Abstract
Mast syndrome is an autosomal recessive, complicated form of hereditary spastic paraplegia with dementia that is present at high frequency among the Old Order Amish. Subtle childhood abnormalities may be present, but the main features develop in early adulthood. The disease is slowly progressive, and cerebellar and extrapyramidal signs are also found in patients with advanced disease. Patients have a thin corpus callosum and white-matter abnormalities, as seen on magnetic resonance imaging. Using an extensive Amish pedigree, we have mapped the Mast syndrome locus (SPG21) to a small interval of chromosome 15q22.31 that encompasses just three genes. Sequence analysis of the three transcripts revealed that all 14 affected cases were homozygous for a single base-pair insertion (601insA) in the acid-cluster protein of 33 kDa (ACP33) gene. This frameshift results in the premature termination (fs201-212X213) of the encoded product, which is designated “maspardin” (Mast syndrome, spastic paraplegia, autosomal recessive with dementia), and has been shown elsewhere to localize to intracellular endosomal/trans-Golgi transportation vesicles and may function in protein transport and sorting.
Introduction
The hereditary spastic paraplegias (HSPs) are a clinically diverse group of disorders that share the primary feature of progressive lower-limb spasticity and weakness. These disorders are classified as “pure,” when spasticity occurs in isolation, and as “complicated,” when additional neurological or other manifestations are present (McDermott et al. 2000). In addition to the marked clinical variability, HSP is genetically heterogeneous, with 11 autosomal dominant, 7 autosomal recessive, and 3 X-linked forms of the disease mapped (HUGO Gene Nomenclature Committee). HSP pathology involves degeneration of the longest corticospinal and dorsal column axons, which is maximal in their distal portions. Despite the identification of genes for 9 of the 21 forms of the condition currently mapped, the pathogenic basis of the HSPs remains uncertain. Although the function of these genes appears to be divergent, there is accumulating evidence to support aberrant subcellular transportation and protein sorting as a common mechanism for a number of forms of motor neuron degeneration (Crosby and Proukakis 2002).
Mast syndrome (MIM 248900) is a complicated form of HSP in which progressive spastic paraparesis is associated in more advanced cases with dementia and other CNS abnormalities (Cross and McKusick 1967). Mast syndrome occurs with high frequency in the Old Order Amish, a population that constitutes a genetic isolate in which a number of disorders have been shown to arise from an ancestral founder mutation, including Troyer syndrome, another complicated form of HSP (Patel et al. 2002). In the current study, we present clinical and radiological findings from 14 patients with Mast syndrome and the mapping and identification of the causative mutation in the ACP33 gene (National Center for Biotechnology Information [accession number NM_016630]), the protein product of which has been shown to associate with intracellular transportation vesicles.
Methods
Linkage Analysis
The genomewide linkage analysis was performed with the ABI LMS, version 2.5, with an ABI 3100 sequence analyzer and Genotyper software, v3.7. Marker saturation analysis was performed using existing and novel microsatellite markers. Alleles were size fractionated with 8% polyacrylamide gels, and DNA was visualized by silver staining. Multipoint LOD scores were generated using GENEHUNTER, v2.1, and the pedigree was deconstructed to facilitate computational efficiency. An autosomal recessive mode of inheritance with complete penetrance and a disease gene frequency of 0.039 were assumed.
Mutation Analysis
Intronic primers were designed flanking each of the nine exons of the ACP33 gene, as well as those of the other genes located within the critical interval. PCR was performed using 50 ng genomic DNA and 5 pmol for each primer. Amplified products were examined on 2% agarose gels and purified for sequencing with a GENECLEAN Turbo for PCR Kit (Q-BIOgene). PCR products were sequenced with the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems) and run on an ABI3100 sequence analyzer.
For SSCP analysis, PCR products from exon 4 of the ACP33 gene were electrophoresed, using 0.55×MDE (National Diagnostics) gels for 18 h at 10°C (constant 250 V), with BioRad Protean II equipment. DNA was visualized by silver staining.
Structure Modeling
Sequences were retrieved through ESTHER (Cousin et al. 1998), and 3D-PSSM (Kelley et al. 2000) was used to predict structural features. The model was built by side-chain replacement with SCWRL (Bower et al. 1997), and the figure was prepared using SwissModel (Guex and Peitsch 1997).
