Abstract
We describe a novel gene family that forms clusters in subtelomeric regions of Trypanosoma brucei chromosomes and partially accounts for the observed clustering of retrotransposons. The ingi and ribosomal inserted mobile element (RIME) non-LTR retrotransposons share 250 bp at both extremities and are the most abundant putatively mobile elements, with about 500 copies per haploid genome. From cDNA clones and subsequently in the T. brucei genomic DNA databases, we identified 52 homologous gene and pseudogene sequences, 16 of which contain a RIME and/or ingi retrotransposon inserted at exactly the same relative position. Here these genes are called the RHS family, for retrotransposon hot spot. Comparison of the protein sequences encoded by RHS genes (21 copies) and pseudogenes (24 copies) revealed a conserved central region containing an ATP/GTP-binding motif and the RIME/ingi insertion site. The RHS proteins share between 13 and 96% identity, and six subfamilies, RHS1 to RHS6, can be defined on the basis of their divergent C-terminal domains. Immunofluorescence and Western blot analyses using RHS subfamily-specific immune sera show that RHS proteins are constitutively expressed and occur mainly in the nucleus. Analysis of Genome Survey Sequence databases indicated that the Trypanosoma brucei diploid genome contains about 280 RHS (pseudo)-genes. Among the 52 identified RHS (pseudo)genes, 48 copies are in three RHS clusters located in subtelomeric regions of chromosomes Ia and II and adjacent to the active bloodstream form expression site in T. brucei strain TREU927/4 GUTat10.1. RHS genes comprise the remaining sequence of the size-polymorphic “repetitive region” described for T. brucei chromosome I, and a homologous gene family is present in the Trypanosoma cruzi genome.
African trypanosomes, including Trypanosoma brucei, are unicellular protists that are responsible for diseases affecting humans and livestock. The nuclear chromosomes of T. brucei can be grouped into three size classes based on their migration in pulsed-field gel electrophoresis: 11 pairs of diploid megabase chromosomes (1 to 6 Mb) that contain the housekeeping genes and represent about 80% of the nuclear DNA content, a few intermediate-sized chromosomes (200 to 900 kb), and an undetermined number of minichromosomes (in the range of 100 that were 50 to 150 kb in size) (27, 44, 68). The intermediate and minichromosomes, whose ploidy is uncertain, play a role in antigenic variation. The genome sizes of different T. brucei isolates can vary by up to 30% (30, 32, 44, 45). T. brucei TREU927/4 GUTat10.1, the reference strain for genome sequencing, contains an estimated 62-Mb diploid nuclear genome, including 53.4 Mb of diploid megabase chromosomal DNA (44).
T. brucei has a life cycle that alternates between the tsetse fly and the mammal. In the bloodstream of their mammalian hosts, the parasites evade the immune response by antigenic variation, a continual switching of the variant surface glycoprotein (VSG) that constitutes the surface coat. Although each bloodstream trypanosome has a single VSG species on its surface, the parasite genome has a repertoire of several hundred to 1,000 different VSG genes that are expressed in a mutually exclusive manner from about 20 potential bloodstream form expression sites (B-ESs), invariantly located near telomeres (see references 5, 10, 19, 53, and 66 for recent reviews). Only one B-ES at a time is activated by an unknown mechanism. These expression sites are long polycistronic transcription units in which the VSG is cotranscribed with several intervening expression site-associated genes (ESAGs) from a promoter located about 45 to 60 kb upstream (34, 54) and are separated from the rest of the chromosome by a 10- to 40-kb region of 50-bp repeats. VSGs are also expressed during the metacyclic stage of the life cycle in the salivary glands of the tsetse fly as a preadaptation to life in the mammal. The genome contains 20 to 30 telomere-linked metacyclic expression sites (M-ESs) containing VSGs that are transcribed into monocistronic precursor RNAs from a proximal promoter located within 2 kb upstream (see references 4 and 22 for recent reviews).
The genome is highly plastic, as revealed by pulsed-field gel electrophoresis (PFGE) and analysis of the recombination events associated with VSG switching. It also contains a large number of putative non-long terminal repeat (LTR) retrotransposons: ingi's and ribosomal inserted mobile elements (RIMEs) (29, 33, 47). Non-LTR retrotransposons, exemplified by the human short interspersed nucleotide elements (SINE) and long interspersed nucleotide elements (LINE), are replicating retroelements of a type that are ubiquitous in nature and may constitute as much as 14% of host genomes (60). Retroelements replicate by copying their RNA transcript into DNA by using a reverse transcriptase. The DNA copy then integrates into the genome (35). All the non-LTR retroelements are flanked by target site duplications of variable length, have variable length poly(A) or A-rich 3" tails, and are devoid of the LTRs present in retroviruses and LTR retrotransposons. As reported for mammals (60) and plants (58), the non-LTR retrotransposons constitute the most abundant repeat elements described for the genome of T. brucei (ingi, RIME, and SLACS) (3, 47). The ingi elements (5.2 kb) has the characteristics of LINE elements, while the RIME (500-bp) elements are similar to the nonautonomous SINE elements. ingi's are composed of a 4.7-kb fragment bordered by two separate halves of RIME and, if their reading frames are not mutated to possess termination codons, they may encode a single large protein containing a central reverse transcriptase domain, a C-terminal DNA-binding domain (52), and an N-terminal apurinic-apyrimidinic-like endonuclease domain (48). SLACS are site-specific retroelements found only in the spliced leader RNA genes (3), but ingi's and RIMEs were previously thought to be randomly distributed in the host genome (47). Individual ingi and RIME are associated with rRNA genes (29) and tubulin gene arrays (1) and precede or are within most of the B-ESs and M-ESs characterized so far (4, 7, 13, 36, 39, 43, 54, 55, 59).
Recently, Melville et al. showed that a large region (about 200 kb) of uncharacterized repeated sequences is present upstream of the 50-bp repeats preceding the B-ES of chromosome I (ChrI) (43). Interestingly, this region also contains a high number of ingi's and RIMEs and is very size polymorphic between strains, and similar sequences are present in many of the megabase chromosomes of T. brucei (43; unpublished data). We have characterized a novel, large multigene family (about 128 copies per haploid nonminichromosomal genome) encoding mainly nuclear proteins, multiple copies of which are also located in the RIME/ingi-rich region. Approximately 60% of the identified members of this gene family are pseudogenes. The gene family can be divided into six subfamilies, called RHS1 through RHS6 (for retrotransposon hot spot), based on deduced amino acid sequences. About one-third of the RHS (pseudo)genes contain RIME and/or ingi retroelement(s) inserted in frame and at exactly the same relative nucleotide position. Analysis of the ChrIa sequence indicates that the RHS genes are clustered upstream of the 50-bp repeats preceding the bloodstream expression site (B-ES). They account for most of the unknown sequences present in the RIME/ingi-rich repeated region described previously (43).
MATERIALS AND METHODS
Trypanosomes.
Cells of the bloodstream form of T. brucei AnTat1 were used to infect rats and then were isolated by ion exchange chromatography (37). Procyclic form of T. brucei EATRO1125, TREU927/4, and 427 were cultured at 27°C in SDM-79 medium (15) containing 10% fetal calf serum and 5 mg of hemin liter−1.
Construction and screening of genomic and cDNA libraries.
λZAP II clones containing cDNA-004, cDNA-005, cDNA-040, and cDNA-132 (accession numbers AF403385, AF403388, AF403386, and AF403387, respectively) were randomly isolated from a T. brucei AnTat1 cDNA library (derived from the bloodstream form). The cDNA was synthesized from poly(A)+ mRNA as described previously (12) and was inserted into the EcoRI site of λZAP II cloning vector (Stratagene). Recombinant pBluescript II plasmids containing cDNA fragments were excised from the λZAP II clones according to the manufacturer's instructions (Stratagene). The genomic DNA library of the T. brucei AnTat1 strain was constructed in the c2X75 cosmid vector (17). Large DNA fragments generated by Sau3A partial digestion of genomic DNA were inserted into the BamHI site of the vector as previously described (14), and the cosmid library (20,000 clones) was screened with α-32P-labeled cDNA-132. We have selected and partially sequenced five cosmid clones, three containing a full-length and apparently functional RHS1 gene (Cos-02, Cos-03, and Cos-17 with the accession numbers AY046893, AY046894, and AY046895, respectively) and two containing one RHS1 pseudogene inactivated by a RIME/ingi insertion (Cos-12 [accession number AY046896] and Cos-23 [accession numbers AY046897S1 and AY046897S2]).
DNA sequencing, alignments, and phylogenetic analysis.
Inserts of recombinant pBluescript II plasmids and c2X75 cosmids were sequenced by the dideoxynucleotide chain termination method, using AmpliTaq DNA polymerase, as described by the manufacturer (ABI PRISM, Perkin-Elmer). DNA and amino acid sequences were analyzed using the DNA STRIDER and Artemis programs (The Wellcome Trust Sanger Institute), and database searches were done with BLAST. Multiple alignments of amino acid sequences were obtained using MacVector 6.0.1. For the phylogenetic analysis, multiple alignments of DNA and amino acid sequences were obtained using CLUSTAL W version 1.6 (64). For DNA alignments, all the available full-length RHS1 or RHS2 (pseudo)gene sequences located downstream of the RIME/ingi insertion site were used. For amino acid alignments, the full-length protein sequence encoded by functional genes and pseudogenes (ϕRHS1c, ϕRHS3b, and ϕRHS3c), corrected to remove frame shifts or premature stop codons, were analyzed. The phylogenetic trees were constructed using version 3.5c of the PHYLIP program package of J. Felsenstein (CLUSTAL W and PHYLIP were obtained through Bisance and Infobiogen facilities). The matrix of pairwise sequence distances were calculated by the Dayhoff's method using DNADIST or PROTDIST. The unrooted phylogenetic trees were constructed from the distance matrix using the neighbor or Fitch methods and were drawn with TREEVIEW version 1.3 (50). The statistical robustness of the resulting phylogenetic trees was assessed with the SEQBOOT program by bootstrap resampling analysis generating 100 reiterated data sets. The resulting bootstrap values were added manually at each corresponding node.
Southern blot analysis.
Approximately 2.5 μg of genomic DNA from T. brucei TREU927/4 and 427, extracted as described elsewhere (8), was subjected to endonuclease digestion (HincII for RHS1 and RHS5, ClaI for RHS2 and RHS6. AseI for RHS3, and HpaII and KpnI for RHS4), electrophoresed in 0.6% agarose gel, blotted onto neutral membrane (Quantum-Appligene), and hybridized with α-32P-labeled RHS-specific probes at 65°C in 6× SSPE (1× SSPE is 0.18 mM NaCl, 10 mM NaH2PO4, 1 mM EDTA, pH 7.0)-0.1% sodium dodecyl sulfate (SDS). The probes specific for each RHS1 to RHS6 multigene subfamily were obtained by PCR from the most divergent 3" region of the (pseudo)genes, which corresponds to box 2 in Fig. 2. The membranes were washed at 65°C using 0.1× SSPE-0.1% SDS, before autoradiography. Probes were removed by boiling in a solution of 0.5% SDS, before rehybridizing blots.
