Abstract
Nance-Horan syndrome (NHS) is an X-linked disorder characterized by congenital cataracts, dental anomalies, dysmorphic features, and, in some cases, mental retardation. NHS has been mapped to a 1.3-Mb interval on Xp22.13. We have confirmed the same localization in the original, extended Australian family with NHS and have identified protein-truncating mutations in a novel gene, which we have called “NHS,” in five families. The NHS gene encompasses ∼650 kb of genomic DNA, coding for a 1,630–amino acid putative nuclear protein. NHS orthologs were found in other vertebrates, but no sequence similarity to known genes was identified. The murine developmental expression profile of the NHS gene was studied using in situ hybridization and a mouse line containing a lacZ reporter-gene insertion in the Nhs locus. We found a complex pattern of temporally and spatially regulated expression, which, together with the pleiotropic features of NHS, suggests that this gene has key functions in the regulation of eye, tooth, brain, and craniofacial development.
Introduction
Nance-Horan syndrome (NHS [MIM 302350]) was first described independently in 1974, in Australia (Horan and Billson 1974) and the United States (Nance et al. 1974), as an X-linked syndrome involving congenital cataract and dental anomalies. Ophthalmological findings in affected males include bilateral severe congenital cataract involving the fetal nucleus and posterior Y suture with variable zonular extensions into the posterior cortex, usually leading to profound visual loss and requiring surgery. Microcornea, nystagmus, and microphthalmia have also been reported in some pedigrees (Lewis et al. 1990; Stambolian et al. 1990; Walpole et al. 1990). Dental abnormalities include screwdriver blade–shaped incisors, supernumerary maxillary incisors (mesiodens), and diastema (Walpole et al. 1990). Approximately 30% of affected males display mental retardation and behavioral disturbance (Walpole et al. 1990; Toutain et al. 1997a), and several families were reported to have lateral brachymetacarpalia (Lewis et al. 1990). Characteristic dysmorphic facial features, which may be subtle, include large anteverted pinnae; long, narrow facies; and prominent nose and nasal bridge (Lewis et al. 1990; Walpole et al. 1990). The disorder appears to be inherited in a codominant fashion, with heterozygous females often manifesting similar but less severe features than affected males, including posterior Y-sutural cataracts, with little or no loss of vision, and characteristic dental abnormalities (Walpole et al. 1990; Zhu et al. 1990).
NHS was mapped to Xp21.1-p22.3 (Lewis et al. 1990; Stambolian et al. 1990) and, more recently, was refined to a critical region of 1.3 Mb between STR markers DXS1195 and DXS999 (Toutain et al. 2002). We have independently confirmed the same minimal genetic interval by detailed analysis of the original Australian family with NHS. A truncating mutation in a novel gene, which we have called “NHS” (accession number AY436752), was found in this pedigree, and, subsequently, different truncating mutations have been found in four other unrelated pedigrees with NHS. Here, we present detailed expression data, in human and mouse, confirming a developmental expression pattern consistent with a role in the pleiotropic features of NHS.
Subjects and Methods
Affected Individuals and Families Tested
Approval for this study was obtained from the Human Research Ethics Committees of the Royal Children’s Hospital, Melbourne, the Royal Victorian Eye and Ear Hospital, Melbourne, and the University of Tasmania, Hobart, and the study adhered to the tenets of the Declaration of Helsinki. Written informed consent was obtained from all participating individuals or their guardians. All available family members were examined by one or more ophthalmologists (J.E.C., I.R.-E., M.G.W., D.A.M., J.E.E., A.N., or M.C.). DNA was extracted from whole blood or buccal mucosa swabs through use of the PureGene DNA Isolation Kit (Gentra Systems).
Database Analysis
Nucleotide and protein database searches were performed, through the Web site of the National Center for Biotechnology Information, by BLAST algorithms BLASTN, BLASTX, and TBLASTX, against the nonredundant dbEST, high-throughput genomic sequences, and species-specific databases. The putative promoter was identified using the Neural Network Promoter Prediction tool (Reese 2001), and subcellular localization of the putative NHS protein has been predicted using PSORTII (PSORT II Prediction Web site). Zebrafish data have been accessed via the Danio rerio Sequencing Project Web page.
Genotyping and Linkage Analysis
All individuals were genotyped at seven microsatellite loci within a 10-cM region on Xp22.31-p22.13 previously reported as linked to NHS (tel-DXS1224-DXS1053-DXS1195-DXS418-DXS999-DXS365-DXS989-cen) (Toutain et al. 1997b). An additional four microsatellite markers—labeled as markers 1, 2, 3, and 4 in our data—were extracted from the draft genome sequence and were genotyped in family 1 (primers available on request). Two-point linkage analysis was performed with MLINK, part of the FASTLINK package (Cottingham et al. 1993), and multipoint and haplotype analyses were performed with X-linked GeneHunter Plus (Kong and Cox 1997).
Mutation Analysis
Coding regions of all characterized genes in the critical region (SCML1, RAI2, SCML2, STK9, RS1, and PPEF1) were directly sequenced using an ABI PRISM 310 Genetic Analyzer (Applied Biosystems), after PCR amplification and purification with the Ultraclean PCR cleanup kit (MoBio). The NHS candidate gene was screened using intronic primers for the amplification of all identified exons (primer sequences available on request). Where possible, we confirmed familial segregation of the NHS mutation. We tested 200 control chromosomes for each mutation by restriction enzyme analysis or by sequencing of both strands of the PCR product.