Results
Clinical and Radiological Findings
History and Clinical Examination
We investigated 29 family members, of whom 14 were affected with Mast syndrome and 1 had been part of the original study (Cross and McKusick 1967) (table 1). The ages of patients examined had a range of 31–62 years (mean age 51.79 years). The condition was clearly progressive in all 29 family members, leading to akinetic mutism in the most severely affected subjects. A slight delay in milestones was reported in those for whom information was available. Problems were usually noted in childhood, with difficulties at school, although most were able to complete 8th grade. Motor difficulties were also noted during childhood in some cases, with mild incoordination and awkward running. Patients were perceived as relatively normal in teenage years and early adulthood, and several were married, had children, obtained driving licenses, and held jobs. It is difficult to precisely define the onset of decline in walking and mental function, but, for many, this was already evident by the early 20s; in others, there is no clear history of a decline before the late 30s or early 40s. Speech declined progressively, but the character of speech was not reported as significantly affected. Early subtle personality disturbances were reported in several individuals who were always shy, nervous, or “not very talkative,” and, in three individuals, clear psychotic episodes had occurred (bipolar disorder, paranoid ideation, or hearing voices). Seizures were reported in two cases but had subsided spontaneously. Progressive difficulties in swallowing arose, often necessitating percutaneous enterogastrostomy feeding.
Table 1.
Clinical Features of Patients with Mast Syndrome[Note]
|
Patient Number |
||||||||||||||
| Characteristic | 3 | 4 | 8 | 9 | 14 | 17 | 20 | 23 | 26 | 27 | 31 | 32 | 36 | 39 |
| Age at exam (years) | 64 | 62 | 48 | 52 | 50 | 51 | 43 | 58 | 62 | 60 | 37 | 39 | 48 | 31 |
| Milestone delay | NK | NK | + | + | ++ | + | − | + | NK | + | + | +/− | + | +/− |
| Personality/psychiatric problems | + | +++ | ++ | + | − | − | + | + | − | + | + | +++ | − | +++ |
| Walking difficultiesa | +++ | +++ | ++ | ++ | +++ | +++ | + | +++ | +++ | +++ | + | + | +++ | + |
| Dementia severity | +++ | +++ | ++ | ++ | +++ | +++ | + | +++ | +++ | ++ | +++ | + | ++ | + |
| Dysphagia | +++ | ++ | − | − | + | +++ | − | + | ++ | ++ | − | − | + | +/− |
| Brisk jaw jerk | ++ | + | − | NK | + | + | − | + | ++ | NK | − | − | +/− | − |
| Pyramidal signs (ULb) | +++ | + | − | NK | + | + | + | +++ | + | + | + | + | + | + |
| Pyramidal signs (LLc) | +++ | +++ | ++ | NK | +++ | +++ | ++ | +++ | +++ | +++ | ++ | ++ | ++ | ++ |
| Primitive reflexes | +++ | ++ | ++ | NK | ++ | ++ | − | ++ | ++ | + | − | − | + | − |
| Cerebellar signs | NK | NK | ++ | + | NK | NK | − | NK | NK | NK | NK | + | +++ | + |
| Extrapyramidal movements | − | − | − | − | − | + | − | − | + | ++ | − | − | − | − |
| MMTSd | 0 | 0 | … | … | … | 0 | … | … | … | … | 0 | 9 | 6 | 14 |
Note.— Findings are graded by relative severity: − = absent; +/− = equivocal; + = mild; ++ = moderate; +++ = severe. NK = not known (either owing to inadequate historical information or owing to inability to comply with exam).
+ = can walk without aid; ++ = can walk with walker; +++ = unable to walk.
UL = upper limbs.
LL = lower limbs.
MMTS = mini mental test score (of 30).
Examination revealed clear pyramidal signs in all subjects, with hypertonicity, brisk reflexes, and extensor plantars; these were more severe in the lower than in the upper limbs. In the most advanced cases, legs were rigid, with no spontaneous movements and very little passive flexion, and the patients were bedbound. Higher mental function was significantly impaired in all patients, and even those who were able to answer certain questions were apathetic, with paucity of facial expression, and were very difficult to engage in conversation. Mini mental test score was 0/30 in advanced cases and was only 14/30 in the youngest affected individual. For all individuals studied, speech was very limited where present, but it sounded only mildly dysarthric and was quite easy to understand. Primitive reflexes were found in patients with more advanced disease. Cerebellar dysfunction (incoordination and disdiadochokinesia but no nystagmus) was demonstrated in subjects with mild-to-moderate disease; assessment was not possible in the most severely affected subjects. Extrapyramidal movements (oromandibular dyskinesia and athetoid movements of the fingers or entire limbs) were seen in three advanced cases. Sensation was normal where it could be tested, but diminished ankle jerks in two subjects with advanced disease indicated that a peripheral neuropathy could not be excluded in later stages.
