Abstract
Mutations in the ASPM gene at the MCPH5 locus are expected to be the most common cause of human autosomal recessive primary microcephaly (MCPH), a condition in which there is a failure of normal fetal brain development, resulting in congenital microcephaly and mental retardation. We have performed the first comprehensive mutation screen of the 10.4-kb ASPM gene, identifying all 19 mutations in a cohort of 23 consanguineous families. Mutations occurred throughout the ASPM gene and were all predicted to be protein truncating. Phenotypic variation in the 51 affected individuals occurred in the degree of microcephaly (5–11 SDs below normal) and of mental retardation (mild to severe) but appeared independent of mutation position.
Human autosomal recessive primary microcephaly (MCPH [MIM 251200]; appendix A) (fig. 1) is a rare disorder in which the head circumference is reduced because of a congenital deficiency of fetal brain growth, particularly affecting the cerebral cortex (Bundey 1997; Mochida and Walsh 2001). The MCPH brain, although small, is structurally normal, and the phenotypic result is mental retardation but no other neurological defect (Aicardi 1998). Genetic heterogeneity has been shown, with five loci (MCPH1–5) reported to date (Jackson et al. 1998, 2002; Jamieson et al. 1999, 2000; Roberts et al. 1999; Moynihan et al. 2000; Pattison et al. 2000), all of which are clinically indistinguishable disorders (Roberts et al. 2002). Linkage analysis has identified MCPH5 on chromosome 1q31 (Jamieson et al. 2000; Pattison et al. 2000) as potentially the most common MCPH locus, accounting for 43% (24/56) of MCPH families in the northern Pakistani population that was studied (Roberts et al. 2002).
Figure 1.
A northern Pakistani individual with MCPH. This individual has the sloping forehead typical of those with MCPH. He and other affected members of his family have an ASPM mutation described in this report.
The ASPM gene (abnormal, spindle-like, microcephaly-associated [MIM 650481]), the human orthologue of the Drosophila abnormal, spindle gene (asp) (Ripoll et al. 1985), was recently identified as the MCPH5 gene (Bond et al. 2002). ASPM is composed of 28 exons with a 10,434-bp ORF (NCBI GenBank accession number AF509326) and encodes 3,477 amino acids. Collectively, exon 3 (1,486 bp) and exon 18 (4,755 bp) contain most of the coding region of the gene. ASPM is predicted to contain an amino terminal microtubule binding domain (Saunders et al. 1997), a calponin homology domain, 74 isoleucine-glutamine (IQ) domains (which potentially bind calmodulin) (Craig and Norbury 1998; Bond et al. 2002), and a carboxy-terminal region with no identified domains. The predicted ASPM protein is conserved between human, mouse, Drosophila, and worm, with a consistent correlation of nervous-system complexity and protein length, principally involving an increase in the number of encoded IQ domains (Bond et al. 2002). Drosophila asp is associated with the minus ends of microtubules at spindle poles in mitosis and meiosis (Ripoll et al. 1985; Gonzalez et al. 1990). Drosophila asp mutants are embryonic lethal, and in the CNS many neuron progenitors are unable to complete asymmetric cell division, arresting in metaphase (Gonzalez et al. 1989).
We have previously reported four homozygous ASPM mutations in four consanguineous northern Pakistani families linked to the MCPH5 locus (Bond et al. 2002). We now report our comprehensive mutation analysis of a further 23 consanguineous families: 17 from northern Pakistan (a Pakistani individual with MCPH is shown in fig. 1), 2 white Dutch families (1 of which has been previously described as part of “family seven” in the work of Van den Bosch [1959]), 2 families from Yemen, 1 Jordanian Bedouin family, and 1 Saudi Arabian family. The 23 families encompass a total of 51 affected individuals, all of whom are homozygous for the MCPH5 region and excluded from linkage to the MCPH1–4 loci. The study was approved by the Ethical Committee of the Combined Leeds Health Care Trusts and informed consent was obtained from all subjects involved or, in the case of subjects under 18 years of age, from their parents.
