Abstract
Migraine is a common form of headache and has a significant genetic component. Here, we report linkage results from a study in Iceland of migraine without aura (MO). The study group comprised patients with migraine recruited by neurologists and from the registry of the Icelandic Migraine Society, as well as through the use of a questionnaire sent to a random sample of 20,000 Icelanders. Migraine diagnoses were made and confirmed using diagnostic criteria established by the International Headache Society. A genomewide scan with multipoint allele-sharing methods was performed on 289 patients suffering from MO. Linkage was observed to a locus on chromosome 4q21 (LOD=2.05; P=.001). The locus reported here overlaps a locus (MGR1) reported elsewhere for patients with migraine with aura (MA) in the Finnish population. This replication of the MGR1 locus in families with MO indicates that the gene we have mapped may contribute to both MA and MO. Further analysis indicates that the linkage evidence improves for affected females and, especially, with a slightly relaxed definition of MO (LOD=4.08; P=7.2×10-6).
Introduction
Migraine (MIM 157300) is a common headache disorder that usually begins in early adulthood and affects ∼18% of females and ∼6% of males every year (Stewart et al. 1992). It is characterized by recurrent, unilateral, pulsating headaches of moderate-to-severe intensity, aggravated by physical activity; the headaches are of 4- to 72-h duration. Other manifestations of the disease include nausea, vomiting, photophobia, and phonophobia. The diagnostic criteria most commonly used are the standardized guidelines of the International Headache Society (IHS) (Headache Classification Committee of the International Headache Society [HCCIHS] 1988).
Migraine without aura (MO) or “common migraine,” affects ∼58% of migraineurs (Russell et al. 1995b). Migraine with aura (MA) or “classic migraine” is the form of migraine with headache attacks preceded by neurologic manifestations, called “aura,” and affects ∼28% of migraineurs (Russell et al. 1995b). Both subtypes of migraine can occur within the same family (Nyholt et al. 1998a, 1998b; Carlsson et al. 2002; Lea et al. 2002) or in the same individual (Rasmussen and Olesen 1992; Launer et al. 1999; Kallela et al. 2001b). A recent questionnaire-based clinical study in Finland indicates that the migraine subtype breakdown for families with four or more migraineurs is as follows: 23% have only MO, 41% experience both MA and MO episodes, 11% have only MA, 20% have unclassified aura, and 3% have migraine aura without headache (Kallela et al. 2001b). In a population-based twin registry, the number of discordant twins with MA or MO did not indicate that these phenotypes co-occur more frequently than by chance, which suggests that they are distinct entities, inherited independently (Russell et al. 2002). Some epidemiology studies have further emphasized differences between MA and MO, with the former commonly having unilateral headaches and photophobia and the latter having headaches of greater duration and more nausea (Kallela et al. 1999).
Twin studies of migraine with and without aura indicate a genetic component of ∼40%–65% (Honkasalo et al. 1995; Larsson et al. 1995; Gervil et al. 1999a, 1999b; Ulrich et al. 1999a, 1999b). This is further supported by studies of familial clustering of migraine (Mochi et al. 1993; Russell et al. 1995a; Stewart et al. 1997).
The genetic component of the two major subtypes of migraine, MO and MA, was evaluated in a Danish twin study (Ulrich and Gervil 2000). The probandwise concordance rates were significantly higher for MZ twins than for DZ twins for both MO and MA, indicating a genetic component for susceptibility to migraine. Their data were also consistent with several studies indicating greater relative risks for MA than for MO (Russell and Olesen 1995; Stewart et al. 1997). The results of a Swedish twin study have suggested that the genetic component in migraine is more prominent for females than for males (Larsson et al. 1995).
A rare Mendelian subtype of MA is called “familial hemiplegic migraine” (FHM), which is characterized by unilateral motor weakness during the aura and by hemicranial headaches (HCCIHS 1988). Mutations at the FHM1 locus (MIM 141500), in the calcium channel gene CACNA1A (MIM 601011) on 19p13.1, have been shown to cause 30%–50% of the FHM cases that have been studied (Joutel et al. 1993; Ophoff et al. 1996; Ducros et al. 2001). Mutations have also been found in this gene that cause other phenotypes, such as episodic ataxia type 2 (EA2 [MIM 108500]) (Ophoff et al. 1996) and spinocerebellar ataxia type 6 (SCA6 [MIM 183086]) (Zhuchenko et al. 1997). Another FHM locus (FHM2 [MIM 602481]) has been mapped to chromosome 1q21-q23 (Ducros et al. 1997) and mutations identified in a gene encoding a subunit of a sodium/potassium pump (ATP1A2 [MIM 182340]) (De Fusco et al. 2003). Finally, an FHM3 locus (MIM 607516) has been identified on chromosome 1q31 (Gardner et al. 1997), which may be the same as the typical migraine locus MGR6 (Lea et al. 2002).