Northern Blot Analysis
Northern blot analysis was performed on Human Multiple Tissue Northern filters (human 12-lane MTN blot Clontech and human Fetal II MTN blot Clontech), by following the manufacturer’s protocol. The blots were hybridized with three independent cDNA probes derived from exon 6, exons 6–8, and the 3′ UTR of the NHS gene and were radiolabeled with α[32P]-dATP by random priming (details available on request). The mouse northern blot (Seegene) contained 20 μg of total RNA per lane, from mouse brain at various stages of development. The probe was generated from the 3′ end of the mouse Nhs gene. Hybridization was performed in ExpressHyb solution (Clontech) at 68°C, as per the manufacturer’s instructions.
RT-PCR
RT-PCR was performed on oligo-dT–primed first-strand cDNA from human adult and fetal brain, lens, retina, retinal pigment epithelium, placenta, lymphocytes, and fibroblasts. A 206-bp fragment was amplified with the forward primer RT-6 (5′-GAGACCCAAGGAAATGTGGA-3′) and the reverse primer RT-8 (5′-ATGTCCCCGGAATCTTTTCT-3′), designed to amplify a region of cDNA corresponding to the region from the end of exon 6 to the beginning of exon 8. PCR was performed using Hot Star Taq (Qiagen) at 95°C for 15 min, followed by 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s. Amplification was performed on both RT+ and RT− templates.
Mouse In Situ Hybridization
Nonradioactive mouse embryo in situ hybridization was performed as described elsewhere (Dunwoodie et al. 1997). Mouse embryonic and neonatal sections were hybridized with the sense and antisense RNA probes generated from regions of Nhs in exon 1 (genomic sequences AL672082 and AC097354, Nhs5 probe; primers forward [5′-GCCTTTCGCCAAGCGGATCG-3′] and reverse [5′-GCCTCCTGCTTGGGGTCCAAC-3′]; amplicon of 549 bp) and exon 6 (genomic sequences AL732391 and AC093447, Nhs4 probe; primers Fw [5′-CTCAGCTAGCAGCAATCTTCCAG-3′], and Rev [5′-CAATGAAGTCTCGTCCATACTTCC-3′]; amplicon of 511 bp). Radioactive in situ hybridization using [33P]-labeled probes 5′ (Nhs5) and 3′ (Nhs4) to the lacZ insertion site was performed as described elsewhere (Thomas et al. 2000).
Generation and Analysis of a Mouse Nhs lacZ Insertion Mutant
The mouse line 414, carrying a lacZ reporter gene in the Nhs locus, was generated in a gene-trap screen (Voss et al. 1998) through use of the splice-acceptor gene-trap construct pGT1.8geo (Skarnes et al. 1995). The insertion event occurred in the first intron. The Nhs exon 1 upstream of the gene trap insertion was identified by 5′ RACE (Voss et al. 1998) and was confirmed by northern analysis (data not shown). The gene trap construct contained a lacZ reporter gene, enabling the study of Nhs promoter activity. β-galactosidase staining was performed as described elsewhere (Voss et al. 1998).
Results
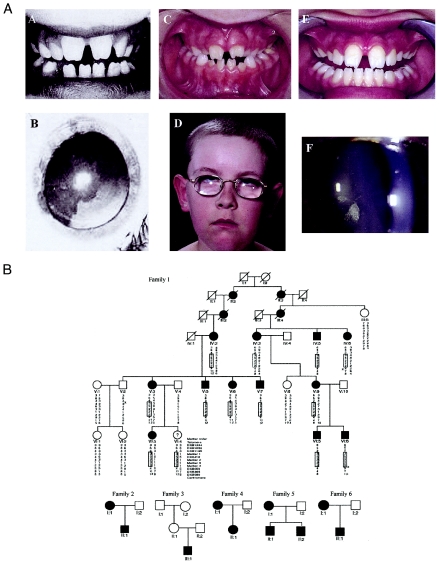
We investigated the original Australian kindred with NHS (Horan and Billson 1974) and a further five smaller pedigrees with NHS. Clinical features from a range of affected individuals in the present study are shown in figure 1A. A further branch of the original kindred with NHS was discovered, enabling the study of a six-generation family with 12 living affected individuals (family 1 in fig. 1B). Two of the affected males have severe intellectual impairment. We confirmed the linkage to Xp22.13 through the genotyping of 11 microsatellite markers in the region (including 4 novel markers) and through use of the MLINK and X-Linked GeneHunter Plus programs. Recombination events refined the critical region to a 1.3-Mb interval between markers DXS1195 and DXS999 (figs. 1B and 2A), in agreement with another recent study (Toutain et al. 2002). All exons of the six characterized genes in the region (SCML1, SCML2, RAI2, RS1, STK9, and PPEF1) were directly sequenced in three affected individuals and in one unaffected individual. No segregating mutations were detected.
Figure 1.