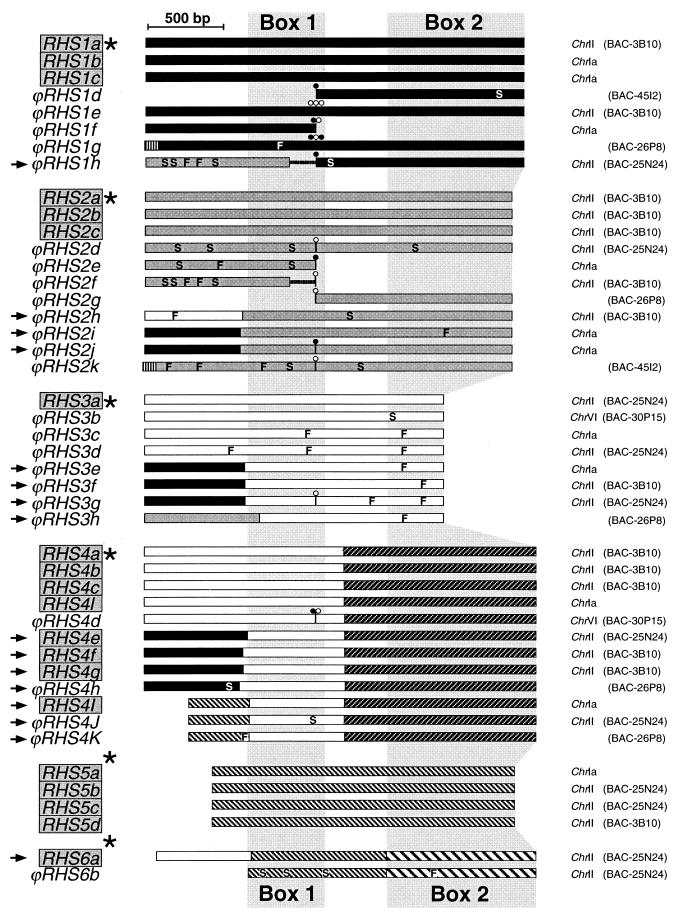
FIG. 2.
Schematic representation of RHS (pseudo)genes present in T. brucei (TREU927/4) database. The name of each gene or pseudogene is on the left; those in a grey box are potentially functional, while the unboxed names correspond to nonfunctional pseudogenes. Arrows, chimeric sequences; ∗, sequences (RHS genes and conserved flanking regions) present in the database under accession numbers AY046887 (RHS1a), AY046888 (RHS2a), AY046889 (RHS3a), AY046890 (RHS4a), AY046891 (RHS5a), and AY046892 (RHS6a). The amino acid sequences of box 1 encoded by the RHS1a to RHS6a genes which are labeled by an asterisk are compared in Fig. 3. Chromosome and/or BAC clones containing the sequence are indicated on the right. The color code for each subfamily is as follows: RHS1 ( ), RHS2 (
), RHS2 ( ), RHS3 (▭), RHS4 (
), RHS3 (▭), RHS4 ( ), RHS5 (
), RHS5 ( ), and RHS6 (
), and RHS6 ( ). In the coding sequences, the positions of frame shifts (F), premature stop codons (S), and RIME (○) and/or ingi (•) insertions are indicated. Multiple retroelement insertions are shown by a corresponding number of open and filled circles. Horizontal lines in the middle of ϕRHS1h and ϕRHS2f coding sequences represent a deletion of a part of the RHS coding sequence. The most conserved (box 1) and most divergent (box 2) coding regions between RHS subfamilies are shaded.
). In the coding sequences, the positions of frame shifts (F), premature stop codons (S), and RIME (○) and/or ingi (•) insertions are indicated. Multiple retroelement insertions are shown by a corresponding number of open and filled circles. Horizontal lines in the middle of ϕRHS1h and ϕRHS2f coding sequences represent a deletion of a part of the RHS coding sequence. The most conserved (box 1) and most divergent (box 2) coding regions between RHS subfamilies are shaded.
Estimation of RHS1 to RHS6 (pseudo)gene and RIME/ingi copy numbers.
The copy numbers per haploid nonminichromosomal genome (T. brucei TREU927/4) of each RHS subfamily and the RIME and ingi retrotransposons were estimated by BLAST analysis of a Genome Survey Sequence (GSS) database and hydridization to a P1 genomic DNA library. For the BLAST analysis, the calculation of the copy number per haploid nonminichromosomal genome (CN) includes the number of GSSs (GSS) homologous to the probe, the size of the probe (GS), the size of the haploid nonminichromosomal genome (HGS = 30 Mb), and the number of GSSs contained in the library (TGSS), using the following equation: CN = (GSS × HGS)/(TGSS × GS). For P1 library hybridization, the copy number per haploid nonminichromosomal genome (CN) is calculated from the number of positive P1 clones (PC), the total number of P1 clones (TC = 1,819 clones), the average size of the P1 DNA inserts (PIS = 65 kb), and the size of the haploid genome without mini and intermediate chromosomes (HGS = 26.7 Mb): CN = (PC × HGS)/(TC × PIS). Since all complete ingi retroelements contain a full-length RIME sequence, the ingi copy number was deduced from the RIME copy number.
Production of recombinant proteins in Escherichia coli and antibody production.
PCR fragments encoding the C-terminal subfamily-specific domain of RHS1 (372 amino acids [aa]), RHS2 (260 aa), RHS4 (289 aa), RHS5 (285 aa), and RHS6 (286 aa), preceded by a methionine and six histidine residues, were obtained using the respective 5" primers (5"-GCCTCA CATATGcaccatcaccatcaccatTTGAAGGATTTGGAAGCCA-3", 5"-AATTTA CATATGcatcaccatcaccatcacGAAGAATGCAGAAACAGAGC-3", 5"-TATTTA CATATGcatcaccatcaccatcacCGAGATGCCGGAGAGAGCGT-3", 5"-AATTTA CATATGcatcaccatcaccatcacAAAGCTCGAGAAGGAAACT-3" and 5"-AATTTA CATATGcatcaccatcaccatcacGTACCTCACTCTGAATCCAT-3") and 3" primers (5"-TCCTTC GGATCCCTATGCATTGTTACCACC-3", 5"-TTTATT GGATCCTCAGTCAGCGGGGCCACCAG-3", 5"-AATTAA GGATCCTCACCCTCCTTGCGCTCCCG-3", 5"-TTTAAA GGATCCTTACCTTCGGCCCGCAGCAG-3" and 5"-TTTAAA GGATCCTTATTCGTTATTCGCCACTT-3"). The 5" primers contain an NdeI restriction site (italicized), a start codon (italicized and bold), and six histidine codons (lower case). The 3" primers contain a BamHI restriction site (italicized) and a stop codon (bold). DNA isolated from Cos-02 (RHS1) and BAC-25N24 (RHS2, RHS4, RHS5, and RHS6) clones was used as template for PCR. The resulting DNA fragments were cloned into the pET3a expression vector (Novagen) and expressed in E. coli BL21 cells. Expression and affinity purification of the recombinant proteins were performed as described by the manufacturer (Novagen). The affinity-purified recombinant proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE), electroeluted, and emulsified with complete (first injection) or incomplete Freund adjuvants. Antisera were raised in rabbits (RHS1) or rats (RHS2, RHS4, RHS5, and RHS6) by five injections at 2-week intervals by using 100 or 30 μg of protein per injection, respectively.
Western blot analysis.
Total extracts of trypanosomes were boiled for 5 min in 2% (wt/vol) SDS. Sample preparation, migration in SDS-8% PAGE, immunoblotting on Immobilon-P membranes (Millipore), and immunodetection using as secondary antibody goat anti-rabbit or anti-goat antibody conjugated to horseradish peroxidase (SIGMA) were achieved as previously described (28, 57). The antisera were diluted 1:100 in phosphate-buffered saline (PBS)-0.05% (vol/vol) Tween 20 containing 5% (wt/vol) nonfat milk, and blots were developed with 3,3"-diaminobenzidine.
Immunolocalization of RHS proteins.
For immunofluorescence microscopy, trypanosomes were fixed in PBS-1% (vol/vol) formaldehyde for 30 min, permeabilized for 10 min by adjusting the solution to 0.1% (vol/vol) Triton X-100, and finally 0.1 M glycine was added for 10 min to neutralize active aldehyde groups. Cells were washed once in PBS, and trypanosomes were resuspended in PBS and allowed to adhere to glass slides until completely dry before incubation with antibodies. Rabbit or rat antisera raised against the RHS recombinant proteins were diluted 1:100, whereas secondary goat anti-rabbit fluorescein isothiocyanate (FITC) or anti-rat FITC were used at a 1:10,000 or 1:1,000 dilution, respectively. All incubations were carried out for 30 min at room temperature, and all dilutions were performed with PBS containing 0.1% (vol/vol) Triton X-100 and 0.1% (wt/vol) bovine serum albumin. At the end of the immunofluorescence assay, cells were incubated for 5 min with PBS containing 1 μg of the fluorescent DNA dye DAPI (4",6"-diamino-2-phenylindole; SIGMA) ml−1. Observations were made after mounting in Vectashield (Valbiotech) mounting medium using a Zeiss epifluorescence microscope fitted with FITC and UV filters. Images were captured by camera (Princeton) and MetaView software (Universal Imaging Corporation) and were processed in Adobe Photoshop (Adobe Systems, Mountain View, Calif.) on a Macintosh iMac computer.
Nucleotide sequence accession numbers.
The sequences have been deposited in GenBank and assigned accession numbers as follows: cDNA-004, AF403385; cDNA-005, AF403388; cDNA-040, AF403386; cDNA-132, AF403387; Cos-02, AY046893; Cos-03, AY046894; Cos-12, AY046896; Cos-17, AY046895; Cos-23, AY046897S1 and AY046897S2; RHS1a, AY046887; RHS2a, AY046888; RHS3a, AY046889; RHS4a, AY046890; RHS5a, AY046891; RHS6a, AY046892.
RESULTS
Characterization of new multigene family containing potential retroelement insertion site.