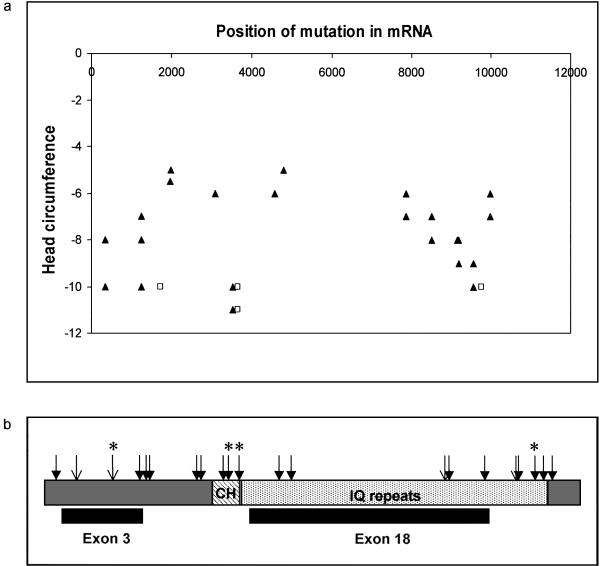
Within our 23 consanguineous families, we identified a greater phenotypic spectrum than previously described. Of the 23 families studied, 19 displayed head circumferences of 5–11 SDs below the mean (corrected for age and sex) and had mild to moderate mental retardation (fig. 2a). The remaining four families (two northern Pakistani families, each with four affected members, and two Yemeni families, each with one affected member) all had severe mental retardation and head circumferences of 10–11 SDs below the mean (shown as squares in fig. 2a). These individuals were unable to read or write, displayed primitive language skills, and could perform only the most basic tasks. No affected individual had seizures, spasticity, dementia, or psychiatric disease.
Figure 2.
The position of ASPM mutations identified in families with MCPH5. a, ASPM mutation position compared to the degree of microcephaly. On the x axis, the position of the ASPM gene mutations (listed in table C1) is shown, with the gene sequence aligned 5′→3′. The head circumference of individuals with MCPH5 in whom a mutation was found in this study is shown on the y axis. The head circumference is shown as a SD from the mean (corrected for age and sex). Some points represent more than one affected individual. Those individuals who were judged to exhibit severe mental retardation are shown as unfilled squares, whereas those with mild or moderate mental retardation are represented as filled triangles. There is no relationship between mutation position and head circumference or the degree of mental retardation. b, The ASPM protein as a schematic with the major protein domains depicted; CH (diagonally striped box) depicts the calponin homology domain, and the IQ domain repeats (light gray hatched box) are represented. The position of the large nonterminal exons 3 and 18 are indicated beneath the corresponding region they encode. Novel mutations identified in this study are shown as filled arrows. Unfilled arrows indicate mutations reported in the work of Bond et al. (2002). Mutations found in more than one family are denoted with an asterisk.
To aid mutation detection, the 17 northern Pakistani families were initially categorized by genotyping novel and published microsatellite repeats in a 0.6-Mb region centered upon the ASPM gene (1pter-D1S2816-AL139418TG18-AL353809TA20-AL353809GA13-[ASPM]-AL356315TG19-AL356315CA20-1qter). Within these 17 families, 11 haplotypes (A–K) were identified, of which 1 haplotype was common to 4 families and 3 haplotypes were each common to 2 families (appendix B, table B1). The genomic structure of ASPM had previously been determined, and intronic primers had been designed to amplify and sequence across the ASPM coding regions, including splice and polyadenylation sites (Bond et al. 2002). The ASPM gene was directly sequenced from genomic DNA in one affected individual from each northern Pakistani haplotype and from each of the other families.
