Abstract
The successful dispersal of transposons depends on the critical balance between the fitness of the host and the ability of the transposon to insert into the host genome. One method transposons may use to avoid the disruption of coding sequences is to target integration into safe havens. We explored the interaction between the long terminal repeat retrotransposon Tf1 and the genome of the yeast Schizosaccharomyces pombe. Using techniques that were specifically designed to detect integration of Tf1 throughout the genome and to avoid bias in this detection, we generated 51 insertion events. Although 60.2% of the genome of S. pombe is coding sequence, all but one of the insertions occurred in intergenic regions. We also found that Tf1 was significantly more likely to insert into intergenic regions that included polymerase II promoters than into regions between convergent gene pairs. Interestingly, 8 of the 51 insertion sites were isolated multiple times from genetically independent cultures. This result suggests that specific sites in intergenic regions are targeted by Tf1. Perhaps the most surprising observation was that per kilobase of nonrepetitive sequence, Tf1 was significantly more likely to insert into chromosome 3 than into one of the other two chromosomes. This preference was found not to be due to differences in the distribution or composition of intergenic sequences within the three chromosomes.
Long terminal repeat (LTR) retrotransposons are retrovirus-like elements that encode Gag, protease (PR), reverse transcriptase (RT), and integrase (IN) proteins. Because of their close relationship to retroviruses, the propagation of LTR retrotransposons is dependent on a life cycle that is similar to that of retroviruses (9). Insertion of the cDNA into the host DNA is a critical event required for the propagation of both LTR retrotransposons and retroviruses. The similarity between retroviruses and LTR retrotransposons extends to the mechanism of integration. IN mediates insertion by creating staggered nicks in the genomic DNA of the host and attaching the 3" ends of the cDNA to the 5" ends of the target site (10). The result of this integration reaction is the insertion of full-length cDNA that is flanked by short target site duplications.
A wealth of information has been generated that documents the biochemical behavior of the INs of retroviruses and LTR retrotransposons under in vitro conditions (1). However, much less is known about the integration sites that LTR retroelements choose when influenced by the full spectrum of host factors. Ultimately, the impact of host factors on integration may best be observed in vivo. The selection of insertion sites in the genome of the host is of the highest importance. The disruption of coding sequences in the host genome can reduce the fitness of the host and, as a result, lower the ability of the transposon to propagate. It is therefore critical that a balance be struck between the fitness of the host and the ability of the transposon to integrate into the host genome.
The integration of the Ty elements into the genome of Saccharomyces cerevisiae serves as a model for how LTR elements populate a host genome without disrupting coding sequences. Perhaps the most specific targeting mechanism is that of Ty3. This element inserts one to four nucleotides upstream of polymerase III (pol III) promoter initiation sites, such as those responsible for tRNA transcription (11). This strategy allows Ty3 to amplify its copy number without risk of damaging coding sequences. In recent work, this mechanism was attributed to interactions with transcription factor IIIB (22, 41). In similar types of analyses, Ty1 was found to integrate within a window of 75 to 700 nucleotides upstream of the start sites of pol III promoters (14, 19). Here, too, this strategy avoids the disruption of coding sequences, since this integration window is gene poor (E. C. Bolton and J. D. Boeke, unpublished data). It is interesting that although Ty1 and Ty3 are unrelated transposons, they have converged on the same type of target selection. On the other hand, both Ty1 and Ty5 are members of the copia family, yet they use very different targeting mechanisms. Ty5 specifically inserts into regions of silent chromatin, such as telomeres and the silent sequences of the mating type cassette (43, 44). This targeting behavior is due to the ability of the IN to recognize the SIR complex of silent chromatin (42). The insertion of Ty5 into regions of silent chromatin is yet another effective method for avoiding the disruption of important host genes.
Tf1 is an active LTR retrotransposon found in the genome of the fission yeast Schizosaccharomyces pombe. Tf1 contains a single open reading frame encoding 1,331 amino acids that constitute the capsid, PR, RT, and IN proteins (3, 28, 29). These proteins are functional and are required for efficient transposition in vivo (2, 25-27). Analysis of the conserved residues in RT has demonstrated that Tf1 belongs to the gypsy group of LTR retroelements, as does Ty3 of S. cerevisiae (15, 39, 40). The similarity of Tf1 to Ty3 suggests the interesting possibility that Tf1 may also integrate into specific types of target sites.