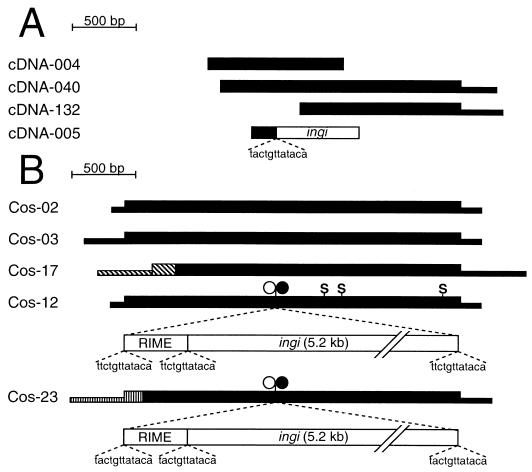
We previously observed that cDNA can be synthesized from T. brucei mRNA by self-priming, probably through the presence of a poly(U) stretch located a few dozen nucleotides upstream of the poly(A) tail (11, 12). With the aim of identifying novel genes, we produced sequence tags from both ends of 114 new cDNAs derived from a library of T. brucei (AnTat1) bloodstream form cDNAs and ranging from 0.3 to 2.2 kb. Among the 40 sequenced pairs that did not have any significant matches in databases, cDNA-004, cDNA-040, cDNA-132, and the 5" end of cDNA-005 shared an overlap with over 80% sequence identity. Only the 5" end of cDNA-005 is similar to the three other cDNA sequences, due to the presence of a non-LTR retrotransposon (ingi) sequence (Fig. 1A). These four cDNA sequences are sufficiently divergent to indicate that they may originate from four different genes and could be members of a new multigene family.
FIG. 1.
New multigene family containing a hot spot for retroelement insertion. (A) Map of homologous cDNAs (cDNA-004, cDNA-005, cDNA-040, and cDNA-132 with accession numbers AF403385, AF403388, AF403386, and AF403387, respectively) isolated from a T. brucei AnTat1 library. The large and small black boxes represent coding and noncoding sequences, respectively, and the white box shows the 5" end of a non-LTR retrotransposon ingi. The 12-bp sequence upstream of the retrotransposon is shown below the cDNA-005 map. (B) Schematic map of the RHS genes and pseudogenes sequenced from five different cosmid clones of AnTat1 genomic DNA (Cos-02, -03, -12, -17, and -23 with accession numbers AY046893, AY046894, AY046896, AY046895, and AY046897S1, and AY046897S2, respectively). Coding and noncoding sequences are represented by large and small boxes, respectively. The black boxes correspond to sequences presenting at least 80% identity, while the hatched boxes correspond to unrelated sequences. Premature stop codons (S) and retroelement RIME (○) or ingi (•) insertion are indicated above the maps. The RIME and ingi retroelements inserted in the RHS pseudogene of Cos-12 and Cos-23 clones are shown below the maps. The 12-bp repetitive duplication sequences flanking the retroelements are shown below the RIME/ingi map.
To obtain full-length sequences of members of this novel, putative multigene family, we screened a cosmid library of AnTat1 genomic DNA with an α-32P-labeled cDNA-132 fragment. Of 18,000 cosmid clones, the probe hybridized to 319 (1.8%). Comparison of the nucleotide sequences of five genes isolated from different cosmid clones revealed a high degree of conservation (from 63 to 87% identity). However, the 5" coding sequences of two of these genes, in Cos-17 and Cos-23, are unrelated to the same regions in the three other sequences (Fig. 1B). In addition, both the Cos-12 and Cos-23 coding sequences are interrupted by two tandemly arranged non-LTR retrotransposons (a RIME followed by ingi) inserted at exactly the same relative position in each sequence (Fig. 1B). These elements are flanked by short duplicated sequences as shown in Fig. 1B and described previously (29, 33, 47). In addition, the ingi retroelement present in cDNA-005 is inserted at the same relative position with identical flanking sequences (Fig. 1A), suggesting that members of this new multigene family, called RHS for retrotransposon hot spot, may contain a hot spot for non-LTR retrotransposon insertion.
Identification of six RHS multigene subfamilies.
To further our analyses of this multigene family, we studied the T. brucei (TREU927/4 GUTat10.1 strain) sequence databases that contain the 1.1-Mb ChrIa sequence (http://www.sanger.ac.uk/Projects/T_brucei/ [The Wellcome Trust Sanger Institute]) and about 30 sequenced bacterial artificial chromosome (BAC) clones containing genomic DNA fragments of ca. 140 kb (http://www.tigr.org/tdb/mdb/tbdb/index.shtml [The Institute of Genome Research {TIGR}]). Ten of these BAC sequences have been assembled to generate a contig covering chromosome II (ChrII) (unpublished data). To date, the T. brucei databases contain about 5 Mb of large, fully sequenced genomic DNA fragments (about 17% of the 30-Mb haploid nonminichromosomal genome). A BLAST computer search performed on these databases, using the RHS amino acid sequence of Cos-02 as the query, identified 52 different sequences. Schematic maps of these sequences, omitting seven that are highly degenerate, are presented in Fig. 2. Among these 52 related sequences, 31 are pseudogenes (60%) due to the presence in the coding sequence of RIME and/or ingi (16 sequences [∼31%]), frame shift(s), unexpected stop codon(s), and/or deletion(s), whereas the other 21 gene sequences (40%) may code for functional proteins. Interestingly, RIME and ingi were invariably inserted at exactly the same relative nucleotide position in the 16 different RHS pseudogenes analyzed, strongly suggesting that the (pseudo)genes contain a site-specific hot spot for insertion of non-LTR retrotransposons.
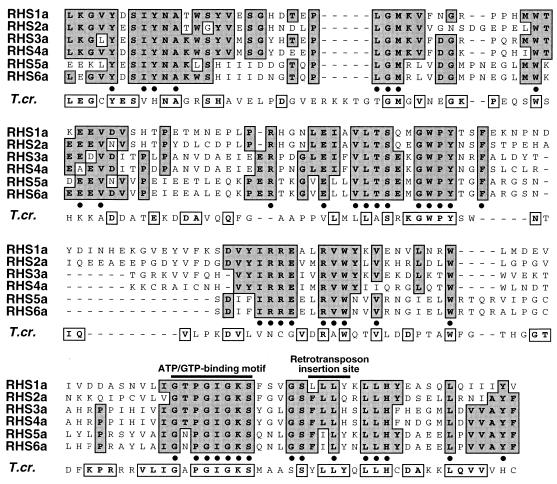
The N-terminal halves of the proteins encoded by these genes contain a highly conserved domain (box 1 in Fig. 2 and Fig. 3) whose coding region includes the retrotransposon insertion site. In contrast, the C-terminal sequences are very divergent (Fig. 2, box 2) and constitute the best criterion to tentatively group these proteins: six RHS subfamilies have been defined and named RHS1 to RHS6 (Fig. 2). These groups were formed to maximize the number of genes contained within each group; however, sequence analysis of the N-terminal regions detected 16 sequences that are the result of chimeric formations between four different sequences constituting RHS1, RHS2, RHS3/4, and RHS5. To determine if the proposed subdivision is significant, we compared three full-length and nonchimeric proteins of each subfamily, with the exception of RHS6, which presently has only one full-length member. The construction of a phylogenetic tree clearly showed six clusters corresponding to the six RHS subfamilies (Fig. 4A). Calculation of the amino acid identity between all members of the same or different subfamilies supports this analysis: the intragroup alignments reveal a high level of conservation (81 to 97% identity), while the intergroup alignments show 13 to 39% identity with the exception of the RHS3/RHS4 (43%) and RHS5/RHS6 (50%) comparisons, which overlap by about one-half and one-third, respectively (Fig. 2). The four cDNA and five cosmid clones containing RHS (pseudo)genes isolated from the AnTat1 strain (Fig. 1) all belong to the RHS1 subfamily.
FIG. 3.
Amino acid alignment of box 1 domain from RHS1a-6a proteins and related T. cruzi protein. The aligned box 1 sequences indicated by an asterisk in the left margin of Fig. 2 are representative of each RHS subfamily. Dashes were introduced to maximize the alignment. Identical amino acids are shaded and in bold. The positions of the ATP/GTP-binding motif and the duplicated insertion site sequence associated with non-LTR retroelement (RIME and/or ingi) insertion are indicated above the alignment. The last row (T.cr.) represents the chimeric RHS-related protein identified in the T. cruzi database, as shown in Fig. 10. All boxed and bold residues of the T. cruzi sequence are identical to residues located at the same relative position in the T. brucei RHS proteins.
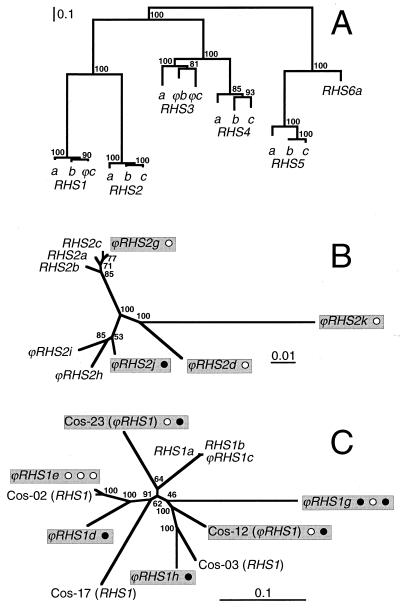
FIG. 4.
Phylogenetic analysis of RHS proteins and RHS (pseudo) genes. (A) The phylogenetic tree was constructed with three full-length RHS amino acid sequences (a, b, and c) specific for each RHS subfamily except RHS6, for which only a single full-length sequence has been identified. Premature stop codons and frame shifts present in three RHS pseudogenes (ϕRHS1c, ϕRHS3b, and ϕRHS3c) were corrected to obtain a chimeric full-length RHS protein. The two other phylogenetic trees were constructed using the full-length RHS2 (B) or RHS1 (C) (pseudo)gene sequences located downstream of the RIME/ingi insertion site. The grey boxes include RHS (pseudo)genes with RIME (○) and/or ingi (•) retroelement(s). In each panel, the scale bar represents a genetic distance of 0.1 or 0.01 amino acid or nucleotide substitutions per site, and numbers given beside the nodes represent the percentage of bootstrap replicas yielding these trees.
A BLAST search revealed no significant homology between any deduced RHS sequences and any sequences in the SWISS-PROT database. However, using the Profile program (Infobiogen), they all contain a predicted ATP/GTP-binding motif specified by a coding sequence located five codons upstream of the RIME/ingi insertion site (Fig. 3).
To investigate the presence of conserved sequences associated with RHS (pseudo)genes, the flanking regions of all identified RHS copies were compared (data not shown). The organization of conserved regions upstream of RHS (pseudo)genes, ranging from 0.4 to 20 kb, is complex. However, two main groups of 1.4- and 0.4-kb sequence tracts located upstream of the initiation codon are associated with RHS1 and RHS2 and with RHS3 to RHS6 N-terminal coding sequences, respectively. In contrast, the sequence downstream of RHS (pseudo)genes has a less complicated organization. These sequences are 0.8 to 5.2 kb in length depending on the subfamily, they are specific to each RHS subfamily, and they lack interfamily conserved sequences. No gene has been identified in the conserved RHS flanking regions upstream or downstream (up to 7 kb) of the RHS genes.
RHS (pseudo)genes contain hot spot of homologous recombination.