Investigations
Magnetic resonance imaging (MRI) brain scans were available for patient 8 at age 46 years (fig. 1), patient 17 at age 47 years, and patient 32 at age 31 years. All MRIs showed a thin corpus callosum, cerebral and cerebellar atrophy, and white-matter hyperintensity compatible with demyelination. Patient 17 also had a frontal lesion, likely to be an incidental meningioma. She had been previously diagnosed as having a leucodystrophy, but very-long-chain fatty acids and arylsulfatase A were normal. Nerve-conduction studies performed at age 35 years had revealed no abnormality.
Figure 1.
MRI of patient 17, showing thin corpus callosum and cerebral atrophy (A) and periventricular white-matter hyperintensity (B)
Mapping of the Mast Syndrome Locus (SPG21) to Chromosome 15q22
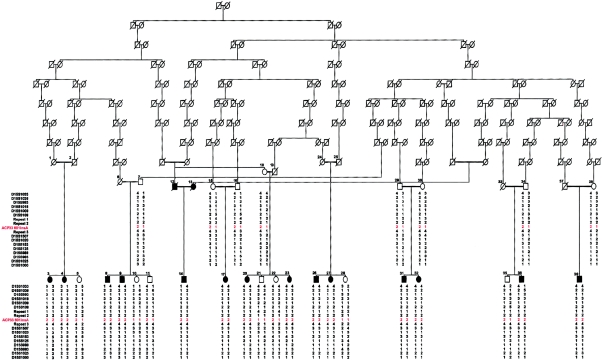
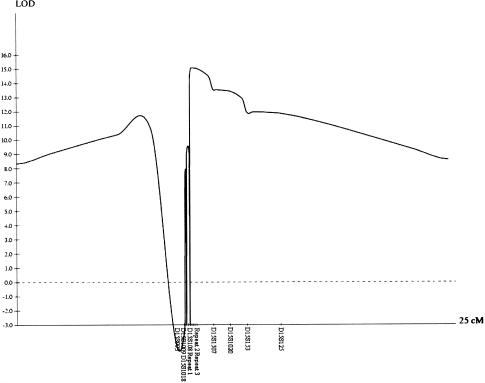
After exclusion of the known autosomal recessive HSP loci on chromosomes 3q27-q28, 8p12-q13, 14q22-q24, 15q13-q15, and 16q24.3 by linkage analysis (data not shown), DNA from 8 of the 14 affected individuals with Mast syndrome was used to conduct a genomewide screen for linkage, under the assumption that a founder mutation was responsible for the condition. Genotyping with the LMS2 marker set revealed only a single significant region of homozygosity, located on chromosome 15q in the region of marker D15S153. Subsequent analyses—by utilization of a total of 16 novel and existing microsatellite markers and incorporation of the remaining affected subjects and unaffected parents and siblings—identified the presence of an ancestral chromosome on which the causative mutation resides (table 2). When examined in parallel, recombination events in the affected cases identify a single block of homozygosity that encompasses markers repeat 1, repeat 2, and repeat 3 (table 2). The apparent homozygosity of these markers in unaffected individuals 7, 11, and 35 (fig. 2) can be clearly explained by extended haplotype analysis, which shows that the homozygosity results from partial informativity of these markers and that they are not identical by descent. This result was further confirmed by mutation analysis (see below). Recombination events in the ancestral chromosome were identified proximally in individuals 3, 4, 8, 14, and 36—with individual 3 being delimiting—and distally in individuals 3, 4, 8, 9, 20, 23, and 39—with individuals 3 and 4 being delimiting (table 2). Taken together, the haplotype analysis indicates that the Mast syndrome locus resides within a 156-kb interval situated between markers D115S108 and D15S1507. Multipoint linkage analysis—by use of GENEHUNTER, v2.1, with 100% penetrance and a trait-allele frequency of 0.039—conclusively confirmed linkage to this region, yielding a maximum LOD score of 15.1 (fig. 3).
Table 2.