Families studied. A, Clinical features in individuals with NHS from the current study. Family 1, V-5: dentition (A.A), and cataract (A.B); family 5, II-2: dentition (A.C), and facial features (A.D); family 4, II-1: dentition (A.E); and family 4, I-1: cataract (A.F). Dental anomalies in the cases illustrated are diastema and notched incisors, showing striking similarity in three unrelated individuals. B, Pedigrees of the families with NHS. Xp22.13 haplotypes are shown for family 1, with the common haplotype boxed and recombinations indicated with an “x.” Blackened boxes denote affected males, blackened circles denote females with confirmed features of NHS, and unblackened boxes and circles denote unaffected males and females, respectively. ? = not available for examination.
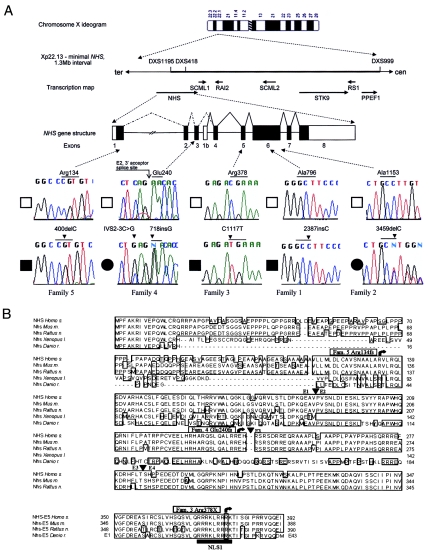
We investigated a novel candidate gene, LOC90334, predicted by Genescan (Burge and Karlin 1997) within the NHS minimal genetic interval and supported by clustered ESTs (NCBI UniGene cluster numbers Hs. 444940, Hs. 282164, and Hs. 21470) from multiple tissues, including fetal eye, brain, kidney, and colon. These UniGene clusters, as well as ESTs AL926872 (Danio rerio) and BF776631 (Bos taurus), suggested that the gene contained additional upstream exon(s). Characterization of this novel gene has established that it consists of nine exons transcribed as two mRNA isoforms, A (8.7 kb encoding a putative 1,630–amino acid protein; the major isoform) and B (7.7 kb encoding a putative N-terminus truncated 1,335–amino acid protein, with translation beginning in exon 4; the minor isoform) (fig. 2A). Exon 1 was detected in genomic sequence AL845433, ∼350 kb upstream of exon 2, through use of the EST AL926872 (D. rerio) and the TBLASTX algorithm. Exons 2 and 3 (genomic sequence Z93242) of isoform A were identified from EST BF776631 (B. taurus). Exons 1–3 were confirmed by RT-PCR from human brain RNA. The 5′ end of isoform B (exon 1b in fig. 2A) was identified by 5′ RACE. Exons 4–8 were identified by a combination of Genescan, clustered ESTs (Hs. 444940, Hs. 282164, and Hs. 21470), and RT-PCR. Intron-exon boundaries are given in table 1.
Figure 2.
Localization and characterization of the NHS gene. A, Schematic map of the Xp22.13 region, indicating positions of flanking markers and positions of candidate genes excluded, together with the structure of the NHS gene. Exon 1b defines a new putative NHS transcription start site. Note that markers DXS1195 and DXS418 are intragenic, residing in introns 1 and 5 of the NHS gene, respectively. Blackened segments indicate the ORF. Sequence chromatograms and positions of five truncation mutations identified in families 1–5 are shown. Blackened boxes next to chromatograms indicate affected males, blackened circles indicate affected females, and unblackened boxes indicate unaffected controls. B, Partial ClustalW multiple-protein alignment of NHS orthologs. Protein sequence encoded by exons 1–4 (upper panel) and exon 5 (lower panel) are shown. Mutations are indicated by arrows, and the nuclear localization sequence NLS1 is underlined. Conserved amino acid residues are boxed. Family numbers correspond to those in fig. 1B. Exon/exon boundaries are indicated with arrowheads and the corresponding exon numbers. The mouse protein sequence was based on mouse genomic sequences (AL672082, AC097354, AL732391, and AC093447) and mouse partial mRNAs similar to Nhs (XM_142285 and XM_112126).
Table 1.
Intron/Exon Boundaries of the NHS Gene[Note]
| Exon | 3′ Acceptor Splice Site | 5′ Donor Splice Site | Exon Size(bp) |
| 1 | GAGGCAGTGCgtgagtaccc |
903 | |
| 2 | tgtcttgcagCCGTCTCCAA |
CTGCGCAGAGgtgacagatc |
153 |
| 3 | tccttctcagAACACCGGAG |
CTTTTTAACGgtaagtttgg |
134 |
| 1b | CTCTGACAAGgtaagtaaat |
138 | |
| 4 | ttgtttgcagTCCCATCCCC |
AATGTTACTGgtatcgttct |
193 |
| 5 | tgggttgcagGAGTTGGCTT |
CAAGAAATAGgtgtgatatc |
132 |
| 6 | tgtgccctagATTCTGATGA |
TCATTGAAAGgtcagtcact |
2,982 |
| 7 | ttattttaagAATCATCACC |
TCATTCACAGgtgaggcaac |
127 |
| 8 | tttctcaaagATCCAAGAGG |
GCAAATAAACGTGACTGCA | 4,134 |
Note.— Exons are indicated in capital letters, and introns are indicated in lowercase letters. Consensus ag/gt intron splice sites are underlined.