The availability of a transposition system in S. pombe provides the unique opportunity to compare the interactions between LTR retrotransposons and the host genome in two yeasts that diverged 109 years ago (38). This is a particularly important comparison, because much of what is known about the selection of target sites by LTR retroelements is derived from studies of S. cerevisiae. An understanding of target selection in S. pombe will reveal whether the types of target selection in S. cerevisiae are general. However, not much is known about the target site preferences of Tf1. What is known is the result of an analysis of 27 insertion events that were identified by screening for cells of S. pombe that become resistant to G418 due to the insertion of Tf1 tagged with neo (Tf1-neo) (6). Behrens et al. (6) reported an interesting pattern of integration into intergenic sequences near the 5" ends of genes. Although this is an intriguing observation, the technique used to isolate the insertion events did not guard against the possibility that transposition might specifically target regions of the genome that would block the expression of neo. This situation would bias the detection of target sites. In fact, what is known about the insertion preferences of LTR retrotransposons is the result of studies that rely on identification within the host genome of the insertion of transposons tagged with genetic markers. Should transposition occur in a class of target sites that block the transcription of the marker gene, these sites would go undetected.
We report a genome-wide study of Tf1 transposition in vivo that was designed to avoid any biases due to the transcription environment of the target sites. Our results confirmed the preference of Tf1 integration for intergenic sequences that was reported by Behrens et al. (6). Interestingly, 8 of the 51 insertion sites were isolated multiple times from genetically independent cultures, suggesting that specific sites in intergenic regions are targeted by Tf1. Perhaps the most significant result was that the analysis of our data and the reexamination of those reported by Behrens et al. (6) led to the surprising observation that Tf1 integration has a significant preference for integration into chromosome 3.
MATERIALS AND METHODS
Media.
The S. pombe minimal liquid and plate media were composed of Edinburgh minimal medium (EMM) (32). Selective plates contained EMM and 2 g of dropout mix (35), which included all the amino acids except leucine and adenine. A final concentration of 10 μM vitamin B1 (thiamine) was added to EMM to repress the nmt1 promoter, as needed. 5-Fluoroorotic acid (5-FOA) was used at 1 mg/ml as a selective agent in EMM, along with a final concentration of 50 μg of uracil and 250 μg of leucine per ml (8). The rich medium (YES) contained 5 g of yeast extract (Difco), 30 g of glucose, and 2 g of complete dropout mix per liter. YES-5-FOA-G418 plates were made with YES containing 5-FOA and G418 at final concentrations of 1 g/liter and 500 μg/ml, respectively. All yeast strains were incubated at 32°C unless indicated otherwise. The medium selective for bacteria was Luria-Bertani medium supplemented with kanamycin at 60 μg/ml and ampicillin at 100 μg/ml. Nonselective SOB-minus-magnesium medium was composed of 2% Bacto-Tryptone, 0.5% Bacto-Yeast Extract, 10 mM NaCl, and 2.5 mM KCl (17). All bacterial strains were incubated at 37°C.
Construction of strains and plasmids.