Linkage scans have been made in attempts to map migraine genes. Linkage scans with typical migraine (MO or MA) as the phenotype have reported loci on chromosome 1q31 (MGR6 [MIM 607516]) (Lea et al. 2002), 6p21.1-p12.2 (MGR3 [MIM 607498]) (Carlsson et al. 2002), and Xq (MGR2 [MIM 300125]) (Nyholt et al. 1998a, 2000). Linkage studies show that MA maps to 4q24 (MGR1 [MIM 157300]) (Wessman et al. 2002) and 19p13 (MGR5 [MIM 607508]) (Jones et al. 2001), although typical migraine has also been mapped to the 19p13 locus (Nyholt et al. 1998b; Terwindt et al. 2001). MO has been mapped to 14q21.2-q22.3 (MGR4 [MIM 607501]) (Soragna et al. 2003). Follow-up association analysis has been reported only for the locus on chromosome 19p13, with support for a role for the insulin receptor gene (INSR [MIM 147670]) (McCarthy et al. 2001).
We describe here the results of a genomewide scan for MO genes in Icelandic families and the mapping of a major gene to a locus on chromosome 4q21. The mapping of MO to the same locus, MGR1, as recently reported for MA, indicates that the same gene may have a role in both forms of migraine.
Materials and Methods
Patient Material
deCODE Genetics is conducting a large study of migraine in Iceland. Individuals suffering from headache have been recruited from three sources: (1) a list of patients provided by two neurologists (401 potential participants), (2) responses to an advertisement in the newsletter of the Icelandic Migraine Society (137 participants), and (3) responses to a brief screening questionnaire mailed to a random sample of 20,000 Icelanders, aged 18–50 years and living in the Reykjavik area (including 2,481 respondents who report recurrent headache attacks). From these sources, 2,611 patients with likely migraine and 1,865 of their close relatives have been recruited for a genetic study. All recruits were asked to answer a comprehensive migraine questionnaire (deCODE Genetics) before giving blood for the study. The questionnaire was patterned after the IHS criteria (HCCIHS 1988). Diagnoses of migraine through a questionnaire, as done in this study, have been shown to be highly effective (Kallela et al. 2001a). We have genotyped 1,943 patients with headache and 1,510 unaffected relatives, using 1,000 microsatellite markers. Of the genotyped headache-afflicted patients, 596 meet the IHS criteria for MO and did not report any aura (138 males and 458 females). There were an additional 702 patients, possibly having MO and reporting some aura symptoms, who were not included in this analysis, except as relatives with unknown affection status. Using our unique Icelandic genealogy database and clustering software (Gulcher and Stefánsson 1998; Gulcher et al. 2000), we identified 103 families with 289 MO-afflicted patients who have a relative with MO at or within 5 meioses (i.e., 5 meioses separate first cousins, once removed). This is our primary linkage cohort for the results presented in this study. This left 307 patients who were unrelated to other patients with MO within a distance of 5 meioses; thus, they were not included in the linkage analyses of the families with MO. Subsequently, in our investigation of the contribution of phenotypes to an identified locus, we attempted to increase the number of patients and families that have no reported aura by slightly relaxing the IHS criteria for MO, by dropping either pain criteria C or D but not both (appendix A). This increased the linkage cohort to 117 families with 351 affected patients. Thus, the two phenotype models are MO and a less strict version of MO, MO2. We also considered subsets of these models that consisted of only male affected members or only female affected members. The family sets, including the number of relative pairs at various meiotic distances, are described in table 1. This study was approved by the National Bioethics Committee and the Data Protection Commission of Iceland. Informed consent was obtained from all participants.
Table 1.
