Abstract
There is a critical need to understand why missense mutations are deleterious. The deleterious effects of missense mutations are commonly attributed to their impact on primary amino acid sequence and protein structure. However, several recent studies have shown that some missense mutations are deleterious because they disturb cis-acting splicing elements—so-called “exonic splicing enhancers” (ESEs). It is not clear whether the ESE-related deleterious effects of missense mutations are common. We have evaluated colocalization of pathogenic missense mutations (found in affected individuals) with high-score ESE motifs in the human mismatch-repair genes hMSH2 and hMLH1. We found that pathogenic missense mutations in the hMSH2 and hMLH1 genes are located in ESE sites significantly more frequently than expected. Pathogenic missense mutations also tended to decrease ESE scores, thus leading to a higher propensity for splicing defects. In contrast, nonpathogenic missense mutations (polymorphisms found in unaffected individuals) and nonsense mutations are distributed randomly in relation to ESE sites. Comparison of the observed and expected frequencies of missense mutations in ESE sites shows that pathogenic effects of ⩾20% of mutations in hMSH2 result from disruption of ESE sites and disturbed splicing. Similarly, pathogenic effects of ⩾16% of missense mutations in the hMLH1 gene are ESE related. The colocalization of pathogenic missense mutations with ESE sites strongly suggests that their pathogenic effects are splicing related.
Missense mutations—nucleotide substitutions that change an amino acid in a protein—are among the most common types of mutations underlying inherited human diseases. The deleterious effects of missense mutations are usually attributed to their effects on protein function. However, recent studies of normal and alternative splicing suggest that the deleterious effects of nucleotide substitutions might, in fact, be splicing related when they are located in exonic splicing enhancers (ESEs) (Cartegni and Krainer 2002; Cartegni et al. 2002; Fackenthal et al. 2002; Moseley et al. 2002; Pollard et al. 2002). ESEs are discrete, degenerate motifs of 6–8 nts located inside exons (Liu et al. 1998; Blencowe 2000). The study of normal splicing suggests that most exons contain at least one functional ESE site (Blencowe 2000; Hastings and Krainer 2001; Cartegni et al. 2002). ESEs are target sequences for the family of conserved essential splicing factors—the serine- and arginine-rich (SR) proteins (Stojdl and Bell 1999; Graveley 2000; Hastings and Krainer 2001). ESEs play an important role in exon recognition. Nucleotide substitutions in ESEs can result in failure of SR proteins to bind to the ESE, which leads to failure of splisosome machinery to recognize the sequence as exonic and causes exon skipping (Ars et al. 2000; Cartegni et al. 2002; Fackenthal et al. 2002; Moseley et al. 2002). Each SR protein recognizes specific, albeit degenerate and partially redundant, sequence motifs. ESE motifs for four members of the SR family (SF2/ASF, SRp40, SRp55, and SC35) have been identified (Liu et al. 1998; Stojdl and Bell 1999; Graveley 2000; Liu et al. 2000). To identify the ESE motifs that are recognized by individual SR proteins, a PCR-based approach called “SELEX” (systematic evolution of ligands by exponential) enrichment was used. In this approach, a natural splicing enhancer in a minigene is replaced by short, random sequences derived from an oligonucleotide library. The generated pool of minigenes is transfected into cultured cells, and spliced mRNAs are amplified by RT-PCR and sequenced (Liu et al. 1998, 2000). On the basis of the frequencies of the individual nucleotides at each position, a score matrix for each nucleotide in each position was calculated. This score matrix can be used to predict SR protein–specific ESEs (ESEfinder).
We studied the association of pathogenic missense mutations (found in affected kindreds), nonpathogenic missense mutations (polymorphic mutations and sequence variants found in nonaffected individuals), nonsense mutations, and frameshifts (here restricted to 1- or 2-nt deletions and insertions) with ESE sites in hMSH2 and hMLH1, human mismatch repair genes that are related to human nonpolyposis colon cancer (HNPCC [MIM 114500]) (Peltomaki and Vasen 1997). We used published and our own data on pathogenic and nonpathogenic missense mutations in the hMSH2 and hMLH1 genes (tables A, B, C, D, and E [online only]). Only mutations found in independent families were used. We excluded multiple reported mutations found in the same family. The numbers of different types of mutations analyzed are shown in table 1.
Table A.
Pathogenic Missense Mutations Found in the hMSH2 Gene[Note]
| Nucleotide Position in ORF | ESE | Exon | Codon | Nucleotide | Nucleotide Change | Consequence | Type of Amino Acid Substitutiona | Geographic Origin | Family ID | Reference |
| 4 | 0 | 1 | 2 | 4 | G→A | Ala→Thr | C | None specified | … | Myriad et al. |
| 182 | 1 | 1 | 61 | 182 | A→C | Gln→Pro | C | Western Europe | … | Myriad et al. |
| 308 | 1 | 2 | 103 | 308 | A→G | Tyr→Cys | R | None specified | … | Myriad et al. |
| 319 | 0 | 2 | 107 | 319 | G→C | Ala→Prol | C | Czech Republic | PT523 | A. Krepelova (unpublished) |
| 380 | 0 | 3 | 127 | 380 | A→G | Asn→Ser | C | None specified | … | Myriad et al. |
| 380 | 0 | 3 | 127 | 380 | A→G | Asn→Ser | C | Africa | … | Myriad et al. |
| 380 | 0 | 3 | 127 | 380 | A→G | Asn→Ser | C | Africa | … | Myriad et al. |
| 380 | 0 | 3 | 127 | 380 | A→G | Asn→Ser | C | None specified | … | Myriad et al. |
| 380 | 0 | 3 | 127 | 380 | A→G | Asn→Ser | C | None specified | … | Myriad et al. |
| 435 | 1 | 3 | 145 | 435 | T→G | Ile→Met | C | Czech Republic | Patient 338 | A. Krepelova (unpublished) |
| 435 | 1 | 3 | 145 | 435 | T→G | Ile→Met | C | None specified | … | A. M. Deffenbaugh et al. (unpublished) |
| 435 | 1 | 3 | 145 | 435 | T→G | Ile→Met | C | Western European | … | Myriad et al. |
| 446 | 0 | 3 | 149 | 446 | G→A | Gly→Asp | R | None specified | … | A. M. Deffenbaugh et al. (unpublished) |
| 505 | 1 | 3 | 169 | 505 | A→G | Ile→Valb | C | Asia | … | A. M. Deffenbaugh et al. (unpublished) |
| 593 | 1 | 3 | 198 | 593 | GAA→GGA | Glu→Gly | R | Sweden | 199 | Unpublished |
| 595 | 0 | 3 | 199 | 595 | T→C | Cys→Arg | R | Hong Kong | [Turcot] | Yuen et al., Am J Pathol 153:1181–1188 (1998) |