The yeast and bacterial strains and plasmids used in this study are described in Table 1. The oligonucleotides used are shown in Table 2. Yeast strain YHL6488, derived from strainYHL5661, is a diploid strain carrying the assay plasmid pTS1559-9. Plasmid pHL411, containing Tf1 under the control of an nmt1 promoter, was constructed as described previously (28) and was the backbone of all assay plasmids. PCR primers HL423 and HL424 produced an 850-bp product that contained the bacterial p15A origin of replication (ori) amplified from pACYC184 (36). The 850-bp product was digested with BamHI/BglII and ligated into a unique BglII site of pHL411 in a noncoding region of Tf1 in both orientations, generating pHL1555-7 and pHL1555-4. pHL1558-14 and pHL1558-10 were generated by insertion of a 1-kb neo-containing BamHI fragment upstream of the ori gene into the BglII site of pHL1555-4 in the direct and reverse orientations, respectively. pHL1559-14 and pHL1559-9 were generated by insertion of a 1-kb neo-containing BamHI fragment downstream of the ori gene into the BglII site of pHL1555-7 in the direct and reverse orientations, respectively.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype | Parent/plasmid | Description | Reference or source |
|---|---|---|---|---|
| Strains | ||||
| Yeast | ||||
| YHL1282 | h−ura4-294 leu1-32 | YHL912/pHL449-1 | Wild-type Tf1-neoAI | 25 |
| YHL1554 | h−ura4-294 leu1-32 | YHL912/pHL476-3 | Tf1-neoAI with frameshift in beginning of IN | 25 |
| YHL1836 | h−ura4-294 leu1-32 | YHL912/pHL490-80 | Tf1-neoAI with frameshift in PR | 26 |
| YHL5661 | Diploid ura4-294 ade6-m210/ade6-m216 leu1-32/leu1-32::nmt1-lacZ-leu1 | Homozygosed mating type locus | This study | |
| YHL6488 | See YHL5661 | YHL5661/pHL1559-9 | Homozygosed mating type locus | This study |
| Bacterial, DH10B | ||||
| Plasmids | ||||
| pHL411-62 | Tf1 fused to nmt1 promoter | 25 | ||
| pHL414-2 | Tf1-neo fused to nmt1 promoter | 25 | ||
| pHL765 | neo in pBSII at BamHI site | This study | ||
| pHL1559-9 | Tf1-ori/neo fused to nmt1 promoter | This study | ||
| pACYC184 | Contains p15A ori | 36 |
TABLE 2.
Oligonucleotides
| Oligonucleotide | Sequence (5" to 3") | Use |
|---|---|---|
| HL423 | AACCATCAGTGGATCCGAAGTGCTTCATGTGGCAGG | 5" Oligonucleotide for PCR of bacterial ori from pACYC184 |
| HL424 | AACCATCAGTAGATCTGGCTGACTTCAGGTGCTACATT | 3" Oligonucleotide for PCR of bacterial ori from pACYC184 |
| HL273 | ATATTTTTATCTTGTGCAATG | DNA sequencing primer that hybridizes to 3" neo region to generate sequence downstream from 3" LTR |
| JB54.3 | ATCACAAGAGTTCAGTTA | DNA sequencing primer that hybridizes to 3" neo region to generate sequence downstream from 3" LTR |
| HL781 | AAACGGATCGATTAATGTCGTACAA | 5" Oligonucleotide for PCR of empty target site in parent strain for clone pTS117 |
| HL782 | GTACGATTAAACGGCGAAGATATACT | 3" Oligonucleotide for PCR of empty target site in parent strain for clone pTS117 |
| HL783 | TCCTACATGCTCAAGGTATCAAT | 5" Oligonucleotide for PCR of empty target site in parent strain for clone pTS118 |
| HL784 | CCTAATCACAGGCATTCATCTGCT | 3" Oligonucleotide for PCR of empty target site in parent strain for clone pTS118 |
| HL785 | CCTTGTGGGACCTAATGGCACATTC | 5" Oligonucleotide for PCR of empty target site in parent strain for clone pTS512 |
| HL786 | AACAGTGCATTATGTCAGATAATGAG | 3" Oligonucleotide for PCR of empty target site in parent strain for clone pTS512 |
| HL787 | TCCCGTTGAATATGGCTCATAACAC | For sequencing flanks from 3" end of neo region |
| HL788 | ATGGACGCTATTTTCTTTAATTTCCAG | For sequencing flanks from gag region |
Transposition assay.
All assay plasmids with various orientations and orders of ori and neo were tested for transposition and G418 resistance in S. pombe. S. pombe strains containing pTS1558-10, pTS1558-14, pTS1559-9, and pTS1559-14 were first grown as patches on EMM plates lacking uracil in the presence of vitamin B1 to repress the nmt1 promoter and propagate the assay plasmids. Patches were replica printed onto EMM plates lacking uracil in the absence of vitamin B1 to induce the nmt1 promoter. After 4 days, the cells were replica printed onto 5-FOA-containing EMM plates to select against cells retaining the assay plasmids which possess a URA3 marker gene (8). Cells were subsequently replica printed onto YES-5-FOA-G418 plates to detect copies of Tf1-neo transposed into the genome. The presence of the neo gene allowed S. pombe to grow in G418.
