Abstract
Charcot-Marie-Tooth disease type 4C (CMT4C) is a childhood-onset demyelinating form of hereditary motor and sensory neuropathy associated with an early-onset scoliosis and a distinct Schwann cell pathology. CMT4C is inherited as an autosomal recessive trait and has been mapped to a 13-cM linkage interval on chromosome 5q23-q33. By homozygosity mapping and allele-sharing analysis, we refined the CMT4C locus to a suggestive critical region of 1.7 Mb. We subsequently identified mutations in an uncharacterized transcript, KIAA1985, in 12 families with autosomal recessive neuropathy. We observed eight distinct protein-truncating mutations and three nonconservative missense mutations affecting amino acids conserved through evolution. In all families, we identified a mutation on each disease allele, either in the homozygous or in the compound heterozygous state. The CMT4C gene is strongly expressed in neural tissues, including peripheral nerve tissue. The translated protein defines a new protein family of unknown function with putative orthologues in vertebrates. Comparative sequence alignments indicate that members of this protein family contain multiple SH3 and TPR domains that are likely involved in the formation of protein complexes.
Introduction
Charcot-Marie-Tooth disease (CMT) comprises a group of clinically and genetically heterogeneous hereditary motor and sensory neuropathies (HMSNs). With an overall population prevalence of 1 in 2,500, CMT constitutes the most common inherited neuromuscular disorder (Skre 1974). The clinical picture includes progressive distal muscle weakness and atrophy, foot deformities, and distal sensory loss. Two major types have been discerned by combined clinical, electrophysiological, and nerve biopsy studies (Dyck et al. 1993). The demyelinating form (CMT1 or HMSN I [summarized in MIM 118200 for CMT1B]) is characterized by reduced nerve-conduction velocities (NCVs) with values <38 m/s for the motor median nerve, segmental demyelination and remyelination, and onion bulb formation in nerve biopsies. These changes differentiate CMT1 from the axonal type (CMT2 or HMSN II [summarized in MIM 118210 for CMT2A]), in which NCVs are near normal and nerve pathology shows signs of axonal degeneration. Both forms are usually inherited as an autosomal dominant trait, but autosomal recessive and X-linked inheritance also occurs. In recent years, considerable advances have been made in understanding the molecular genetics of CMT neuropathies. (Mutations of genes that cause CMT are listed in the Inherited Peripheral Neuropathies Mutation database.)
Autosomal recessive CMT neuropathies are, in general, much less common than the dominant forms. Skre (1974) estimated the frequency of autosomal recessive CMT to be 1.4 per 100,000 in western Norway. However, in communities with a high prevalence of consanguineous marriages, autosomal recessive inheritance is likely to account for 30%–50% of all forms of CMT and may even be the most common form (European CMT-Consortium–ENMC 1999). The clinical presentation is similar to dominant CMT, but it is usually more severe and has an earlier onset age (Harding and Thomas 1980; Thomas 2000). The distinction in demyelinating and axonal forms is also valid for the recessive CMTs, with the demyelinating forms named “CMT4” (summarized in MIM 214400 for CMT4A) and the axonal types designated “ARCMT2” (MIM 605588 for CMT2B1; MIM 605589 for CMT2B2). Autosomal recessive CMT is genetically heterogeneous, with ⩾11 chromosomal loci involved. One form of recessive demyelinating CMT, CMT4C (MIM 601596), was mapped to chromosome 5q23-q33 in two Algerian inbred kindreds (LeGuern et al. 1996). Additional families compatible with linkage to CMT4C were reported from the Mediterranean basin and in a cohort of Dutch families (Gabreëls-Festen et al. 1999; Guilbot et al. 1999; Nelis et al. 2001). It has been suggested that CMT4C is associated with a peculiar peripheral-nerve pathology characterized by a combination of extended formation of Schwann cell processes and onion bulbs consisting only of basal membranes. Clinically, CMT4C is complicated by an early-onset, severe scoliosis that contrasts with the relatively mild peripheral neuropathy symptoms (Kessali et al. 1997; Gabreëls-Festen et al. 1999).
Since the first recognition of the CMT4C locus in 1996, seven genes responsible for recessive CMT subtypes have been identified (Warner et al. 1998; Bolino et al. 2000; Kalaydjieva et al. 2000; Boerkoel et al. 2001; Guilbot et al. 2001b; Baxter et al. 2002; Cuesta et al. 2002; De Sandre-Giovannoli et al. 2002; Azzedine et al. 2003; Senderek et al. 2003), but the CMT4C gene has remained unknown. In this study, we report the fine mapping of the CMT4C locus and the identification of a novel gene that, when mutated, causes CMT4C.
Subjects and Methods
Patients
We obtained family data and blood samples for isolation of genomic DNA, after receiving informed consent from the patients with CMT and their healthy relatives, according to the Declaration of Helsinki and protocols approved by the review boards for medical ethics at the different institutions involved in this study. A series of 14 families with CMT (including 21 affected individuals) was available for the initial search for the CMT4C gene. Diagnostic criteria were (i) sensorimotor neuropathy with onset in the 1st or 2nd decade, (ii) absence of clinical symptoms and neurophysiological signs in the parents, and (iii) parental consanguinity or at least one other affected sibling (European CMT-Consortium–ENMC 1999). Most of the patients were diagnosed with a demyelinating neuropathy. After revealing the identity of the CMT4C gene, we screened a set of 55 families with recessive CMT (including 72 patients) for compatibility with linkage to the CMT4C region. Those families compatible with linkage underwent mutation analysis. We also sequenced the novel CMT4C gene in 21 patients whose DNAs were received for routine diagnostic testing for demyelinating CMT. These patients were mainly single affected children whose parents were reported to be nonconsanguineous and healthy. Six of the patients had severe, early-onset scoliosis.
Refinement of the CMT4C Locus
Haplotype analysis at the CMT4C locus was performed with STR polymorphism markers D5S658, D5S2011, D5S1360, D5S2090, D5S413, D5S2015, D5S636, and D5S673 (Genome Database and November 2002 release of the public genome assembly at the University of California–Santa Cruz [UCSC]). For fine mapping and allele-sharing analysis, 10 additional STRs were retrieved from public databases, and 15 new repeat polymorphisms were identified by using the Repeat-Masker program on genomic DNA from the critical interval. Primers for PCR amplification of known STRs are reported in the Genome Database, and primers for new STRs were designed by the Primer3 program (table A [online only]). Sense primers were labeled with FAM fluorophores (MWG Biotech) for analysis on an ABI 377 DNA sequencer (Applied Biosystems). Data on 12 SNPs from the minimal candidate interval were retrieved from the NCBI dbSNP database and ABI SNP genotyping repository (table B [online only]). Sequences were PCR amplified and assayed by DNA sequencing with ABI dye terminator technology (Applied Biosystems) for electrophoresis on an ABI 310 genetic analyzer (Applied Biosystems).
Table A.