Summary of Linkage Families with Count of Affected Relative Pairs at Various Meiotic Distances[Note]
|
Meiotic Distance |
||||||
| Phenotype | No. of Families | No. of Those Affected | No. of Those Unaffected | 2 | 3 | ⩾4 |
| MO: | ||||||
| All patients | 103 | 289 | 518 | 67 | 36 | 150 |
| Females only | 64 | 167 | 356 | 36 | 20 | 79 |
| Males only | 14 | 31 | 70 | 6 | 3 | 8 |
| MO2: | ||||||
| All patients | 117 | 351 | 553 | 90 | 48 | 186 |
| Females only | 77 | 203 | 406 | 46 | 25 | 98 |
| Males only | 18 | 39 | 85 | 10 | 4 | 8 |
Note.— The family counts include only genotyped persons.
Microsatellite Markers and Genotyping
Genotypes were obtained for a marker set developed at deCODE Genetics that contains markers from the ABI Linkage marker (v.2) screening and intercalating sets and 500 custom-made markers. The set has 1,000 fluorescently labeled primers, with an average spacing of 3.6 cM. The set has been extensively tested for multiplex PCR results. PCR reactions were set up by Zymark SciClone (ALH-500) robots, and PCR amplifications were made on MJ Research machines (PTC-25). The reaction volume was 5 μl for each PCR reaction, and 20 ng of genomic DNA was amplified in the presence of 2 pmol of each primer, 0.25 U AmpliTaq Gold, 0.2 mM dNTPs, and 2.5 mM MgCl2 (buffer was supplied by Applera). Cycling conditions were: 95°C for 10 min, followed by 37 cycles of 94°C for 15 s, annealing for 30 s at 55°C, and 1-min extension at 72°C. The PCR products were supplemented with the internal size standard, and the pools were separated and detected on 3700 Sequencers using Genescan (v3.0) peak calling software (Applera). The genotypes, for a total of 866 markers from the genomewide set, were used for the linkage analysis, resulting in an average intermarker distance of 4.2 cM. Alleles were automatically called using DAC, an allele-calling program developed at deCODE Genetics (Fjalldal et al. 2001), and the program DecodeGT was used to fractionate called genotypes, according to quality, and to edit, when necessary (Pálsson et al. 1999).
Statistical Methods for Linkage Analysis
To evaluate linkage, we used multipoint, affected-only allele-sharing methods. The program Allegro (Gudbjartsson et al. 2000) was used for calculating nonparametric linkage and LOD scores, as well as for producing information measures on identity-by-descent (IBD) sharing. We employed the Spairs scoring function (Whittemore and Halpern 1994; Kruglyak et al. 1996) and the exponential allele-sharing model (Kong and Cox 1997) to generate the relevant 1-df test statistic. Family scores were combined to obtain an overall score, by use of a weighting scheme that is halfway on the logarithm scale between weighting the families equally (Kruglyak et al. 1996) and by weighting the affected pairs equally; with the resulting weight equal to the geometric mean of the two weights. This scheme gives weights similar to those proposed by Weeks and Lange (1988) as an extension of the scheme Hodge designed for sibships (Hodge 1984). To increase our confidence in the evidence for linkage to a particular region, additional microsatellite markers were genotyped to increase the information on IBD sharing to ⩾85% in that region. For the families in this study, to achieve this level of information required an average spacing of markers of ∼1 every 1.5 cM. The marker order and positions used in linkage analysis are from our high-resolution genetic map (Kong et al. 2002). All reported locations are in Kosambi centimorgans.
Results
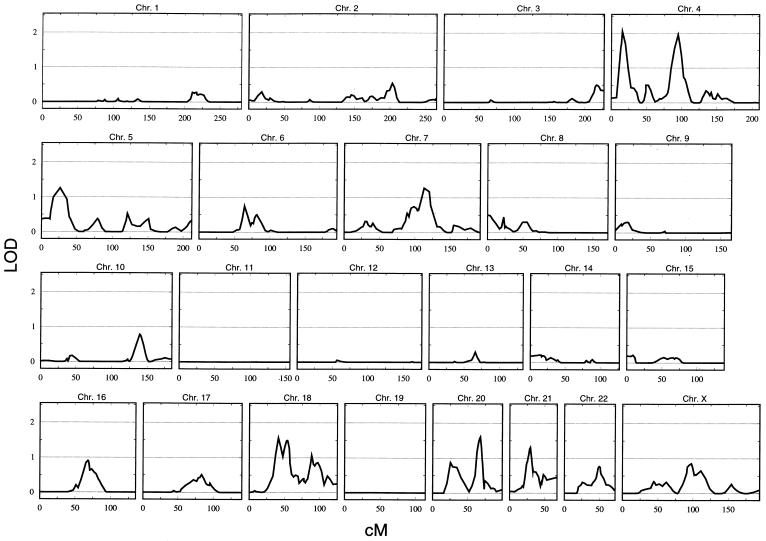
A genomewide scan was made on 103 families containing 289 genotyped patients with MO and 518 of their genotyped relatives (fig. 1). Five locations had a LOD score ⩾1.5. They are on chromosomes 4p (LOD=2.05 at D4S394), 4q (LOD=1.96 at D4S2361), 18p (LOD=1.57 at D18S453), 18q (LOD=1.50 at D18S877), and 20q (LOD=1.60 at D20S96).