Oligonucleotide primers were designed to PCR amplify and directly sequence the nine exons of the gene, along with the flanking intron/exon boundaries and the 5′ and 3′ UTRs. A single-nucleotide insertion mutation, 2387insC, was identified in exon 6 and segregated with the NHS phenotype in family 1 (figs. 1B and 2A). This mutation causes a frameshift in the reading frame, Ala796fs, and is predicted to result in a premature stop codon following the addition of 35 amino acids. A further five families (fig. 1B) with typical NHS were recruited, and truncation mutations were also detected in four of these families. A single-nucleotide deletion, 3459delC (Ala1153fs), was found in a second Australian family with NHS (Walpole et al. 1990) (figs. 1B and 2A). It is interesting that a de novo nonsense mutation, 1117C→T (Arg378X), was identified in an isolated male infant with NHS (figs. 1B and 2A). The index case of family 4 (fig. 1B) displayed an unusually severe ocular phenotype for a female with NHS. Two sequence variations were identified: 718insG (Glu240fs) at the first base of exon 3, leading to a frameshift predicted to result in a premature stop codon following the addition of 16 amino acids; and a base substitution, IVS2−3C→G, at the 3′ acceptor splice site (fig. 2A). The significance of the latter change is uncertain, but it was not detected in 200 control chromosomes analyzed. The affected mother of the proband displays the same genotype. A single-base deletion, 400delC (Arg134fs), was identified in family 5 (figs. 1B and 2A). It is notable that this mutation is telomeric with regard to (and de facto, outside of) the minimal critical region as determined in family 1 (the same critical interval was also determined by an independent group, Toutain et al. [2002]). The marker DXS1195 lies within the large, ∼350-kb intron 1 of the NHS gene (fig. 2A). All five mutations detected to date would result in a substantial C-terminal truncation of the predicted NHS protein and were not detected in 200 control chromosomes. No coding region or splice-site nucleotide changes were detected in family 6.
Database searches revealed NHS orthologs in other vertebrate species, including mouse, rat, cow, chicken, zebrafish, and frog. Protein similarity between the NHS orthologs is relatively high, with the human protein 76%, 76%, 50%, and 46.5% identical to the mouse, rat, zebrafish (partial), and frog (partial) proteins, respectively (fig. 2B). The NHS protein shows no significant similarity to any known protein or class of proteins. Four conserved putative monopartite nuclear localization signals (Robbins et al. 1991) were detected at amino acid positions 371–379 (RRRKLRRRK), 438–444 (PSRRRIR), 822–825 (RKPK), and 1026–1034 (PGGSKRKPK), suggesting a functional role in the cell nucleus (fig. 2B and results not shown).
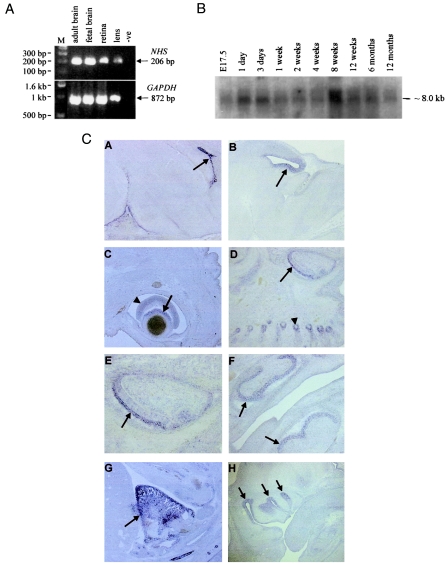
To study the expression of the NHS gene, human adult and fetal multiple-tissue northern blot filters (Clontech) were hybridized with three independent NHS probes. Two major transcripts, of ∼8.7 kb and ∼7.7 kb, were detected at low levels in all tissues analyzed (results not shown). RT-PCR analysis of RNA from human adult and fetal brain, lens, retina, retinal pigment epithelium, placenta, lymphocytes, and fibroblasts revealed the presence of NHS transcript in all tissues, although the transcript levels in retinal pigment epithelium, placenta, lymphocytes, and fibroblasts were very low (fig. 3A and data not shown). To assess the developmental expression pattern of the Nhs gene, a mouse northern blot (Seegene) containing brain RNA from different developmental stages was probed. As shown in figure 3B, the mouse Nhs gene is broadly expressed throughout brain development and in the adult mouse brain, from E17.5 until age 12 mo, although the transcript levels vary. In situ hybridization of mouse embryo sections showed that Nhs expression was developmentally regulated in a range of embryonic tissues (fig. 3C). The Nhs5 probe, generated from exon 1 (see the “Subjects and Methods” section), detected expression in the midbrain, the lens and retina, the tooth primordia, the olfactory epithelium, and the whisker follicles. The Nhs4 probe, generated from exon 6, detected expression in all these domains, as well as in the choroid plexus and the heart (fig. 3C).
Figure 3.