| 742 | 0 | 4 | 248 | 742 | A→G | Lys→Glu | C | Western Europe | … | Myriad et al. |
| 792 | 1 | 4 | 264 | 792 | G→C | Gln→His | R | Western Europe | … | Myriad et al. |
| 806 | 1 | 5 | 269 | 806 | C→T | Ser→Leu | R | Latin America/Caribbean | … | A. M. Deffenbaugh et al. (unpublished) |
| 815 | 1 | 5 | 272 | 815 | C→T | Ala→Val | C | None specified | 0108 | MDACCc |
| 913 | 1 | 5 | 305 | 913 | G→A | Ala→Thr | C | Netherlands | NL-38 | Wijnen et al. (unpublished) |
| 965 | 1 | 6 | 322 | 965 | G→Ab | Gly→Asp | C | Russia | 1 | Maliaka et al. Hum Genet 97:251–255 (1996) |
| 965 | 1 | 6 | 322 | 965 | G→Ab | Gly→Asp | C | Germany | 186 | Brieger et al. Clinical Chemistry 45: 1564–1567 (1999) |
| 965 | 1 | 6 | 322 | 965 | G→Ab | Gly→Asp | C | United States | 0205 | MDACCc |
| 997 | 0 | 6 | 333 | 997 | T→C | Cys→Arg | R | Nigeria | 114-I-OL | A. de la Chapelle et al. (unpublished) |
| 998 | 0 | 6 | 333 | 998 | G→A | Cys→Tyr | R | Australia | IMS5 | S. E. Bennett (unpublished) |
| 1012 | 1 | 6 | 338 | 1012 | G→A | Gly→Arg | R | None specified | … | A. M. Deffenbaugh et al. (unpublished) |
| 1082 | 0 | 7 | 361 | 1082 | A→G | Arg→Ser | R | Western Europe | … | Myriad et al. |
| 1319 | 1 | 8 | 440 | 1319 | T→C | Leu→Pro | R | Lithuania | O3 | E. Avizienyte et al. (unpublished) |
| 1508 | 0 | 9 | 503 | 1508 | T→C | Leu→Pro | R | None specified | … | Myriad et al. |
| 1516 | 1 | 10 | 506 | 1516 | G→T | Asp→Tyr | R | Korea | SNU-YC13 | Han et al., J. Natl. Cancer Inst. 88: 1317–1319 (1996) |
| 1571 | 1 | 10 | 524 | 1571 | G→C | Arg→Pro | R | None specified | Patient 2774 with ovarian cancer | Orth et al., Cold Spring Harbor Symp Quant Biol LIX: 349 (1994) |
| 1600 | 1 | 10 | 534 | 1600 | C→T | Arg→Cys | R | United States | 3080 | MDACCc |
| 1774 | 1 | 12 | 592 | 1774 | A→G | Met→Val | C | None specified | … | Myriad et al. |
| 1807 | 0 | 12 | 603 | 1807 | G→A | Asp→Asn | R | Finland | Patient 138 | Salovaara et al., J Clin Oncol 18: 2193–2200 (2000) |
| 1864 | 0 | 12 | 622 | 1864 | C→A | Missense | R | Argentina | CRE001 | Chialina et al. (unpublished) |
| 1865 | 0 | 12 | 622 | 1865 | C→T | Pro→Leu | R | New Zealand | J | Liu et al., Cancer Res. 54: 4590–4594 (1994) |
| 1906 | 1 | 12 | 636 | 1906 | G→Cb | Ala→Pro | C | France | CHI-S6351 | B. Bressac-de Paillerets, unpublished |
| 1906 | 1 | 12 | 636 | 1906 | G→Cb | Ala→Pro | C | United States | 54 | A. de la Chapelle (unpublished) |
| 1906 | 1 | 12 | 636 | 1906 | G→C | Ala→Pro | C | Ashkenazi Jewish | MONO7 (y) | Yuan et al. J Med Genet 36:790–793 (1999) |
| 1906 | 1 | 12 | 636 | 1906 | G→C | Ala→Pro | C | None specified | … | Myriad et al. |
| 2090 | 0 | 13 | 697 | 2090 | G→T | Cys→Phe | R | Germany | 56 | Raedle et al. Gastroenterology 116:489A (1999) |
| 2090 | 0 | 13 | 697 | 2090 | G→T | Cys→Phe | R | Germany | 62 | Wehner et al., Hum Mut 10:241–244 (1997) |
| 2164 | 1 | 13 | 722 | 2164 | G→A | Val→Ile | R | United States | 0024 | MDACCc |
| 2251 | 0 | 14 | 751 | 2251 | G→A | Gly→Arg | R | France | BIL-IGR1924 | B. Bressac-de Paillerets |
| 2500 | 0 | 15 | 834 | 2500 | G→A | Ala→Thr | C | Netherlands | NL57 | J. T. Wijnen et al. (unpublished) |
| 2714 | 1 | 16 | 905 | 2714 | C→G | Thr→Arg | R | United Kingdom | C015 | Froggatt et al., J. Med. Genet. 33:726–730 (1996) |
| 2714 | 1 | 16 | 905 | 2714 | C→T | Thr→Ile | R | Central/eastern Europe | … | Myriad et al. |
| 2790 | 0 | 16 | 930 | 2790 | A→Gb | Ile→Met | C | Spain | GE008 | P. Hutter (unpublished) (1999) |
Note.— The table enlists missense mutations from the HNPCC mutation database (accessed March 2003) and mutation data obtained by M.L.F. at The University of Texas M. D. Anderson Cancer Center (MDACC).
C = conservative; R = radical.
Of unknown pathogenicity.
The University of Texas M. D. Anderson Cancer Center.
Table B.
Pathogenic Missense Mutations Found in the hMLH1 Gene[Note]
| Exon | ORF Position | ESE | Exonic Position | Initial Nucleotide | Mutated Nucleotide | Initial Amino Acid | Mutated Amino Acid | Type of Amino Acid Substitutiona | Family ID | Reference |
| 1 | 69 | 0 | 69 | A | T | Glu | Asp | C | … | Myriad et al. |
| 1 | 73 | 0 | 73 | A | T | Ile | Phe | R | 166 | W. Weber and M. Rodriguez-Bigas (unpublished) |
| 1 | 74 | 0 | 74 | T | C | Ile | Thr | R | 40547 | G. Norbury et al. (unpublished) |
| 1 | 83 | 1 | 83 | C | T | Pro | Leu | R | 52 | Wehner et al., Hum Mut 10:241–244 (1997) |
| 1 | 85 | 0 | 85 | G | T | Ala | Ser | C | … | Myriad et al. |
| 1 | 104 | 0 | 104 | T | G | Met | Arg | C | 3 | Tannergard et al., Cancer Res 55:6092–6096 (1995) |
| 1 | 116 | 0 | 116 | G | A | Cys | Ter | R | MDACC3014 | MDACC |
| 2 | 184 | 1 | 68 | C | A | Gln | Lys | C | 498 | Wehner et al., Hum Mut 10:241–244 (1997) |
| 2 | 191 | 1 | 75 | A | G | Asm | Ser | C | 2104 | J. T. Wijnen et al. (unpublished) |
| 2 | 199 | 1 | 83 | G | A | Gly | Arg | C | MDACC3011 | MDACC |
| 2 | 199 | 1 | 83 | G | A | Gly | Arg | C | MDACC3202 | MDACC |
| 2 | 199 | 1 | 83 | G | A | Gly | Arg | C | 304 | Tannergard et al., Cancer Res 55:6092–6096 (1995) |
| 2 | 199 | 1 | 83 | G | A | Gly | Arg | C | 1652 | Tannergard et al., Cancer Res 55:6092–6096 (1995) |
| 2 | 199 | 1 | 83 | G | A | Gly | Arg | C | 68 | Heinimann et al., Cancer 85 (12):2512–2518 (1999) |
| 2 | 199 | 1 | 83 | G | A | Gly | Arg | C | JPN-28 | Sasaki et al., Hum Mut 9:164 (1977} |
| 2 | 199 | 1 | 83 | G | A | Gly | Arg | C | MFI | Herfarth et al., Genes Chrom Cancer 18:42 (1997) |
| 2 | 199 | 1 | 83 | G | A | Gly | Arg | C | VSO15 | Hutter et al., Int J Cancer 78:680–684 (1998) |
| 2 | 200 | 1 | 84 | G | A | Gly | Glu | R | MDACC413075 | MDACC |
| 2 | 200 | 1 | 84 | G | A | Gly | Glu | R | MDACC380548 | MDACC |
| 2 | 203 | 0 | 87 | T | A | Ile | Asn | R | 18 | Tannergard et al., Cancer Res 55:6092–6096 (1995) |
| 3 | 230 | 0 | 23 | G | A | Cys | Tyr | R | IMS2 | S. E. Bennet (unpublished) |
| 3 | 250 | 0 | 43 | A | G | Lys | Glu | R | 171 | Lamberti et al., Gut 44:839–843 (1999) |
| 3 | 277 | 0 | 70 | A | G | Ser | Gly | R | 1 | Quaresima et al., Hum Mut 12; 433 (1998) |
| 3 | 299 | 1 | 92 | G | C | Arg | Pro | C | … | Myriad et al. |
| 3 | 304 | 1 | 97 | G | A | Glu | Lys | R | 61 | |
| 3 | 306 | 1 | 99 | G | T | Glu | Asp | R | … | Myriad et al. |
| 4 | 320 | 1 | 14 | T | G | Ile | Arg | R | 28/51/67 | Nyström-Lahti et al., Hum Mol Genet 5:763–769 (1996) |
| 4 | 350 | 1 | 44 | C | T | Thr | Met | R | … | A. M. Deffenbaugh et al. (unpublished) |