STR Markers from the CMT4C Interval
|
Primer Sequencesa (5′→3′) |
||||||
| Markera | Position in Mapb(Mb) | Calculated heterozygositya(%) | Repeat Sequencea | Forward | Reverse | Product Sizea(bp) |
| D5S658 | 140.4 | 84 | CA | TTTGAAGGGCACTACGAAGATCCTC | ATTCTCTATTGGAGCCAAGCCAAG | 266–282 |
| D5S2011 | 141.2 | 86 | CA | GCTTGGAACACAAGAGTGG | TCAGCCTCACCCAGAGA | 138–154 |
| D5S1360 | 144.9 | 70 | ATA | ACAAACAAAACCAAGAGTGC | TGGCTCATGTATCCCTATGT | 132–147 |
| D5S643 | 145.0 | 49 | CA | TGGGCGACAGAGCCATC | TGTGGTGTGCCATTTATTGACT | 134–168 |
| D5S436 | 145.2 | 87 | CA | TATGTGCCAGGCATTACGCT | GTCTCCACCCACAGCAGG | 234–254 |
| 11359 | 145.3 | ND | GT | CATGACTTAGATCCCCAGCC | CAGCAATAGAAGGGGGACAA | 279–285 |
| D5S2099 | 145.3 | 55 | CA | ATTGCACAGAAATGGTAAAGG | TATCAGCATAAATGCCTCCC | 236–244 |
| 5216 | 145.4 | ND | CA | TATAGCTCTGGGAGCTGGGA | TTGTTCCCAGAATGCTACCC | 170–202 |
| 11396 | 145.7 | ND | AC | TGCGCTCGCTCTCTTTTTAT | AATGAAATTACGGTGGTGCC | 231–239 |
| 8728 | 145.9 | ND | GT | CCTAAACAGGAAGCAGCCTT | TGCCTTTTCTCAGCCATAAGA | 210–238 |
| D5S2033 | 145.9 | 65 | CA | TAGCTTTTCCATGTTATCTAGCA | TTCCAGTTTGGGTCCTTAT | 215–229 |
| D5S638 | 146.3 | 63 | CA | GGAAACACTCAGTAGTATCATTGCC | TTTAGAGAAACACACGCACCAGTA | 137–145 |
| D5S2490 | 147.0 | ND | ATA | TTTGGCATCCTTGCTATTTC | GTTCTGCGCATATATCCCAG | 119–134 |
| D5S463 | 147.1 | ND | CA | AAGCACACCCATTTCCCAAG | GATCGGGGTTAAGGCTTCAA | 178–186 |
| D5S2090 | 147.2 | 83 | CA | CATGGGCATGTTTCAAAAT | AGTACCTCCTTAGTAACTCTGGGC | 189–205 |
| D5S434 | 147.3 | 71 | CA | CTTGAAATGTTCCCAACACA | TGCACAGATGAGGAAACTGA | 246–258 |
| 91948 | 147.7 | 87c | GT | TTTCTCAGGCCGTACTGGTT | TTGCCTCAGCAGTAAATGGA | 226–252 |
| 8627 | 148.0 | ND | GT | TCTTAAAGGCGAACTGAGTTGTT | TGCTTTTCCCAATGAATTACAC | 183–187 |
| 116312A | 148.3 | 84c | GT | TGAGAAGCAGGGTATCTGACC | ATACACACTCAGGCCCGTTC | 184–214 |
| D5S413 | 148.4 | 70c | CA | AGTCACCTTCTCTGTCTCCA | AGGCCTCATTCAAAATCTGT | 264–276 |
| 91940 | 148.5 | 65c | CA | ATTCCAGGAGGGGAATGAAC | CCGAAGGTGGAAGTGAGGTA | 147–157 |
| 12613 | 148.6 | 57c | CA | GCCCAAGCAACTGGTACATT | TCTCAGGCAGTGAGTGGTCA | 157–179 |
| 8681 | 148.9 | 83c | CA | ATCTCACAGCACCTCCATCC | GCCTCAAATGACACCCTACTTT | 165–185 |
| 11636 | 149.0 | ND | CA | GTGGCCAAATGTTCCTGAAG | CAGGGATGCTTGTCCTCTTT | 206–212 |
| D5S812 | 149.0 | ND | TAAG | TACCACAGCAACCACAAAGA | GAGGAAAGCAAAGACCATGA | 154–162 |
| 221 | 149.0 | ND | CA | AGTCTGTGGCCAAATGTTCC | TCAGGGATGCTTGTCCTCTT | 215–221 |
| 8545 | 149.2 | 42c | CA | TCTGATTTGCTGTTGATTGC | CTGGGACAACAGCCTCCTTA | 237–253 |
| 11382 | 149.5 | ND | CA | CTCCACTAGCTCCCACTTGC | ATGAGATCCTGGGCTGAAGA | 168–180 |
| D5S2015 | 149.6 | 70 | CA | TTGGCTAATGGGAGGCAACA | GCTACCTAAAGAACACAGTCATGGC | 171–185 |
| D5S2013 | 149.6 | 76 | CA | AATGGTCTCCAGAAAAGCA | AGCCAGGTGAGTCTCGTC | 150–162 |
| 11372 | 149.7 | ND | CA | CTTCCCAGGAGGATGCAATA | CTTCCACCACCTCATCCACT | 243–249 |
| D5S636 | 149.9 | 78 | CA | AAGGCATATGGGAAATATCTGT | CCACACCATTATGACATTTTCT | 130–152 |
| D5S673 | 151.6 | 82 | CA | TGGAGAGATAGTGTCCCG | GGTCTGAGTTCCACATCAT | 260–288 |
Data on published markers (beginning with “D5S”) are according to the Genome Database. Allele sizes on ABI 377 differed by 1–2 bp. For new markers (plain numerals), allele sizes observed in families with CMT are given.
Map positions are according to the UCSC November 2002 freeze.
For some markers, heterozygosity rates and the distribution of alleles were determined by genotyping 23 Turkish control individuals. ND = not determined.
Table B.
SNP Markers from the CMT4C Interval
|
Primer Sequences (5′→3′) |
||||||
| dbSNP Numbera | Position in Mapa,b(Mb) | Variationa | Minor-Allele Frequencya(%) | Forward | Reverse | Product Size(bp) |
| rs1984789 | 147.8 | C/T | ND | TTGGTGCTGTGCTGTCTACC | TGGCCAGAAGAGGTAAATGC | 165 |
| rs3734119 | 147.9 | C/T | C: 27 | TCCAATCCTCCACTTTCTTCC | TAGGGCTTGTTGACCATGAA | 198 |
| rs1042718 | 148.2 | C/A | A: 40 | ATCGCAGTGGATCGCTACTT | GATCACCAGGGGAACGTAGA | 265 |
| rs1432793 | 148.4 | C/T | C: 43 | GGTTGCTCAACTTTGCCTGT | CCTGACCAGTGCTTAGGTCC | 233 |
| rs9512 | 148.6 | C/T | C: 40c | TGGCCAAAAAGAGAAGTTGC | AGGCAGAAAGTGATCGGAGA | 244 |
| rs783777 | 148.7 | C/T | ND | CGACCACGAAGCTTTATCCT | TACCATTGGGCATTTGAACA | 243 |
| rs2242376 | 148.7 | C/T | C: 40c | CTCCCTGCCTGAGGTCATAG | TGTCAGATGAAGCGAGAGGA | 199 |
| rs3733847 | 148.9 | A/T | T: 13c | TGAAAAAGGCAGCATGAAAA | GGGCAACAAGAGCAAAACAT | 226 |
| rs3756505 | 148.9 | C/T | C: 18 | ACTATTGTCGTGGGCTACGC | GAAAAATGCCAAATTAGTCCCA | 237 |
| rs30825 | 149.3 | C/T | T: 47 | GGGATTTACTGCATGCCAAC | TTGAGAAACTTGAGGCAGCTT | 165 |
| rs2276982 | 149.4 | A/G | G: 08 | CAGTCCATGCTGTTTCCTGT | GACCACATGGAGAGAGAAAGAT | 197 |
| rs246388 | 149.5 | C/T | T: 38 | AGCCCCCTACAGGAAGCTAT | GCTCCACCCTGAATGAAGTC2 | 191 |
All data are according to the NCBI dbSNP repository or the Applied Biosystems SNP genotyping resources.
Map positions are according to the UCSC November 2002 freeze.
For some markers, allele frequencies were determined by genotyping 23 Turkish control individuals. ND = not determined.
Candidate Gene Analysis
For selection of candidate genes, we used the November 2002 freeze of the UCSC human genome assembly. The UCSC map provided known proteins and genes (as curated by Swiss-Prot and the NCBI RefSeq project), aligned mRNAs, ESTs, Unigene clusters, Ensembl gene builds, Genscan and FGENESH gene predictions, and alignments of other genomes by similarity. Selection of positional candidate genes and anonymous transcripts was performed on the basis of neural expression, especially in spinal cord and peripheral nerve. We queried public data repositories for EST matches and SAGE (serial analysis of gene expression) tags and evaluated microarray data on the temporal expression pattern of genes during sciatic nerve myelination (Verheijen et al. 2003). To test for expression of candidate genes by RT-PCR, total human sciatic nerve and spinal cord RNAs were extracted using the RNeasy kit (Qiagen) and were reverse transcribed by oligo-dT primers and AMV reverse transcriptase (Promega). RT-PCR-products were generated using gene-specific exonic primer sets.
Five transcripts (SP329/FLJ13962 [GenBank accession numbers AF177339 and AK024024], KIAA1985 [GenBank accession numbers AB075865, BQ421935, BQ213149, W86370, BM552239, BE026883, and BE302610; HUGE Protein Database], KIAA0843 [GenBank accession number AB020650; HUGE Protein Database], FLJ23713 [GenBank accession number AK074293], and CSNK1A1 [GenBank accession number NM_001892]) were screened for mutations. Primers flanking exons were used to amplify genomic DNA, and products were subjected to DNA sequencing on ABI 310 or 3730 DNA analyzers (Applied Biosystems) (for KIAA1985 primers, see table C [online only]). Sequence variants were identified by visual inspection of electropherograms. The DNAs of parents and siblings were sequenced to test for segregation of mutations. Control samples were screened by restriction enzyme digestion or by WAVE denaturing high-performance liquid chromatography (Transgenomic).
Table C.