Different homozygous mutations were identified in all 11 northern Pakistani haplotypes, as well as the Jordanian family, the Saudi Arabian family, and the two Yemeni families (table 1). Both of the Dutch families displayed the same mutation. To confirm the segregation of each mutation, genomic DNA from members of each family was assessed for the respective alteration. In 20 families, 16 mutations segregated correctly. However, three of the families from northern Pakistan (two families with haplotype B and one family with haplotype C) did not exhibit the expected mutations. Upon screening of an individual from each of these families, three different mutations were identified, two on the haplotype B background and one on the haplotype C background. These mutations segregated with the disease within the respective families. Therefore, although analysis of genotype data from the six markers flanking ASPM identified 11 separate haplotypes, it failed to discriminate between all mutation-specific haplotypes. This may be because of the paucity of polymorphic microsatellite markers in the ASPM region, but it more likely reflects mutational heterogeneity, even in a seemingly ethnically and geographically defined population.
Table 1.
Mutations in ASPM Causing Premature Protein Truncation
| Mutation | Haplotype | Location | Alterationa | ASPM Sizeb | Familiesc | Population |
| 349C→T | A | Exon 2 | Immediate stop | 116 | 1 | Northern Pakistani |
| 1258delTCTCAAGd | B1e | Exon 3 | Frameshift, ORF incorporating 31 aa then stop | 450 | 2 | Northern Pakistani |
| 9190C→T | B2e | Exon 21 | Immediate stop | 3,063 | 1 | Northern Pakistani |
| IVS9+5G→T (2936+5G→T) | B3e | Intron 7 | Removes splice-donor site, additional 2 aa then stop | 981 | 1 | Northern Pakistani |
| 1990C→T | C1f | Exon 4 | Immediate stop | 663 | 1 | Northern Pakistani |
| 8508delGA | C2f | Exon 18 | Frameshift, ORF incorporating 33 aa then stop | 2,869 | 1 | Northern Pakistani |
| 3082G→A | D | Exon 11 | Removes splice-donor site, additional 3 aa then stop | 1,030 | 1 | Northern Pakistani |
| 4581delA | E | Exon 18 | Frameshift, ORF incorporating 23 aa then stop | 1,550 | 1 | Northern Pakistani |
| 4795C→T | F | Exon 18 | Immediate stop | 1,598 | 1 | Northern Pakistani |
| 7895C→T | G | Exon 18 | Frameshift, ORF incorporating 15 aa then stop | 2,634 | 1 | Northern Pakistani |
| 9159delAd | H | Exon 21 | Frameshift, ORF incorporating 4 aa then stop | 3,056 | 1 | Northern Pakistani |
| 9557C→G | I | Exon 23 | Immediate stop | 3,185 | 2 | Northern Pakistani |
| IVS25+1G→T (9984+1G→T) | J | Intron 25 | Removes splice-donor site, additional 29 aa then stop | 3,357 | 1 | Northern Pakistani |
| 3663delGg | K | Exon 15 | Frameshift, ORF incorporating 12 aa then stop | 1,223 | 2 | Northern Pakistani |
| 3811C→T | Exon 16 | Immediate stop | 1,270 | 2 | Dutch | |
| 3527C→G | Exon 14 | Immediate stop | 1,175 | 1 | Jordanian | |
| 1959delCAAA | Exon 4 | Frameshift, ORF incorporating 13 aa then stop | 666 | 1 | Saudi Arabian | |
| 1727delAGg | Exon 3 | Frameshift, ORF incorporating 32 aa then stop | 607 | 1 | Yemeni | |
| 9754delAg | Exon 24 | Frameshift, ORF incorporating 9 aa then stop | 3,260 | 1 | Yemeni |
Mechanism of truncation caused by mutation.
Predicted size in amino acids of the truncated protein product.
Number of families with mutation.
Mutations previously reported in other northern Pakistani families (Bond et al. 2002).
Three mutations were associated with the haplotype B background.
Two mutations were associated with the haplotype C background.
Mutations associated with severe mental retardation.