Figure 1.
Genomewide linkage analysis of patients with MO. This analysis includes 103 families, with 289 affected and 518 relatives. The multipoint allele-sharing LOD score is shown on the vertical axis, and the distance (in Kosambi cM) from the chromosomal p terminus is on the horizontal axis.
Given that the prevalence of MO in women is three times that in men, we decided to investigate families restricted to either male patients or female patients. For males with MO, the LOD scores on chromosomes 4 and 18 dropped to <0.3, whereas the peak on chromosome 20 dropped to <1.0. No LOD score >1.5 was observed. For females with MO, however, whereas the 4p, 18p, 18q, and 20q peaks dropped to <1.5, the LOD score at chromosome 4q rose to 2.68 at D4S2361. Figure 2 shows a comparison of linkage scans on chromosome 4 in males and females with MO. A region on chromosome 21q also achieved a LOD score >1.5 for females with MO (LOD=1.74 between D21S1896 and D21S1442).
Figure 2.
Multipoint allele-sharing LOD scores for chromosome 4 of females with MO (solid line) and males with MO (dashed line). The multipoint allele-sharing LOD score is shown on the vertical axis, and the distance (in Kosambi cM) from the chromosomal p terminus is on the horizontal axis.
The 4q peak is interesting, since it overlaps with the MGR1 locus that was reported by a Finnish group as being linked to MA (Wessman et al. 2002). We decided to genotype 15 additional markers at this locus to increase the information on IBD sharing of alleles by affected relatives and to determine if that would support genetic sharing in the region. After this higher-resolution mapping, the MO phenotype gave a LOD score of 2.05 (P=.001) at D4S1534 (93.19 cM), thus confirming the MGR1 locus. The run for males with MO continued to show no evidence for linkage to this region, and the run for females with MO produced a slightly lower LOD score than for the framework mapping (LOD=2.33 at D4S1534). A summary of the fine-mapping results is displayed in table 2.
Table 2.
Summary of Fine-Mapping Results at the Chromosome 4q21 Locus
| Phenotype | Peak Marker | Position(cM) | LOD |
| MO: | |||
| All patients | D4S1534 | 93.19 | 2.05 |
| Females only | D4S1534 | 93.19 | 2.33 |
| Males onlya | … | … | … |
| MO2: | |||
| All patients | D4S1534 | 93.19 | 2.87 |
| Females only | D4S2409 | 94.23 | 4.08 |
| Males only | D4S2361 | 92.30 | .79 |
In the results for only males with MO, the LOD score is <.10 throughout the region.
We then expanded our study cohort by including patients who scored positive for all MO diagnostic criteria except either criteria C or D but not both (appendix A). For the 117 families containing 351 genotyped patients who were diagnosed according to this MO2 criterion, we observed a LOD score of 2.87, again at D4S1534. The males with MO2 showed some linkage, with a LOD score of 0.66, but the females with MO2 gave a very striking result (LOD=4.08; P=7.2×10-6; D4S2409; 94.23 cM). The region corresponding to a drop of 1 in LOD is flanked by markers D4S2627 (91.86 cM) and D4S423 (100.16 cM). Figure 3 shows the high-resolution mapping of chromosome 4 for males and females with MO2.
Figure 3.
Multipoint allele-sharing LOD scores for chromosome 4 of females with MO2 (solid line) and males with MO2 (dashed line), with a higher-resolution map. With an average intermarker distance of 1.5 cM in the linked region, a multipoint LOD score of 4.08 was detected at marker D4S2409 (94.23 cM). The multipoint allele-sharing LOD score is shown on the vertical axis, and the distance (in Kosambi cM) from the chromosomal p terminus is on the horizontal axis.