Haplotype Analysis of Affected Individuals in Large Amish Pedigree[Note]
|
Haplotypes in Affected Individual |
||||||||||||||
| Location | 3 | 4 | 8 | 9 | 14 | 17 | 20 | 23 | 26 | 27 | 31 | 32 | 36 | 39 |
| D15S1033 | 3, 5 | 3, 1 | 1, 4 | 4, 4 | 1, 4 | 4, 4 | 4, 4 | 4, 4 | 4, 4 | 4, 4 | 4, 4 | 4, 4 | 4, 1 | 4, 4 |
| D15S1036 | 2, 1 | 2, 3 | 2, 2 | 2, 2 | 1, 2 | 2, 2 | 2, 2 | 2, 2 | 2, 2 | 2, 2 | 2, 2 | 2, 2 | 2, 5 | 2, 2 |
| D15S993 | 1, 3 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 2 | 1, 1 |
| D15S1018 | 3, 1 | 3, 3 | 3, 3 | 3, 3 | 3, 3 | 3, 3 | 3, 3 | 3, 3 | 3, 3 | 3, 3 | 3, 3 | 3, 3 | 3, 3 | 3, 3 |
| D15S1009 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 |
| D15S108 | 2, 5 | 2, 2 | 2, 2 | 2, 2 | 2, 2 | 2, 2 | 2, 2 | 2, 2 | 2, 2 | 2, 2 | 2, 2 | 2, 2 | 2, 2 | 2, 2 |
| Repeat 1 |
1, 1 |
1, 1 |
1, 1 |
1, 1 |
1, 1 |
1, 1 |
1, 1 |
1, 1 |
1, 1 |
1, 1 |
1, 1 |
1, 1 |
1, 1 |
1, 1 |
| Repeat 2 |
2, 2 |
2, 2 |
2, 2 |
2, 2 |
2, 2 |
2, 2 |
2, 2 |
2, 2 |
2, 2 |
2, 2 |
2, 2 |
2, 2 |
2, 2 |
2, 2 |
| ACP33 601insA |
1, 1 |
1, 1 |
1, 1 |
1, 1 |
1, 1 |
1, 1 |
1, 1 |
1, 1 |
1, 1 |
1, 1 |
1, 1 |
1, 1 |
1, 1 |
1, 1 |
| Repeat 3 |
4, 4 |
4, 4 |
4, 4 |
4, 4 |
4, 4 |
4, 4 |
4, 4 |
4, 4 |
4, 4 |
4, 4 |
4, 4 |
4, 4 |
4, 4 |
4, 4 |
| D15S1507 | 1, 3 | 3, 1 | 3, 3 | 3, 3 | 3, 3 | 3, 3 | 3, 3 | 3, 3 | 3, 3 | 3, 3 | 3, 3 | 3, 3 | 3, 3 | 3, 3 |
| D15S1020 | 3, 1 | 1, 3 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 |
| D15S153 | 5, 3 | 3, 5 | 3, 3 | 3, 3 | 3, 3 | 3, 3 | 3, 4 | 3, 4 | 3, 3 | 3, 3 | 3, 3 | 3, 3 | 3, 3 | 3, 3 |
| D15S125 | 3, 1 | 1, 3 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 2 | 1, 2 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 |
| D15S988 | 2, 1 | 1, 2 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 2 | 1, 2 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 |
| D15S983 | 8, 1 | 1, 8 | 1, 2 | 1, 2 | 1, 1 | 1, 1 | 1, 3 | 1, 3 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 |
| D15S1025 | 2, 2 | 2, 2 | 2, 3 | 2, 3 | 2, 2 | 2, 2 | 2, 2 | 2, 2 | 2, 2 | 2, 2 | 2, 2 | 2, 2 | 2, 2 | 2, 3 |
| D15S1000 | 3, 1 | 1, 3 | 1, 3 | 1, 3 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 | 1, 1 |
Note.— Analysis reveals the mutation-bearing ancestral chromosome (shown in boldface italics) and recombination events delimiting the common region of homozygosity (underlined). Repeat 1 is a GATA repeat, and repeat 2 is a TA repeat; both were identified in BAC clone AC069368. Repeat 3 is an AAAT repeat located in BAC clone AC103691.
Figure 2.
Pedigree of the large Amish family with Mast syndrome, with consanguinity dating to the 17th century, showing haplotypes for markers across the SPG21 interval
Figure 3.