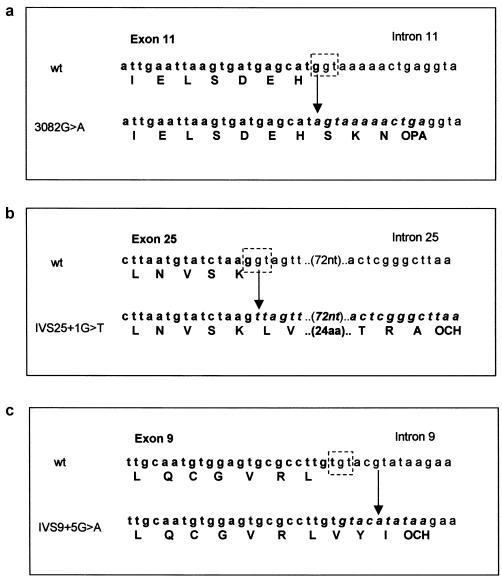
In total, 19 homozygous ASPM mutations (fig. 2b) and 12 nonpathogenic SNPs (appendix C, table C1) were identified in our 23 families. Of these, 17 mutations were novel and 2 mutations, 1258delTCTCAAG and 9159delA, had previously been reported in the northern Pakistani population (Bond et al. 2002). All mutations were predicted to cause premature protein truncation. In nine of the mutations, deletions of one to seven consecutive nucleotides resulted in the addition of 4 to 33 amino acids downstream of the mutation (translations performed using the INFOBIOGEN program), terminating because of the generation of a stop signal (table 1). Single-base changes caused 10 mutations, 7 of which introduced immediate-stop codons. The three remaining point mutations occurred at splice-donor sites as follows: 3082G→A, the last base of exon 11, removed the splice-donor site exon/intron 11, G/gt→A/gt (using the Berkeley Drosophila Genome Project splice prediction program, donor-motif probability was reduced from 0.89 to <0.1), potentially allowing transcription read-through for a further three amino acids until an alternative stop signal was reached (fig. 3a); IVS25+1G→T (9984+1G→T), an intronic mutation at exon/intron 25, G/gt→G/tt (donor-motif probability reduced from 0.76 to <0.1), incorporated 29 amino acids into the protein before a stop signal was reached (fig. 3b); IVS9+5G→A (2936+5G→A) was an intronic mutation in exon/intron 9, T/gtacg→T/gtaca. The wild-type sequence (T/gtacg) was predicted to be a poor splice-donor site, but we had previously sequenced human fetal brain ASPM cDNA (Bond et al. 2002) and had confirmed splicing to occur at nucleotide 2936 of ASPM. The mutation IVS9+5G→A reduced the donor-motif probability from 0.21 to <0.01, allowing the incorporation of two amino acids into the protein before a stop signal was reached (fig. 3c).
Figure 3.
Mutations in ASPM splice-donor motifs. The splice-donor site (the last guanine of the exon in boldface and the guanine-thymidine of the intron in plain text) is shown as a broken boxed area drawn onto the wild-type genomic sequence. In each panel, the upper genomic sequence is wild type, and the lower contains the mutation, indicated by an arrow. Bases that are exonic are shown in boldface, and those that are potentially translated intronic sequence are shown in italicized boldface. The normal and novel amino acids introduced by the failure to splice and the translation read-through are indicated beneath the wild-type and mutation sequences, respectively. The predicted premature stop codons caused by the mutations are indicated as OPA (opal) and OCH (ochre). a, The exon 11 splice-donor site is removed by a G→A mutation at base 3082, thus allowing the incorporation of an additional three amino acids into the protein before translation termination. b, The point mutation G→T at IVS25 +1 eliminates the splice-donor site, allowing translation to continue until a stop signal is reached 29 codons downstream. c, The point mutation G→A at IVS9+5 is predicted to remove the splice-donor site, therefore translation continues for a further two amino acids until a translation-termination motif is reached.