Discussion
Our linkage analysis places a gene for MO in Icelandic families on chromosome 4q21. Our data indicate that this MO locus overlaps with a known MA locus, MGR1, that is reported by a Finnish study (Wessman et al. 2002). This locus for MA, with peak marker D4S1647 (103.77 cM), was identified using a parametric two-point linkage model, assuming a dominant mode of inheritance. The Finnish group also reported its best multipoint parametric and nonparametric linkage for MA between markers D4S2409 and D4S2380, slightly centromeric to marker D4S1647. We map our peak marker for MO, D4S1534, to within 1.1 cM of D4S2409, which is near the peak of the reported MGR1 multipoint locus. This locus is most prominent for female patients with MO, especially when including headache patients who were diagnosed with slightly relaxed criteria for MO (dropping IHS criteria class C or D) (appendix A). We note that D4S2409 is also in the center of our most prominent peak for females with MO2. Since D4S2409 maps to the same physical location as cytogenetic band 4q21, we suggest that 4q21 may be a better localization for the MGR1 locus than 4q24.
The peak that we observe on chromosome 18q, at marker D18S877 in our strict MO linkage run (fig. 1), is also in the Finnish linkage data (Wessman et al. 2002). Their peak has the same top marker. However, this linkage decreases when the MO2 phenotype model is analyzed.
An MO locus has been mapped to chromosome 14q21.2-q22.3 in a large Italian pedigree that has a dominant inheritance pattern (Soragna et al. 2003). We have no evidence for linkage in this region. Also, no linkage >1 in LOD score is observed for typical migraine loci on chromosome 1q31 (MGR6) (Lea et al. 2002), 6p21.1-p12.2 (MGR3) (Carlsson et al. 2002), 19p13 (MGR5) (Nyholt et al. 1998b), or Xq (MGR2) (Nyholt et al. 1998a, 2000).
The MGR1 locus reported elsewhere is based on 50 Finnish families that have three or more first-degree relatives with MA, with a seemingly dominant mode of inheritance (Wessman et al. 2002). In contrast, the Icelandic cohort with MO is defined as “MO patients who do not report any symptoms that can be interpreted as aura.” Headache patients who report only one aura episode but otherwise five or more MO attacks are not included in our cohort with MO. We are currently evaluating the Icelandic MA material and intend to report our findings on it and other phenotype breakdowns as soon as they become available.
Our data show that the chromosome 4q21 locus is more significant when only females are analyzed. This may relate to the stronger genetic effect that has been reported for females (Larsson et al. 1995). Epidemiologic studies (Rasmussen and Olesen 1992; Russell et al. 1996; Launer et al. 1999) and population-based twin studies (Kallela et al. 1999; Ulrich et al. 1999a; Svensson et al. 2002) show that MO is more frequent in females than males. Our cohort with MO has, similarly, more affected females than males (458 females and 138 males). The preponderance of females with MO could relate to a stronger genetic component. We note, however, that the smaller number of male patients in our study decreases the power of detecting linkage for males, and the affect of a gene at this locus on susceptibility in males may bear further investigation.
On the basis of our data and the Finnish work, the MGR1 locus may be best characterized as “containing a gene predisposing to typical migraine that contributes to both MA and MO,” and the type may be decided by other gene(s) or environmental factors. However, further work is necessary to support this conclusion, since no linkage study has been published on MO in the Finnish population or on MA in the Icelandic populations.
Acknowledgments
We thank the participating patients and their families and Hjördís Jóhannesdóttir, Elísabet Grétarsdóttir, Halldóra Gröndal, and Hjördís Pálsdóttir, for the collection of samples.
Appendix A: Migraine without Aura: International Headache Society Criteria
Previously used terms: common migraine, hemicrania simplex.
Diagnostic criteria:
-
1.
At least five attacks fulfilling items B–D.
-
2.
Headache lasting 4–72 hours (untreated or unsuccessfully treated).
-
3.
Headache with at least two of the following characteristics:
-
A.
Unilateral location.
-
B.
Pulsating quality.
-
C.
Moderate or severe intensity (inhibits or prohibits daily activities).
-
D.
Aggravation by walking stairs or similar routine physical activity.
-
A.
-
4.
During headache, at least one of the following:
-
A.
Nausea and/or vomiting.
-
B.
Photophobia and phonophobia.
-
A.
-
5.
At least one of the following:
-
A.
History and physical and neurologic examinations do not suggest one of the disorders listed in groups 5–11 (organic disorders).
-
B.
History and/or physical and/or neurologic examinations suggest such disorder, but it is ruled out by appropriate investigations.