About one-third of the RHS genes analyzed (16 out of 49 copies) are chimeric between copies belonging to the different subfamilies defined above, such as the two, three, and four (pseudo)genes of the RHS2, RHS3, and RHS4 subfamilies, respectively, which contain the 5" extremity of RHS1 (pseudo)-genes (Fig. 2). These chimera probably result from homologous recombination between two copies from different subfamilies, the crossing over taking place in the N-terminal region upstream of the retroelement insertion site. The probable site of homologous recombination was determined by comparing chimeric RHS nucleotide sequences with the corresponding RHS (pseudo)genes to find the region of overlap (data not shown). Analysis of the 16 chimeric RHS (pseudo)genes reveals eight different sites of recombination clustered in a 140-bp fragment that includes the 5" end of conserved box 1 (Fig. 2), suggesting that there is one or more hot spots of homologous recombination in this region.
Insertion of RIME and/or ingi elements to form RHS pseudogenes.
The analysis of the TREU927/4 databases showed that about one-third of the RHS (pseudo)genes (16 out of 52 copies) contain a RIME and/or ingi retrotransposon inserted at exactly the same relative position. This observation suggests that RHS (pseudo)genes contain a hot spot for retroelement insertion. However, at this stage of the analysis, we cannot rule out the possibility that most, if not all, of the RHS pseudogenes containing RIME/ingi retroelement(s) are derived by gene duplication from a common RHS ancestor inactivated by random insertion of a retroelement. This hypothesis implies that RHS (pseudo)genes should form separate monophylogenetic groups, depending on the presence or the absence of retroelement(s). The phylogenetic analysis of the RHS1-6 proteins shows that each RHS subfamily forms a monophylogenetic group, suggesting that the retroelement insertions in RHS1, RHS2, and RHS3-4 subfamilies occurred after the differentiation of each subfamily from a common ancestor (Fig. 4A). To further address this question for the RHS1 and RHS2 subfamilies, we constructed phylogenetic trees with the RHS1 or RHS2 (pseudo)gene nucleotide sequences located downstream of the retroelement insertion site (12 and 9 sequences, respectively). This analysis shows that ϕRHS2g and ϕRHS2j, which contain one retroelement, are more closely related to several RHS2 (pseudo)genes without RIME/ingi than to ϕRHS2d and ϕRHS2k, which also contain one retrotransposon (Fig. 4B). Similarly, RHS1 (pseudo)genes appear to be randomly distributed in the tree regardless of the presence or absence of retrotransposons (Fig. 4C). Furthermore, based on the number, the type (RIME or ingi), and the organization of the inserted retroelement(s) (Fig. 1 and 2), at least half of the retroelement-containing RHS pseudogenes were generated by independent retroelement insertion. In summary, these data show that the RHS (pseudo)genes do indeed contain a hot spot for retroelement insertion.
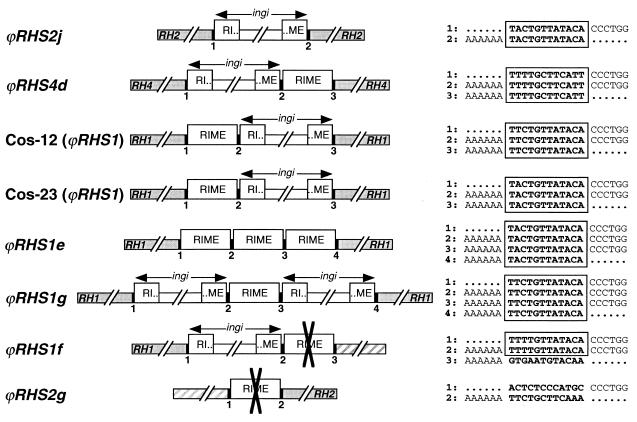
The insertion of retrotransposons, such as RIMEs or ingi's, generates a duplication of the target site sequence to form a direct repeat of a few base pairs flanking the inserted retroelement. This is exemplified in the ϕRHS2j pseudogene, which contains an ingi retroelement flanked by 12 conserved nucleotides (Fig. 5). The analysis of RHS pseudogenes inactivated by retrotransposon insertion shows that ingi's are often associated with a RIME to form a doublet of retroelements. The RIME may follow (ϕRHS4d) or precede (ϕRHS1 in Cos-23 and Cos-12) ingi (Fig. 5). In the case of Cos-12 ϕRHS1, Cos-23 ϕRHS1, and ϕRHS4d, the 12-bp sequence located between the RIME and ingi is identical to the 12-bp duplicated target site. This strict conservation of length and sequence is unlikely to occur by chance and may be involved in the mechanism of retrotransposon insertion. Thus, we propose that after the insertion of a first retroelement (RIME or ingi), a second one (RIME or ingi) recognizes the same target site (the original one or the duplicated one) for insertion upstream or downstream of the first retrotransposon, resulting in the formation of another copy of the target site. The presence of RHS pseudogenes inactivated by insertion of three tandemly arranged retroelements flanked by 12-bp conserved sequences, such as ϕRHS1e and ϕRHS1g (Fig. 5), supports this hypothesis of multiple insertion events at the same target site. Alternatively, we cannot rule out the possibility that tandem arrangement of retroelements is the result of a single event by an unknown mechanism.
FIG. 5.
Comparison of 12-bp sequences flanking RIME or ingi elements inserted into RHS pseudogenes. The RIME and ingi retroelements (□), the RHS flanking pseudogenes (░⃞), and the unknown flanking region (▨) are schematically represented. The 12 bp located at the junction between the RIME/ingi retroelements and the RHS/unknown flanking regions, and also between retroelements, are indicated by numbered black boxes, and the corresponding sequence is indicated on the right. The nucleotides in bold correspond to the duplicated region associated with the RIME/ingi insertion, and the boxes define the conserved region. The AAAAAA and CCCTGG sequences correspond to the end and the beginning of the RIME/ingi elements, respectively, and dots represent the RHS or unknown sequences. The crosses (✖) in the middle of retroelements indicate in which RIME or ingi element homologous recombination probably occurred. The accession numbers of ϕRHS2j, ϕRHS4d, and ϕRHS1 in Cos-12 and Cos-23, ϕRHS1e, ϕRHS1g, ϕRHS1f, and ϕRHS2g are AL359782 (ChrIa), AC008146 (BAC-30P15), AY046896, AY046897S1 and AY046897S2, AC079606 (BAC-3B10), AC087701 (BAC-26P8), AL359782 (ChrIa), and AC087701 (BAC-26P8), respectively.
The two other RHS pseudogenes (ϕRHS1f and ϕRHS2g) inactivated by retrotransposon insertion differ in that one duplicated 12-bp sequence is different from the other(s). It is possible that genetic exchange by homologous recombination between retrotransposons occurred to generate chimeric transposons flanked by unrelated regions (Fig. 5). This hypothesis was previously considered to explain the loss of a conserved region located upstream of a VSG gene family member, due to the presence of an ingi 22 bp upstream of the VSG gene without a flanking repeat sequence (59). The presence of unknown sequences (not related to RHS [pseudo]genes) downstream or upstream of the divergent 12-bp duplicated sequence in ϕRHS1f and ϕRHS2g, respectively, supports this hypothesis.
Expression and subcellular localization of RHS proteins.
A BLAST search with RHS subfamily-specific DNA fragments detected a total of 14 matches among ca. 4,500 T. brucei expressed sequence tags (ESTs) (http://www.ebi.ac.uk/blast2/parasites.html) (21, 23), suggesting that some of the RHS genes may be expressed (data not shown). Interestingly, as was observed for cDNA-005 (Fig. 1), one EST (AQ657854) homologous to RHS2 (pseudo)genes contains the 5" extremity of a RIME/ingi sequence, with the boundary between the RHS2 and RIME sequence corresponding exactly to the insertion site described above. Northern blot analyses performed with RHS subfamily-specific probes on RNA from T. brucei bloodstream (AnTat1) and procyclic (EATRO1125) forms indicate that the RHS multigene subfamilies are constitutively transcribed (data not shown). The mRNA detected by the RHS probes range between 3 and 3.5 kb, depending on the probe, which is consistent with the size of RHS (pseudo)genes (1.8 to 2.5 kb).
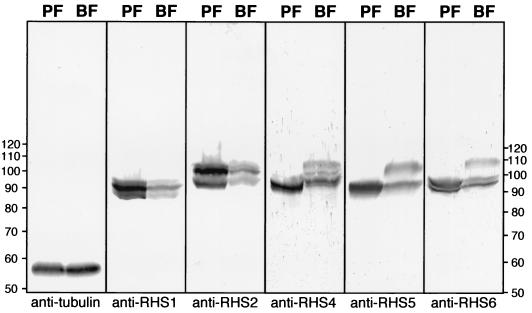
To study the expression of RHS protein, we raised antibodies against each RHS protein subfamily C-terminal domain, defined as the RHS subfamily-specific domain. The specificity of each immune serum was determined by testing the absence of cross-reaction with the other RHS recombinant proteins. All were found to be specific for the corresponding RHS recombinant protein except for the anti-RHS3 immune serum, which did not recognize the RHS3 recombinant protein, probably due to the weak immunogenicity of the 10-kDa recombinant protein (data not shown). Western blot analysis showed that all RHS proteins are constitutively expressed, although they are more abundant in the procyclic form than in the bloodstream form of T. brucei (Fig. 6). In addition, the different anti-RHS immune sera produced different protein profiles, confirming the absence of cross-reactivity. The detected proteins ranged from 85 to 110 kDa, which corresponds to the molecular mass calculated from the RHS genes. RHS proteins appear to be present in both life cycle stages with the exception of the highest band (110 kDa), which is only detected in the bloodstream form with the anti-RHS4, anti-RHS5, and anti-RHS6 immune sera (Fig. 6).
FIG. 6.
Western blot analysis of RHS proteins. Lysates (4 × 107 cells) of T. brucei procyclic form EATRO1125 (PF) and bloodstream form AnTat1 (BF) were analyzed by Western blotting with the immune sera specific for tubulin, RHS1, RHS2, RHS4, RHS5, and RHS6. The positions of the molecular mass markers (in kilodaltons) are indicated on the left and right, and the names of the immune sera is given under each blot.
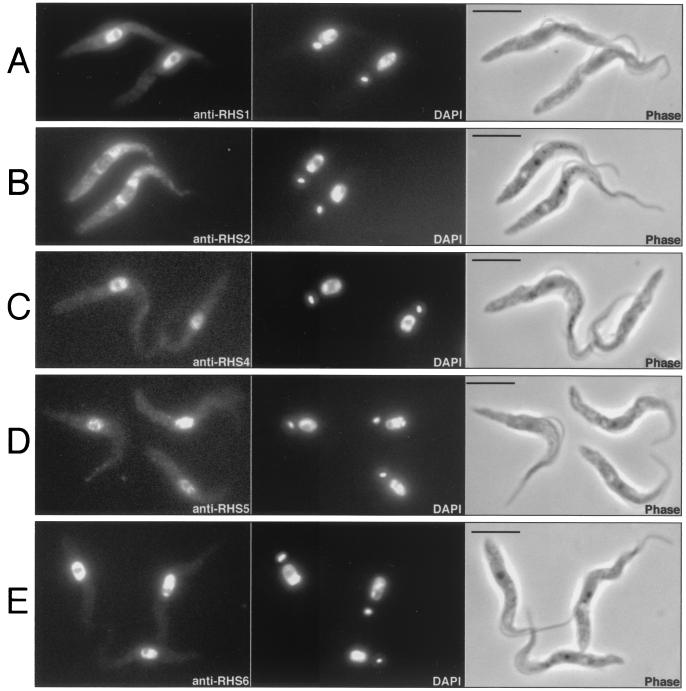
Immunofluorescence analysis of the T. brucei procyclic form (Fig. 7) and bloodstream form (data not shown) revealed that the RHS1, RHS4, RHS5, and RHS6 proteins colocalize with the DAPI-stained nuclear DNA with no visible label in the nucleolus (Fig. 7A and C to E). In contrast, the anti-RHS2 immune serum showed a perinuclear signal, whereby fluorescence intensity was higher around the nucleus (Fig. 7B).