Primer Sets Used to Amplify KIAA1985 Exons for Mutation Analysis
|
Primer Sequences (5′→3′) |
||||
| PrimerSet | Exon(s) Covered | Forward | Reverse | Product Size (bp) |
| 1 | 1 | GCTGTAGCCGGGATATCAAA | AGCACAATCATCTCCCCATC | 337 |
| 2 | 2 | CCATGCCACTAATCCTGGAA | TCATCAAGAGGGAAAGAGGG | 318 |
| 3 | 3 | CACCAGTTGAGGGTTCAGGT | GGACCTACTTTGTTCCAGATGC | 338 |
| 4 | 4 | TGACGTAAAAATCAATGCCA | TAATTACCAGAGCCAGGCCA | 320 |
| 5 | 5 | GGTCTGGGAATCAAGCTCAG | ACAGGTGGGTTCATTTGTGA | 376 |
| 6 | 6 | CCAGGAAGAAAAGTACTCCAGC | TCCACACTATGGATGCCCA | 413 |
| 7 | 7 | TTCTCAATAAACTTTTGCCATCA | TGGCTTCCCTAAATCCAGTT | 348 |
| 8 | 8 | AGTCTGGGAACCCCTCATTT | GTGCTTTTCTGGCTCCATGT | 416 |
| 9 | 9 | ATTTGCGGGAAGTTGGTTT | CATGGCCACCCAAATTCAT | 378 |
| 10 | 10, 11 | TGAATGCCAAGGCTCCTAAG | TCCAGGTAGGCCACAAACTC | 629 |
| 11A | 11 | ACTGGTCAGGAGGAGGAGG | GGAGGGTGTCCAGAGAGGA | 629 |
| 11B | 11 | AAGCATGAACTCGACGTGGT | TAAGAGATAGGCCCATGCCA | 597 |
| 11C | 11 | GCCCTGTGTCTCATCCTTTC | GATGCCTTAAGCCAAACAGC | 569 |
| 11D | 11, 12 | CCTGCAGGCTGTACGACTCT | CTTCCTTTGGCTGATGAGGA | 692 |
| 13 | 13 | GCTGTTCCTGCTCAGAGCTT | TCCACAGGCTTAGGGTGAAC | 367 |
| 14 | 14 | GGGGGACTTTAGCAGAGGAT | AGGAGAAGAGGGACTCAGGC | 331 |
| 15 | 15 | TTTTAGTCCACTGCCGTTCC | CACACAGTCTGACTCCAGGG | 364 |
| 16 | 16 | CAGCACAGTCCCTGACATCT | CTGGTTTTCTCAGCTCCACC | 415 |
| 17 | 17 | ATGTTGGGCAAACAAACAAA | AGGTAAGGACTCGGACCCTC | 416 |
Expression Study
A 535-bp cDNA fragment encompassing KIAA1985 exons 15–17 was amplified by RT-PCR and was cloned in the pCRII-TOPO vector (Invitrogen). A 693-bp cDNA fragment corresponding to exons 1–6 was obtained by KpnI and BsmI digestion from the IMAGE cDNA clone 6048901 obtained through RZPD Deutsches Ressourcenzentrum für Genomforschung. Inserts were gel purified, were [32P]-labeled with a Multiprime kit (Amersham Biosciences), and were hybridized to a human adult multiple-tissue northern blot (BD Bioscience). Hybridization was performed for 1 h at 68°C with Express Hyb solution (BD Bioscience). Membranes were washed for 3×30 min in 2×SSC, 0.05% SDS, at room temperature, followed by 3×30 min washes in 0.1×SSC, 0.1% SDS at 50°C. Filters were exposed to autoradiography film for 24–36 h at −70°C. Human β-actin probes were used for control hybridizations.
For RT-PCR, total RNA and cDNA from human adult sciatic nerve, spinal cord, brain, and skeletal muscle were prepared as described above. Overlapping RT-PCR fragments covering the KIAA1985 cDNA were sequenced (table D [online only]). Expression levels were assessed by using β2-microglobulin–specific primers in control reactions. For the expression analysis of the mouse orthologue D430044G18Rik (GenBank accession numbers AK052534, BC037000, BI414954, and NM_172628), we amplified exons 4–11 in cDNA from brain, ventral horn (VH) of spinal cord, and dorsal root ganglia (DRG) of E13 mouse embryos, and we cloned the products in the pCRII-TOPO vector with the TOPO TA Cloning kit (Invitrogen). After one-shot electroporation, 20 colonies were selected for each tissue, and the inserts were sequenced.
Table D.
Primer Pairs Used to Amplify KIAA1985 Fragments from cDNA
|
Primers (5′–3′) |
||||
| PrimerSet | Position in cDNA | Forward | Reverse | Product Size(bp) |
| C1 | −53–900 | ACTCTTCTGGCCTGTGTTCC | AAAGCCGATGATCTCAATGC | 953 |
| C2 | 342–1413 | GACCATGGAGGAAATCTGGA | CAATATGGGGGCGAAGTTCT | 1,072 |
| C3 | 1029–2424 | TGAGGAGAGATGCTCCCTGTT | TAAGAGATAGGCCCATGCCA | 1,396 |
| C4 | 2272–3522 | GCCCTGTGTCTCATCCTTTC | TGTAGCCAGGCGGTGAAAGG | 1,251 |
| C5 | 2933–3899 | AGGCATGCATCACCTACCATG | TGCCAAATGTCTAGAGACAG | 967 |
| C23a | 574–1267 | GAGAAGGAAGGGGAATGCTT | CAGAGCTGCTGGACTGTCTG | 694 |
| C5ab | 3365–3899 | TGAGAACTGAGCTCCGGATT | TGCCAAATGTCTAGAGACAG | 535 |
Used for analysis of putative KIAA1985 splice variants.
Used for expression studies by RT-PCR and for generation of the 3′ probe for northern blotting.
Results
Refinement of the CMT4C Interval
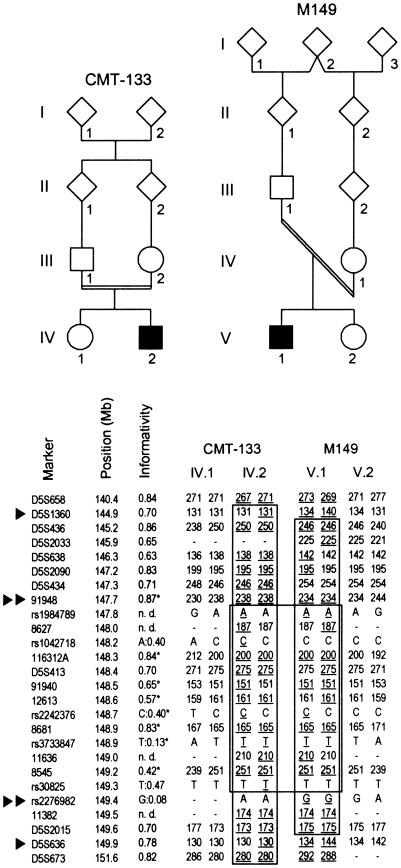
The initial 13-cM CMT4C linkage interval between markers D5S658 and D5S673 (LeGuern et al. 1996) could not be refined in subsequent studies (Gabreëls-Festen et al. 1999; Guilbot et al. 1999, 2001a). To establish compatibility with linkage to CMT4C in further kindreds, we selected 14 families with CMT with multiple affected children or parental consanguinity. Haplotype analysis was compatible with linkage to CMT4C in five families: two German nonconsanguineous multiplex families (M983 and M1494) and three consanguineous Turkish nuclear families (CMT-133, M149, and M2045). Family M149 yielded recombination events by homozygosity by descent, reducing the CMT4C critical region, within suggestive borders, to 5.6 cM between markers D5S1360 and D5S636 (fig. 1). Genotyping of additional STRs from the reduced region confirmed homozygosity at the CMT4C locus in all three Turkish families. Moreover, in families CMT-133 and M149, we observed a shared disease haplotype between STRs 91948 and 11382, suggesting further refinement of the CMT4C region to 1.8 Mb. Genotyping of SNPs from this interval showed an ancestral recombination event, under the assumption of a common haplotype, for rs2276982 and the gene (fig. 1). The shared 20-marker haplotype spanned a 1.7-Mb interval and contained rare alleles for STRs 116312A, D5S413, 12613, 8681, and 8545 and for SNP rs3733847. This haplotype was not detected in 23 Turkish control individuals, and both affected individuals were heterozygous for STR markers at other recessive CMT loci (data not shown).
Figure 1.
Homozygosity mapping in CMT4C-affected families (CMT-133 and M149) with chromosome 5q32 markers. A subset of markers used in this study is given in cen-qter orientation. The physical map positions are according to the UCSC November 2002 freeze. The informativity of each marker is shown as a calculated heterozygosity rate for microsatellite markers (“D5S” locus numbers and plain numerals) and as a minor allele frequency for SNPs (“rs” numbers). Values determined in the Turkish population are marked by an asterisk (*). Haplotypes are shown for affected and healthy offspring. Paternal haplotypes are shown at left; maternal haplotypes are at right. Informative alleles in the affected individuals are underlined. Regions of homozygosity and shared haplotypes are boxed. The recombination events suggested from regions of homozygosity in patient M149.V.1 support a 5.6-Mb critical interval flanked by markers D5S1360 and D5S636 (single arrowheads). Further refinement was inferred through identical haplotypes shared by patients CMT-133.IV.2 and M149.V.1 between markers 91948 and rs2276982 (double arrowheads). This narrowed the critical genetic region to a suggestive interval of 1.7 Mb.