-
C.
Such disorder is present, but migraine attacks do not occur for the first time in close temporal relation to the disorder.
-
A.
Electronic-Database Information
URLs for data presented herein are as follows:
- deCODE Genetics, http://www.decode.com/migraine/questionnaire (for the English translation of the Icelandic, deCODE migraine questionnaire used in this study)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for MGR1, MGR2, MGR3, MGR4, MGR5, MGR6, FHM1, FHM2, FHM3, EA2, SCA6, CACNA1A, ATP1A2, and INSR)
References
- Carlsson A, Forsgren L, Nylander PO, Hellman U, Forsman-Semb K, Holmgren G, Holmberg D, Holmberg M (2002) Identification of a susceptibility locus for migraine with and without aura on 6p12.2-p21.1. Neurology 59:1804–1807 [DOI] [PubMed] [Google Scholar]
- De Fusco M, Marconi R, Silvestri L, Atorino L, Rampoldi L, Morgante L, Ballabio A, Aridon P, Casari G (2003) Haploinsufficiency of ATP1A2 encoding the Na+/K+ pump α2 subunit associated with familial hemiplegic migraine type 2. Nat Genet 33:192–196 [DOI] [PubMed] [Google Scholar]
- Ducros A, Denier C, Joutel A, Cecillon M, Lescoat C, Vahedi K, Darcel F, Vicaut E, Bousser MG, Tournier-Lasserve E (2001) The clinical spectrum of familial hemiplegic migraine associated with mutations in a neuronal calcium channel. N Engl J Med 345:17–24 [DOI] [PubMed] [Google Scholar]
- Ducros A, Joutel A, Vahedi K, Cecillon M, Ferreira A, Bernard E, Verier A, Echenne B, Lopez de Munain A, Bousser MG, Tournier-Lasserve E (1997) Mapping of a second locus for familial hemiplegic migraine to 1q21-q23 and evidence of further heterogeneity. Ann Neurol 42:885–890 [DOI] [PubMed] [Google Scholar]
- Fjalldal JG, Benediktsson K, Sigurdsson J, Ellingssen LM (2001) Automated genotyping: combining neural networks and decision trees to perform robust allele calling. Proc Int Joint Conf Neural Networks A1–A6 [Google Scholar]
- Gardner K, Barmada MM, Ptacek LJ, Hoffman EP (1997) A new locus for hemiplegic migraine maps to chromosome 1q31. Neurology 49:1231–1238 [DOI] [PubMed] [Google Scholar]
- Gervil M, Ulrich V, Kaprio J, Olesen J, Russell MB (1999a) The relative role of genetic and environmental factors in migraine without aura. Neurology 53:995–999 [DOI] [PubMed] [Google Scholar]
- Gervil M, Ulrich V, Kyvik KO, Olesen J, Russell MB (1999b) Migraine without aura: a population-based twin study. Ann Neurol 46:606–611 [DOI] [PubMed] [Google Scholar]
- Gudbjartsson DF, Jónasson K, Frigge ML, Kong A (2000) Allegro, a new computer program for multipoint linkage analysis. Nat Genet 25:12–13 [DOI] [PubMed] [Google Scholar]
- Gulcher J, Stefánsson K (1998) Population genomics: laying the groundwork for genetic disease modeling and targeting. Clin Chem Lab Med 36:523–527 [DOI] [PubMed] [Google Scholar]
- Gulcher JR, Kristjánsson K, Gudbjartsson H, Stefánsson K (2000) Protection of privacy by third-party encryption in genetic research in Iceland. Eur J Hum Genet 8:739–742 [DOI] [PubMed] [Google Scholar]
- Headache Classification Committee of the International Headache Society (1988) Classification and diagnostic criteria for headache disorders, cranial neuralgias and facial pain. Cephalalgia Suppl 7:1–96 [PubMed] [Google Scholar]
- Hodge SE (1984) The information contained in multiple sibling pairs. Genet Epidemiol 1:109–122 [DOI] [PubMed] [Google Scholar]
- Honkasalo ML, Kaprio J, Winter T, Heikkila K, Sillanpaa M, Koskenvuo M (1995) Migraine and concomitant symptoms among 8167 adult twin pairs. Headache 35:70–78 [DOI] [PubMed] [Google Scholar]
- Jones KW, Ehm MG, Pericak-Vance MA, Haines JL, Boyd PR, Peroutka SJ (2001) Migraine with aura susceptibility locus on chromosome 19p13 is distinct from the familial hemiplegic migraine locus. Genomics 78:150–154 [DOI] [PubMed] [Google Scholar]