Expression analysis of the NHS gene. A, RT-PCR from human adult brain, fetal brain, retina, and lens tissues. RNA was reverse transcribed and amplified using primers from exons 6 and 8. RT-minus controls were negative for each tissue. B, Mouse northern blot (Seegene), containing 20 μg of total RNA per lane from mouse brain at various developmental stages. The probe was generated from the 3′ end of the mouse Nhs gene. C, Mouse Nhs is developmentally regulated and is expressed in the brain, eye, and tooth primordia. C.A, Midsagittal section from E16.5 mouse embryo, showing Nhs expression in the choroid plexus of the fourth ventricle. C.B, Parasagittal section of E14.5 mouse embryo, showing Nhs expression in the midbrain. C.C, Sagittal section from E16.5 mouse embryo, showing expression in lens cell bodies (arrow) and retina (arrowhead). C.D, Sagittal section from E16.5 mouse embryo, showing Nhs expression in the primordium of upper right incisor tooth (arrow) and the whisker follicles (arrowhead). C.E, Detail of panel D, showing Nhs expression in the primordium of upper right incisor tooth (arrow). C.F, Parasagittal section from E16.5 mouse embryo, showing Nhs expression in the primordia of the upper and lower right molar teeth (arrows). C.G, Midsagittal section from E14.5 mouse embryo, showing Nhs expression in the right ventricle of the heart (arrow). C.H, Parasagittal section from E14.5 mouse embryo, showing NHS expression in olfactory epithelium (arrows). The Nhs4 probe was used in panels A and G, and the Nhs5 probe was used in panels B, C, D, E, F, and H.
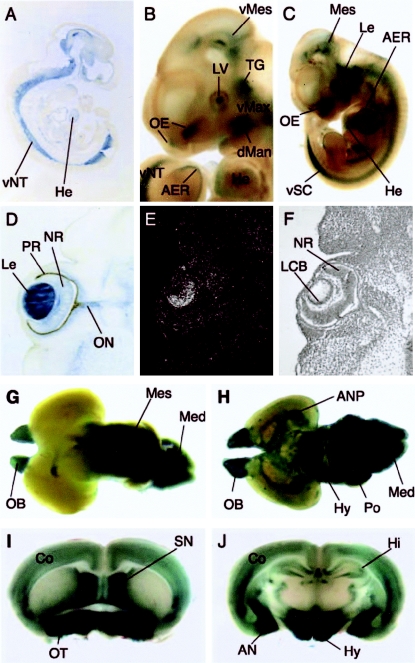
To further study the expression profile of Nhs throughout development, we studied a mouse line containing a lacZ reporter-gene cassette inserted in the Nhs locus. The gene-trap insertion in intron 1 did not prevent transcription and normal splicing of Nhs exons 3′ to the insertion site. Therefore, this insertion event did not completely disrupt the synthesis of a full-length Nhs mRNA (data not shown). The Nhs gene (as traced by lacZ expression) showed a complex pattern of expression. It was first visible in the ventral neural tube at E9.5 (not shown). At E10.5, the expression in the ventral neural tube became stronger, and additional domains of expression—including the heart—appeared (fig. 4A). At E11.5 and E12.5, expression was seen in the olfactory epithelium, in the ventral aspects of the maxillary process, and in the dorsal aspects of the mandibular process (fig. 4B and 4C). Reporter-gene expression was most striking in the lens tissue, in which cataract is a hallmark of the NHS phenotype. Expression in the lens vesicle was first visible at E11.5 (fig. 4B), became stronger at E12.5 (fig. 4C), and persisted until at least E16.5 (fig. 3C.C). β-galactosidase protein was seen throughout the primary lens fibers (fig. 4D). Reporter-gene activity was verified as a true reflection of the endogenous gene activity by parallel in situ hybridization using probes 5′ (Nhs5) and 3′ (Nhs4) of the reporter-gene insertion at one embryonic, one fetal, and one postnatal stage. The reporter-gene expression was found to associate with the lens fibers; however, the endogenous Nhs mRNA is restricted to the cell bodies, as expected (fig. 4E and 4F). Sense probes did not show any signal above background (data not shown).
Figure 4.
Expression analysis of the Nhs locus, using the lacZ reporter gene. Shown is β-galactosidase staining of a parasagittal section of an E10.5 embryo (A), a whole-mount E11.5 embryo (B), a whole-mount E12.5 embryo (C), and a coronal section of an E12.5 eye (D); radioactive in situ hybridization of a coronal section of E12.5 eye using the Nhs5 probe (E) and bright field of E (F); β-galactosidase staining of whole-mount PN0 brain (dorsal aspect) (G), whole-mount PN0 brain (ventral aspect) (H), adult brain (slice at frontal level) (I), and adult brain (slice at parietal level) (J). AER = apical ectodermal ridge; AN = amygdaloid nuclei; ANP = amygdaloid nuclei primordium; Co = cortex; dMan = dorsal mandibular process; He = heart; Hi = hippocampus; Hy = hypothalamus; Le = lens; LCB = lens cell bodies; LV = lens vesicle; Med = medulla; Mes = mesencephalon; NR = neural retina; OB = olfactory bulb; OE = olfactory epithelium lining the olfactory pit; ON = optic nerve; OT = olfactory tubercle; Po = pons; PR = pigmented retina; SN = septal nuclei; TG = trigeminal ganglion; vMax = ventral maxillary process; vMes = ventral mesencephalon; vNT = ventral neural tube; vSC = ventral spinal cord.