Multipoint LOD scores for markers situated across the SPG21 locus, flanked by markers D15S108 and D15S1507, producing a peak score of 15.1 for marker repeat 3.
Mutation Analysis
The 156-kb interval of band 15q22.31 contains three nonoverlapping transcriptional units listed from centromere to telomere: the ankyrin domain-containing protein LOC348094, an acid-cluster protein of 33 kDa (ACP33), and the mitochondrial methionyl-tRNA formyltransferase (MtFMT) (fig. 4). For all three transcripts, primers were positioned to incorporate each exon and its associated splice signals and were used to sequence genomic DNA from an affected individual and an unaffected parent. We identified a single nucleotide insertion (601insA) in exon 7 of the ACP33 gene (fig. 5), which is predicted to produce a frameshift causing substitution of the following 12 amino acids and premature termination of translation (fs201-212X213) of the protein product maspardin (Mast syndrome, spastic paraplegia, autosomal recessive with dementia). Sequence analysis revealed that this mutation cosegregates perfectly with the disease in this population, since unaffected siblings are either homozygous wild-type or heterozygous carriers. This mutation was not detected in 236 normal control chromosomes of mixed white (80%), Asian (10%), and African (10%) descent, and only one carrier of the mutation was identified in 64 chromosomes originating from the same Amish community in which this condition is present at high frequency.
Figure 4.
A schematic transcript map of the SPG21 locus, located within contig NT_010265 and spanning BAC clones AC069368, AC013691, and AC013553 (NCBI). The relative positions of the markers used for haplotyping, transcripts (and direction of transcription) located within and around the critical region, and the 601insA mutation in ACP33 are shown.
Figure 5.
Sequence chromatograms of the region around the exon 7 601insA mutation (arrow) of ACP33 in a wild-type homozygote (A), a homozygous affected individual (B), and a carrier parent (C).
Comparative Protein Alignment and Structural Predictions
To determine whether maspardin is likely to possess enzymatic function, we have conducted alignment of maspardin with a typical hydrolase molecule (chloroperoxidase, PRXC_PSEFL). Although it is clear from this alignment that maspardin is a member of this superfamily (fig. 6), the nucleophile-acid-histidine triad required for catalytic function is not present, which suggests that maspardin is unlikely to be an enzyme. This is further confirmed by modeling of maspardin on the basis of the results of 3D-PSSM, with 1A8S as a template (fig. 7). This reveals that only one of the residues required for catalysis is present (ser109) and that Asp 226 (located adjacent to Gln 227) points in the wrong orientation.
Figure 6.
Alignment of maspardin with pseudomonas fluorescens chloroperoxidase (SwissProt: O31158 PRXC_PSEFL, PDB:1A8S). Line 1 shows the predicted secondary structure (“C3” loop, “E” strand, and “H” helix); lines 2 and 3 show the sequence of maspardin and chloroperoxydase, respectively; line 4 shows the known secondary structure of PRXC_PSEFL; and line 5, CORE, is a measure of the burial and number of contacts made by each residue, on a scale between 0 (not buried/few contacts) and 9 (very buried/many contacts). Amino acids of the catalytic triad of PRXC_PSEFL are shown in bold and are marked by an asterisk (*).
Figure 7.
Model of the structure of maspardin on the basis of the results of 3D-PSSM, with 1A8S as a template, which indicates that two of the three residues (Gln227 and Asn255) do not conform to the α/β hydrolase catalytic triad and that Asp226 is in the opposite orientation. Alpha helices are shown in yellow, and beta strands are shown in blue.
Discussion
Mast syndrome (SPG21) is an autosomal recessive complicated form of HSP associated with dementia and mild developmental, pseudobulbar, cerebellar, and extrapyramidal abnormalities, first described at high frequency in an Old Order Amish deme (Cross and McKusick 1967). Mild early milestone delay and subtle cognitive and motor difficulties in childhood are often reported, indicating a neurodevelopmental problem. Symptom progression usually commences in early adulthood and is slow but relentless, suggesting ongoing neurodegeneration affecting several different areas of the CNS. Although there is no evidence of peripheral nerve involvement, brain MRI reveals a thin corpus callosum, together with gray-matter atrophy and white-matter demyelination (fig. 1). The clinical pattern and radiological findings suggest a pattern of degeneration more widespread than that of the axons of the corticospinal tracts and the posterior columns that degenerate in pure HSP.