FIG. 7.
Immunolocalization of RHS proteins. T. brucei procyclic cells (EATRO1125) were stained with anti-RHS1 (A), anti-RHS2 (B), anti-RHS4 (C), anti-RHS5 (D), and anti-RHS6 (E) immune sera (first column) and with DAPI (second column). Respective phase contrast (phase) images are shown in the third column. Bar = 5 μm.
RHS gene copy number.
In Trypanosoma cruzi, GSS databases proved to be an extremely powerful and accurate tool to study repeated sequences and particularly to estimate their copy number per genome (2). We used the same approach to estimate the copy number of each RHS subfamily in the T. brucei genome. A BLAST analysis was performed on the T. brucei GSS sequences (http://www.ebi.ac.uk/blast2/parasites.html [TIGR and The Wellcome Trust Sanger Institute]) using specific 500-bp sequences located in the 3" end of each multigene subfamily. The GSS represent about 1.8-fold coverage of the haploid nonminichromosomal genome (ca. 30 Mb in strain TREU927/4). The gene copy number per haploid genome was found to range between 6 and 31 depending on the subfamily, with a total of 128 copies for the RHS (pseudo)gene family. The RHS1 (28 copies), RHS2 (26 copies), RHS3 (31 copies), and RHS4 (27 copies) (pseudo)genes are the most abundantly represented, while RHS5 (10 copies) and RHS6 (6 copies) are less abundant. The same BLAST computer analysis was conducted with ingi and RIME sequences. The ingi and RHS (pseudo)gene copy numbers are similar (140 versus 128 copies per haploid nonminichromosomal genome), while the RIME copy number is about two to three times higher (380 copies). A previous Southern blot analysis estimated the ingi copy number in the range of 200 per haploid total genome (47).
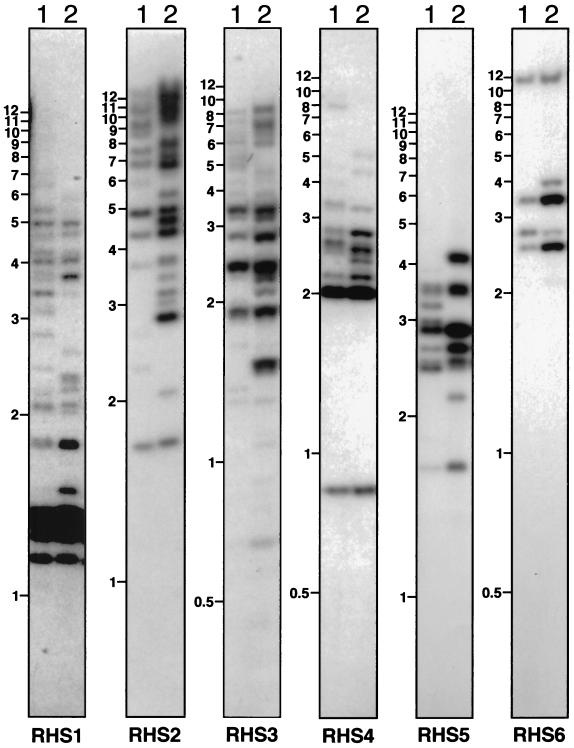
After comparing the available RHS (pseudo)genes and their conserved flanking regions, a Southern blot analysis of genomic DNA was conducted using restriction enzymes selected for their capacity to generate (i) relatively small DNA fragments that separate on a 0.6% agarose gel, (ii) a single DNA fragment for each RHS (pseudo)gene (the enzymes do not cleave the DNA fragments hybridizing with the probes), and (iii) size-polymorphic DNA fragments due to restriction site polymorphism in the different subfamily members. Using subfamily-specific probes (Fig. 2, box 2), ca. 100 different bands were detected in the genome of T. brucei TREU927/4 for the whole RHS multigene family (Fig. 8), indicating that at least 50 RHS (pseudo)genes are present in the haploid genome. This value is two to three times lower than obtained by GSS database analysis due to the comigration of fragments, as is clear from the variable intensity of hybridization to restriction fragments on the Southern blot (Fig. 8). Comparison of five different T. brucei strains showed a moderate DNA fragment polymorphism but the overall copy number for each RHS multigene subfamily appeared to be in the same range (Fig. 8 and data not shown).
FIG. 8.
Southern blot analysis of RHS (pseudo)genes. Genomic DNA from T. brucei TREU927/4 (lanes 1) and 427 (lanes 2) was digested to completion with HincII (RHS1 and RHS5), ClaI (RHS2 and RHS6), AseI (RHS3), or HpaII and KpnI (RHS4) and was analyzed by hybridization with the RHS1- to RHS6-specific probes to a Southern blot. The names of the probes and molecular size markers (in kilobases) are indicated below and on the left of each panel, respectively.
RHS (pseudo)genes are clustered in genome.
A P1 library of T. brucei (TREU927/4) genomic DNA containing average inserts of 65 kb (46) was screened with RHS subfamily-specific probes. Only 134 P1 clones (7.4% of the P1 library) are recognized by RHS probe(s), representing approximately 2 Mb of the haploid genome and yielding an estimated RHS1 to RHS6 copy number of only 35 copies per haploid nonminichromosomal genome. The latter figure is 3.3 times lower than that obtained by BLAST computer analysis of the GSS databases, suggesting that positive P1 clones contain several RHS (pseudo)genes. Indeed, more than 70% of the RHS-positive P1 clones contain at least two representatives of different subfamilies, since 29, 38, 21, 14, 15, and 17 P1 clones are recognized by one, two, three, four, five, and six different RHS probes, respectively. However, the estimated copy numbers of the less abundant RHS subfamilies (RHS5 and RHS6) are about the same using both approaches (10 versus 10 and 10 versus 6, respectively).
To determine the extent of RHS (pseudo)gene clustering, we analyzed several fully or almost fully sequenced BAC clones containing TREU927/4 genomic DNA fragments. Among the 30 sequenced BACs, we found clones containing 2 (BAC-45I2 and BAC-30P15), 5 (BAC-26P8), 12 (BAC-25N24), or 16 (BAC-3B10) RHS (pseudo)genes (Fig. 2). BAC-3B10 (163 kb) and BAC-25N24 (115 kb) constitute one end of ChrII (unpublished data) and contain a 250-kb region mainly composed of 28 RHS (pseudo)genes and their conserved flanking regions, as defined above, with some RIME or ingi retroelements inserted in the RHS1 to RHS4 coding sequences. This clearly indicates that RHS (pseudo)genes and their conserved flanking regions are often tandemly arranged. BAC-30P15 and BAC-26P8 clones contain only two and five RHS (pseudo)genes, respectively, which are also tandemly arranged (Fig. 9B and data not shown).
FIG. 9.
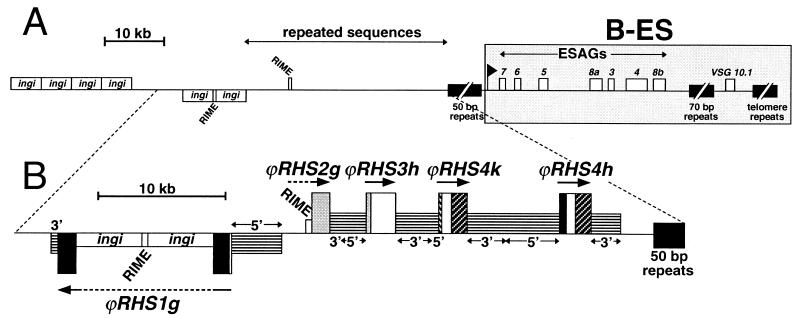
Gene organization of the BAC-26P8 clone which contains the B-ES of VSG 10.1. (A) Map of the B-ES of VSG 10.1 and upstream regions in TREU927/4 GUTat10.1, as previously described (36). The locations of the genes and retrotransposons (RIME and ingi) in BAC-26P8 are shown (□). Genes shown above the line are oriented towards the telomere, whereas those shown below the line are oriented away from the telomere. ▪, 50-bp, 70-bp, and telomere repeats. Expressions site-associated genes (ESAGs) are numbered 1 to 8 and the black flag indicates the position of the B-ES promoter. The B-ES, starting at the promoter, is highlighted by a large grey box. The region containing uncharacterized repeated sequences located upstream of the B-ES is indicated. (B) Detailed analysis of the region containing RHS pseudogenes in BAC-26P8. Sequences encoding RHS1 to RHS4 pseudogenes (large boxes) are shown using the same color code as in Fig. 2. The name and orientation of each RHS pseudogene are indicated above or below the boxes. The upstream (5") and downstream (3") sequences conserved between the different RHS (pseudo)genes of the same or different subfamilies are indicated by intermediate size boxes containing horizontal lines. All the RIME and ingi retroelements that are inserted into RHS genes are indicated by small white boxes.
RHS (pseudo)genes are clustered upstream of B-ES.
VSGs are expressed in T. brucei bloodstream forms in 1 of about 20 VSG expression sites (B-ESs) located upstream of the telomere repeats. Upstream of the B-ES promoter there is a large array of 50-bp repeats (up to 15 kb) that is specific to B-ES (44, 51). In ChrI, and probably in most of the other megabase chromosomes containing a B-ES, the 50-bp repeats are preceded by a large region (100 to 300 kb) composed of RIME/ingi retroelements and previously uncharacterized repeats (43). Analysis of the ChrIa sequence (1.1 Mb), recently completed by The Wellcome Trust Sanger Institute (data not shown), revealed the presence of 15 RHS (pseudo)genes clustered upstream of the B-ES in a 150-kb DNA region corresponding to the RIME/ingi-rich region (43). The RIME/ingi-rich region of ChrIa contains only five full-length and probably functional RHS genes (21%), and of the 10 RHS pseudogenes, four are highly degenerate. As was observed for ChrII, all the RIME/ingi retroelements present in the RHS-rich region of ChrIa (seven elements) are inserted into RHS-related (pseudo)genes, suggesting that the RIME/ingi richness of this repetitive region is directly related to the presence of the RHS (pseudo)genes. RIME, ingi, and the RHS (pseudo)genes with their conserved flanking regions constitute most (if not all) of the repetitive sequences in this region.