Isolation of DNA with insertion sites.
Large patches of cells (YHL6488) were grown for 4 days on agar containing EMM and dropout mix but lacking vitamin B1. These patches were then replica printed onto plates containing EMM and 5-FOA. After 2 days of growth on EMM-5-FOA plates, the cells were scraped off and inoculated into separate 1-liter cultures of 5-FOA-EMM containing uracil and leucine but lacking vitamin B1. These cultures were allowed to reach an optical density at 600 nm of 2.0. Cells were collected at 4,000 rpm for 10 min and resuspended in 0.1 M NaCl. After pelleting, cells were resuspended in 20 ml of solution I (1 M sorbitol, 50 mM Na2PO4 [pH 7.5]). Beta-mercaptoethanol (14 mM; Sigma) and 3 mg of Zymolyase (100T; ICN) per g of cells were added and incubated at 30°C for 2 h. Spheroplasts were collected for 10 min at 3,000 rpm and gently resuspended in 20 ml of solution II (50 mM Tris-HCl [pH 8.0], 50 mM EDTA [pH 8.0], 1% Sarkosyl). An RNase A solution (10 mg/ml; 20 μl) was added and incubated for 1 h at 37°C. A proteinase K solution (10 mg/ml; 500 μl) was added and incubated for 1 h at 65°C. Extensive extractions (two chloroform-isoamyl alcohol [24:1], one phenol-chloroform-isoamyl alcohol, and one chloroform-isoamyl alcohol) were gently performed. Genomic DNA was precipitated by using 2× ethanol and was resuspended in 1 mM EDTA-10 mM Tris-HCl buffer.
Gel fractionation of genomic DNA with transposon insertions.
The genomic DNA (10 μg) was digested with either BamHI/BglII, SpeI/NheI, or BglII and gel fractionated. Digested DNA was allowed to migrate through a 0.5% low-melting-temperature agarose gel in Tris-acetate-EDTA at 4°C for 16 h at 22 V. Fragments within the 7- to 23-kb size range were excised and purified by using beta-agarase (New England Biolabs). Beta-agarase-digested fragments were allowed to migrate through a second 0.5% low-melting-temperature gel, and fragments within the 7- to 23-kb size range were excised, digested with beta-agarase, and extracted by using phenol-chloroform-isoamyl alcohol (25:24:1).
Recovery and DNA sequencing of Tf1 targets.
The restriction-digested and gel-purified DNA (50 ng) was allowed to self-ligate for 16 h at 12°C and was transformed into competent DH10B bacterial cells (17) by electroporation with a BRL electroporation apparatus. Cells were electroporated by using the parameters 1.75 V, 25 μF, and 100 Ω. Transformed cells were selected for kanamycin resistance. Because of the presence of endogenous Tf2 elements and the probability of recombination events between Tf1 and Tf2, kanamycin-resistant colonies were screened by bacterial colony hybridization with an EcoRI fragment that was specific for the Gag sequence of Tf1 (29). Clones were analyzed by digestion with SnaBI and EcoRI to identify intact Tf1 elements and any deletions, rearrangements, or recombination events. Primers JB54.3 (anneals just 3" to the 5" LTR) and HL273 (anneals just 5" to the 3" LTR) were used to sequence the genomic DNA flanking Tf1-ori/neo. The flanking sequence was then used to search the Sanger Centre S. pombe sequence database to identify the positions of the insertions. The sequence from the database was compared to that of the isolated DNA to determine whether the introduction of Tf1-ori/neo was due to true integration events or recombination into preexisting transposon sequences. Flanking sequence information was also used to design primers for PCR amplification of the empty target site in parental strain YHL5661.
PCR.