The corpus callosum abnormality, which is part of many inherited syndromes (Dobyns 1996), is particularly interesting and may represent partial agenesis, atrophy, or a combination of both. A developmental abnormality would explain the slight delay in milestones and the subtle problems in intellect and coordination seen early in life. White-matter abnormalities and a thin corpus callosum have only rarely been reported in some forms of HSP, including SPG11 (McDermott et al. 2000), an autosomal recessive form of the disease (Martinez Murillo et al. 1999). HSP complicated by dementia is also relatively unusual, although a mild intellectual impairment, possibly progressive, has been reported in HSP caused by SPG4 (spastin) mutations (Byrne et al. 2000). Conversely, there are reports of adult-onset dementias accompanied by spastic paraparesis, including Alzheimer disease, caused by presenilin-1 mutations (Houlden et al. 2000), and a variant of the Gerstmann-Sträussler syndrome, an inherited prion disease (Kitamoto et al. 1993). However, these conditions differ from Mast syndrome in that they lack a developmental phenotype, and Mast syndrome is almost certain to have a pathology distinct from both of them.
The identification of the 601insA mutation in the ACP33 gene is expected to produce a frameshift, generating a premature stop codon, which is likely to result in a loss of function of the polypeptide product assigned the more phenotypically descriptive alias “maspardin.” The facts that the remaining genes located in the Mast syndrome interval contained no disease-associated mutations and that we did not detect the 601insA mutation in a large number of controls strongly suggest that this is the causative mutation. To date, only one functional study relating to maspardin is available in which the molecule was identified through its association with CD4, a cell-surface glycoprotein involved in the cellular immune response; therefore, the study focused specifically on this interaction (Zeitlmann et al. 2001). The subcellular localization of the protein was investigated, using a monoclonal antibody directed against the complete polypeptide in HUT78 (CD4-positive T cell line) cells and using a chimeric fusion protein (Zeitlmann et al. 2001). These results revealed that both endogenous and transiently expressed proteins are cytosolic and that they partially associate with particulate structures. Subsequent studies revealed that the vesicular structures marked by maspardin partially coincided with those marked for transferrin—which is confined to the early endosomal recycling pathway—and with acidic organelles, suggesting that maspardin is partitioned between the cytosol and vesicles of the endosomal/trans-Golgi network. Taken together, it appears that maspardin may be involved in the sorting and/or trafficking of molecules in HUT78 cells (Zeitlmann et al. 2001).
Although our BLAST homology searches indicate no known subcellular localization signals, leader sequence, or transmembrane regions, they did reveal a clear but limited similarity to the α/β hydrolases that mainly comprise enzymes that catalyze a diverse range of reactions (Holmquist 2000). Previous studies have indicated that the α/β hydrolases can be delineated by the presence of three essential structural features required for catalysis: the presence of at least five parallel β strands, a catalytic triad in a specific order (nucleophile-acid-histidine), and a nucleophile elbow (Shaw et al. 2002). Although our alignment studies indicate that maspardin is clearly related to members of this superfamily and possesses a nucleophilic elbow and parallel β strands, it does not possess the catalytic triad, which suggests that it is unlikely to possess enzymatic function (figs. 6 and 7). Further support for this is provided by studies of the hydrolase fold of maspardin. The hydrolase fold comprises a nucleophile elbow with a nucleophilic residue at its center (Holmquist 2000), which corresponds to residue 109 (serine) of maspardin. A single point mutation disrupting the nucleophilic elbow (serine109alanine) was sufficient to completely abolish interaction with CD4 (Zeitlmann et al. 2001). Consequently, it appears that the hydrolase-related domain of maspardin has evolved from an ancient enzymatic function and is now likely to serve as an intracellular protein-protein interaction module. Two families of noncatalytic α/β hydrolase fold proteins have been described elsewhere: the electrotactins—which include the neural cell adhesion proteins gliotactin, neurotactin, and neuregulin (Botti et al. 1998)—and the N-myc downstream regulated (NDRG) proteins NDRG1–NDRG4 (Qu et al. 2002; Shaw et al. 2002). Of particular interest in the context of Mast syndrome is the identification of a nonsense mutation in NDRG1, shown elsewhere to underlie hereditary motor and sensory neuropathy-Lom (HMSNL), an autosomal recessive peripheral neuropathy with early axonal involvement possibly resulting from impaired axonal-glial interactions (Kalaydjieva et al. 2000). HMSNL shows no features suggestive of upper motor neuron involvement (Kalaydjieva et al. 1998), and, conversely, there was no clear evidence of lower motor neuron involvement in Mast syndrome. Mutations in similar molecules causing other pure forms of HSP, an upper motor neuron disease, and HMSN, a lower motor neuron disease, have been described elsewhere in the kinesins microtubule motor proteins: KIF5A mutations lead to a pure form of HSP (Reid et al. 2002), and KIF1Bβ mutations lead to HMSNIIA (Zhao et al. 2001). Consequently, the identification of mutations in two nonenzymatic hydrolases, associated with two motor neuron degenerative phenotypes, suggests a related function of these molecules that is essential for neuronal survival and highlights these molecules as candidates for other inherited forms of neurodegeneration.