Recently, LaCount et al. sequenced a BAC genomic DNA insert (BAC-26P8) containing the active B-ES expressing the VSG10.1 gene of strain TREU927/4 GUTat10.1 and the region (90 kb) upstream of the 50-bp repeats (36). As previously observed for the B-ES flanking region of ChrIa, this 90-kb DNA region is RIME/ingi-rich (six ingis and two RIMEs) and contains uncharacterized repeats (Fig. 9A). We found that this 90-kb repeated region is composed of five RHS pseudogenes (ϕRHS2g and ϕRHS1g contain one and three RIME/ingi retroelements, respectively) and conserved flanking sequences, as shown in Fig. 9B. The extent of the RHS (pseudo)gene cluster flanking VSG10.1-ES may be longer since the region upstream of the BAC-26P8 is not yet sequenced.
To determine if the presence of an RHS (pseudo)gene cluster is a general feature of the regions upstream of B-ESs, we studied the locations of RHS (pseudo)genes and B-ES-associated sequences in the T. brucei (TREU927/4) P1 library. All B-ESs described to date contain ESAG7 and ESAG6 genes downstream of the B-ES-specific promoter region and are separated from the remainder of the chromosome by a 50-bp repeat cluster (Fig. 9). All these sequences are considered to be B-ES specific (44, 51). Interestingly, 72, 73, and 86% of the P1 clones recognized by the 50-bp repeat, B-ES promoter and/or ESAG6/7 probes, respectively, contain RHS sequences. These data strongly support the hypothesis that most, if not all, B-ESs are preceded by RHS sequences.
Presence of RHS-related genes in T. cruzi.
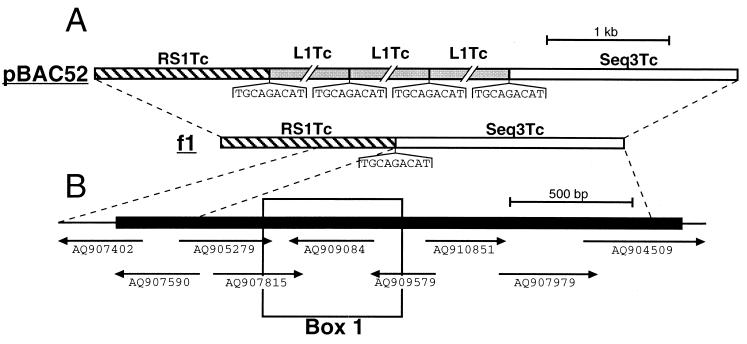
A BLAST search of the trypanosomatid databases (http://www.ebi.ac.uk/blast2/parasites.html) revealed that RHS-related sequences are also abundant in the T. cruzi genome. Recently, Olivares et al. showed that the non-LTR retrotransposon L1Tc, the T. cruzi homologue of ingi, is frequently inserted between a RS1Tc (1.5-kb) and Seq3Tc (1.9-kb) fragment (49) (Fig. 10A). We have determined that these could encode RHS-related proteins. Since, the RS1Tc and Seq3Tc sequences contain numerous frame shifts and stop codons, a chimeric full-length RHS-related gene was assembled in silico using the T. cruzi GSS database (http://www.ebi.ac.uk/parasites/paratable.html) (Fig. 10B). This database contains 11,459 sequences and represents about 10% of the 40-Mb haploid genome of the T. cruzi CL strain (2). The T. cruzi chimeric protein has 16.5 to 23.4% identity with the different T. brucei RHS proteins, and most of the residues conserved between the T. brucei RHS proteins, including the ATP/GTP-binding motif, are also conserved in the T. cruzi protein (Fig. 3). The RS1Tc and Seq3Tc sequences detected 133 and 167, respectively, significantly similar DNA fragments in the T. cruzi GSS database by BLAST analysis, which corresponds to 348 and 304 copies per haploid genome, respectively. Furthermore, we detected 49 sequences related to the Seq3Tc sequence among approximately 5,000 T. cruzi ESTs (http://www.genpat.uu.se/tryp/tryp.html [Uppsala University]). In summary, these computer analyses indicate that T. cruzi contains an expressed RHS-related mutigene family, which may also contain a hot spot for retroelement insertion.
FIG. 10.
Characterization of an RHS-related multigene family in T. cruzi. (A) Map of the pBAC52 and f1 genomic clones from T. cruzi CL-Brener and Maracay strains, respectively, as previously reported by Olivares et al. (49). Grey, hatched, and white boxes represent the L1Tc retrotransposons (5 kb) and the RS1Tc (1.4 kb) and Seq3Tc (1.9 kb) repetitive elements, respectively. The conserved duplicated 9-bp sequences (TGCAGACAT) flanking the L1Tc elements and the corresponding L1Tc insertion site in the f1 sequences are shown under the map. (B) Chimeric RS1Tc/Seq3Tc sequence coding for an RHS-related protein. To generate an RS1Tc/Seq3Tc-related sequence containing a large open reading frame (black box on the map), a selection of nine representative T. cruzi GSS were assembled. The relative positions and accession numbers of the selected GSSs are indicated under the map. Box 1 indicates the amino acid sequence, aligned with the T. brucei RHS proteins in Fig. 3.
DISCUSSION
We have characterized a new, large multigene family encoding nuclear and perinuclear proteins in T. brucei. We analyzed a total of 61 different RHS genes and pseudogenes detected in four cDNA clones, two BAC clones from ChrII, the contiguous sequence of ChrIa, and three BACs and five cosmids of unknown genomic location. Analysis of the C-terminal DNA sequence allowed us to subdivide the family into six multigene subfamilies, RHS1 to RHS6. More than half of the RHS copies described here are pseudogenes. To estimate the number of RHS (pseudo)genes in the nuclear genome of strain TREU927/4, we took advantage of the T. brucei GSS databases at TIGR and The Wellcome Trust Sanger Institute, which provide about 1.8-fold coverage of the haploid DNA (excluding minichromosomes). We estimate that there are 128 RHS (pseudo)gene fragments per nonminichromosomal haploid genome. RHS (pseudo)genes also appear to be present in a subset of minichromosomes (hybridization data not shown).
The computational analysis of DNA sequences from TIGR and The Wellcome Trust Sanger Institute, selected cosmids and cDNAs revealed that this multigene family contains a hot spot for insertion of the RIME and ingi retrotransposons: (i) approximately one-third of the RHS (pseudo)genes contain RIME and/or ingi retrotransposons (16 out of 51 copies), (ii) the retroelements are always inserted at exactly the same relative position in the RHS pseudogenes, even though these genes display up to 50% variation in nucleotide sequence in the vicinity of the insertion site (data not shown), (iii) of the 16 RHS pseudogenes containing RIME/ingi element(s), 25% contain two or three retroelements while only 1 of the 10 non-RHS sequences in the databases containing RIME/ingi retroelements has tandemly arranged elements (data not shown), (iv) a phylogenetic analysis shows that most were generated by independent insertion events, and (v) among the 10 RIME/ingi retroelements present in the sequenced ChrIa of strain TREU927/4, 7 are inserted into RHS pseudogenes. Many eukaryotes contain site-specific non-LTR retrotransposons (3, 9, 16, 26, 38, 63, 67). Also, non-LTR retrotransposons that appear to be randomly distributed in the host genome in fact show a bias of recognition for insertion sites, as exemplified by the TTAAAA sequence of human LINEs (31). The exact site specificity of retroelement insertion into RHS genes leads to the observed tandem arrays of elements. Interestingly, the tandem arrangement of the T. brucei (RIME and ingi) and T. cruzi (L1Tc) non-LTR retrotransposons is unique since, to our knowledge, none of the site-specific or randomly distributed retroelements show this organization in other organisms.
It appears that all the RIME/ingi elements present in RHS genes are inserted in frame with the RHS gene. When the retroelement is unmutated, this results in the generation of long open reading frames encoding putative chimeric proteins composed of the RHS N-terminal half followed by a peptide encoded by the retroelement. However, it is noteworthy that only a few ingi elements contain a single long open reading frame encoding a putative multifunctional protein (data not shown). Most, including those originally described (33, 47), are probably not able to encode functional mRNAs due to the presence of frame shifts or premature stop codons. Consequently, the putative RHS/ingi chimeric proteins may exhibit an important size and sequence polymorphism due to the ingi polymorphism. At least seven different chimeric proteins formed between cellular and mobile element genes are expressed in humans (24, 56, 60, 65). Thus, it is tempting to consider that some of the RHS/ingi chimeric proteins may be expressed and that the proteins may have a cellular role. This would provide a functional raison d'être for the presence and conservation of a RIME/ingi insertion hot spot within the RHS genes. This hypothesis is supported by the characterization of RHS/retrotransposon chimeric cDNA molecules in which the boundary between the RHS pseudogene and the RIME sequence corresponds exactly to the conserved RIME/ingi insertion site observed in genomic DNA. Production of antibodies against the N-terminal region of the ingi products will allow us to determine if the RHS/ingi chimeric proteins are expressed.
Analysis of the T. cruzi databases revealed that the genome of T. cruzi also contains polymorphic repeated sequences that potentially code for proteins homologous to the T. brucei RHS proteins. Interestingly, these DNA sequences were initially characterized as non-LTR retrotransposon (L1Tc) flanking sequences (49), suggesting that such elements also frequently insert into the putative T. cruzi RHS-like genes. In contrast, a BLAST analysis of the Leishmania GSS and cosmid sequence databases, which contain at least as many sequences as the T. brucei databases, does not reveal the presence of any RHS homologue. The absence of these sequences is probably correlated with the apparent absence of mobile elements, including retrotransposons, as revealed by the ongoing sequence analysis of this highly related genome (http://www.ebi.ac.uk/parasites/leish.html).
Comparison of ChrI homologues in different T. brucei strains indicates that the large RIME/ingi-rich repetitive region presents a polymorphism with an important size (43). The RIME/ingi richness observed for this large section of ChrIa in TREU927/4 (43), but also in ChrII and BAC-26P8, is entirely due to insertion into the clustered RHS (pseudo)genes. Detailed analysis of the RHS multigene family shows that they are subject to frequent homologous recombination. Where this occurs within and between nonhomologous chromosomes may explain not only the size of the polymorphism of the RIME/ingi-rich repetitive area (43) but also the variation in number and location of B-ESs observed in different strains (43, 44, 45). Our analysis reveals that among 23 retroelements present in 14 RHS pseudogenes within the large clusters described here, 8 are flanked by one RHS sequence and one unknown sequence. The latter were probably generated by homologous recombination between two retroelements, one inserted in an RHS pseudogene and another inserted into the unknown sequence. In addition, approximately one-third of the RHS (pseudo)-genes studied are chimeric, and we suggest that these probably result from homologous recombination in conserved regions of RHS copies belonging to different subfamilies. These suspected homologous recombination events are probably the tip of the iceberg, since numerous undetectable events probably occur between the abundant homologous sequences clustered in large sections of multiple chromosomes.