| 4 | 350 | 1 | 44 | C | T | Thr | Met | R | 431 | Krpelova (unpublished) |
| 4 | 350 | 1 | 44 | C | T | Thr | Met | R | S.B.-2575 | Stachow-Kurzawski (unpublished) |
| 4 | 350 | 1 | 44 | C | T | Thr | Met | R | 26 | Raedle et al. Gastro 116:A-489 (1999) |
| 4 | 350 | 1 | 44 | C | T | Thr | Met | R | LG | Liu et al., Nature Med 2:169–174 (1996) |
| 4 | 350 | 1 | 44 | C | T | Thr | Met | R | h83 | D. J. Bunyan (unpublished) |
| 4 | 350 | 1 | 44 | C | T | Thr | Met | R | BES-SG624 | B. Bressac-de Paillerets (unpublished) |
| 4 | 350 | 1 | 44 | C | T | Thr | Met | R | 84 | A. de la Chapelle (unpublished) |
| 4 | 350 | 1 | 44 | C | T | Thr | Met | R | 5 | Maliaka et al., Hum Genet 97:251–255 (1996) |
| 4 | 350 | 1 | 44 | C | T | Thr | Met | R | 434 | Buerstedde et al., J Med Genet 32:909–912 (1995) |
| 4 | 350 | 1 | 44 | C | G | Thr | Arg | C | … | Myriad et al. |
| 5 | 382 | 1 | 3 | G | C | Ala | Pro | C | 338 | V. Pensotti et al. (unpublished) |
| 6 | 479 | 1 | 25 | C | T | Ala | Val | R | … | A. M. Deffenbaugh et al. (unpublished) |
| 7 | 554 | 0 | 9 | T | G | Val | Gly | R | OZ-1 | A. M. Deffenbaugh et al. (unpublished) |
| 7 | 554 | 0 | 9 | T | G | Val | Gly | R | … | Kohonen-Corish et al., Am J Hum Genet 59:818–824 (1996) |
| 7 | 577 | 0 | 32 | T | C | Ser | Pro | C | NA-G.C. | P. Izzo et al. (unpublished) |
| 8 | 595 | 0 | 7 | G | C | Glu | Gln | R | 34 | T. Liu et al. (unpublished) |
| 8 | 637 | 0 | 49 | G | A | Val | Met | C | 39356 | Myriad et al. |
| 8 | 637 | 0 | 49 | G | A | Val | Met | C | … | Myriad et al. |
| 8 | 637 | 0 | 49 | G | A | Val | Met | C | … | G. Norbury et al. (unpublished) |
| 8 | 637 | 0 | 49 | G | A | Val | Met | C | … | Myriad et al. |
| 8 | 649 | 0 | 61 | C | T | Arg | Cys | R | 9 | Miyaki et al., J Mol Med 73:515–520 (1995) |
| 8 | 649 | 0 | 61 | C | T | Arg | Cys | R | SNU-H1006 | Han et al., J Natl Cancer Inst 88:1317–1319 (1996) |
| 8 | 677 | 1 | 89 | G | A | Arg | Gln | R | … | A. M. Deffenbaugh et al. (unpublished) |
| 8 | 677 | 1 | 89 | G | A | Arg | Gln | C | 6 | Maliaka et al., Hum Genet 97:251–255 (1996) |
| 8 | 677 | 1 | 89 | G | A | Arg | Gln | R | NLH-20 | Wijnen et al., Am J Hum Genet 58:300–307 (1996) |
| 9 | 731 | 1 | 55 | G | A | Gly | Asp | R | 311 | V. Pensotti et al. (unpublished) |
| 10 | 791 | 0 | 1 | A | G | Arg | Asp | R | 333633 | G. Norbury et al. (unpublished) |
| 10 | 793 | 1 | 3 | C | T | Arg | CyS | R | 198 | P. Hutter (unpublished) (1999) |
| 10 | 793 | 1 | 3 | C | T | Arg | CyS | R | VD0004 | T. Liu et al. (unpublished) |
| 10 | 794 | 1 | 4 | G | A | Arg | His | C | A-PD1 | Viel et al., Genes Chromosomes Cancer 18:8–18 (1997) |
| 10 | 803 | 0 | 13 | A | G | Glu | Gly | R | 80 | Liu et al., Clin Genet 53:131–135 (1998) |
| 10 | 814 | 0 | 24 | T | G | Leo | Val | R | … | Myriad et al. |
| 10 | 842 | 1 | 52 | C | T | Ala | Val | R | … | A. M. Deffenbaugh et al. (unpublished) |
| 11 | 977 | 1 | 94 | T | C | Val | Ala | R | 3273 | Myriad et al. |
| 11 | 977 | 1 | 94 | T | C | Val | Ala | R | 1515 | Liu et al., Nature Med 2:169–174 (1996) |
| 11 | 977 | 1 | 94 | T | C | Val | Ala | R | … | Buerstedde et al., J Med Genet 32:909–912 (1995) |
| 11 | 986 | 1 | 103 | A | C | His | Pro | C | 96 | Lamberti et al., Gut 44:839–843 (1999) |
| 12 | 1166 | 1 | 129 | G | A | Arg | Gln | C | … | A. M. Deffenbaugh et al. |
| 12 | 1217 | 0 | 180 | G | A | Ser | Asn | C | 209 | Cunningham et al., Am J Hum Genet 69:780–790 (2001) |
| 12 | 1321 | 1 | 284 | G | A | Ala | Thr | R | 466 | Cunningham et al., Am J Hum Genet 69:780–790 (2001) |
| 12 | 1321 | 1 | 284 | G | A | Ala | Thr | R | … | A. Krepelova (unpublished) |
| 13 | 1421 | 0 | 13 | G | A | Arg | Gln | R | DES-SG407 | B. Bressac-de Paillerets (unpublished) |
| 13 | 1474 | 1 | 66 | G | A | Ala | Thr | R | A4T | Möslein et al., Hum Mol Genet 5:1245–1252 (1996) |
| 13 | 1517 | 0 | 109 | T | C | Val | Ala | R | 3118 | Liu et al., Nature Med 2:169–174 (1996) |
| 13 | 1517 | 0 | 109 | T | C | Val | Ala | R | MDACC387858 | MDACC |
| 13 | 1517 | 0 | 109 | T | C | Val | Ala | R | … | A. M. Deffenbaugh et al. (unpublished) |
| 14 | 1569 | 0 | 12 | G | T | Glu | Asp | R | … | Myriad et al. |
| 14 | 1625 | 1 | 68 | A | T | Gln | Leu | R | SNUH-H2 | Han et al., Hum Mol Genet 4:237–242 (1995) |
| 14 | 1646 | 0 | 89 | T | C | Leo | Pro | R | SNU-H14 | Han et al., J. Natl. Cancer Inst. 88:1317–1319 (1996) |
| 14 | 1652 | 1 | 95 | A | T | Asp | Thr | R | VS012 | Hutter et al., Int J Cancer 78:680–684 (1998) |
| 15 | 1693 | 0 | 27 | A | T | Ile | Phe | R | VD001 | Hutter et al., Int J Cancer 78:680–684 (1998) |
| 15 | 1721 | 1 | 55 | T | C | Leo | Pro | R | SNUH-H1 | Han et al., Hum Mol Genet 4:237–242 (1995) |
| 16 | 1733 | 0 | 2 | A | G | Glu | Gly | R | 21 | A. de la Chapelle et al. (unpublished) |
| 16 | 1744 | 0 | 13 | C | G | Leu | Val | C | JPN-1 | Han et al., Hum Mol Genet 4:237–242 (1995) |
| 16 | 1771 | 0 | 40 | A | G | Glu | Gly | R | END0003 | Hutter et al., Int J Cancer 78:680–684 (1998) |
| 16 | 1853 | 0 | 122 | A | C | Lys | Thr | R | US-6 | T. Caldes (unpublished) |
| 17 | 1958 | 1 | 62 | T | G | Leu | Arg | R | … | Han et al., Hum Mol Genet 14:237–242 (1995) |
| 17 | 1961 | 1 | 65 | C | T | Pro | Leu | R | 38469 | G. Norbury et al. (unpublished) |
| 17 | 1963 | 1 | 67 | A | G | Ile | Val | C | … | Myriad et al. |
| 17 | 1976 | 1 | 80 | G | C | Arg | Pro | C | NL56 | Nyström-Lahti et al., Hum Mol Genet 5:763–769 (1996) |
| 17 | 1976 | 1 | 80 | G | C | Arg | Pro | C | 7 | A. de la Chapelle et al. (unpublished) (also mut. MLH1 exon 16, codon 618) |
| 17 | 1976 | 1 | 80 | G | A | Arg | Gla | C | 51 | J. T. Wijnen et al. (unpublished) |
| 17 | 1988 | 1 | 92 | A | G | Glu | Gly | R | CHI-SG277 | B. Bressac-de Paillerets (unpublished) |
| 18 | 2027 | 0 | 38 | T | G | Leu | Arg | R | … | Myriad et al. |
| 18 | 2041 | 0 | 52 | G | A | Ala | Thr | R | CO10 | Kurzawska and G. Kurzawski (unpublished) |
| 18 | 2041 | 0 | 52 | G | A | Ala | Thr | R | B.T.-1881 | Froggatt et al., J. Med. Genet.33: 726–730 (1996) |
| 19 | 2146 | 1 | 44 | G | A | Val | Meth | C | GE111 | Myriad et al. |
| 19 | 2146 | 1 | 44 | G | A | Val | Meth | C | … | Hutter et al., Int J Cancer 78:680–684 (1998) |
| 19 | 2152 | 0 | 50 | C | T | His | Tyr | R | CHO-SG632 | D. J. Bunyan (unpublished) |
Note.— The table enlists missense mutations from the HNPCC mutation database (accessed March 2003) and mutation data obtained by M.L.F. at MDACC.
C = conservative; R = radical.
Table C.