There is now increasing direct and circumstantial evidence to support the hypothesis that defective protein sorting and trafficking underlies a range of neurodegenerative conditions (Crosby 2003) and that defective trafficking may be a final pathway common to a number of genes involved in HSP (Crosby and Proukakis 2002). Neurons possess a unique morphology, typically characterized by an extensive branching dendritic tree and a single long axon (spinal motor neuron axons reach 1 m in length), and these cell processes typically contain the bulk of the cytoplasm of the cell; yet protein manufacture, packaging, and sorting is essentially restricted to the cell body. In consequence, these factors place tremendous demands on intracellular transport systems, which may constitute a “weak link” susceptible to various insults, resulting in neurodegenerative diseases. Although the role of maspardin is not clear and now warrants detailed investigation, the studies described above are consistent with an involvement in vesicle-mediated trafficking and protein sorting. Therefore, Mast syndrome may represent an example of another condition in which disruption of these mechanisms leads to neurodegeneration.
Acknowledgments
This work was supported by the Birth Defects Foundation (United Kingdom), the Wellcome Trust, and Research to Prevent Blindness. We thank the Amish families for their support, help, and generosity over the duration of this project, and we are grateful to Dr. A. Valentine for reviewing the MRI scans.
Electronic-Database Information
Accession number and URLs for data presented herein are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/
- HUGO Gene Nomenclature Committee, http://www.gene.ucl.ac.uk/nomenclature/
- National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/ (for ACP33 [accession number NM_016630], BAC clones [accession numbers AC069368, AC013691, and AC013553], and SPG21 [contig NT_010265])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for Mast syndrome)
References
- Botti SA, Felder CE, Sussman JL, Silman I (1998) Electrotactins: a class of adhesion proteins with conserved electrostatic and structural motifs. Protein Eng 11:415–420 [DOI] [PubMed] [Google Scholar]
- Bower MJ, Cohen FE, Dunbrack RL Jr (1997) Prediction of protein side-chain rotamers from a backbone-dependent rotamer library: a new homology modeling tool. J Mol Biol 267:1268–1282 [DOI] [PubMed] [Google Scholar]
- Byrne PC, McMonagle P, Webb S, Fitzgerald B, Parfrey NA, Hutchinson M (2000) Age-related cognitive decline in hereditary spastic paraparesis linked to chromosome 2p. Neurology 54:1510–1517 [DOI] [PubMed] [Google Scholar]
- Cousin X, Hotelier T, Giles K, Toutant JP, Chatonnet A (1998) aCHEdb: the database system for ESTHER, the alpha/beta fold family of proteins and the cholinesterase gene server. Nucleic Acids Res 26:226–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby A (2003) Are road blocks in the cellular highways a common cause of neurodegeneration. Lancet Neurology 2:311–316 [DOI] [PubMed] [Google Scholar]
- Crosby AH, Proukakis C (2002) Is the transportation highway the right road for hereditary spastic paraplegia? Am J Hum Genet 71:1009–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross HE, McKusick VA (1967) The Mast syndrome: a recessively inherited form of presenile dementia with motor disturbances. Arch Neurol 16:1–13 [DOI] [PubMed] [Google Scholar]
- Dobyns WB (1996) Absence makes the search grow longer. Am J Hum Genet 58:7–16 [PMC free article] [PubMed] [Google Scholar]
- Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723 [DOI] [PubMed] [Google Scholar]