Mutation Analysis
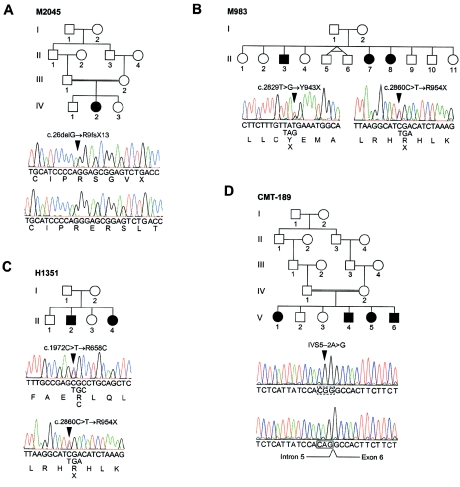
Eight known genes and eight anonymous transcripts mapped within the 1.7-Mb CMT4C region (represented by GenBank human genomic contig NT_006859), none of which was an apparent candidate gene for a peripheral neuropathy (fig. 2). Experimental and in silico expression studies did not allow a clear categorization of positional candidates by likelihood of being the disease gene. We performed mutation analysis on DNAs of the affected individuals from the five putative families with CMT4C. No pathogenic mutations were found in CSNK1A1, SP329/FLJ13962, KIAA0843, and FLJ23713 (data not shown). However, in KIAA1985, sequence variations were detected in four of the five families (CMT-133, M149, M983, and M2045) predicted to result in three truncating changes and one missense mutation (tables 1 and 2; fig. 3). All affected individuals were identified with mutations on both alleles; the parents carried these mutations in the heterozygous state, whereas unaffected siblings also carried one mutation or were homozygous for the wild-type alleles. Consistent with the common flanking haplotype in families CMT-133 and M149, we found a shared seven-marker intragenic SNP haplotype (table E [online only]) and the same c.1586G→A transition causing an R529Q exchange. Among 160 Turkish control subjects, we found one heterozygous individual for the c.1586G→A mutation. That person also shared the flanking and intragenic haplotype found in families CMT-133 and M149. The c.1586G→A transition was not found in the western European control population (600 chromosomes). The mutations in families M983 and M2045 were absent from 120 control chromosomes matched for ethnic background.
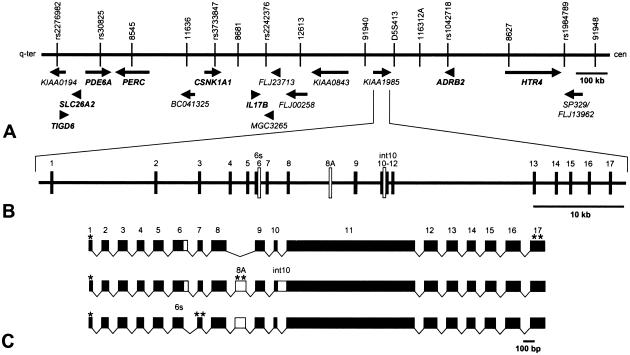
Figure 2.
Transcript map of the CMT4C region on 5q32, genomic organization of the CMT4C gene, and alternative splicing of its transcripts. A, Partial physical and transcript map of the 1.7-Mb CMT4C linkage region delimited by the centromeric marker 91948 and telomeric marker rs2276982. Known genes (shown in bold text) and predicted genes (shown in plain text) are indicated by arrows in the direction of transcription. B, Genomic structure of the CMT4C gene (KIAA1985). The gene covers 62 kb of genomic sequence and consists of ⩾18 variably spliced exons. Exons are indicated by vertical hatches and are numbered. Empty bars represent alternatively spliced sequences. The sizes of introns are shown relative to each other. C, Alternative splice products of KIAA1985. The transcript encoding the putative full-length protein is given above the products. Coding regions of exons are drawn to scale. Use of exon 6s (instead of 6), presence of exon 8A, and retention of intron 10 predict shorter translation products. An asterisk (*) indicates predicted translation-initiation site; a double asterisk (**) indicates predicted translation-termination signal.
Table 1.
Clinical, Electrophysiologic, and Morphological Data in CMT4C
|
NCV (m/s)h |
||||||||||||||||||||||||
| Motor NCV |
Sensory NCV |
Nerve Biopsy (EM)i |
||||||||||||||||||||||
| Family and Patient | Sex | Ethnic Origin | Mutationa | Consanguinity | Age at Walking(mo) | Age at Diagnosis(years) | Age at Last Examination(years) | Distal Weaknessb | Distal Muscle Atrophyb | Involvement of Proximal Musclesc | Foot Deformityd | Walking Aids (since age in years) | Distal Sensory Losse | Scoliosis (age at Onsetin years)f | Reflexesg | Median | Ulnar | Peroneal | Tibial | Median | Sural | BLOB | Branching of SC | Other(age at onset in years) |
| CMT-133: | ||||||||||||||||||||||||
| CMT-133.IV.2 | M | Turkish | R529Q (hom) | + | 30 | Infancy | 8 | + | + | + (legs) | − | − | + | − | (+)/− | 27 | 37 | 24 | NR | − | + | |||
| M149: | ||||||||||||||||||||||||
| M149.V.1 | M | Turkish | R529Q (hom) | + | 11 | 7 | 8.5 | + | + | − | − | − | + | − | +/− | 24.1 | 25.7 | 23.4 | 29.9 | 32.2 | ND | |||
| M983: | ||||||||||||||||||||||||
| M983.II.3 | M | German | Y943X (pat) | − | 18 | 11 | 17 | +++ | + | − | ++ | − | + | + (11) | +/− | 13.9 | 9.5 | NR | − | − | ||||
| M983.II.7 | F | R954X (mat) | 19 | 5 | 26 | ++ | + | + (legs) | ++ | Peroneal braces | + | ++ | (+)/− | 25 | 20.9 | 13.8 | 6.7 | 37.8 | NR | − | + | |||
| M983.II.8 | F | 18 | 7 | 11 | + | − | − | + | − | + | + | +/− | 38 | 24 | ND | |||||||||
| M2045: | ||||||||||||||||||||||||
| M2045.IV.2 | F | Turkish | R9fsX13 (hom) | + | 12 | 8 | 8.5 | ++ | − | − | − | − | + | − | (+)/(+) | 33 | 28 | 30 | + | + | ||||
| CMT-189: | ||||||||||||||||||||||||
| CMT-189.V.1 | F | Italian | IVS5-2A→G (hom) | + | 14 | Infancy | 45 | ++ | ++ | − | + | − | ++ | ++ (infancy) | −/− | 21.6 | 23.7 | NR | ND | Hypoacusis | ||||
| CMT-189.V.4 | M | 16 | Infancy | 39 | +++ | ++ | + (legs) | + | − | ++ | ++ | −/− | 19.5 | 17.6 | NR | NR | + | − | Deafness | |||||
| CMT-189.V.5 | F | 24 | Infancy | 15 | +++ | +++ | ++ | ++ | Chairbound (15) | + | ++ (7) | −/− | ND | Respiratory insufficiency (30) | ||||||||||
| CMT-189.V.6 | M | 15 | 4–5 | 30 | ++ | ++ | + | + | − | ++ | ++ | −/− | 21.1 | 26.9 | NR | ND | Hypoventilation (28); hypoacusis | |||||||
| PN-1289: | ||||||||||||||||||||||||
| PN-1289.1 | F | Iranian | S831fs (hom) | + | >18 | Infancy | 17 | + | + | − | + | − | + | +++ (12) | −/− | 32 | 38 | 27 | NR | ND | Congenital nystagmus | |||
| CMT-219: | ||||||||||||||||||||||||
| CMT-219.1 | F | Turkish | G583fs (hom) | + | 11 | 29 | +++ | +++ | + | + | Chairbound | + | −/− | 13 | 12 | ND | Diabetes mellitus | |||||||
| CMT-225: | ||||||||||||||||||||||||
| CMT-225.1 | M | Turkish | P1114fs (hom) | + | 12 | 25 | +++ | ++ | + | + | ++ | −/− | 4 | ND | Three affected brothers | |||||||||
| CMT-234: | ||||||||||||||||||||||||
| CMT-234.1 | M | Turkish | E657K (hom) | + | Delayed | 2–3 | 26 | +++ | +++ | + | + | Walks with aid | + | −/− | 12 | ND | Nystagmus; palpable great auricular nerve | |||||||
| CMT-235: | ||||||||||||||||||||||||
| CMT-235.1 | F | Turkish | G583fs (hom) | + | 18 | Infancy | 14 | ++ | − | − | ++ | − | + | + | −/− | 26 | 21 | 34 | NR | ++ | + | Asymmetry of pareses | ||
| AC70: | ||||||||||||||||||||||||
| AC70.II.1 | F | Greek | R954X (pat) Q1201X (mat) | − | 21 | Infancy | 8.5 | ++ | ++ | +++ | − | Chairbound (7.5) | + | ++ (4) | −/− | 27 | 16 | 16 | NR | ++ | ++ | CPAP treatment required at night | ||
| H1351: | ||||||||||||||||||||||||
| H1351.II.2 | M | German | R658C (pat) | − | Delayed | 12 | 33 | ++ | ++ | − | +++ | − | + | − | (+)/− | 28 | NR | 30 | 39 | NR | ND | |||
| H1351.II.4 | F | R954X (mat) | 27 | ++ | + | − | + | − | + | − | (+)/− | 37 | 12 | ND | ||||||||||
hom = homozygous alleles; pat = paternal allele; mat = maternal allele.
− = not affected; + = mild in the lower extremities; ++ = marked in the lower extremities; +++ = also affecting the hands and forearms.
− = not affected; + = mild; ++ = severe.
− = no deformity; + = pes cavus and hammer toes; ++ = clubfoot deformity; +++ = surgery required.
+ = decreased sensibility; ++ = absent sensibility.