Database on hMSH2: Intragenic Polymorphisms and Sequence Variants[Note]
| Exon | Codon | Nucleotide Change | Consequence | ESE | Allele Frequency | Geographic Origin | Family ID | Reference |
| 2 | 110 | A→G at 329 | Lys→Arg | 0 | .02 | United States | HNPCC kindred | W. Weber and M. Rodriguez-Bigas (unpublished) |
| 2 | 113 | G→A at 339 | Lys→Lys | 0 | … | Czech Republic | Patient 435 | A. Krepelova (unpublished) |
| 2 | 113 | AAG→AAA | Lys→Lys | 0 | .0076 | … | Liu et al. Clin Genet 53:131–135 (1998) | |
| 3 | 133 | C→T at 399 | Asp→Asp | 0 | … | African American | Patient 31 | Johnson (unpublished) |
| 3 | 153 | C→T at 459 | Ser→Ser | 1 | … | None specified | … | Möslein et al., Hum Mol Genet 5:1245–1252 (1996) |
| 3 | 127 | A→G at 380 | Asn→Ser | 0 | .17 | Blood donors | A. de la Chapelle et al. (unpublished) | |
| 6 | 322 | G→A at 965 | Gly→Asp | 1 | … | Czech Republic | Patients 338, 376, 461, 468, 491 | A. Krepelova (unpublished) |
| 6 | 328 | GCC→GCT | Ala→Ala | 0 | … | France | Gue-SG618 | A. Lindblom et al. (unpublished) (1999) |
| 10 | 521 | T→C at 1563 | Tyr→Tyr | 0 | … | Czech Republic | Patient 424 | A. Krepelova (unpublished) |
| 11 | 556 | T→C at 1666 | Leu→Leu | 0 | .01 | Germany | 70 | Wehner et al., Hum Mut 10:241–244 (1997) |
| 11 | 579 | A→G at 1737 | Lys→Lys | 0 | .01 | Netherlands | NL22 | Wijnen et al., Am J Hum Genet 56:1060–1066 (1995) |
| 11 | 585 | T→C at 1755 | Ser→Ser | 1 | .005 | Finland | Finnish kindreds | Wahlberg et al. 74:134–137 (1997) |
| 12 | 596 | A→G at 1787 | Asn→Ser | 0 | <.004 | Italy | R-MD4 | Viel et al., Genes Chrom Cancer 18:8–18 (1997) |
| 13 | 713 | G→C at 2139 | Gly→Gly | 1 | .04 | Finland | Finnish kindreds | Nyström-Lahti et al., Hum Mol Genet 5:763–769 (1996) |
| 13 | 718 | A→G at 2154 | Gln→Gln | 0 | … | Lithuania | 03 | E. Avizienyte et al. (unpublished) |
Note.— The table enlists missense mutations from the HNPCC mutation database (accessed March 2003).
Table D.
Database on hMLH1: Intragenic Polymorphisms and Sequence Variants[Note]
| Exon | Codon | Nucleotide Change | Consequence | ESE | Allele Frequency | Geographic Origin | Family ID | Reference |
| 2 | 66 | C→T at 198 | Thr→Thr | 0 | .01 | Germany | 7 | Wehner et al., Hum Mut 10:241–244 (1997) |
| 8 | 219 | A→G at 655 | Ile→Val | 1 | .33 | Italy | Italian kindreds | A. Piepoli (unpublished) |
| 8 | 219 | A→C at 655 | Ile→Leu | 1 | .31 | None specified | … | Möslein et al., Hum Mol Genet 5:1245–1252 (1996) |
| 12 | 406 | G→A at 1217 | Ser→Asn | 0 | .015 | Finland | Finnish patients with colorectal cancer | Wu et al., Genes Chrom Cancer 4:269–278 (1997) |
| 16 | 618 | AA→GC at 1852, 1853 | Lys→Ala | 0 | .001 | None specified | … | A. M. Deffenbaugh et al. (unpublished) |
| 17 | 653 | G→T at 1959 | Leu→Leu | 1 | … | United Kingdom | 36902 | G. Norbury et al. (unpublished) (1999) |
| 18 | 676 | C→T at 2028 | Leu→Leu | 0 | … | Czech Republic | Patient 386 | A. Krepelova (unpublished) |
| 19 | 718 | C→T at 2152 | His→Tyr | 0 | .144 | African American | … | Kowalski et al., Genes Chromosomes Cancer 18:219–227 (1997) |
Note.— The table enlists missense mutations from the HNPCC mutation database (accessed March 2003).
Table E.
ESE Motifs Found in the hMSH2 and hMLH1 Genes
| SF2/ASF ESE Motifs |
SC35 ESE Motifs |
SRp40 ESE Motifs |
SRp55 ESE Motifs |
|||||||||||||
| Position in ORF |
Position in ORF |
Position in ORF |
Position in ORF |
|||||||||||||
| Gene | Left | Right | Motif | Score | Left | Right | Motif | Score | Left | Right | Motif | Score | Left | Right | Motif | Score |
| hMSH2 | 10 | 16 | CAGCCGA | 5.3 | 90 | 97 | GACCACCA | 4.5 | 67 | 73 | TTTCAGG | 5.0 | 8 | 13 | TGCAGC | 4.7 |
| hMSH2 | 14 | 20 | CGAAGGA | 3.2 | 121 | 128 | GACTTCTA | 4.7 | 95 | 101 | CCACAGT | 3.6 | 39 | 44 | CGCGGC | 4.5 |
| hMSH2 | 41 | 47 | CGGCCGA | 4.9 | 149 | 156 | CGCTGCTG | 3.3 | 111 | 117 | CGACCGG | 3.4 | 61 | 66 | CGCTTC | 3.0 |
| hMSH2 | 44 | 50 | CCGAGGT | 3.7 | 203 | 210 | GGCCGGCA | 3.5 | 277 | 283 | CTTCTGG | 4.4 | 101 | 106 | TGCGCC | 3.5 |
| hMSH2 | 96 | 102 | CACAGTG | 3.1 | 346 | 353 | GATTGGTA | 3.0 | 306 | 312 | TTATAAG | 3.1 | 129 | 134 | TACGGC | 4.7 |
| hMSH2 | 134 | 140 | CGCACGG | 5.4 | 366 | 373 | GGCTTCTC | 3.2 | 368 | 374 | CTTCTCC | 3.2 | 136 | 141 | CACGGC | 3.9 |
| hMSH2 | 159 | 165 | CGCCCGG | 4.4 | 369 | 376 | TTCTCCTG | 3.5 | 371 | 377 | CTCCTGG | 3.0 | 196 | 201 | TACATG | 3.2 |
| hMSH2 | 164 | 170 | GGGAGGT | 3.4 | 456 | 463 | GTCCGCAG | 3.9 | 422 | 428 | TGTCAGC | 3.6 | 224 | 229 | TGCAGA | 4.3 |
| hMSH2 | 179 | 185 | CCCAGGG | 3.5 | 469 | 476 | GGCCAGAG | 4.0 | 431 | 437 | CCATTGG | 3.3 | 236 | 241 | TGCTTA | 3.5 |
| hMSH2 | 207 | 213 | GGCAGGA | 4.0 | 498 | 505 | GGATTCCA | 3.4 | 475 | 481 | AGACAGG | 4.5 | 255 | 260 | TGAATC | 3.4 |
| hMSH2 | 459 | 465 | CGCAGTT | 3.1 | 520 | 527 | GGACTGTG | 3.8 | 507 | 513 | ACAGAGG | 3.6 | 284 | 289 | TTCGTC | 3.8 |
| hMSH2 | 476 | 482 | GACAGGT | 4.1 | 531 | 538 | ATTCCCTG | 3.5 | 586 | 592 | CCAAAGG | 4.4 | 420 | 425 | TATGTC | 4.0 |
| hMSH2 | 508 | 514 | CAGAGGA | 5.7 | 549 | 556 | GTTCTCCA | 3.4 | 601 | 607 | TTACCCG | 4.0 | 495 | 500 | TGTGGA | 3.4 |
| hMSH2 | 587 | 593 | CAAAGGA | 3.7 | 584 | 591 | GACCAAAG | 3.3 | 614 | 620 | AGACTGC | 3.4 | 506 | 511 | TACAGA | 3.7 |
| hMSH2 | 606 | 612 | CGGAGGA | 5.3 | 615 | 622 | GACTGCTG | 5.0 | 671 | 677 | TCACAGA | 3.9 | 561 | 566 | TGAGGC | 3.2 |
| hMSH2 | 654 | 660 | AAGAGGA | 3.2 | 662 | 669 | GAATTCTG | 4.1 | 698 | 704 | CCACAAA | 3.0 | 758 | 763 | TGAATA | 3.0 |
| hMSH2 | 672 | 678 | CACAGAA | 3.4 | 669 | 676 | GATCACAG | 4.0 | 700 | 706 | ACAAAAG | 3.1 | 801 | 806 | TTCATC | 3.2 |
| hMSH2 | 754 | 760 | CAGATGA | 3.8 | 718 | 725 | GACCTCAA | 3.9 | 712 | 718 | TATCAGG | 3.0 | 912 | 917 | TGCAGC | 4.7 |