The 52 RHS (pseudo)genes identified so far in the T. brucei (TREU927/4) databases are located in five different clusters that are almost exclusively composed of RHS copies and their large conserved flanking regions: 28 copies (15 genes, 13 pseudogenes) in a 250-kb area of ChrII (unpublished data), 15 copies (5 genes, 10 pseudogenes) in the 150-kb RIME/ingi-rich region in ChrIa (42, 43), five pseudogenes in BAC-26P8 (36), two pseudogenes in BAC-45I2 (36), and two pseudogenes in BAC-30P15 (unpublished data). The three largest RHS clusters are located upstream of the TTAGGG telomere repeats (ChrII) or upstream of a 45- to 60-kb B-ES that is adjacent to the telomere repeats (ChrIa and BAC-26P8). The tandemly arranged RHS pseudogenes in BAC-45I2 are located 30 kb upstream of a region with the characteristics of a telomeric M-ES. Similarly, the M-ES active in T. brucei rhodesiense WRATat1.1-MVAT5 (41) and present in T. brucei AnTat1 (13) is preceded by a RHS1 pseudogene (Cos-12) located 10 kb upstream of the telomere repeats (unpublished data). Although the chromosomal positions of the DNA sequences derived from the other BACs and the cosmids are not known, it appears from this analysis that the RHS (pseudo)genes are located in subtelomeric regions of chromosomes, upstream of ESs (B-ESs or M-ESs) or directly adjacent to the telomere repeats. However, in the fully sequenced ChrI and ChrII, the large clusters are found only at one end, indicating that not all telomeres are separated from the central coding regions by RHS clusters. Nevertheless, it is interesting that the P1 genomic library analysis showed that most of the B-ESs, maybe all of them, are flanked by RHS (pseudo)genes.
The subtelomeric localization of the RHS (pseudo)genes may be related to their function. In most eukaryotes, subtelomeric regions are large and repetitive, and poorly transcribed sequences are located at both ends of chromosomes and directly adjacent to the short telomere repeats (69). Although subtelomeres are essentially composed of noncoding sequences, expressed genes are found embedded in subtelomeric repeats, such as the PAU, SUC, MAL, and MEL multigene families in yeast (40), and surface antigen gene families in Plasmodium (6, 18, 20, 61, 62). Apparently there is a selective advantage for the Plasmodium surface antigen genes, which are involved in antigenic variation, to be located within subtelomeric regions. The high recombination frequencies in subtelomeric domains seem to create a favorable environment for the rapid generation of novel genes encoding surface proteins (25). Interestingly, in Plasmodium vivax, a large cluster of 35 vir genes and pseudogenes encoding immunovariant surface proteins is located directly upstream of the telomere repeats (20), exactly as observed for the RHS cluster in ChrII. In addition, T. brucei VSGs are expressed in the telomeric ESs (B-ESs and M-ESs) and homologous recombination is required to mediate antigenic variation. These observations suggest that the diversity observed for the RHS multigene family, probably generated by the high rate of recombination in subtelomeric regions, may be advantageous for the parasite. Our experiments indicate that the RHS proteins are located inside the cell, not on the cell surface, and it is now a priority to investigate the function of this diverse and potentially rapidly evolving gene family.
In summary, we describe for the first time a gene family with conserved flanking regions that constitutes about 5% of the T. brucei genome. This multigene family is associated with the most abundant putative mobile elements (about 5% of the genome content) and may be undergoing rapid evolution by recombination and sequence divergence. The RHS genes are clustered in defined regions of chromosomes in T. brucei and are probably always found upstream of B-ESs, although also present on chromosomes not carrying B-ESs. A homologous family is present in T. cruzi, and for both of these organisms the data presented here will be very significant to the finishing stages of the genome sequencing projects.
Acknowledgments
The contributions of the two first coauthors, F.B. and N.B., are equivalent.
We are grateful to D. Baltz and A. Ambit for technical help and T. Heidmann, S. Litvak, M. Pages and D. R. Robinson for critical reading of the manuscript.
This work was supported by the CNRS, the Conseil Régional d'Aquitaine, the GDR Parasitologie (CNRS), the Ministère de l'Education Nationale de la Recherche et de la Technologie (Action Microbiologie), the Programme Alliance Franco-Britannique 2001, UNDP/World Bank/WHO-TDR T. brucei Genome Project and the Wellcome Trust Beowulf Genomics Initiative.
REFERENCES
- 1.Affolter, M., L. Rindisbacher, and R. Braun. 1989. The tubulin gene cluster of Trypanosoma brucei starts with an intact beta-gene and ends with a truncated beta-gene interrupted by a retrotransposon-like sequence. Gene 80:177-183. [DOI] [PubMed] [Google Scholar]
- 2.Aguero, F., R. E. Verdun, A. C. Frasch, and D. O. Sanchez. 2000. A random sequencing approach for the analysis of the Trypanosoma cruzi genome: general structure, large gene and repetitive DNA families, and gene discovery. Genome Res. 10:1996-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aksoy, S., T. M. Lalor, J. Martin, L. H. Van der Ploeg, and F. F. Richards. 1987. Multiple copies of a retroposon interrupt spliced leader RNA genes in the African trypanosome Trypanosoma gambiense. EMBO J. 6:3819-3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barry, J. D., S. V. Graham, M. Fotheringham, V. S. Graham, K. Kobryn, and B. Wymer. 1998. VSG gene control and infectivity strategy of metacyclic stage Trypanosoma brucei. Mol. Biochem. Parasitol. 91:93-105. [DOI] [PubMed] [Google Scholar]
- 5.Barry, J. D., and R. McCulloch. 2001. Antigenic variation in trypanosomes: enhanced phenotypic variation in a eukaryotic parasite. Adv. Parasitol. 49:1-70. [DOI] [PubMed] [Google Scholar]
- 6.Baruch, D. I., B. L. Pasloske, H. B. Singh, X. Bi, X. C. Ma, M. Feldman, T. F. Taraschi, and R. J. Howard. 1995. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell 82:77-87. [DOI] [PubMed] [Google Scholar]
- 7.Beals, T. P., and J. C. Boothroyd. 1992. Genomic organization and context of a trypanosome variant surface glycoprotein gene family. J. Mol. Biol. 225:961-971. [DOI] [PubMed] [Google Scholar]
- 8.Bernards, A., L. H. Van der Ploeg, A. C. Frasch, P. Borst, J. C. Boothroyd, S. Coleman, and G. A. Cross. 1981. Activation of trypanosome surface glycoprotein genes involves a duplication-transposition leading to an altered 3" end. Cell 27:497-505. [DOI] [PubMed] [Google Scholar]
- 9.Besansky, N. J., S. M. Paskewitz, D. M. Hamm, and F. H. Collins. 1992. Distinct families of site-specific retrotransposons occupy identical positions in the rRNA genes of Anopheles gambiae. Mol. Cell. Biol. 12:5102-5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borst, P., W. Bitter, P. A. Blundell, I. Chaves, M. Cross, H. Gerrits, F. van Leeuwen, R. McCulloch, M. Taylor, and G. Rudenko. 1998. Control of VSG gene expression sites in Trypanosoma brucei. Mol. Biochem. Parasitol. 91:67-76. [DOI] [PubMed] [Google Scholar]
- 11.Bringaud, F., D. Baltz, and T. Baltz. 1998. Functional and molecular characterization of a glycosomal PPi-dependent enzyme in trypanosomatids: pyruvate, phosphate dikinase. Proc. Natl. Acad. Sci. USA 95:7963-7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bringaud, F., and T. Baltz. 1992. A potential hexose transporter gene expressed predominantly in the bloodstream form of Trypanosoma brucei. Mol. Biochem. Parasitol. 52:111-121. [DOI] [PubMed] [Google Scholar]
- 13.Bringaud, F., N. Biteau, J. E. Donelson, and T. Baltz. 2001. Conservation of metacyclic variant surface glycoprotein expression sites among different trypanosome isolates. Mol. Biochem. Parasitol. 113:67-78. [DOI] [PubMed] [Google Scholar]
- 14.Bringaud, F., C. Vedrenne, A. Cuvillier, D. Parzy, D. Baltz, E. Tetaud, E. Pays, J. Venegas, G. Merlin, and T. Baltz. 1998. Conserved organization of genes in trypanosomatids. Mol. Biochem. Parasitol. 94:249-264. [DOI] [PubMed] [Google Scholar]
- 15.Brun, R., and M. Schonenberger. 1979. Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 36:289-292. [PubMed] [Google Scholar]
- 16.Burke, W. D., F. Muller, and T. H. Eickbush. 1995. R4, a non-LTR retrotransposon specific to the large subunit rRNA genes of nematodes. Nucleic Acids Res. 23:4628-4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campbell, D. A. 1989. c2X75, a derivative of the cosmid vector c2XB. Nucleic Acids Res. 17:458.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng, Q., N. Cloonan, K. Fischer, J. Thompson, G. Waine, M. Lanzer, and A. Saul. 1998. stevor and rif are Plasmodium falciparum multicopy gene families which potentially encode variant antigens. Mol. Biochem. Parasitol. 97:161-176. [DOI] [PubMed] [Google Scholar]
- 19.Cross, G. A., L. E. Wirtz, and M. Navarro. 1998. Regulation of vsg expression site transcription and switching in Trypanosoma brucei. Mol. Biochem. Parasitol. 91:77-91. [DOI] [PubMed] [Google Scholar]
- 20.del Portillo, H. A., C. Fernandez-Becerra, S. Bowman, K. Oliver, M. Preuss, C. P. Sanchez, N. K. Schneider, J. M. Villalobos, M. A. Rajandream, D. Harris, L. H. Pereira da Silva, B. Barrell, and M. Lanzer. 2001. A superfamily of variant genes encoded in the subtelomeric region of Plasmodium vivax. Nature 410:839-842. [DOI] [PubMed] [Google Scholar]
- 21.Djikeng, A., C. Agufa, J. E. Donelson, and P. A. Majiwa. 1998. Generation of expressed sequence tags as physical landmarks in the genome of Trypanosoma brucei. Gene 221:93-106. [DOI] [PubMed] [Google Scholar]
- 22.Donelson, J. E., K. L. Hill, and N. M. El-Sayed. 1998. Multiple mechanisms of immune evasion by African trypanosomes. Mol. Biochem. Parasitol. 91:51-66. [DOI] [PubMed] [Google Scholar]