- Holmquist M (2000) Alpha/Beta-hydrolase fold enzymes: structures, functions and mechanisms. Curr Protein Pept Sci 1:209–235 [DOI] [PubMed] [Google Scholar]
- Houlden H, Baker M, McGowan E, Lewis P, Hutton M, Crook R, Wood NW, Kumar-Singh S, Geddes J, Swash M, Scaravilli F, Holton JL, Lashley T, Tomita T, Hashimoto T, Verkkoniemi A, Kalimo H, Somer M, Paetau A, Martin JJ, Van Broeckhoven C, Golde T, Hardy J, Haltia M, Revesz T (2000) Variant Alzheimer’s disease with spastic paraparesis and cotton wool plaques is caused by PS-1 mutations that lead to exceptionally high amyloid-beta concentrations. Ann Neurol 48:806–808 [PubMed] [Google Scholar]
- Kalaydjieva L, Gresham D, Gooding R, Heather L, Baas F, de Jonge R, Blechschmidt K, Angelicheva D, Chandler D, Worsley P, Rosenthal A, King RHM, Thomas PK (2000) N-myc downstream-regulated gene 1 is mutated in hereditary motor and sensory neuropathy–Lom. Am J Hum Genet 67:47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaydjieva L, Nikolova A, Turnev I, Petrova J, Hristova A, Ishpekova B, Petkova I, Shmarov A, Stancheva S, Middleton L, Merlini L, Trogu A, Muddle JR, King RH, Thomas PK (1998) Hereditary motor and sensory neuropathy–Lom, a novel demyelinating neuropathy associated with deafness in Gypsies: clinical, electrophysiological and nerve biopsy findings. Brain 121:399–408 [DOI] [PubMed] [Google Scholar]
- Kelley LA, MacCallum RM, Sternberg MJ (2000) Enhanced genome annotation using structural profiles in the program 3D-PSSM. J Mol Biol 299:499–520 [DOI] [PubMed] [Google Scholar]
- Kitamoto T, Amano N, Terao Y, Nakazato Y, Isshiki T, Mizutani T, Tateishi J (1993) A new inherited prion disease (PrP-P105L mutation) showing spastic paraparesis. Ann Neurol 34:808–813 [DOI] [PubMed] [Google Scholar]
- Martinez Murillo F, Kobayashi H, Pegoraro E, Galluzzi G, Creel G, Mariani C, Farina E, Ricci E, Alfonso G, Pauli RM, Hoffman EP (1999) Genetic localization of a new locus for recessive familial spastic paraparesis to 15q13-15. Neurology 53:50–56 [DOI] [PubMed] [Google Scholar]
- McDermott C, White K, Bushby K, Shaw P (2000) Hereditary spastic paraparesis: a review of new developments. J Neurol Neurosurg Psychiatry 69:150–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel H, Cross H, Proukakis C, Hershberger R, Bork P, Ciccarelli FD, Patton MA, McKusick VA, Crosby AH (2002) SPG20 is mutated in Troyer syndrome, an hereditary spastic paraplegia. Nat Genet 31:347–348 [DOI] [PubMed] [Google Scholar]
- Qu X, Zhai Y, Wei H, Zhang C, Xing G, Yu Y, He F (2002) Characterization and expression of three novel differentiation-related genes belong to the human NDRG gene family. Mol Cell Biochem 229:35–44 [DOI] [PubMed] [Google Scholar]
- Reid E, Kloos M, Ashley-Koch A, Hughes L, Bevan S, Svenson IK, Graham FL, Gaskell PC, Dearlove A, Pericak-Vance MA, Rubinsztein DC, Marchuk DA (2002) A kinesin heavy chain (KIF5A) mutation in hereditary spastic paraplegia (SPG10). Am J Hum Genet 71:1189–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw E, McCue LA, Lawrence CE, Dordick JS (2002) Identification of a novel class in the alpha/beta hydrolase fold superfamily: the N-myc differentiation-related proteins. Proteins 47:163–168 [DOI] [PubMed] [Google Scholar]
- Zeitlmann L, Sirim P, Kremmer E, Kolanus W (2001) Cloning of ACP33 as a novel intracellular ligand of CD4. J Biol Chem 276:9123–9132 [DOI] [PubMed] [Google Scholar]
- Zhao C, Takita J, Tanaka Y, Setou M, Nakagawa T, Takeda S, Yang HW, Terada S, Nakata T, Takei Y, Saito M, Tsuji S, Hayashi Y, Hirokawa N (2001) Charcot-Marie-Tooth disease type 2A caused by mutation in a microtubule motor KIF1Bbeta. Cell 105: 587–597 [DOI] [PubMed] [Google Scholar]