| hMSH2 | 812 | 818 | CTGCGGT | 3.3 | 728 | 735 | GGTTGTTG | 3.0 | 720 | 726 | CCTCAAC | 3.5 | 989 | 994 | TGAATA | 3.0 |
| hMSH2 | 842 | 848 | CAGATGA | 3.8 | 759 | 766 | GAATAGTG | 3.5 | 734 | 740 | TGAAAGG | 3.9 | 1107 | 1112 | TGCAGA | 4.3 |
| hMSH2 | 871 | 877 | CTGACTA | 3.1 | 792 | 799 | GGTTGCAG | 3.7 | 758 | 764 | TGAATAG | 3.1 | 1115 | 1120 | TGAGGC | 3.2 |
| hMSH2 | 1030 | 1036 | CAGTGGA | 3.3 | 803 | 810 | CATCACTG | 3.0 | 805 | 811 | TCACTGT | 3.7 | 1139 | 1144 | TACTTC | 3.2 |
| hMSH2 | 1120 | 1126 | CAGACTT | 3.9 | 847 | 854 | GATTCCAA | 3.8 | 809 | 815 | TGTCTGC | 3.4 | 1142 | 1147 | TTCGTC | 3.8 |
| hMSH2 | 1282 | 1288 | CACCAGA | 3.1 | 873 | 880 | GACTACTT | 3.1 | 872 | 878 | TGACTAC | 3.8 | 1494 | 1499 | TGCAGC | 4.7 |
| hMSH2 | 1550 | 1556 | CACAGTT | 3.5 | 883 | 890 | GACTTCAG | 4.1 | 885 | 891 | CTTCAGC | 3.8 | 1570 | 1575 | CGTGTA | 3.4 |
| hMSH2 | 1633 | 1639 | CAGAAGA | 3.8 | 906 | 913 | GGATATTG | 3.0 | 920 | 926 | TCAGAGC | 3.8 | 1598 | 1603 | TTCGTA | 3.5 |
| hMSH2 | 1718 | 1724 | CCCAGGA | 4.3 | 942 | 949 | GGGTTCTG | 3.2 | 937 | 943 | TTTCAGG | 5.0 | 1762 | 1767 | TATGTA | 3.6 |
| hMSH2 | 1826 | 1832 | CTCACGT | 5.1 | 957 | 964 | TACCACTG | 3.6 | 959 | 965 | CCACTGG | 5.7 | 1775 | 1780 | TGCAGA | 4.3 |
| hMSH2 | 1984 | 1990 | CAGATGT | 3.5 | 964 | 971 | GGCTCTCA | 3.4 | 973 | 979 | TCTCTGG | 5.0 | 1830 | 1835 | CGTGTC | 3.8 |
| hMSH2 | 2016 | 2022 | GGGAGGT | 3.4 | 971 | 978 | AGTCTCTG | 3.4 | 1006 | 1012 | CCTCAAG | 4.4 | 1855 | 1860 | TATGTA | 3.6 |
| hMSH2 | 2099 | 2105 | CAGAAGT | 3.5 | 1148 | 1155 | GATTCCCA | 4.2 | 1029 | 1035 | CCAGTGG | 4.0 | 1905 | 1910 | AGCATC | 3.6 |
| hMSH2 | 2206 | 2212 | CTCAGGT | 4.5 | 1311 | 1318 | GACTCCTC | 3.6 | 1047 | 1053 | TCTCATG | 3.3 | 1993 | 1998 | CACATC | 4.1 |
| hMSH2 | 2253 | 2259 | AAGAGGA | 3.2 | 1515 | 1522 | GGACCCTG | 5.4 | 1066 | 1072 | ATAGAGG | 3.2 | 2091 | 2096 | TGAGTC | 4.0 |
| hMSH2 | 2406 | 2412 | CACAGCA | 3.4 | 1539 | 1546 | GGATTCCA | 3.4 | 1084 | 1090 | TTAGTGG | 4.0 | 2112 | 2117 | TGTGGA | 3.4 |
| hMSH2 | 2699 | 2705 | CAGAAGA | 3.8 | 1540 | 1547 | GATTCCAG | 4.3 | 1126 | 1132 | TTACAAG | 5.4 | 2119 | 2124 | TGCATC | 5.5 |
| hMSH2 | 1638 | 1645 | GAATGGTG | 3.0 | 1186 | 1192 | AGACAAG | 3.9 | 2196 | 2201 | TGCTTC | 3.8 | ||||
| hMSH2 | 1722 | 1729 | GGATGCCA | 3.2 | 1201 | 1207 | TTACAAG | 5.4 | 2295 | 2300 | TATATC | 3.4 | ||||
| hMSH2 | 1849 | 1856 | GTTCCATA | 3.5 | 1222 | 1228 | TATCAGG | 3.0 | 2332 | 2337 | TGCATG | 3.8 | ||||
| hMSH2 | 2005 | 2012 | GGCCCCAA | 5.0 | 1252 | 1258 | ATACAGG | 5.0 | 2399 | 2404 | TACATG | 3.2 | ||||
| hMSH2 | 2064 | 2071 | GGCCCAAA | 3.7 | 1310 | 1316 | TGACTCC | 4.1 | 2401 | 2406 | CATGTC | 3.1 | ||||
| hMSH2 | 2078 | 2085 | GTTTTGTG | 3.3 | 1319 | 1325 | TTACTGA | 3.4 | 2406 | 2411 | CACAGC | 3.3 | ||||
| hMSH2 | 2106 | 2113 | GTCCATTG | 3.6 | 1338 | 1344 | CTTCTCC | 3.2 | 2490 | 2495 | TGCAGA | 4.3 | ||||
| hMSH2 | 2139 | 2146 | GGCTGGTG | 4.4 | 1348 | 1354 | TTTCAGG | 5.0 | 2707 | 2712 | AACATC | 3.0 | ||||
| hMSH2 | 2210 | 2217 | GGTCTGCA | 3.1 | 1458 | 1464 | TGACTTG | 3.2 | 2799 | 2804 | TACGTG | 3.8 | ||||
| hMSH2 | 2211 | 2218 | GTCTGCAA | 3.2 | 1544 | 1550 | CCAGTGC | 3.2 | ||||||||
| hMSH2 | 2282 | 2289 | GGTTAGCA | 3.2 | 1632 | 1638 | CCAGAAG | 3.7 | ||||||||
| hMSH2 | 2319 | 2326 | GATTGGTG | 3.5 | 1754 | 1760 | CTTCAGG | 4.6 | ||||||||
| hMSH2 | 2415 | 2422 | CACCACTG | 3.6 | 1771 | 1777 | CCAATGC | 3.3 | ||||||||
| hMSH2 | 2481 | 2488 | GATTCATG | 4.0 | 1815 | 1821 | TGTCAGC | 3.6 | ||||||||
| hMSH2 | 2657 | 2664 | AGTTCCTG | 3.9 | 1900 | 1906 | TTAAAAG | 3.9 | ||||||||
| hMSH2 | 1992 | 1998 | CCACATC | 3.1 | ||||||||||||
| hMSH2 | 2000 | 2006 | TTACTGG | 5.8 | ||||||||||||
| hMSH2 | 2145 | 2151 | TGACAGT | 3.1 | ||||||||||||
| hMSH2 | 2156 | 2162 | TGAAAGG | 3.9 | ||||||||||||
| hMSH2 | 2205 | 2211 | CCTCAGG | 4.9 | ||||||||||||
| hMSH2 | 2360 | 2366 | TTACTGC | 4.9 | ||||||||||||
| hMSH2 | 2398 | 2404 | CTACATG | 3.6 | ||||||||||||
| hMSH2 | 2405 | 2411 | TCACAGC | 5.5 | ||||||||||||
| hMSH2 | 2412 | 2418 | ACTCACC | 3.1 | ||||||||||||
| hMSH2 | 2414 | 2420 | TCACCAC | 3.2 | ||||||||||||
| hMSH2 | 2417 | 2423 | CCACTGA | 3.3 | ||||||||||||
| hMSH2 | 2443 | 2449 | TATCAGG | 3.0 | ||||||||||||
| hMSH2 | 2578 | 2584 | TCGCAAG | 3.1 | ||||||||||||
| hMSH2 | 2650 | 2656 | ATTCAGG | 3.9 | ||||||||||||
| hMSH2 | 2687 | 2693 | TTACTGA | 3.4 | ||||||||||||
| hMSH2 | 2698 | 2704 | TCAGAAG | 4.0 | ||||||||||||
| hMSH2 | 2711 | 2717 | TCACAAT | 3.4 | ||||||||||||
| hMSH2 | 2722 | 2728 | TTAAAAC | 3.0 | ||||||||||||
| hMSH2 | 2731 | 2737 | CTAAAAG | 3.5 | ||||||||||||
| hMSH2 | 2780 | 2786 | TTTCACG | 4.7 | ||||||||||||
| hMSH2 | 2795 | 2801 | TTACTAC | 4.4 | ||||||||||||
| hMLH1 | 12 | 18 | GGCAGGG | 3.2 | 48 | 55 | GAACCGCA | 3.1 | 20 | 26 | TTATTCG | 3.1 | 23 | 28 | TTCGGC | 3.0 |
| hMLH1 | 59 | 65 | CGGCGGG | 3.6 | 163 | 170 | GGCCTGAA | 3.3 | 40 | 46 | ACAGTGG | 3.3 | 52 | 57 | CGCATC | 4.7 |
| hMLH1 | 132 | 138 | CACAAGT | 3.7 | 180 | 187 | GATCCAAG | 3.2 | 131 | 137 | CCACAAG | 5.4 | 57 | 62 | CGCGGC | 4.5 |
| hMLH1 | 195 | 201 | CACCGGG | 4.3 | 219 | 226 | GGATATTG | 3.0 | 139 | 145 | ATTCAAG | 3.3 | 77 | 82 | AGCGGC | 3.4 |
| hMLH1 | 392 | 398 | CAGATGG | 3.1 | 237 | 244 | GTTCACTA | 4.3 | 188 | 194 | ACAATGG | 3.5 | 289 | 294 | TATGGC | 3.2 |
| hMLH1 | 478 | 484 | GCCACGA | 3.3 | 258 | 265 | GTCCTTTG | 3.3 | 231 | 237 | TGAAAGG | 3.9 | 303 | 308 | TGAGGC | 3.2 |
| hMLH1 | 698 | 704 | GTGAGGA | 3.0 | 292 | 299 | GGCTTTCG | 3.1 | 239 | 245 | TCACTAC | 4.7 | 316 | 321 | AGCATA | 3.3 |
| hMLH1 | 842 | 848 | CAGCCTA | 3.2 | 312 | 319 | GGCCAGCA | 4.0 | 242 | 248 | CTACTAG | 4.8 | 327 | 332 | TGTGGC | 3.8 |