An intricate pattern of Nhs promoter activity was seen in the early postnatal brain. Reporter-gene activity was detected in the ventral neural tube and in its derivatives from the level of the midbrain extending to the tail (fig. 4A, 4C, and 4H). Strong expression was found in the olfactory epithelium (fig. 4B and 4C) and in the olfactory bulbs and medulla (fig. 4G and 4H). Particularly striking were strong domains of reporter-gene activity in aspects of the limbic system of the adult brain. These included expression in the septum, as well as in the amygdala and the hypothalamus (fig. 4H, 4I, and 4J). Lower levels of expression were seen in the cortex and hippocampus (fig. 4J). Reporter-gene activity was also noted in the heart (fig. 4A, 4B, and 4C), the apical ectodermal ridge (fig. 4B and 4C), and the olfactory tubercle (fig. 4I).
Discussion
We used a positional cloning strategy to delineate a 1.3-Mb critical interval, between markers DXS1195 and DXS999, confining the gene for NHS, and we subsequently characterized a large novel gene, without significant homology to other genes, that resides predominantly within the critical interval. The disease interval was entirely determined from recombination events in one very large pedigree. Since a branch of this family formed the basis for one of the two original studies of NHS (Horan and Billson 1974), we felt confident that the disease gene would indeed be confined between these two markers. A recent study (Toutain et al. 2002) confirmed the same disease interval through use of recombinations in different families. It was interesting to note that, after characterizing the NHS candidate gene and finding mutations in four pedigrees, a fifth family with features very typical of NHS had no identified mutation. Further detailed analysis of the gene structure revealed the presence of another exon, ∼350 kb upstream. A truncating mutation was detected in this exon, which is telomeric to marker DXS1195 (present in intron 1). Consequently, the causative mutation in this family lies outside the minimal genetic interval as determined in family 1 and in the study by Toutain et al. (2002), thus illustrating the potential limitations of critical intervals for those involved in positional cloning studies.
The affected individuals from all families involved in the present study had features typical of NHS. The presence of a de novo mutation in a male infant with NHS indicates that mutations in the NHS gene may cause congenital cataract, despite the absence of a positive family history. Mutations were found in a total of five of six families with features typical of NHS. Despite detailed analysis of family 6, no mutation has been identified in the coding sequence, intron-exon boundaries, or the 5′ and 3′ UTR. This family may have an intronic or noncoding regulatory mutation that has not been identified as yet. Alternatively, there may be genetic heterogeneity in NHS.
It is noteworthy that all of the mutations described led to significant truncation of the carboxy terminus of the predicted protein. The mutations found in the families with NHS are located throughout the gene, and there was no obvious genotype/phenotype correlation between mutation position and severity of disease. It should also be noted that a wide variation in the degree of severity of ocular manifestations and mental retardation occurred within family 1, raising the possibility of genetic modifiers. The mutations identified in families 4 and 5, located in exons 3 and 1, respectively, indicate that loss of the major NHS isoform A is sufficient to cause disease. The phenotype in families 4 and 5 is similar to that in other families with NHS, indicating that there is no obvious correlation between the loss of only isoform A and severity of disease. Similarly, we found no obvious differences in the phenotype of individuals in whom mutations remove all four nuclear localization signals (families 4 and 5) and those in whom these sequences are not affected (family 2). Therefore, it is likely that the phenotype is caused, at least in part, by loss of function, either through mRNA degradation or protein truncation, rather than simply by loss of subcellular localization.
NHS is a large gene of unknown function that encodes a predicted 1,630–amino acid protein lacking sequence similarity to any other known protein or protein classes. The identification of nuclear localization signals, along with the high serine and proline content (24%) and the pleiotropic features of the NHS phenotype, support a regulatory role for the NHS protein in the development of ocular, craniofacial, and neural tissue. Conservation in other vertebrate species, such as mouse, rat, and zebrafish, supports an important role for NHS in development. We have performed detailed studies of the expression pattern of NHS, in fetal and adult derived tissue from human and mice. These indicated low levels expression in all tissues analyzed, including human adult lens tissue. The finding of different expression levels in the developing mouse brain at different developmental stages led us to further investigate the broader developmental expression profile in mice, through use of in situ hybridization studies. These studies suggested an intricate pattern of spatially and developmentally regulated expression, particularly as expected in tissues known to be affected in NHS, such as the lens, brain, craniofacial mesenchyme, and dental primordia.
To further study the developmental expression pattern, we analyzed a mouse line containing a lacZ reporter-gene cassette inserted in the Nhs locus. This enabled sensitive detection of expression driven by the Nhs promoter. Striking expression in the developing lens was demonstrated, consistent with a role in lens development. In the developing and early postnatal brain, the presence of relatively high levels of expression in some tissues—in particular, the olfactory bulbs, olfactory epithelium, and limbic system—was striking. These findings raise the possibility that NHS is of importance in development of the limbic system, which is of interest given the range of neuropsychological abnormalities reported in some males affected with NHS. These abnormalities include (in addition to the well documented incidence of mental retardation) autism, aggression, anxiety, stereotypical behavior, mood disturbance (e.g., unprovoked laughter), and sleep-wake cycle abnormalities (Toutain et al. 1997a). Our finding of Nhs expression in the heart is also of interest, since a family was recently reported as having X-linked cataract associated with cardiac anomalies mapping to this region of chromosome Xp22 (Francis et al. 2002). It has been postulated that isolated X-linked cataract and NHS may be allelic, because of the similarity in the cataract phenotype and the overlapping critical regions on the X chromosome (Francis et al. 2002). The mouse ortholog of NHS is a strong candidate for the Xcat mouse phenotype, which maps to the syntenic region of chromosome Xp (Favor and Pretsch 1990). The Xcat mouse displays only an ocular phenotype of bilateral total lens opacity in males, with varying severity of nuclear and cortical opacity in heterozygous females. Further analysis is also required to determine whether NHS has a role in isolated X-linked mental retardation, which is known to be highly heterogeneous and, at least in some cases, to map to Xp22 (Chelly and Mandel 2001).