− = none; + = mild; ++ = severe; +++ = surgery required.
Upper/lower extremities; + = normal; (+) = decreased; − = absent.
Normal values: motor-median and ulnar nerve >45 m/s; motor-tibial and peroneal nerve >40 m/s; sensory median nerve >45 m/s; sural nerve >40 m/s. NR = not recordable.
Light microscopy indicated a demyelinating neuropathy in all investigated cases. Electron microscopy (EM) findings: BLOB = basal lamina onion bulbs; SC = Schwann cell; − = not observed; + = present; ++ = prominent finding. ND = no biopsy done.
Table 2.
Mutations Detected in Patients with CMT4C
| Exon/Intron | Nucleotide Change | Effect on Coding Sequence | Families with CMT4Ca | Ethnic Origin |
| 1 | c.26delG | R9fsX13 | M2045 (hom) | Turkish |
| IVS5 | IVS5-2A→G | Exon skipping supposed | CMT-189 (hom) | Italian |
| 11 | c.1586G→Ab | R529Qb | CMT-133c (hom), M149c (hom) | Turkish |
| 11 | c.1747_1748delAG | G583fsX586 | CMT-235c (hom), CMT-219c (hom) | Turkish |
| 11 | c.1969G→A | E657K | CMT-234 (hom) | Turkish |
| 11 | c.1972C→T | R658C | H1351 (het) | German |
| 11 | c.2491_2492delAG | S831fsX839 | PN-1289 (hom) | Iranian |
| 11 | c.2829T→G | Y943X | M983 (het) | German |
| 11 | c.2860C→T | R954X | M983c (het), H1351c (het), AC70c (het) | German, Greek |
| 15 | c.3341delC | P1114fsX1115 | CMT-225 (hom) | Turkish |
| 16 | c.3601C→T | Q1201X | AC70 (het) | Greek |
hom = homozygous; het = heterozygous.
The R529Q mutation was also found in 1 of 320 Turkish control chromosomes. The same flanking and intragenic polymorphisms as those in CMT-133 and M149 were observed.
Disease chromosomes carried the same flanking and intragenic polymorphisms.
Figure 3.
KIAA1985 mutations in four families with CMT4C. Arrowheads in the electropherograms indicate the disease-causing mutations. A, Homozygous c.26delG mutation in family M2045, resulting in the R9fsX13 frameshift mutation. The wild-type sequence is shown in the lower chromatogram. B, Compound heterozygote c.2829T→G and c.2860C→T base substitutions in family M983, resulting in Y943X and R954X nonsense mutations. C, Compound heterozygote c.1972C→T and c.2860C→T mutations in family H1351, resulting in the R658C missense and the R954X nonsense mutations. D, Homozygous acceptor splice site IVS5-2A→G mutation in family CMT-189. The wild-type sequence is shown in the lower chromatogram.
Table E.
KIAA1985: Observed Intragenic SNPs
| Exon/Intron | Nucleotide Change | Effect on Coding Sequence | Minor Allele Frequency | dbSNP Number |
| 11 | c.1194T→C | No change of G398 | C: .43 | rs1432793 |
| 11 | c.1402G→T | A468S | ND | |
| IVS11 | IVS11+37A→G | ND | ||
| IVS12 | IVS12+103G→A | G: .22a | ||
| IVS14 | IVS14+69C→T | ND | rs1025476 | |
| 15 | c.3472G→A | V1158I | ND | |
| 16 | c.3594A→C | No change of P1198 | A: .40a |
The distribution of some alleles was determined by genotyping 23 Turkish control individuals. ND = not determined.
When we screened a cohort of 55 families with autosomal recessive CMT by using closely linked, highly informative markers 91948, 116312A, and 8681, we discovered 15 families compatible with CMT4C. KIAA1985 sequencing revealed homozygous mutations in six families, CMT-189, CMT-219, CMT-225, CMT-234, CMT-235, and PN-1289 (tables 1 and 2; fig. 3). We also screened 21 patients with early-onset demyelinating neuropathy, who requested DNA diagnosis for CMT. In two patients, one with an affected sibling (family H1351) and another with sporadic disease (patient AC70.II.1), we found compound heterozygous sequence variations in KIAA1985 (tables 1 and 2). In the case of missense mutations, 220 control chromosomes from a similar ethnic background were analyzed and found to be negative for the mutations. Corresponding segregation of mutations was found in every family.
Expression Analysis and Genomic Organization
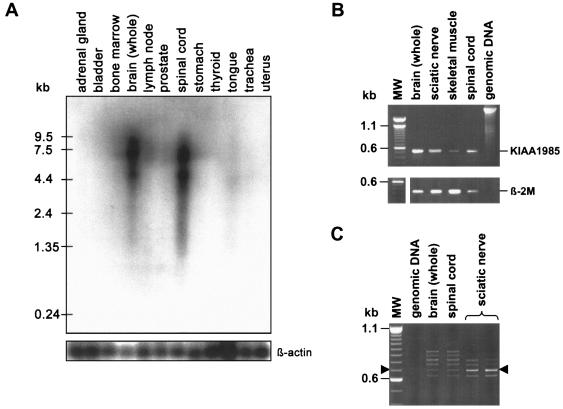
On human adult multiple-tissue northern blots, we found expression of KIAA1985 to be strong in brain and spinal cord and to be very weak in striated muscle. Two bands at ∼7.5 kb and ∼4.5 kb were observed with 5′ and 3′ KIAA1985 probes. RT-PCR experiments revealed equal levels of expression in spinal cord and sciatic nerve (fig. 4). The reported KIAA1985 mRNA corresponded to the size of the 7.5-kb band on the northern blot. An alternative polyadenylation signal was predicted by the Polyadq algorithm at ∼4.5 kb and may give rise to the smaller band. We confirmed the KIAA1985 mRNA sequence by alignment with human EST sequences by querying the NCBI dbEST database and by RT-PCR amplification on mRNA from sciatic nerve, brain, and spinal cord (fig. A [online only]). The putative ATG translation-initiation codon is surrounded by an incomplete Kozak consensus sequence (ACACACATGG) (Kozak 1989) and is preceded by an inframe stop codon 15 bp upstream. The longest continuous ORF (GenBank accession number AY341075) predicts a protein of 1,288 amino acids, with a calculated molecular mass of 144.7 kDa (fig. B [online only]). Sequence alignments of the confirmed KIAA1985 cDNA with genomic sequences on 5q32 were performed with the BLAT tool implemented on the UCSC Web site. The KIAA1985 gene is flanked by the centromeric marker D5S413 and the novel telomeric marker 91940. The gene is transcribed from telomere to centromere and spans 62 kb of genomic sequence. We identified a total of 18 exons; the longest putative ORF is encoded by a 17-exon transcript (fig. 2). All exon-intron boundaries followed the canonical GT/AG rule. The size of exons had a range of 42–1,695 bp, and the length of introns was 0.1–14 kb.
Figure 4.
Expression analysis of KIAA1985. A, Human adult multiple-tissue northern blot. Upper panel, Membrane probed with a radioactively labeled cDNA fragment corresponding to KIAA1985 exons 15–17. Two strong signals at ∼7.5 kb and ∼4.5 kb are seen in brain and spinal cord, whereas striated muscle (tongue) gives only faint signals. All other tissues are negative for KIAA1985 expression by northern blotting. Comparable results were obtained when using a 5′ probe (exons 1–6). Lower panel, Membrane hybridized with radioactively labeled β-actin cDNA fragment for an internal control. B, Expression analysis by RT-PCR. Upper panel, Amplification of a 535-bp KIAA1985 fragment corresponding to exons 15–17. Lower panel, β2-microglobulin (β-2M)–specific primers were used in control reactions. KIAA1985 expression levels in sciatic nerve and spinal cord seem similar. Weak signals are obtained on skeletal muscle mRNA. Genomic DNA was used as a control to test for specific amplification from cDNA targets. (MW = DNA size standard) C, Expression of different KIAA1985 splice variants in human brain, spinal cord, and sciatic nerve. RT-PCR with primers positioned in exons 6 and 11 yielded a 694-bp product from transcripts encoding the longest continuous ORF (arrowheads). The additional bands arise from alternative usage of exons 6s or 8A or retention of intron 10, which would be predicted to result in truncated forms of the KIAA1985 protein (also see fig. 2). The intact mRNA is the dominant transcript in sciatic nerve, whereas spinal cord and brain predominantly express alternative splice products.
Figure A.
KIAA1985 cDNA sequence encoding the longest potential reading frame (GenBank accession number AY341075). Bases appearing in bold text and italics establish the exon-exon boundaries. The translation start site and the stop codon are in bold text and underlined.
Figure B.
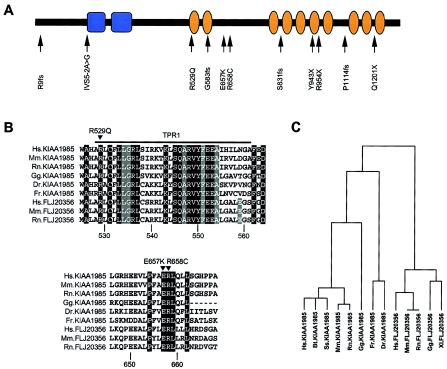
Amino acid sequence of KIAA1985 encoded by the longest ORF. Blue denotes SH3 domains; orange denotes TPR motifs.