| hMLH1 | 922 | 928 | CACCCCA | 3.0 | 330 | 337 | GGCTCATG | 4.9 | 262 | 268 | TTTGAGG | 3.2 | 372 | 377 | TGCATA | 5.2 |
| hMLH1 | 950 | 956 | TGCACGA | 3.2 | 440 | 447 | GGACCCAG | 4.4 | 281 | 287 | TTTCTAC | 3.3 | 376 | 381 | TACAGA | 3.7 |
| hMLH1 | 1141 | 1147 | CACCAGA | 3.1 | 614 | 621 | GGACACTA | 4.5 | 287 | 293 | CCTATGG | 3.1 | 627 | 632 | TGCCTC | 3.8 |
| hMLH1 | 1144 | 1150 | CAGATGG | 3.1 | 639 | 646 | GGACAATA | 3.2 | 344 | 350 | TTACAAC | 4.6 | 652 | 657 | TCCATC | 3.2 |
| hMLH1 | 1238 | 1244 | CAGAGGA | 5.7 | 794 | 801 | GTCTGGTA | 3.3 | 387 | 393 | TTACTCA | 3.1 | 733 | 738 | TACATA | 4.6 |
| hMLH1 | 1264 | 1270 | GGCAGGG | 3.2 | 869 | 876 | CATTCCTG | 3.3 | 404 | 410 | TGAAAGC | 3.1 | 766 | 771 | TGCATC | 5.5 |
| hMLH1 | 1376 | 1382 | CAGAGAA | 3.2 | 895 | 902 | AGTCCCCA | 3.0 | 468 | 474 | TTACAAC | 4.6 | 781 | 786 | TTCATC | 3.2 |
| hMLH1 | 1383 | 1389 | GAGAGGA | 4.2 | 896 | 903 | GTCCCCAG | 4.8 | 616 | 622 | ACACTAC | 3.6 | 840 | 845 | TGCAGC | 4.7 |
| hMLH1 | 1485 | 1491 | CCCCCGG | 3.1 | 962 | 969 | GCATCCTG | 3.2 | 623 | 629 | CCAATGC | 3.3 | 877 | 882 | TACCTC | 3.2 |
| hMLH1 | 1486 | 1492 | CCCCGGA | 3.3 | 998 | 1005 | AGCTCCTG | 4.5 | 629 | 635 | CCTCAAC | 3.5 | 906 | 911 | TGTGGA | 3.4 |
| hMLH1 | 1507 | 1513 | CTCACTA | 3.3 | 1006 | 1013 | GGCTCCAA | 4.7 | 671 | 677 | TTAGTCG | 3.7 | 961 | 966 | AGCATC | 3.6 |
| hMLH1 | 1561 | 1567 | CTCCGGG | 3.1 | 1060 | 1067 | GGCCCCTC | 4.2 | 725 | 731 | TGAATGG | 3.7 | 977 | 982 | TGCAGC | 4.7 |
| hMLH1 | 1622 | 1628 | CACAGCA | 3.4 | 1104 | 1111 | GTCTTCTA | 4.4 | 740 | 746 | CCAATGC | 3.3 | 985 | 990 | CACATC | 4.1 |
| hMLH1 | 1720 | 1726 | CTCAGGT | 4.5 | 1110 | 1117 | TACTTCTG | 3.1 | 775 | 781 | TTACTCT | 3.1 | 1027 | 1032 | TACTTC | 3.2 |
| hMLH1 | 1793 | 1799 | CAGAGGA | 5.7 | 1131 | 1138 | GGTCTATG | 3.8 | 793 | 799 | CGTCTGG | 3.9 | 1101 | 1106 | CTCGTC | 3.0 |
| hMLH1 | 1868 | 1874 | CAGACTA | 4.2 | 1149 | 1156 | GGTTCGTA | 4.3 | 862 | 868 | ACACACC | 4.1 | 1110 | 1115 | TACTTC | 3.2 |
| hMLH1 | 1997 | 2003 | GGGACGA | 4.3 | 1159 | 1166 | GATTCCCG | 4.6 | 893 | 899 | TCAGTCC | 3.2 | 1151 | 1156 | TTCGTA | 3.5 |
| hMLH1 | 2075 | 2081 | CTGAGGA | 4.6 | 1193 | 1200 | AGCCTCTG | 3.9 | 926 | 932 | CCACAAA | 3.0 | 1155 | 1160 | TACAGA | 3.7 |
| hMLH1 | 2177 | 2183 | CACACAT | 3.7 | 1205 | 1212 | AACCCCTG | 4.3 | 942 | 948 | TCACTTC | 3.2 | 1190 | 1195 | TGCAGC | 4.7 |
| hMLH1 | 2202 | 2208 | CACAGAA | 3.4 | 1220 | 1227 | AGCCCCAG | 3.7 | 1000 | 1006 | CTCCTGG | 3.0 | 1392 | 1397 | TACTTC | 3.2 |
| hMLH1 | 2204 | 2210 | CAGAAGA | 3.8 | 1227 | 1234 | GGCCATTG | 4.3 | 1017 | 1023 | CTCCAGG | 3.3 | 1434 | 1439 | TGTGGA | 3.4 |
| hMLH1 | 2260 | 2266 | GAGAGGT | 3.8 | 1298 | 1305 | AACTCCCA | 3.0 | 1029 | 1035 | CTTCACC | 3.5 | 1473 | 1478 | TGCAGC | 4.7 |
| hMLH1 | 1305 | 1312 | AGCCCCTG | 4.7 | 1045 | 1051 | CTACCAG | 3.4 | 1520 | 1525 | TGAGTC | 4.0 | ||||
| hMLH1 | 1320 | 1327 | GGCTGCCA | 4.2 | 1055 | 1061 | TTGCTGG | 3.2 | 1574 | 1579 | TGCATA | 5.2 | ||||
| hMLH1 | 1347 | 1354 | GGATACAA | 3.3 | 1064 | 1070 | CCTCTGG | 4.7 | 1601 | 1606 | TGAATC | 3.4 | ||||
| hMLH1 | 1362 | 1369 | GACTTCAG | 4.1 | 1085 | 1091 | CCACAAC | 4.6 | 1622 | 1627 | CACAGC | 3.3 | ||||
| hMLH1 | 1453 | 1460 | GATTCCCG | 4.6 | 1088 | 1094 | CAACAAG | 3.1 | 1625 | 1630 | AGCATC | 3.6 | ||||
| hMLH1 | 1470 | 1477 | GACTGCAG | 3.9 | 1112 | 1118 | CTTCTGG | 4.4 | 1824 | 1829 | TGAATA | 3.0 | ||||
| hMLH1 | 1523 | 1530 | GTCTCCAG | 4.6 | 1140 | 1146 | CCACCAG | 3.7 | 1866 | 1871 | TGCAGA | 4.3 | ||||
| hMLH1 | 1558 | 1565 | GTTCTCCG | 3.8 | 1186 | 1192 | TTTCTGC | 3.9 | 1958 | 1963 | TGCCTA | 3.5 | ||||
| hMLH1 | 1717 | 1724 | GTTCTCAG | 3.5 | 1222 | 1228 | CCCCAGG | 3.6 | 2053 | 2058 | TCCATC | 3.2 | ||||
| hMLH1 | 1775 | 1782 | GTCCAGAG | 3.4 | 1235 | 1241 | TCACAGA | 3.9 | 2068 | 2073 | TACATA | 4.6 | ||||
| hMLH1 | 1790 | 1797 | GGACAGAG | 3.0 | 1237 | 1243 | ACAGAGG | 3.6 | 2145 | 2150 | TGTGGA | 3.4 | ||||
| hMLH1 | 1804 | 1811 | GGTCCCAA | 4.4 | 1256 | 1262 | TTTCTAG | 4.2 | 2186 | 2191 | TGCCTC | 3.8 | ||||
| hMLH1 | 1805 | 1812 | GTCCCAAA | 3.1 | 1259 | 1265 | CTAGTGG | 3.7 | 2222 | 2227 | TGCAGC | 4.7 | ||||
| hMLH1 | 1838 | 1845 | AGTTTCTG | 3.1 | 1301 | 1307 | TCCCAGC | 3.1 | ||||||||
| hMLH1 | 1980 | 1987 | AGCCACTG | 4.2 | 1332 | 1338 | TCAGAGC | 3.8 | ||||||||
| hMLH1 | 2095 | 2102 | GGCCAGCA | 4.0 | 1349 | 1355 | ATACAAC | 3.6 | ||||||||
| hMLH1 | 2128 | 2135 | AACTCCTG | 4.1 | 1354 | 1360 | ACAAAGG | 3.7 | ||||||||
| hMLH1 | 2141 | 2148 | GGACTGTG | 3.8 | 1459 | 1465 | CGAAAGG | 3.6 | ||||||||
| hMLH1 | 2187 | 2194 | GCCTCCTA | 3.7 | 1469 | 1475 | TGACTGC | 4.4 | ||||||||
| hMLH1 | 2242 | 2249 | GATCTATA | 3.0 | 1508 | 1514 | TCACTAG | 5.5 | ||||||||
| hMLH1 | 1525 | 1531 | CTCCAGG | 3.3 | ||||||||||||
| hMLH1 | 1560 | 1566 | TCTCCGG | 3.5 | ||||||||||||
| hMLH1 | 1581 | 1587 | CCACTCC | 4.6 | ||||||||||||
| hMLH1 | 1608 | 1614 | TCAGTGG | 4.4 | ||||||||||||
| hMLH1 | 1629 | 1635 | TCAAACC | 3.6 | ||||||||||||
| hMLH1 | 1647 | 1653 | TCTCAAC | 3.9 | ||||||||||||
| hMLH1 | 1680 | 1686 | CTACCAG | 3.4 | ||||||||||||
| hMLH1 | 1719 | 1725 | TCTCAGG | 5.3 | ||||||||||||
| hMLH1 | 1792 | 1798 | ACAGAGG | 3.6 | ||||||||||||
| hMLH1 | 1915 | 1921 | TTACCCC | 3.1 | ||||||||||||
| hMLH1 | 1929 | 1935 | TGACAAC | 4.1 | ||||||||||||
| hMLH1 | 1975 | 1981 | CGACTAG | 4.3 | ||||||||||||
| hMLH1 | 1982 | 1988 | CCACTGA | 3.3 | ||||||||||||
| hMLH1 | 2019 | 2025 | TGAAAGC | 3.1 | ||||||||||||
| hMLH1 | 2074 | 2080 | TCTGAGG | 3.6 | ||||||||||||
| hMLH1 | 2090 | 2096 | TCTCAGG | 5.3 | ||||||||||||
| hMLH1 | 2130 | 2136 | CTCCTGG | 3.0 | ||||||||||||
| hMLH1 | 2176 | 2182 | TCACACA | 3.6 | ||||||||||||
| hMLH1 | 2201 | 2207 | TCACAGA | 3.9 | ||||||||||||
| hMLH1 | 2259 | 2265 | TGAGAGG | 3.8 | ||||||||||||
Table 1.