The mutations identified in the Dutch, Jordanian, Saudi Arabian, and Yemeni families were not identified in the Pakistani cohort. Three homozygous deletions (3663delG in two Pakistani families; 1727delAG and 9754delA each in a single Yemeni family) were associated with MCPH with severe mental retardation. Interestingly, an outlying cousin from a typical Pakistani family with MCPH (haplotype J) was identified as a compound heterozygote by haplotype analysis. In comparison to her affected cousins, she had a more profound degree of mental retardation and was heterozygous for mutations IVS25+1G→T (seen in her affected, homozygous cousins) and 3663delG (associated with severe mental retardation). Her head circumference was 9 SDs below the mean, whereas the head circumference of haplotype J/IVS25+1G→T was 7 SDs below the mean and that of haplotype K/3663delG was 11 SDs below the mean.
We found no evidence for a heterozygote effect of carrying an ASPM gene mutation; such an effect has been previously postulated (Qazi and Reed 1975). Parents of the affected individuals in this study had normal intellect and head circumference. All were proven to be heterozygotes for the ASPM gene mutation found in their affected child, excepting those who had died prior to our clinical studies and who therefore could not be tested.
In our individuals with MCPH5, the minimum predicted ASPM size was 116 aa, resulting from the most 5′ mutation, 349C→T. We had previously reported loss of the C-terminal 425 amino acids to be the minimum truncated region to cause MCPH (Bond et al. 2002), but in this study the mutation IVS25+1G→T redefined the minimum common truncated region to be the C-terminal 149 amino acids. The remaining mutations were found throughout the gene (fig. 3). Despite containing 45% of the ASPM coding sequence, the AT-rich exon 18 displayed a relative paucity of mutations but a high proportion of nonpathogenic SNPs.
We have identified 19 mutations in the ASPM gene in families linked to MCPH5, but none of the changes appear to be common. All of the mutations were predicted to lead to protein truncation with the resulting protein ranging in size from 116 aa (349C→T) to 3,357 aa (IVS25+1G→T). The severity of the degree of MCPH and of mental retardation varied considerably within and between families. There appeared to be no correlation between the phenotype and the size of the predicted mutant protein. The MCPH5 phenotype could be due to instability of the truncated ASPM protein, nonsense-mediated decay (NMD) of the mRNA, or the absence of an essential but unknown C-terminal protein domain. As non–protein-truncating pathogenic mutations in ASPM were not observed in the cohort with MCPH5, it is possible that these will cause a discrete phenotype. Our study suggests that mutations leading to premature ASPM protein termination are the major cause of MCPH, given the prevalence of MCPH5 in the northern Pakistani population and the detection of mutations in multiple ethnic groups. It seems most likely that NMD is the mechanism by which mutations in the ASPM gene cause the reduced fetal brain growth that results in MCPH.
Notes added in proof.—A sixth MCPH loci (MCPH6) has recently been reported (Leal et al. 2003). Two further homozygous mutations have been found in families linked to MCPH5, 3663delG in a northern Pakistani family and 1365G→T in a Turkish family (who were previously described in Jamieson et al. 2000).
Acknowledgments
The Wellcome Trust supports J.B., E.R., K.S., S.S., and C.G.W. The West Riding Medical Research Trust Fund of the University of Leeds supported D.J.H. M.J.A. is supported by FRSM, Belgium. G.H.M. is a Howard Hughes Medical Institute Physician postdoctoral fellow. C.A.W. is supported by NINDS and the March of Dimes. We thank the families, and we are grateful to M. Ahmed, G. Karbani, and S. Malik for their valuable input into this project.
Appendix A: Clinical Definition of MCPH
The phenotype of MCPH has been defined as congenital microcephaly of at least 4 SDs below the expected mean for age and sex, with mild/moderate mental retardation but no other neurological, growth, health, or dysmorphic findings, and no discernible prenatal or postnatal syndrome or cause, such as an aberrant chromosome or structural brain anomaly. Seizures are no more frequent than in the general population.