There is evidence that the KIAA1985 gene is alternatively spliced. The skipping of the last 58 bp of exon 6 (exon 6s), the insertion of a 130-bp exon between exons 8 and 9 (exon 8A), and the retention of the 122-bp intron 10 were found in ESTs and in RT-PCR–amplified transcripts from sciatic nerve, spinal cord, and brain (figs. 2 and 4). These variants may account for the slight smearing of the bands in the northern blot but do not yield additional signals, since they alter the size of the transcript by only, at maximum, 5% (fig. 4). The alternative exons all exhibited consensus donor and acceptor splice sites. Alternatively spliced transcripts, if translated, would result in a frameshift generating a stop codon with consecutive extensive truncation of the protein. RT-PCR analysis revealed the dominant expression of alternative transcripts, compared with the intact form in brain and spinal cord. Peripheral nerve, however, mainly expressed the transcript encoding the full-length protein (fig. 4).
Analysis of 20 cDNA clones from brain, VH, and DRG of the mouse orthologue D430044G18Rik identified a deletion of the first 16 bp of exon 6 in one clone from brain and one from DRG. A longer version of exon 6 (retaining 154 bp before exon 6) was discovered in cDNA from brain and VH. In 4 of the 20 DRG clones, exon 9 was deleted, and one DRG clone showed a deletion of the first 68 bp of exon 8. By genomic alignment, all splice junctions followed the canonical GT/AG rule. All alternatively spliced transcripts were predicted to shift the reading frame, resulting in severely truncated forms of the D430044G18Rik protein.
Characterization of the Encoded Protein
No significant sequence similarity to known human cDNA or protein sequences was found with the KIAA1985 protein. SMART and ScanProsite analyses on the human and mouse orthologues indicated the presence of two N-terminal SH3 (Src homology 3) domains (Pfam database accession number PF00018) and 10 TPR (tetratricopeptide repeat) motifs (Pfam database accession number PF00515) arranged in tandem arrays (fig. 5). ScanProsite predicted a putative N-terminal myristylation signal, two bipartite nuclear localization signals, a peroxisomal targeting signal, two sites for glycosylation and sulfatation, and multiple phosphorylation sites. The observed splice variants, if translated into protein, would encode SH3 domain–only polypeptides. Conserved orthologues were detected by alignment of KIAA1985 with EST and genomic sequences of various vertebrates. There was a strong sequence conservation for KIAA1985 during evolution, with 81% amino acid identity of human KIAA1985 with its murine orthologue D430044G18Rik. No orthologues were found in Drosophila melanogaster and Caenorhabditis elegans. In addition, BLAST searches on human genomic and translated sequences identified another gene, FLJ20356 (GenBank accession number AK000363), on chromosome 4p16, the putative translation product of which shares 47% amino acid sequence identity with the KIAA1985 protein. The FLJ20356 gene also has conserved orthologues in vertebrates (fig. 5). EST data suggest that FLJ20356 undergoes differential splicing in a manner similar to that of the KIAA1985 gene.
Figure 5.
Protein prediction and sequence conservation among the KIAA1985 protein family. A, Full-length protein of 1,288 amino acids, showing domains as predicted by SMART (blue = SH3 domains; orange = TPR motifs). The sites of mutations detected in this study are indicated. B, Multiple protein sequence alignment generated with the program ClustalW, using translation from genome assemblies and expressed sequences. The amino acid numbering is according to the human KIAA1985 protein. Sequence comparison shows that the three KIAA1985 missense mutations (R529Q, E657K, and R658C) observed in patients with CMT4C affect amino acids that are identical in KIAA1985 and FLJ20356 and their putative orthologues. (Hs = Homo sapiens; Mm = Mus musculus; Rn = Rattus norvegicus; Gg = Gallus gallus; Dr = Danio rerio; Fr = Fugu rubripes.) Upper panel, Sequence of the first TPR motif. Residues generating the TPR consensus sequence are shown on gray background. Sequence conservation outside these consensus residues (shown on black background) is believed to correlate with functional specialization of the TPR motif (Blatch and Lässle 1999). Lower panel, Residues from one of the interdomain regions. Conserved amino acids are given on a black background. C, Average distance tree of KIAA1985 (residues 1000–1100) aligned with homologous sequences. The average distance tree, with percentage identities, was generated by the program Jalview on the basis of a multiple-sequence alignment generated with the program ClustalW. (Ss = Sus scrofa; Bt = Bos taurus; Xl = Xenopus laevis.) The following GenBank database entries were used for generating the multiple-sequence alignments: KIAA1985 [accession number AY341075], Bt.KIAA1985 [accession numbers BI680206, BF652287, BU239452, AW656867, and BI976766], Ss.KIAA1985 [accession numbers BF442400, BI186309, BI183894, and BI345340], Mm.KIAA1985/D430044G18Rik [accession number AK052534], Rn.KIAA1985 [accession numbers BF567582 and XM_225887], Gg.KIAA1985 [accession numbers BU239452 and BU355692], FLJ20356 [accession number AK000363], Mm.FLJ20356 [accession numbers AK028482 and BC024909], Rn.FLJ20356 [accession numbers XM_223527 and XM_223528], Gg.FLJ20356 [accession numbers BU232586, BU471861, and BU474979], Xl.FLJ20356 [accession number BJ062830]). D. rerio and F. rubripes orthologues were retrieved by translated BLAST searches on unfinished genomes, with “human protein sequence” as a query (Ensembl).
The Phenotypic Spectrum of CMT4C
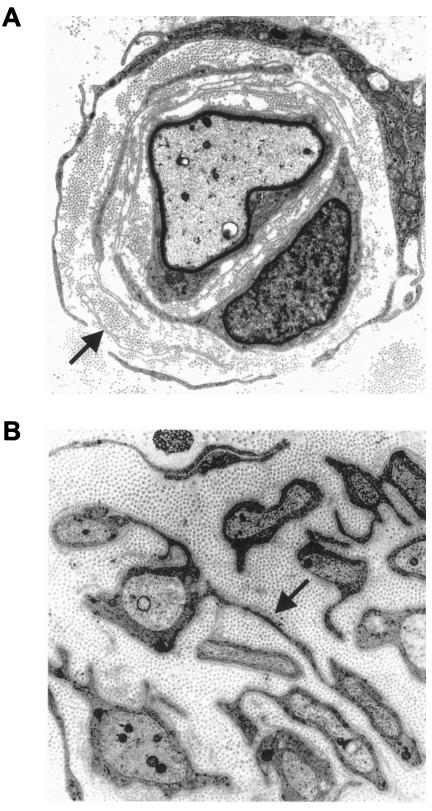
The clinical, electrophysiological, and pathological details of the 18 patients with CMT4C are summarized in table 1. The age at onset peaked during infancy or the late 1st and early 2nd decades of life. Early onset of the disease did not reliably predict a more severe disease or rapid progression. In some patients, CMT4C progressed slowly, allowing independent ambulation up to the 5th decade, whereas others became wheelchair dependent during their teenage years. One patient (AC70.II.1) had severe generalized hypotonia and early-onset respiratory problems, along with a progressive thoracal scoliosis. Altogether, prominent scoliosis was observed in 11 of 18 patients with CMT4C. Scoliosis often preceded apparent pareses, and, in some cases, was the primary reason for medical referral. Other clinical features displayed by the patients with CMT4C were likely unrelated to the CMT (such as diabetes mellitus or nystagmus), represent complications of a neuromuscular disorder (such as respiratory problems arising after a longer course of the disease), or are not uncommon in peripheral neuropathies (such as hypoacusis). Motor and sensory NCVs were reduced, with a mean median motor NCV of 22.6 m/s (range 4–37 m/s). Nerve biopsy findings on the light- and electron-microscopic level displayed a predominantly demyelinating neuropathy in all seven subjects for whom a biopsy specimen was available. The combination of basal lamina onion bulb formation and extended Schwann cell processes was apparent in six cases (table 1; fig. 6).
Figure 6.
Electron micrographs. A, Patient AC70.II.1. A thinly remyelinated axon is surrounded by several layers of basal membranes (arrow) that occasionally contain remnants of Schwann cell cytoplasm (9,000×). B, Patient CMT-133.IV.2. Thin Schwann cell processes (arrow) connect isolated unmyelinated axons and show supernumerary extensions (10,000×).
Discussion
In this study, we refined the CMT4C region on chromosome 5q32 to 1.7 Mb, within suggestive borders, and identified KIAA1985 as the CMT4C gene. In 12 families, we found eight different protein-truncating mutations and three missense mutations affecting evolutionary-conserved amino acid residues. Affected individuals from eight families were homozygous or compound heterozygous for truncating mutations. Segregation analyses of all KIAA1985 mutations were consistent with autosomal recessive inheritance. The absence of mutations in 10 other families compatible with linkage to CMT4C is not unexpected, since linkage in small families can be fortuitous in the setting of genetic heterogeneity. Otherwise, some of these families may represent CMT4C-affected families with KIAA1985 mutations undetectable in the present study (e.g., large genomic alterations or mutations in regulatory sequences). The novel CMT4C gene is expressed in neural tissues, including peripheral nerve, which is consistent with the tissue involved in a demyelinating peripheral neuropathy.