- Joutel A, Bousser MG, Biousse V, Labauge P, Chabriat H, Nibbio A, Maciazek J, Meyer B, Bach MA, Weissenbach J, Lathorp GM, Tournier-Lasserve E (1993) A gene for familial hemiplegic migraine maps to chromosome 19. Nat Genet 5:40–45 [DOI] [PubMed] [Google Scholar]
- Kallela M, Wessman M, Farkkila M (2001a) Validation of a migraine-specific questionnaire for use in family studies. Eur J Neurol 8:61–66 [DOI] [PubMed] [Google Scholar]
- Kallela M, Wessman M, Farkkila M, Palotie A, Koskenvuo M, Honkasalo ML, Kaprio J (1999) Clinical characteristics of migraine in a population-based twin sample: similarities and differences between migraine with and without aura. Cephalalgia 19:151–158 [DOI] [PubMed] [Google Scholar]
- Kallela M, Wessman M, Havanka H, Palotie A, Farkkila M (2001b) Familial migraine with and without aura: clinical characteristics and co-occurrence. Eur J Neurol 8:441–449 [DOI] [PubMed] [Google Scholar]
- Kong A, Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jónsdóttir GM, Gudjónsson SA, Richardsson B, Sigurdardóttir S, Barnard J, Hallbeck B, Másson G, Shlien A, Pálsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefánsson K (2002) A high-resolution recombination map of the human genome. Nat Genet 31:241–247 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Larsson B, Bille B, Pedersen NL (1995) Genetic influence in headaches: a Swedish twin study. Headache 35:513–519 [DOI] [PubMed] [Google Scholar]
- Launer LJ, Terwindt GM, Ferrari MD (1999) The prevalence and characteristics of migraine in a population-based cohort: the GEM study. Neurology 53:537–542 [DOI] [PubMed] [Google Scholar]
- Lea RA, Shepherd AG, Curtain RP, Nyholt DR, Quinlan S, Brimage PJ, Griffiths LR (2002) A typical migraine susceptibility region localizes to chromosome 1q31. Neurogenetics 4:17–22 [DOI] [PubMed] [Google Scholar]
- McCarthy LC, Hosford DA, Riley JH, Bird MI, White NJ, Hewett DR, Peroutka SJ, et al (2001) Single-nucleotide polymorphism alleles in the insulin receptor gene are associated with typical migraine. Genomics 78:135–149 [DOI] [PubMed] [Google Scholar]
- Mochi M, Sangiorgi S, Cortelli P, Carelli V, Scapoli C, Crisci M, Monari L, Pierangeli G, Montagna P (1993) Testing models for genetic determination in migraine. Cephalalgia 13:389–394 [DOI] [PubMed] [Google Scholar]
- Nyholt DR, Curtain RP, Griffiths LR (2000) Familial typical migraine: significant linkage and localization of a gene to Xq24-28. Hum Genet 107:18–23 [DOI] [PubMed] [Google Scholar]
- Nyholt DR, Dawkins JL, Brimage PJ, Goadsby PJ, Nicholson GA, Griffiths LR (1998a) Evidence for an X-linked genetic component in familial typical migraine. Hum Mol Genet 7:459–463 [DOI] [PubMed] [Google Scholar]
- Nyholt DR, Lea RA, Goadsby PJ, Brimage PJ, Griffiths LR (1998b) Familial typical migraine: linkage to chromosome 19p13 and evidence for genetic heterogeneity. Neurology 50:1428–1432 [DOI] [PubMed] [Google Scholar]
- Ophoff RA, Terwindt GM, Vergouwe MN, van Eijk R, Oefner PJ, Hoffman SM, Lamerdin JE, Mohrenweiser HW, Bulman DE, Ferrari M, Haan J, Lindhout D, van Ommen GJ, Hofker MH, Ferrari MD, Frants RR (1996) Familial hemiplegic migraine and episodic ataxia type-2 are caused by mutations in the Ca(2+) channel gene CACNL1A4. Cell 87:543–552 [DOI] [PubMed] [Google Scholar]
- Pálsson B, Pálsson F, Perlin M, Gudbjartsson H, Stefánsson K, Gulcher J (1999) Using quality measures to facilitate allele calling in high-throughput genotyping. Genome Res 9:1002–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen BK, Olesen J (1992) Migraine with aura and migraine without aura: an epidemiological study. Cephalalgia 12:221-228 [DOI] [PubMed] [Google Scholar]