Our findings and further studies to determine the function of NHS will advance understanding of lens development and cataract formation. The embryology of lens development is well characterized, and a number of different mechanisms of cataract formation are known, with the best studied involving structural proteins such as the crystallins (Litt et al. 1997). Mutations in genes encoding proteins involved in ion transport (Shiels and Bassnett 1996) are also known to cause congenital cataract. Disturbance of metabolic pathways in galactosemia are well characterized causes of lens dysfunction resulting in cataract (Stambolian et al. 1995). Aberrations in developmental pathways resulting in cataract are less well understood; however, more recently, mutations in genes such as PAX6, PITX3, and MAF have been described in which congenital cataract is at least one of the features. (Semina et al. 1998; Hanson et al. 1999; Jamieson et al. 2002).
In summary, we present evidence that mutations in a novel gene of as-yet-unknown function, which we have named “NHS,” cause the pleiotropic features of NHS. The lack of sequence similarity between the NHS protein and any other known protein or protein classes suggests that the NHS gene may form a novel component of the genetic program controlling ocular, craniofacial, and CNS development. This work will enable direct DNA diagnosis in families with blindness and mental retardation due to NHS, leading to improvements in genetic counseling and reproductive choices.
Acknowledgments
We thank the families with NHS, for their participation; Dr. J. E. Liebelt, for help with dysmorphology; Dr. J. Poulsen and Dr. J. MacKinnon, for help with patient ascertainment; M. Ring, for help with genealogy; A. Bell, M. Brown, R. Emeny, M. Staeger, V. Salamanca, and G. Weinrich, for technical assistance; W. Skarnes, for providing pGT1.8geo; and A. Nagy, for supplying the R1 ES cell line. T.T. and S.G. were supported by fellowships from the European Molecular Biology Organization. A.K.V. was supported by a fellowship from the Deutsche Forschungsgemeinschaft. P.T. is supported by an R. D. Wright Fellowship from the Australian National Health and Medical Research Council (NHMRC). J.E.C. is supported by an NHMRC Practitioner Fellowship and by the Sylvia and Charles Viertel Charitable Foundation. This work was supported by the Ophthalmic Research Institute of Australia, the Royal Hobart Hospital Research Foundation, the Australian NHMRC, the Jack Brockhoff foundation, the Clifford Craig Medical Research Trust, the Max-Planck-Society, and the Walter and Eliza Hall Institute of Medical Research.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Danio rerio Sequencing Project, The Sanger Institute, http://www.sanger.ac.uk/Projects/D_rerio/wgs.shtml
- NCBI Entrez, http://www.ncbi.nlm.nih.gov/Entrez/ (for putative new gene LOC90334 [the original accession number, XM_030959, has now been withdrawn and partly replaced by AK026164]; human NHS gene sequence [accession number AY436752]; human NHS genomic sequences [accession numbers AL845433 and Z93242]; mouse partial mRNAs similar to Nhs [accession numbers XM_142285 and XM_112126]; mouse Nhs genomic sequences [accession numbers AL672082, AC097354, AL732391, and AC093447]; UniGene EST clusters Hs. 444940, Hs. 282164, and Hs. 21470; and singleton ESTs AL926872 [D. rerio] and BF776631 [B. taurus])
- NCBI BLAST, http://www.ncbi.nlm.nih.gov/BLAST/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for NHS) [PubMed]
- PSORT II Prediction, http://psort.nibb.ac.jp/form2.html
References
- Burge C, Karlin S (1997) Prediction of complete gene structures in human genomic DNA. J Mol Biol 268:78–94 [DOI] [PubMed] [Google Scholar]
- Chelly J, Mandel JL (2001) Monogenic causes of X-linked mental retardation. Nat Rev Genet 2:669–680 [DOI] [PubMed] [Google Scholar]
- Cottingham RW Jr, Idury RM, Schaffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed] [Google Scholar]
- Dunwoodie SL, Henrique D, Harrison SM, Beddington RS (1997) Mouse Dll3: a novel divergent Delta gene which may complement the function of other Delta homologues during early pattern formation in the mouse embryo. Development 124:3065–3076 [DOI] [PubMed] [Google Scholar]
- Favor J, Pretsch W (1990) Genetic localization and phenotypic expression of X-linked cataract (Xcat) in Mus musculus. Genet Res 56:157–162 [DOI] [PubMed] [Google Scholar]