The clinical, electrophysiological, and pathological phenotype of patients with CMT4C was described elsewhere in detail (Kessali et al. 1997; Gabreëls-Festen et al. 1999). With the identification of disease-causing mutations in the CMT4C gene, we have confirmed that patients with CMT4C can have an early-onset scoliosis, relatively well-preserved NCVs, and increased basal lamina production and formation of abnormal Schwann cell processes in their peripheral nerves. However, neuropathic symptoms were not confined to relatively mild disease. The phenotypic spectrum also included infantile neuropathy, with early loss of ambulation and respiratory problems. At present, the limited sample size leaves open any conclusions regarding genotype-phenotype correlations. We found CMT4C to occur in CMT cohorts of various ethnic origins. Together with previous studies (LeGuern et al. 1996; Gabreëls-Festen et al. 1999; Guilbot et al. 1999), our findings indicate that CMT4C represents a comparatively frequent cause of autosomal recessive CMT.
We encountered two aspects that pose challenges to prediction of the consequences of KIAA1985 mutations. First, in one Turkish control individual, we have identified a heterozygous R529Q mutation, the same mutation as in families CMT-133 and M149. The identical rare haplotype found on all R529Q chromosomes supports a common origin of this allele; thus, it is likely that the recurrence of the R529Q mutation is due to a sampling bias of control DNAs arising from linkage disequilibrium or founder effect. Alternatively, R529Q may be a nonpathogenic variant, and an undetected true mutation may reside on the R529Q allele of families CMT-133 and M149. Finally, with regard to genetic heterogeneity in autosomal recessive CMT, linkage data in small families need to be interpreted with caution. Families CMT-133 and M149 may carry a mutation elsewhere in the genome and may share a common 5q32 haplotype by chance. Nevertheless, homozygous and compound heterozygous truncating mutations identified in this study strongly support the notion that KIAA1985 is the CMT4C gene.
Second, we discovered a set of KIAA1985-splicing products that are predicted to have outcomes similar to those of truncating mutations described for the patients’ DNAs. Although some of these transcripts may represent messages that are incompletely spliced or that are not translated into protein, polypeptides lacking major portions of the protein may exist in normal tissues. We suggest two potential explanations: First, all mutations in the CMT4C gene are predicted to alter the sequence of the longest ORF. This may suggest that a critical amount of the full-length protein is necessary for retaining a normal phenotype. It is notable that the highest levels of the transcript with the longest ORF were detected in mRNA from peripheral nerve that is primarily targeted by KIAA1985 mutations. Second, alternative splicing may serve to maintain a critical functional, spatial, or temporal balance between the different protein products. Studies on mouse tissues also showed the presence of alternative transcripts with truncated ORFs, indicating a functional role for this property. A similar splicing diversity, with abundant generation of predicted shorter isoforms, has been described elsewhere in the regulation of genes controlling cell death (Jiang and Wu 1999; Bingle et al. 2000; Horiuchi et al. 2000). Appearance of Schwann cell death with apoptotic morphology has been recently noted in the biopsy of a case of infantile hereditary neuropathy, with evidence of abundant basal lamina onion bulb formation (Fidzianska et al. 2002).
The transcript with the longest ORF is predicted to encode a novel protein of unknown function containing SH3 and TPR motifs, arrayed in a fashion not observed previously in a protein. SH3 domains—found in a great variety of membrane-associated, intracellular, and even extracellular proteins—mediate assembly of protein complexes via binding to proline-rich peptides (Pawson 1995; Stoll et al. 2001; Pfam database [accession number PF00018]). The TPR motif similarly represents a protein-protein interaction module found in many functionally diverse proteins (Blatch and Lässle 1999; Pfam database [accession number PF00515]). Common features in the interaction partners have not been defined, but, in some instances, TPR proteins seem to aggregate to multiprotein complexes. It is conceivable that the KIAA1985 protein acts as an adapter or docking molecule. Therefore, a search for proteins complexed with KIAA1985 might be an effective strategy to shed light on KIAA1985 function and its role in normal and diseased peripheral nerves. BLAST searches on expressed sequences and genome assemblies revealed conserved orthologues in various vertebrates from cattle to zebrafish. In addition, we discovered a second human protein, FLJ20356, with a very similar domain composition. This suggests that the KIAA1985 and FLJ20356 genes and their orthologues in other species belong to a multigene family that encodes putative docking proteins.
In summary, we have identified the gene responsible for CMT4C, a recessive CMT neuropathy linked to chromosome 5q32. The translated protein belongs to a so-far-unrecognized family of putative adapter proteins. Functional studies on the CMT4C protein may add to the knowledge about the pathomechanisms in peripheral neuropathies. In the meantime, the results of our study will aid genetic testing and counseling in families with recessive and sporadic CMTs.
Acknowledgments
We are indebted to the members of the families with CMT for their participation in this study. J.S. was supported by the START program of the medical faculty of Aachen University of Technology. N.V. is a Ph.D. student supported by the Institute for Science and Technology, Belgium. J.M.S. was supported by the Deutsche Forschungsgemeinschaft (DFG). B.R. received grants from the DFG and Deutsche Gesellschaft für Muskelkranke. E.N. is a postdoctoral fellow of the Fund for Scientific Research–Flanders (FWO-Vlaanderen). This research project was supported, in part, by the FWO-Vlaanderen, the Special Research Fund of the University of Antwerp, the Medical Foundation Queen Elisabeth, the Association Belge contre les Maladies Neuro-Musculaires, and the Federal Office for Scientific, Technical and Cultural Affairs, Belgium.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Applied Biosystems SNP genotyping repository, http://myscience.appliedbiosystems.com/navigation/mysciLoginTC.jsp/ (for the publicly accessible section)
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/
- Ensembl Genome Browser, http://www.ensembl.org/Multi/blastview?species=danio_rerio/ and http://www.ensembl.org/Multi/blastview?species=Fugu_rubripes/ (for BLAST searches on D. rerio and F. rubripes genomes, respectively)
- European Bioinformatics Institute Web site, http://www.ebi.ac.uk/index.html (for ClustalW server and Jalview)
- FGENESH, http://www.softberry.com/berry.phtml?topic=fgenesh&group=programs&subgroup=gfind (for ab initio gene prediction)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for human genomic contig [accession number NT_006859], SP329/FLJ13962 [accession numbers AF177339 and AK024024], KIAA1985 [accession numbers AB075865, BQ421935, BQ213149, W86370, BM552239, BE026883, BE302610, and AY341075], KIAA0843 [accession number AB020650], FLJ23713 [accession number AK074293], CSNK1A1 [accession number NM_001892], Bt.KIAA1985 [accession numbers BI680206, BF652287, BU239452, AW656867, and BI976766], Ss.KIAA1985 [accession numbers BF442400, BI186309, BI183894, and BI345340], Mm.KIAA1985/D430044G18Rik [accession numbers AK052534, BC037000, BI414954, and NM_172628], Rn.KIAA1985 [accession numbers BF567582 and XM_225887], Gg.KIAA1985 [accession numbers BU239452 and BU355692], FLJ20356 [accession number AK000363], Mm.FLJ20356 [accession numbers AK028482 and BC024909], Rn.FLJ20356 [accession numbers XM_223527 and XM_223528], Gg.FLJ20356 [accession numbers BU232586, BU471861, and BU474979], and Xl.FLJ20356 [accession number BJ062830])
- Genome Database, http://www.gdb.org/
- Genscan, http://genes.mit.edu/GENSCAN.html (for ab initio gene prediction)
- HUGE Protein Database, http://www.kazusa.or.jp/huge/ (for KIAA1985 and KIAA0843)
- Inherited Peripheral Neuropathies Mutation Database, http://molgen-www.uia.ac.be/CMTMutations/
- NCBI Entrez SNP, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=snp
- NCBI Expressed Sequence Tags Database, http://www.ncbi.nlm.nih.gov/dbEST/index.html (for the dbSNP database)