Types and Numbers of Mutations Analyzed
|
No. of Mutations |
||||
| Gene | Pathogenic Missense | Nonsense and Frameshift | Nonpathogenic Missense | Total Mutations |
| hMSH2 | 50 | 81 | 17 | 148 |
| hMLH1 | 99 | 68 | 8 | 175 |
First, we searched the coding regions of the genes for the presence of ESE motifs with ESEfinder software. To reduce the number of false-positive results, we used a more-stringent-than-recommended threshold value of 3.0 for all four types of ESE motifs. Potential ESE motifs found in the hMSH2 and hMLH1 genes are listed in table E (online only). We excluded ESEs in exon/exon boundaries, accounted for overlap between different ESEs by counting as a single ESE any segment containing two or more ESEs, and estimated the percentage of sequence that consists of ESE motifs for each entire gene and each exon. This estimate provided us with the proportion of mutations expected to be in ESE motifs under the null hypothesis that assumes that there is no association between pathogenic missense mutations and exonic splicing enhancers.
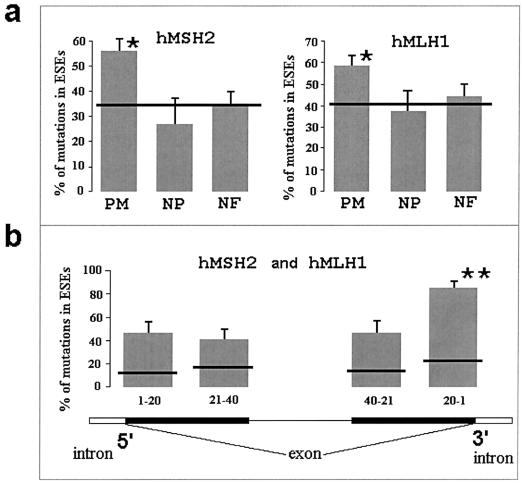
Different nucleotide substitutions in mutation databases for the hMSH2 and hMLH1 genes differ with respect to how many times they are reported in the databases. Some of them are listed only once, whereas others are reported several times (e.g., the C→T transition at position 350 in the hMLH1 gene is reported 11 times). Multiply reported mutations in the mutation databases originate from different families. Counting each reported mutation, we found that missense mutations are colocalized with ESEs (for hMSH2, χ2=11.8, df=1, P<.001; for hMLH1, χ2=7.9, df=1, P<.01) (fig. 1a). Alternatively, we also counted each separate mutation only once, no matter how often it was listed in the database. Again, we found that, in both genes, deleterious missense mutations are located in ESEs more frequently than expected (for hMSH2, χ2=9.4, df=1, P<.001; for hMLH1, χ2=4.3, df=1, P<.05). Counting each mutation only once eliminates all possibility that families are related but probably leads to a downward bias. Nucleotide substitutions can occur at any position in a coding region of a gene; yet only deleterious mutations that disturb important functional sites would lead to cancer and thus have a chance of being detected by screening of cancer-affected families. The more deleterious the mutation, the higher the chance it has of causing disease and of being detected. Therefore, deleterious mutations are expected to be most frequent in mutation databases (Martin et al. 2002; Olivier et al. 2002). Counting multiple mutations only once leads to loss of information and may cause downward bias by reducing the number of observations and by eliminating variation in the number of mutations between different mutant sites.
Figure 1.
a, Frequency of ESE-associated mutations. Horizontal lines show the expected frequencies of mutations calculated as a proportion of the sequence occupied by ESE motifs. Differences between the expected and observed frequencies of pathogenic missense mutations (PM) were significant for both hMSH2 and hMLH1 (marked by an asterisk [*]). Nonpathogenic missense mutations (NP) show a trend to be underrepresented in ESEs. Nonsense and frameshift mutations (NF) did not show preferential association with ESE sites. Bars represent SEs of the frequencies. b, Positions of pathogenic missense mutations with respect to the 5′ and 3′ ends of an exon. The last 20 nts of exons had the highest frequency of mutations in ESEs: 83% of the mutations were in ESE sites (marked by a double asterisk [**]). The difference between the observed and expected numbers of mutations in that region was highly significant (χ2=33.8; df=1; P<.001). When the analysis was limited to short (average size, 80 nts) exons, the colocalization of the pathogenic missense mutations with ESE sites was even higher: 96% of the mutations were in ESEs.
There may be two possible explanations for missense mutations in hMSH2 and hMLH1 genes being preferentially located in ESE motifs: either missense mutations arise more frequently in ESEs, or mutations in ESEs are more pathogenic than mutations outside ESEs and therefore are more likely to be detected during screening of affected individuals. Our analysis favors the second explanation. If a missense mutation becomes pathogenic because it is located in an ESE, then one can expect pathogenic missense mutations to be located in ESE sites more frequently, compared with nonpathogenic ones. We found that 57% (28/49) of pathogenic missense mutations were located in ESEs of the hMSH2 gene versus 24% (4/17) of nonpathogenic mutations (χ21=5.66; P=.02; Fisher's exact test). For the hMLH1 gene, we also found that pathogenic missense mutations were more frequently located in ESE sites than nonpathogenic mutations—58% (57/99) versus 38% (3/8), respectively (χ21=1.21; P=.30; Fisher's exact test)—but there were too few nonpathogenic mutations to draw a meaningful conclusion.
If deleterious effects of nucleotide substitutions located inside ESE motifs result from disruption of splicing, then even missense mutations that are unlikely to affect protein structure (e.g., mutations that do not change the type of amino acid) will have a chance to be deleterious because they disturb ESEs and, therefore, splicing. This idea is supported by stratified analysis. We stratified missense mutations into “conservative” and “radical” (classifying them according to specifications of Dagan et al. [2002]), and we found that, in both genes, missense mutations located outside ESE sites tended to be “radical,” strongly affecting protein-structure mutations, whereas those located in ESE motifs are more likely to be “conservative” mutations that have no or slight effect on protein structure. For hMSH2 genes, the frequency of conservative missense mutations located in ESEs is 0.61±0.09, whereas the frequency of conservative missense mutations outside ESEs is 0.50±0.10. For the hMLH1 gene, we found the same trend: the frequency of conservative missense mutations in ESEs is 0.31±0.06, whereas the frequency of conservative mutations outside ESEs is 0.21±0.10. For both genes, the differences in the proportions of conservative missense mutations located inside and outside of ESE sites are not significant, even after we combine the data for both genes, probably because of the relatively low number of mutations in analysis.