Appendix B: Haplotype Analysis of a 0.6-Mb Region on Chromosome 1q for 17 Northern Pakistani Families with MCPH5
The initial assigned haplotypes are shown, as are the number of families with each haplotype, the polymorphic microsatellite markers used in the study, and the genotyping results used to determine the haplotypes. The markers are arranged cent→1qter, and novel markers have been described elsewhere (Bond et al. 2002).
Table B1.
|
Result for Assigned Haplotypea |
|||||||||||
| Polymorphic MicrosatelliteMarker | A(n=1) | B(n=4) | C(n=2) | D(n=1) | E(n=1) | F(n=1) | G(n=1) | H(n=1) | I(n=2) | J(n=1) | K(n=2) |
| D1S2816 | 3 | 1 | 2 | 3 | 3 | 1 | 1 | 1 | 3 | 2 | 1 |
| AL139418TG18 | 2 | 3 | 1 | 4 | 2 | 2 | 1 | 3 | 1 | 3 | 2 |
| AL353809TA20 | 1 | 4 | 1 | 4 | 3 | 5 | 2 | 4 | 1 | 4 | 5 |
| AL353809GA13 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 |
| AL356315TG19 | 3 | 3 | 3 | 1 | 2 | 3 | 2 | 4 | 4 | 1 | 4 |
| AL356315CA20 | 3 | 3 | 3 | 1 | 2 | 3 | 2 | 4 | 4 | 1 | 3 |
n = No. of families with haplotype.
Appendix C: SNPs in ASPM Identified in 12 Individuals
Of the 12 SNPs identified, 3 altered an amino acid. However, in each of these instances, at least one individual carrying the homozygous SNP exhibited a stop signal downstream from the altered amino acid.
Table C1.
| SNP | Exon | Amino AcidAlteration | Occurrence in12 Individualswith MCPH |
| 2747C→T | 9 | None | 1 |
| 3138G→A | 12 | None | 3 |
| 3714T→C | 15 | None | 1 |
| 4344A→G | 18 | None | 3 |
| 4449G→A | 18 | None | 2 |
| 5856A→G | 18 | None | 1 |
| 5961A→G | 18 | None | 2 |
| 7605G→A | 18 | None | 4 |
| 7674C→T | 18 | None | 3 |
| 7684A→G | 18 | Ser→Gly | 4 |
| 7939C→A | 18 | Leu→Ile | 8 |
| 9395T→G | 22 | Leu→Arg | 3 |
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Berkeley Drosophila Genome Project, http://www.fruitfly.org/seq_tools/splice.html (for splice prediction)
- INFOBIOGEN, http://www.infobiogen.fr/services/analyseq/cgi-bin/traduc_in.pl (for translation of nucleotide sequence to amino acids)
- National Center for Biotechnology Information (NCBI), Entrez search engine, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Nucleotide (for ASPM)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/omim/ (for MCPH [MIM 251200] and ASPM [MIM 650481])
References
- Aicardi J (1998) Malformations of the central nervous system. In: Diseases of the nervous system in childhood, 2nd ed. Mac Keith, London, pp 90–91 [Google Scholar]
- Bond J, Roberts E, Mochida GH, Hampshire DJ, Scott S, Askham JM, Springell K, Mahadevan M, Crow YJ, Markham AF, Walsh CA, Woods CG (2002) ASPM is a major determinant of cerebral cortical size. Nat Genet 32:316–320 [DOI] [PubMed] [Google Scholar]
- Bundey S (1997) Abnormal Mental Development. In: Rimoin DL, Connor JM, Pyeritz RE (eds) Emery and Rimoin’s principles and practice of medical genetics, 3rd ed. Churchill Livingstone, New York, pp 730–731 [Google Scholar]
- Craig R, Norbury C (1998) The novel murine calmodulin-binding protein Sha1 disrupts mitotic spindle and replication checkpoint functions in fission yeast. J Cell Sci 111:3609–3619 [DOI] [PubMed] [Google Scholar]
- Gonzalez C, Molina I, Casal J, Ripoll P (1989) Gross genetic dissection and interaction of the chromosomal region 95E;96F of Drosophila melanogaster. Genetics 123:371–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez C, Saunders RD, Casal J, Molina I, Carmena M, Ripoll P, Glover DM (1990) Mutations at the asp locus of Drosophila lead to multiple free centrosomes in syncytial embryos, but restrict centrosome duplication in larval neuroblasts. J Cell Sci 96:605–616 [DOI] [PubMed] [Google Scholar]