- Francis PJ, Berry V, Hardcastle AJ, Maher ER, Moore AT, Bhattacharya SS (2002) A locus for isolated cataract on human Xp. J Med Genet 39:105–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson I, Churchill A, Love J, Axton R, Moore T, Clarke M, Meire F, van Heyningen V (1999) Missense mutations in the most ancient residues of the PAX6 paired domain underlie a spectrum of human congenital eye malformations. Hum Mol Genet 8:165–172 [DOI] [PubMed] [Google Scholar]
- Horan MB, Billson FA (1974) X-linked cataract and hutchinsonian teeth. Aust Paediatr J 10:98–102 [Google Scholar]
- Jamieson RV, Perveen R, Kerr B, Carette M, Yardley J, Heon E, Wirth MG, van Heyningen V, Donnai D, Munier F, Black GC (2002) Domain disruption and mutation of the bZIP transcription factor, MAF, associated with cataract, ocular anterior segment dysgenesis and coloboma. Hum Mol Genet 11:33–42 [DOI] [PubMed] [Google Scholar]
- Kong A, Cox N (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis RA, Nussbaum RL, Stambolian D (1990) Mapping X-linked ophthalmic diseases. IV. Provisional assignment of the locus for X-linked congenital cataracts and microcornea (the Nance-Horan syndrome) to Xp22.2-p22.3. Ophthalmology 97:110–20; discussion 120–121 [DOI] [PubMed] [Google Scholar]
- Litt M, Carrero-Valenzuela R, LaMorticella DM, Schultz DW, Mitchell TN, Kramer P, Maumenee IH (1997) Autosomal dominant cerulean cataract is associated with a chain termination mutation in the human beta-crystallin gene CRYBB2. Hum Mol Genet 6:665–668 [DOI] [PubMed] [Google Scholar]
- Nance WE, Warburg M, Bixler D, Helveston EM (1974) Congenital X-linked cataract, dental anomalies and brachymetacarpalia. Birth Defects Orig Artic Ser 10:285–291 [PubMed] [Google Scholar]
- Reese MG (2001) Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput Chem 26:51–56 [DOI] [PubMed] [Google Scholar]
- Robbins J, Dilworth SM, Laskey RA, Dingwall C (1991) Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: Identification of a class of bipartite nuclear targeting sequence. Cell 64:615–623 [DOI] [PubMed] [Google Scholar]
- Semina EV, Ferrell RE, Mintz-Hittner HA, Bitoun P, Alward WL, Reiter RS, Funkhauser C, Daack-Hirsch S, Murray JC (1998) A novel homeobox gene PITX3 is mutated in families with autosomal-dominant cataracts and ASMD. Nat Genet 19:167–170 [DOI] [PubMed] [Google Scholar]
- Shiels A, Bassnett S (1996) Mutations in the founder of the MIP gene family underlie cataract development in the mouse. Nat Genet 12:212–215 [DOI] [PubMed] [Google Scholar]
- Skarnes WC, Moss JE, Hurtley SM, Beddington RS (1995) Capturing genes encoding membrane and secreted proteins important for mouse development. Proc Natl Acad Sci USA 92:6592–6596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stambolian D, Ai Y, Sidjanin D, Nesburn K, Sathe G, Rosenberg M, Bergsma DJ (1995) Cloning of the galactokinase cDNA and identification of mutations in two families with cataracts. Nat Genet 10:307–312 [DOI] [PubMed] [Google Scholar]
- Stambolian D, Lewis RA, Buetow K, Bond A, Nussbaum R (1990) Nance-Horan syndrome: localization within the region Xp21.1-Xp22.3 by linkage analysis. Am J Hum Genet 47:13–19 [PMC free article] [PubMed] [Google Scholar]
- Thomas T, Voss AK, Chowdhury K, Gruss P (2000) Querkopf, a MYST family histone acetyltransferase, is required for normal cerebral cortex development. Development 127:2537–2548 [DOI] [PubMed] [Google Scholar]
- Toutain A, Ayrault AD, Moraine C (1997a) Mental retardation in Nance-Horan syndrome: clinical and neuropsychological assessment in four families. Am J Med Genet 71:305–314 [DOI] [PubMed] [Google Scholar]
- Toutain A, Dessay B, Ronce N, Ferrante MI, Tranchemontagne J, Newbury-Ecob R, Wallgren-Pettersson C, Burn J, Kaplan J, Rossi A, Russo S, Walpole I, Hartsfield JK, Oyen N, Nemeth A, Bitoun P, Trump D, Moraine C, Franco B (2002) Refinement of the NHS locus on chromosome Xp22.13 and analysis of five candidate genes. Eur J Hum Genet 10:516–520 [DOI] [PubMed] [Google Scholar]
- Toutain A, Ronce N, Dessay B, Robb L, Francannet C, Le Merrer M, Briard ML, Kaplan J, Moraine C (1997b) Nance-Horan syndrome: linkage analysis in 4 families refines localization in Xp22.31-p22.13 region. Hum Genet 99:256–261 [DOI] [PubMed] [Google Scholar]
- Voss AK, Thomas T, Gruss P (1998) Efficiency assessment of the gene trap approach. Dev Dyn 212:171–180 [DOI] [PubMed] [Google Scholar]
- Walpole IR, Hockey A, Nicoll A (1990) The Nance-Horan syndrome. J Med Genet 27:632–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Alcorn DM, Antonarakis SE, Levin LS, Huang PC, Mitchell TN, Warren AC, Maumenee IH (1990) Assignment of the Nance-Horan syndrome to the distal short arm of the X chromosome. Hum Genet 86:54–58 [DOI] [PubMed] [Google Scholar]