- 23.El-Sayed, N. M., C. M. Alarcon, J. C. Beck, V. C. Sheffield, and J. E. Donelson. 1995. cDNA expressed sequence tags of Trypanosoma brucei rhodesiense provide new insights into the biology of the parasite. Mol. Biochem. Parasitol. 73:75-90. [DOI] [PubMed] [Google Scholar]
- 24.Esposito, T., F. Gianfrancesco, A. Ciccodicola, L. Montanini, S. Mumm, M. D'Urso, and A. Forabosco. 1999. A novel pseudoautosomal human gene encodes a putative protein similar to Ac-like transposases. Hum. Mol. Genet. 8:61-67. [DOI] [PubMed] [Google Scholar]
- 25.Freitas-Junior, L. H., E. Bottius, L. A. Pirrit, K. W. Deitsch, C. Scheidig, F. Guinet, U. Nehrbass, T. E. Wellems, and A. Scherf. 2000. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum. Nature 407:1018-1022. [DOI] [PubMed] [Google Scholar]
- 26.Gabriel, A., T. J. Yen, D. C. Schwartz, C. L. Smith, J. D. Boeke, B. Sollner-Webb, and D. W. Cleveland. 1990. A rapidly rearranging retrotransposon within the miniexon gene locus of Crithidia fasciculata. Mol. Cell. Biol. 10:615-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gottesdiener, K., J. Garcia-Anoveros, M. G. Lee, and L. H. Van der Ploeg. 1990. Chromosome organization of the protozoan Trypanosoma brucei. Mol. Cell. Biol. 10:6079-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harlow, E., and D. Lane (ed.). 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Hasan, G., M. J. Turner, and J. S. Cordingley. 1984. Complete nucleotide sequence of an unusual mobile element from Trypanosoma brucei. Cell 37:333-341. [DOI] [PubMed] [Google Scholar]
- 30.Hope, M., A. MacLeod, V. Leech, S. Melville, J. Sasse, A. Tait, and C. M. Turner. 1999. Analysis of ploidy (in megabase chromosomes) in Trypanosoma brucei after genetic exchange. Mol. Biochem. Parasitol. 104:1-9. [DOI] [PubMed] [Google Scholar]
- 31.Jurka, J. 1997. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc. Natl. Acad. Sci. USA 94:1872-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanmogne, G. D., M. Bailey, and W. C. Gibson. 1997. Wide variation in DNA content among isolates of Trypanosoma brucei ssp. Acta Trop. 63:75-87. [DOI] [PubMed] [Google Scholar]
- 33.Kimmel, B. E., O. K. ole-MoiYoi, and J. R. Young. 1987. Ingi, a 5.2-kb dispersed sequence element from Trypanosoma brucei that carries half of a smaller mobile element at either end and has homology with mammalian LINEs. Mol. Cell. Biol. 7:1465-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kooter, J. M., H. J. van der Spek, R. Wagter, C. E. d'Oliveira, F. van der Hoeven, P. J. Johnson, and P. Borst. 1987. The anatomy and transcription of a telomeric expression site for variant-specific surface antigens in T. brucei. Cell 51:261-272. [DOI] [PubMed] [Google Scholar]
- 35.Labrador, M., and V. G. Corces. 1997. Transposable element-host interactions: regulation of insertion and excision. Annu. Rev. Genet. 31:381-404. [DOI] [PubMed] [Google Scholar]
- 36.LaCount, D. J., N. M. El-Sayed, S. Kaul, D. Wanless, C. M. Turner, and J. E. Donelson. 2001. Analysis of a donor gene region for a variant surface glycoprotein and its expression site in African trypanosomes. Nucleic Acids Res. 29:2012-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lanham, S. M., and D. G. Godfrey. 1970. Isolation of salivarian trypanosomes from man and other mammals using DEAE-cellulose. Exp. Parasitol. 28:521-534. [DOI] [PubMed] [Google Scholar]
- 38.Levis, R. W., R. Ganesan, K. Houtchens, L. A. Tolar, and F. M. Sheen. 1993. Transposons in place of telomeric repeats at a Drosophila telomere. Cell 75:1083-1093. [DOI] [PubMed] [Google Scholar]
- 39.Lodes, M. J., B. L. Smiley, A. W. Stadnyk, J. L. Bennett, P. J. Myler, and K. Stuart. 1993. Expression of a retroposon-like sequence upstream of the putative Trypanosoma brucei variant surface glycoprotein gene expression site promoter. Mol. Cell. Biol. 13:7036-7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Louis, E. J. 1995. The chromosome ends of Saccharomyces cerevisiae. Yeast 11:1553-1573. [DOI] [PubMed] [Google Scholar]
- 41.Lu, Y., T. Hall, L. S. Gay, and J. E. Donelson. 1993. Point mutations are associated with a gene duplication leading to the bloodstream reexpression of a trypanosome metacyclic VSG. Cell 72:397-406. [DOI] [PubMed] [Google Scholar]
- 42.Melville, S. E. 1997. Parasite genome analysis. Genome research in Trypanosoma brucei: chromosome size polymorphism and its relevance to genome mapping and analysis. Trans. R. Soc. Trop. Med. Hyg. 91:116-120. [DOI] [PubMed] [Google Scholar]
- 43.Melville, S. E., C. S. Gerrard, and J. M. Blackwell. 1999. Multiple causes of size variation in the diploid megabase chromosomes of African trypanosomes. Chromosome Res. 7:191-203. [DOI] [PubMed] [Google Scholar]
- 44.Melville, S. E., V. Leech, C. S. Gerrard, A. Tait, and J. M. Blackwell. 1998. The molecular karyotype of the megabase chromosomes of Trypanosoma brucei and the assignment of chromosome markers. Mol. Biochem. Parasitol. 94:155-173. [DOI] [PubMed] [Google Scholar]
- 45.Melville, S. E., V. Leech, M. Navarro, and G. A. Cross. 2000. The molecular karyotype of the megabase chromosomes of Trypanosoma brucei stock 427. Mol. Biochem. Parasitol. 111:261-273. [DOI] [PubMed] [Google Scholar]
- 46.Melville, S. E., N. S. Shepherd, C. S. Gerrard, and R. W. F. Le Page. 1996. Selection of chromosome-specific DNA clones from African trypanosome genomic libraries, p. 257-293. In B. Birren, and E. Lai (ed.), Analysis of non-mammalian genomes. Academic Press, New York, N.Y.
- 47.Murphy, N. B., A. Pays, P. Tebabi, H. Coquelet, M. Guyaux, M. Steinert, and E. Pays. 1987. Trypanosoma brucei repeated element with unusual structural and transcriptional properties. J. Mol. Biol. 195:855-871. [DOI] [PubMed] [Google Scholar]
- 48.Olivares, M., C. Alonso, and M. C. Lopez. 1997. The open reading frame 1 of the L1Tc retrotransposon of Trypanosoma cruzi codes for a protein with apurinic-apyrimidinic nuclease activity. J. Biol. Chem. 272:25224-25228. [DOI] [PubMed] [Google Scholar]
- 49.Olivares, M., M. del Carmen Thomas, A. Lopez-Barajas, J. M. Requena, J. L. Garcia-Perez, S. Angel, C. Alonso, and M. C. Lopez. 2000. Genomic clustering of the Trypanosoma cruzi nonlong terminal L1Tc retrotransposon with defined interspersed repeated DNA elements. Electrophoresis 21:2973-2982. [DOI] [PubMed] [Google Scholar]
- 50.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 51.Pays, E., S. Lips, D. Nolan, L. Vanhamme, and D. Perez-Morga. 2001. The VSG expression sites of Trypanosoma brucei: multipurpose tools for the adaptation of the parasite to mammalian hosts. Mol. Biochem. Parasitol. 114:1-16. [DOI] [PubMed] [Google Scholar]
- 52.Pays, E., and N. B. Murphy. 1987. DNA-binding fingers encoded by a trypanosome retroposon. J. Mol. Biol. 197:147-148. [DOI] [PubMed] [Google Scholar]
- 53.Pays, E., and D. P. Nolan. 1998. Expression and function of surface proteins in Trypanosoma brucei. Mol. Biochem. Parasitol. 91:3-36. [DOI] [PubMed] [Google Scholar]
- 54.Pays, E., P. Tebabi, A. Pays, H. Coquelet, P. Revelard, D. Salmon, and M. Steinert. 1989. The genes and transcripts of an antigen gene expression site from Trypanosoma brucei. Cell. 57:835-845. [DOI] [PubMed] [Google Scholar]
- 55.Pedram, M., and J. E. Donelson. 1999. The anatomy and transcription of a monocistronic expression site for a metacyclic variant surface glycoprotein gene in Trypanosoma brucei. J. Biol. Chem. 274:16876-16883. [DOI] [PubMed] [Google Scholar]
- 56.Robertson, H. M., and K. L. Zumpano. 1997. Molecular evolution of an ancient mariner transposon, Hsmar1, in the human genome. Gene 205:203-217. [DOI] [PubMed] [Google Scholar]
- 57.Sambrook, J., E. F. Fritsch, and T. Maniatis (ed.). 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 58.Schmidt, T. 1999. LINEs, SINEs and repetitive DNA: non-LTR retrotransposons in plant genomes. Plant Mol. Biol. 40:903-910. [DOI] [PubMed] [Google Scholar]
- 59.Smiley, B. L., R. F. Aline, Jr., P. J. Myler, and K. Stuart. 1990. A retroposon in the 5" flank of a Trypanosoma brucei VSG gene lacks insertional terminal repeats. Mol. Biochem. Parasitol. 42:143-152. [DOI] [PubMed] [Google Scholar]
- 60.Smit, A. F. 1999. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 9:657-663. [DOI] [PubMed] [Google Scholar]
- 61.Smith, J. D., C. E. Chitnis, A. G. Craig, D. J. Roberts, D. E. Hudson-Taylor, D. S. Peterson, R. Pinches, C. I. Newbold, and L. H. Miller. 1995. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell 82:101-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Su, X. Z., V. M. Heatwole, S. P. Wertheimer, F. Guinet, J. A. Herrfeldt, D. S. Peterson, J. A. Ravetch, and T. E. Wellems. 1995. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell 82:89-100. [DOI] [PubMed] [Google Scholar]
- 63.Teng, S. C., S. X. Wang, and A. Gabriel. 1995. A new non-LTR retrotransposon provides evidence for multiple distinct site-specific elements in Crithidia fasciculata miniexon arrays. Nucleic Acids Res. 23:2929-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Toth, M., J. Grimsby, G. Buzsaki, and G. P. Donovan. 1995. Epileptic seizures caused by inactivation of a novel gene, jerky, related to centromere binding protein-B in transgenic mice. Nat. Genet. 11:71-75. [DOI] [PubMed] [Google Scholar]
- 66.Vanhamme, L., E. Pays, R. McCulloch, and J. D. Barry. 2001. An update on antigenic variation in African trypanosomes. Trends Parasitol. 17:338-343. [DOI] [PubMed] [Google Scholar]
- 67.Villanueva, M. S., S. P. Williams, C. B. Beard, F. F. Richards, and S. Aksoy. 1991. A new member of a family of site-specific retrotransposons is present in the spliced leader RNA genes of Trypanosoma cruzi. Mol. Cell. Biol. 11:6139-6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weiden, M., Y. N. Osheim, A. L. Beyer, and L. H. Van der Ploeg. 1991. Chromosome structure: DNA nucleotide sequence elements of a subset of the minichromosomes of the protozoan Trypanosoma brucei. Mol. Cell. Biol. 11:3823-3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wellinger, R. J., and D. Sen. 1997. The DNA structures at the ends of eukaryotic chromosomes. Eur. J. Cancer 33:735-749. [DOI] [PubMed] [Google Scholar]