- Russell MB, Iselius L, Olesen J (1995a) Inheritance of migraine investigated by complex segregation analysis. Hum Genet 96:726–730 [DOI] [PubMed] [Google Scholar]
- ——— (1996) Migraine without aura and migraine with aura are inherited disorders. Cephalalgia 16:305–309 [DOI] [PubMed] [Google Scholar]
- Russell MB, Olesen J (1995) Increased familial risk and evidence of genetic factor in migraine. Brit Med J 311:541–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell MB, Rasmussen BK, Thorvaldsen P, Olesen J (1995b) Prevalence and sex-ratio of the subtypes of migraine. Int J Epidemiol 24:612–618 [DOI] [PubMed] [Google Scholar]
- Russell MB, Ulrich V, Gervil M, Olesen J (2002) Migraine without aura and migraine with aura are distinct disorders: a population-based twin survey. Headache 42:332–336 [DOI] [PubMed] [Google Scholar]
- Soragna D, Vettori A, Carraro G, Marchioni E, Vazza G, Bellini S, Tupler R, Savoldi F, Mostacciuolo ML (2003) A locus for migraine without aura maps on chromosome 14q21.2-q22.3. Am J Hum Genet 72:161–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart WF, Lipton RB, Celentano DD, Reed ML (1992) Prevalence of migraine headache in the United States: relation to age, income, race, and other sociodemographic factors. JAMA 267:64–69 [PubMed] [Google Scholar]
- Stewart WF, Staffa J, Lipton RB, Ottman R (1997) Familial risk of migraine: a population-based study. Ann Neurol 41:166–172 [DOI] [PubMed] [Google Scholar]
- Svensson DA, Ekbom K, Larsson B, Waldenlind E (2002) Lifetime prevalence and characteristics of recurrent primary headaches in a population-based sample of Swedish twins. Headache 42:754–765 [DOI] [PubMed] [Google Scholar]
- Terwindt GM, Ophoff RA, van Eijk R, Vergouwe MN, Haan J, Frants RR, Sandkuijl LA, Ferrari MD (2001) Involvement of the CACNA1A gene containing region on 19p13 in migraine with and without aura. Neurology 56:1028–1032 [DOI] [PubMed] [Google Scholar]
- Ulrich V, Gervil M (2000) Twin studies of migraine with and without aura. In: Sydor AM (ed) Frontiers in headache research. Vol 8. Genetics of headache disorders. Lippincott Williams & Wilkins, Philadelphia, pp 27–31 [Google Scholar]
- Ulrich V, Gervil M, Kirsten F, Olesen J, Russell MB (1999a) The prevalence and characteristics of migraine in twins from the general population. Headache 39:173–180 [DOI] [PubMed] [Google Scholar]
- Ulrich V, Gervil M, Kyvik KO, Olesen J, Russell MB (1999b) Evidence of a genetic factor in migraine with aura: a population-based Danish twin study. Ann Neurol 45:242–246 [DOI] [PubMed] [Google Scholar]
- Weeks DE, Lange K (1988) The affected-pedigree-member method of linkage analysis. Am J Hum Genet 42:315–326 [PMC free article] [PubMed] [Google Scholar]
- Wessman M, Kallela M, Kaunisto MA, Marttila P, Sobel E, Hartiala J, Oswell G, Leal SM, Papp JC, Hämäläinen E, Broas P, Joslyn G, Hovatta I, Hiekkalinna T, Kaprio J, Ott J, Cantor RM, Zwart J-A, Ilmavirta M, Havanka H, Färkkilä M, Peltonen L, Palotie A (2002) A susceptibility locus for migraine with aura, on chromosome 4q24. Am J Hum Genet 70:652–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittemore AS, Halpern J (1994) A class of tests for linkage using affected pedigree members. Biometrics 50:118–127 [PubMed] [Google Scholar]
- Zhuchenko O, Bailey J, Bonnen P, Ashizawa T, Stockton DW, Amos C, Dobyns WB, Subramony SH, Zoghbi HY, Lee CC (1997) Autosomal dominant cerebellar ataxia (SCA6) associated with small polyglutamine expansions in the α 1A-voltage-dependent calcium channel. Nat Genet 15:62–69 [DOI] [PubMed] [Google Scholar]