- Jackson AP, McHale DP, Campbell DA, Jafri H, Rashid Y, Mannan J, Karbani G, Corry P, Levene MI, Mueller RF, Markham AF, Lench NJ, Woods CG (1998) Primary autosomal recessive microcephaly (MCPH1) maps to chromosome 8p22-pter. Am J Hum Genet 63:541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AP, Eastwood H, Bell SM, Adu J, Toomes C, Carr IM, Roberts E, Hampshire DJ, Crow YJ, Mighell AJ, Karbani G, Jafri H, Rashid Y, Meuller RF, Markham AF, Woods CG (2002) Identification of microcephalin, a protein implicated in determining the size of the human brain. Am J Hum Genet 71:136–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson CR, Govaerts C, Abramowicz J (1999) Primary autosomal recessive microcephaly: homozygosity mapping of MCPH4 to chromosome 15. Am J Hum Genet 65:1465–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson CR, Fryns J-P, Jacobs J, Matthijs G, Abramowicz MJ (2000) Primary autosomal recessive microcephaly: MCPH5 maps to 1q25–q32. Am J Hum Genet 67:1575–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal GF, Roberts E, Silva EO, Costa SM, Hampshire DJ, Woods CG (2003) A novel locus for autosomal recessive primary microcephaly (MCPH6) maps to 13q12.2. J Med Genet 40:540–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida GH, Walsh CA (2001) Molecular genetics of human microcephaly. Curr Opin Neurol 14:151–156 [DOI] [PubMed] [Google Scholar]
- Moynihan L, Jackson AP, Roberts E, Karbani G, Lewis I, Corry P, Turner G, Mueller RF, Lench NJ, Woods CG (2000) A third novel locus for primary autosomal recessive microcephaly maps to chromosome 9q34. Am J Hum Genet 66:724–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattison L, Crow YJ, Deeble VJ, Jackson AP, Jafri H, Rashid Y, Roberts E, Woods CG (2000) A fifth locus for primary autosomal recessive microcephaly maps to chromosome 1q31. Am J Hum Genet 67:1578–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qazi QH, Reed TE (1975) A possible major contribution to mental retardation in the general population by the gene for microcephaly. Clin Genet 7:85–90 [DOI] [PubMed] [Google Scholar]
- Ripoll P, Pimpinelli S, Valdivia MM, Avila J (1985) A cell division mutant of Drosophila with a functionally abnormal spindle. Cell 41:907–912 [DOI] [PubMed] [Google Scholar]
- Roberts E, Jackson AP, Carradice AC, Deeble VJ, Mannan J, Rashid Y, Jafri H, McHale DP, Markham AF, Lench NJ, Woods CG (1999) The second locus for autosomal recessive primary microcephaly (MCPH2) maps to chromosome 19q13.1–13.2. Eur J Hum Genet 7:815–820 [DOI] [PubMed] [Google Scholar]
- Roberts E, Hampshire DJ, Pattison L, Springell K, Jafri H, Corry P, Mannon J, Rashid Y, Crow Y, Bond J, Woods CG (2002) Autosomal recessive primary microcephaly: an analysis of locus heterogeneity and phenotypic variation. J Med Genet 39:718–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders RD, Avides MC, Howard T, Gonzalez C, Glover DM (1997) The Drosophila gene abnormal spindle encodes a novel microtubule-associated protein that associates with the polar regions of the mitotic spindle. J Cell Biol 137:881–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Bosch J (1959) Microcephaly in the Netherlands: a clinical and genetical study. Ann Hum Genet 23:91–116 [DOI] [PubMed] [Google Scholar]