- NCBI RefSeq project, http://www.ncbi.nih.gov/RefSeq/
- NCBI SAGEmap, http://www.ncbi.nlm.nih.gov/SAGE/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CMT1B, CMT2A, CMT4A, CMT2B1, CMT2B2, and CMT4C)
- Pfam, http://pfam.wustl.edu/index.html (for SH3 domain [accession number PF00018] and TPR motif [accession number PF00515])
- Polyadq algorithm, http://argon.cshl.org/tabaska/polyadq_form.html
- Primer3, http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi
- RepeatMasker program; http://ftp.genome.washington.edu/cgi-bin/RepeatMasker
- ScanProsite, http://us.expasy.org/tools/scanprosite/
- SMART, http://smart.embl-heidelberg.de
- Swiss-Prot, http://www.expasy.org/sprot/
- Unigene, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=unigene
- UCSC Genome Bioinformatics, http://genome.ucsc.edu (for public genome assembly and BLAT alignments)
References
- Azzedine H, Bolino A, Taïeb T, Birouk N, Di Duca M, Bouhouche A, Benamou S, Mrabet A, Hammadouche T, Chkili T, Gouider R, Ravazzolo R, Brice A, Laporte J, LeGuern E (2003) Mutations in MTMR13, a new pseudophosphatase homologue of MTMR2 and Sbf1, in two families with an autosomal recessive demyelinating form of Charcot-Marie-Tooth disease associated with early-onset glaucoma. Am J Hum Genet 72:1141–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter RV, Ben Othmane K, Rochelle JM, Stajich JE, Hulette C, Dew-Knight S, Hentati F, Ben Hamida M, Bel S, Stenger JE, Gilbert JR, Pericak-Vance MA, Vance JM (2002) Ganglioside-induced differentiation-associated protein-1 is mutant in Charcot-Marie-Tooth disease type 4A/8q21. Nat Genet 30:21–22 [DOI] [PubMed] [Google Scholar]
- Bingle CD, Craig RW, Swales BM, Singleton V, Zhou P, Whyte MK (2000) Exon skipping in Mcl-1 results in a bcl-2 homology domain 3 only gene product that promotes cell death. J Biol Chem 275:22136–22146 [DOI] [PubMed] [Google Scholar]
- Blatch GL, Lässle M (1999) The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21:932–939 [DOI] [PubMed] [Google Scholar]
- Boerkoel CF, Takashima H, Stankiewicz P, Garcia CA, Leber SM, Rhee-Morris L, Lupski JR (2001) Periaxin mutations cause recessive Dejerine-Sottas neuropathy. Am J Hum Genet 68:325–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolino A, Muglia M, Conforti FL, LeGuern E, Salih MAM, Georgiou DM, Christodoulou K, Hausmanowa-Petrusewicz I, Mandich P, Schenone A, Gambardella A, Bono F, Quattrone A, Devoto M, Monaco AP (2000) Charcot-Marie-Tooth type 4B is caused by mutations in the gene encoding myotubularin-related protein-2. Nat Genet 25:17–19 [DOI] [PubMed] [Google Scholar]
- Cuesta A, Pedrola L, Sevilla T, Garcia-Planells J, Chumillas MJ, Mayordomo F, LeGuern E, Marin I, Vilchez JJ, Palau F (2002) The gene encoding ganglioside-induced differentiation-associated protein 1 is mutated in axonal Charcot-Marie-Tooth type 4A disease. Nat Genet 30:22–25 [DOI] [PubMed] [Google Scholar]
- De Sandre-Giovannoli A, Chaouch M, Kozlov S, Vallat J-M, Tazir M, Kassouri N, Szepetowski P, Hammadouche T, Vandenberghe A, Stewart CL, Grid D, Lévy N (2002) Homozygous defects in LMNA, encoding lamin A/C nuclear-envelope proteins, cause autosomal recessive axonal neuropathy in human (Charcot-Marie-Tooth disorder type 2) and mouse. Am J Hum Genet 70:726–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyck PJ, Chance P, Lebo R, Carney JA (1993) Hereditary motor and sensory neuropathies. In: Dyck PJ, Thomas PK, Griffin JW, Low PA, Poduslo JF (eds) Peripheral neuropathy. W. B. Saunders, Philadelphia, pp 1094–1136 [Google Scholar]
- European CMT-Consortium–ENMC (1999) 4th workshop of the European CMT-consortium–62nd ENMC international workshop: rare forms of Charcot-Marie-Tooth disease and related disorders 16–18 October 1998, Soestduinen, The Netherlands. Neuromuscul Disord 9:279–287 [DOI] [PubMed] [Google Scholar]
- Fidzianska A, Drac H, Rafalowska J (2002) Phenomenon of Schwann cell apoptosis in a case of congenital hypomyelinating neuropathy with basal lamina onion bulb formation. Brain Dev 24:727–731 [DOI] [PubMed] [Google Scholar]
- Gabreëls-Festen A, van Beersum S, Eshuis L, LeGuern E, Gabreëls F, van Engelen B, Mariman E (1999) Study on the gene and phenotypic characterisation of autosomal recessive demyelinating motor and sensory neuropathy (Charcot-Marie-Tooth disease) with a gene locus on chromosome 5q23-q33. J Neurol Neurosurg Psychiatry 66:569–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilbot A, Ravise N, Bouhouche A, Coullin P, Birouk N, Maisonobe T, Kuntzer T, Vial C, Grid D, Brice A, LeGuern E (1999) Genetic, cytogenetic and physical refinement of the autosomal recessive CMT linked to 5q31-q33. Eur J Hum Genet 7:849–859 [DOI] [PubMed] [Google Scholar]
- Guilbot A, Verny C, Bachelin C, Brice A, Van Evercooren A, LeGuern E (2001a) Progress in the identification of the gene responsible for an autosomal recessive form of demyelinating Charcot-Marie-Tooth disease in 5q32. Acta Myol 20:21–24 [Google Scholar]
- Guilbot A, Williams A, Ravise N, Verny C, Brice A, Sherman DL, Brophy PJ, LeGuern E, Delague V, Bareil C, Megarbane A, Claustres M (2001b) A mutation in periaxin is responsible for CMT4F, an autosomal recessive form of Charcot-Marie-Tooth disease. Hum Mol Genet 10:415–421 [DOI] [PubMed] [Google Scholar]
- Harding AE, Thomas PK (1980) The clinical features of hereditary motor and sensory neuropathy types I and II. Brain 103:259–280 [DOI] [PubMed] [Google Scholar]
- Horiuchi T, Himeji D, Tsukamoto H, Harashima S, Hashimura C, Hayashi K (2000) Dominant expression of a novel splice variant of caspase-8 in human peripheral blood lymphocytes. Biochem Biophys Res Commun 272:877–881 [DOI] [PubMed] [Google Scholar]
- Jiang ZH, Wu JY (1999) Alternative splicing and programmed cell death. Proc Soc Exp Biol Med 220:64–72 [DOI] [PubMed] [Google Scholar]
- Kalaydjieva L, Gresham D, Gooding R, Heather L, Baas F, de Jonge R, Blechschmidt K, Angelicheva D, Chandler D, Worsley P, Rosenthal A, King RHM, Thomas PK (2000) N-myc downstream-regulated gene 1 is mutated in hereditary motor and sensory neuropathy–Lom. Am J Hum Genet 67:47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessali M, Zemmouri R, Guilbot A, Maisonobe T, Brice A, LeGuern E, Grid D (1997) A clinical, electrophysiologic, neuropathologic, and genetic study of two large Algerian families with an autosomal recessive demyelinating form of Charcot-Marie-Tooth disease. Neurology 48:867–873 [DOI] [PubMed] [Google Scholar]
- Kozak M (1989) Context effects and inefficient initiation at non-AUG codons in eucaryotic cell-free translation systems. Mol Cell Biol 9:5073–5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeGuern E, Guilbot A, Kessali M, Ravise N, Tassin J, Maisonobe T, Grid D, Brice A (1996) Homozygosity mapping of an autosomal recessive form of demyelinating Charcot-Marie-Tooth disease to chromosome 5q23-q33. Hum Mol Genet 5:1685–1688 [DOI] [PubMed] [Google Scholar]
- Nelis E, Irobi J, De Vriendt E, Van Gerwen V, Perez Novo C, Topaloglu H, Auer-Grumbach M, Merlini L, Villanova M, Jordanova A, De Jonghe P, Timmerman V (2001) Homozygosity mapping of families with recessive Charcot-Marie-Tooth neuropathies. Acta Myol 20:39–42 [Google Scholar]
- Pawson T (1995) Protein modules and signalling networks. Nature 373:573–580 [DOI] [PubMed] [Google Scholar]
- Senderek J, Bergmann C, Weber S, Ketelsen UP, Schorle H, Rudnik-Schoneborn S, Buttner R, Buchheim E, Zerres K (2003) Mutation of the SBF2 gene, encoding a novel member of the myotubularin family, in Charcot-Marie-Tooth neuropathy type 4B2/11p15. Hum Mol Genet 12:349–356 [DOI] [PubMed] [Google Scholar]
- Skre H (1974) Genetic and clinical aspects of Charcot-Marie-Tooth's disease. Clin Genet 6:98–118 [DOI] [PubMed] [Google Scholar]
- Stoll R, Renner C, Zweckstetter M, Bruggert M, Ambrosius D, Palme S, Engh RA, Golob M, Breibach I, Buettner R, Voelter W, Holak TA, Bosserhoff AK (2001) The extracellular human melanoma inhibitory activity (MIA) protein adopts an SH3 domain-like fold. EMBO J 20:340–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PK (2000) Autosomal recessive hereditary motor and sensory neuropathy. Curr Opin Neurol 13:565–568 [DOI] [PubMed] [Google Scholar]
- Verheijen MH, Chrast R, Burrola P, Lemke G (2003) Local regulation of fat metabolism in peripheral nerves. Genes Dev 17:2450–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner LE, Mancias P, Butler IJ, McDonald CM, Keppen L, Koob KG, Lupski JR (1998) Mutations in the early growth response 2 (EGR2) gene are associated with hereditary myelinopathies. Nat Genet 18:382–384 [DOI] [PubMed] [Google Scholar]