The correlation of different types of mutations with ESE sites can be explained as follows: nonsense and frameshift mutations always produce truncated nonfunctional proteins and therefore always—no matter where they are located with respect to ESEs—are sufficiently damaging to cause disease. Thus, truncating mutations have a high chance of being detected by screening affected families. Missense mutations, especially those located outside important functional domains, may not change protein structure sufficiently to be pathogenic. However, if a nucleotide substitution occurs in a functional ESE site, it could disturb normal splicing and be sufficiently deleterious to cause disease. This could explain why affected individuals are enriched with missense mutations that are located in ESE sites and why polymorphisms found in unaffected individuals are not associated with ESEs and even show a trend not to be localized there.
Different single-nucleotide substitutions can change the ESE score in different directions: some substitutions increase the score, whereas others decrease it. If nucleotide substitutions located in ESE sites are deleterious because they disturb functional ESE sites, then such substitutions are expected mainly to decrease ESE scores. We compared the observed and expected proportion of score-decreasing missense mutations located inside ESE sites. The expected proportion of score-decreasing substitutions was calculated on the basis of all possible substitutions in the ESEs that lead to missense mutations. We found that ESE-located missense mutations reported in hMSH2 and hMLH1 mutation databases decrease ESE scores significantly more frequently than one would expect. For the hMSH2 gene, we found that the expected frequency of score-decreasing mutations is 0.77±0.03, whereas the observed frequency of score-decreasing mutations is 0.96±0.04. The differences are highly significant (χ2=6.5; df=1; P<.01). A similar result was obtained for the hMLH1 gene: the expected frequency of score-decreasing mutations is 0.78±0.02, whereas the observed frequency of score-decreasing mutations is 0.91±0.06 (χ2=5.9; df=1; P<.01).
The excess of pathogenic mutations in ESE sites compared with the expected frequency provides a minimal estimate of the proportion of missense mutations, the pathogenic effects of which are ESE related. As an upper limit for the estimate of the proportion of ESE-related mutations, one can suggest that all pathogenic missense mutations located in ESE sites are deleterious because they disturb functional splicing enhancers. This approach is likely to overestimate the proportion of ESE-related pathogenic mutations. First, not all ESE motifs are actual functional splicing enhancers (Cartegni 2002). Second, not all nucleotide substitutions in functional ESEs disturb their function (Cartegni and Krainer 2002; Fackenthal et al. 2002; Moseley et al. 2002; Pollard et al. 2002; ESEfinder). For the hMSH2 gene, the observed frequency of pathogenic missense mutations in ESEs is 55%, whereas the expected frequency is 36%. This means that 20%–55% of missense mutations in hMSH2 are pathogenic, because they affect ESE sites and therefore disturb normal splicing. A similar reasoning shows that the frequency of ESE-related mutations in the hMLH1 gene is 16%–58%.
Although an exon usually has several ESE motifs, the splicing machinery does not use most of them (Cartegni 2002; ESEfinder). The question is whether the functional ESE sites are distributed randomly. If functional ESEs are preferentially located in some specific regions within an exon, then the association of the pathogenic missense mutations with ESEs will be higher in that region. A study of the molecular mechanisms of splicing suggests that functional ESE sites occupy specific positions relative to the 5′ or 3′ ends of an exon (Blencowe 2000; Hastings and Krainer 2001; Cartegni et al. 2002). We compared the expected and observed frequencies of pathogenic missense mutations in four regions: the first 20 nts located near the 5′ end of an exon, nts 21–40 near the 5′ end of an exon, the first 20 nts near the 3′ end of an exon, and nts 21–40, starting from the 3′ exon region (fig 1b). Because the number of missense mutations located in these specific regions is relatively low, we combined the data on both the hMLH1 and hMSH2 genes. We found that >80% of pathogenic missense mutations that are located in the last 20 nts of exons (especially in short exons—80 nts, on average) strongly colocalize with ESEs (fig. 1b). This finding suggests that functional ESEs are preferentially located near the 3′ ends of exons. However, since we used mostly short exons (∼80 nts), it is noteworthy that, in fact, the functional ESEs are located 60–65 nts from the 5′ ends of exons.
Aside from the SR proteins that we have studied here, other classes of exonic enhancers exist; for example, those driven by hnRNP proteins (Chabot et al. 2003). In future research, studies of the frequency of mutations that affect other ESE sites would be of interest, once algorithms have been developed for evaluating the effect that mutations have on splicing related to these proteins. In summary, our analyses provide compelling evidence that many missense mutations associated with deleterious effects for hMLH1 and hMSH2 affect splicing. Of note, we found that conservative mutations that heretofore may not have had an obvious role in causing HNPCC may disrupt splicing. Further studies to evaluate the isoforms of mRNAs in individuals with missense mutations in ESE sites would help to confirm the role of missense mutations in disease causation and to provide clinical insight into the significance of these mutations.
Acknowledgments
This research was supported, in part, by a cancer-prevention fellowship supported by National Cancer Institute grants R25 CA57730 (Robert M. Chamberlain, Ph.D., principal investigator), CA70759 (to M.L.F.), CA16672, and National Human Genome Research Institute grant HG02275.
Electronic-Database Information
URLs for data presented herein are as follows:
- ESEfinder, http://exon.cshl.org/ESE/index.html
- Mutation Database on Pathogenic Mutations and Polymorphism for the hMLH1 gene, http://www.nfdht.nl/database/database-mlh1.htm
- Mutation Database on Pathogenic Mutations and Polymorphism for the hMSH2 gene, http://www.nfdht.nl/database2/database-msh2.htm
- Mutation Database Polymorphism for the hMLH1 gene, http://www.nfdht.nl/MLH1-poly/poly-mlh1.htm
- Mutation Database Polymorphism for the hMLH2 gene, http://www.nfdht.nl/MSH2-poly/poly-msh2.htm
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for HNPCC) [PubMed]
References
- Ars E, Serra E, Garcia J, Kruyer H, Gaona A, Lazaro C, Estivill X (2000) Mutations affecting mRNA splicing are the most common molecular defects in patients with neurofibromatosis type 1. Hum Mol Genet 9:237–247 [DOI] [PubMed] [Google Scholar]
- Blencowe BJ (2000) Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem Sci 25:106–110 [DOI] [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR (2002) Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 3:285–298 [DOI] [PubMed] [Google Scholar]
- Cartegni L, Krainer AR (2002) Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet 30:377–384 [DOI] [PubMed] [Google Scholar]
- Chabot B, LeBel C, Hutchison S, Nasim FH, Simard MJ (2003) Heterogeneous nuclear ribonucleoprotein particle A/B proteins and the control of alternative splicing of the mammalian heterogeneous nuclear ribonucleoprotein particle A1 pre-mRNA. Prog Mol Subcell Biol 31:59–88 [DOI] [PubMed] [Google Scholar]
- Dagan T, Talmor Y, Graur D (2002) Ratios of radical to conservative amino acid replacement are affected by mutational and compositional factors and may not be indicative of positive Darwinian selection. Mol Biol Evol 19:1022–1025 [DOI] [PubMed] [Google Scholar]
- Fackenthal JD, Cartegni L, Krainer AR, Olopade OI (2002) BRCA2 T2722R is a deleterious allele that causes exon skipping. Am J Hum Genet 71:625–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR (2000) Sorting out the complexity of SR protein functions. RNA 6:1197–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings ML, Krainer AR (2001) Pre-mRNA splicing in the new millennium. Curr Opin Cell Biol 13:302–309 [DOI] [PubMed] [Google Scholar]
- Liu HX, Chew SL, Cartegni L, Zhang MQ, Krainer AR (2000) Exonic splicing enhancer motif recognized by human SC35 under splicing conditions. Mol Cell Biol 20:1063–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HX, Zhang M, Krainer AR (1998) Identification of functional exonic splicing enhancer motifs recognized by indiovidual SR proteins. Genes Dev 12:1998–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AC, Facchiano AM, Cuff AL, Hernandez-Boussard T, Olivier M, Hainaut P, Thornton JM (2002) Integrating mutation data and structural analysis of the TP53 tumor-suppressor protein. Hum Mutat 19:149–164 [DOI] [PubMed] [Google Scholar]
- Moseley CT, Mullis PE, Prince MA, Phillips JA 3rd (2002) An exon splice enhancer mutation causes autosomal dominant GH deficiency. J Clin Endocrinol Metab 87:847–852 [DOI] [PubMed] [Google Scholar]
- Olivier M, Eeles R, Hollstein M, Khan MA, Harris CC, Hainaut P (2002) The IARC TP53 database: new online mutation analysis and recommendations to users. Hum Mutat 19:607–614 [DOI] [PubMed] [Google Scholar]
- Peltomaki P, Vasen HF (1997) Mutations predisposing to hereditary nonpolyposis colorectal cancer: database and results of a collaborative study. The International Collaborative Group on Hereditary Nonpolyposis Colorectal Cancer. Gastroenterology 113:1146–1158 [DOI] [PubMed] [Google Scholar]
- Pollard AJ, Krainer AR, Robson SC, Europe-Finner GN (2002) Alternative splicing of the adenylyl cyclase stimulatory G-protein G α(s) is regulated by SF2/ASF and heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1) and involves the use of an unusual TG 3′-splice site. J Biol Chem 277:15241–15251 [DOI] [PubMed] [Google Scholar]
- Stojdl DF, Bell JC (1999) SR protein kinases: the splice of life. Biochem Cell Biol 77:293–298 [PubMed] [Google Scholar]