To avoid problems related to inadvertent mutations that could arise during PCR, assay plasmids were constructed in duplicate from independent PCRs, and the properties of each plasmid were determined in parallel. The high-fidelity enzyme Pfu DNA polymerase (Stratagene) was used for all cloning PCRs. Taq DNA polymerase (Perkin-Elmer Cetus) was used for all other PCRs. The Pfu cycling conditions were 95°C for 5 min with polymerase added; 95°C for 1 min, 54°C for 2 min, and 72°C for 2 min (30 cycles); and an extension cycle at 72°C for 10 min. PCR products were analyzed on a 1% agarose gel in Tris-borate-EDTA.
RESULTS
Tf1 was modified to allow the identification of integration sites throughout the genome of S. pombe.
To examine the target site preferences of Tf1 on a genome-wide basis, we modified the version of the transposon that is typically used to measure Tf1 activity (4, 13, 28). Methods used to detect Tf1 transposition rely on the genetic selection of phenotypes associated with genes inserted into an active copy of the transposon. We developed a method for identifying insertions into target sites that did not rely on the expression of genes included in the transposon.
The existing assay for Tf1 transposition is based on resistance to G418 that is due to the integration of Tf1-neo. Transposition is induced by the overexpression of Tf1 mRNA from a heterologous promoter, nmt1, located on a high-copy-number plasmid (4, 13). Transposition events were generated in this study by using the same system of overexpressing Tf1 mRNA. However, G418 was not used to select for strains of S. pombe that contained transposed copies of Tf1-neo. Instead, bacterial p15A ori was inserted into Tf1-neo adjacent to neo as shown in Fig. 1A. Insertion sites were identified from pools of genomic DNA extracted from cultures of S. pombe that were induced for transposition. To isolate the sites of insertion, the pools of DNA were digested with restriction enzymes and ligated into circles, and the DNA was transformed into bacteria. Colonies that were resistant to kanamycin contained transposed copies of Tf1-ori/neo flanked by the portions of the S. pombe genome that were disrupted by the insertion.
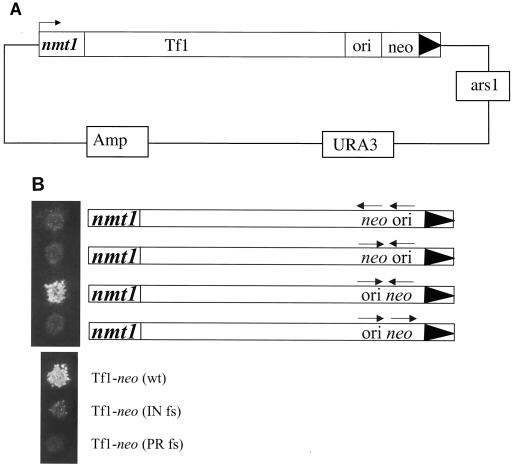
FIG. 1.
Modified version of Tf1 used to detect transposition throughout the genome of S. pombe. (A) An active copy of the transposon was tagged with neo and bacterial plasmid p15A ori. Tf1 was fused to an inducible S. pombe promoter (nmt1) located on a high-copy-number vector. (B) (Top) Growth on agar containing G418 in a standard transposition assay was used to test the ability of the modified elements to transpose. (Bottom) The controls included in this assay were strains containing wild-type (wt) Tf1-neo, Tf1-neo with a frameshift (fs) in IN, and Tf1-neo with a frameshift in PR.
One initial concern with this approach was that the insertion of bacterial p15A ori would reduce the transposition activity of Tf1-neo, possibly by inhibiting reverse transcription. To test this notion, several plasmids containing Tf1 with various orientations of ori and neo were constructed and tested for transposition by using the conventional assay described above. Growth on plates with G418 represents transposition activity and, as shown in Fig. 1B, it indicated that the orientation of the neo gene with respect to the 3" LTR was critical. Transposition was not detected with the Tf1 plasmid in which the neo gene was transcribed in the same orientation as the Tf1 element. The plasmid with neo transcribed in the orientation opposite that of the element and downstream from bacterial p15A ori was able to provide robust resistance to G418. The importance of this configuration may be due to transcription enhancers in the 3" LTR that may contribute to neo expression.
Isolation of target sites for Tf1-ori/neo in the genome of S. pombe.
To avoid a bias for insertion into nonessential genes of S. pombe, all transposition events were carried out with YHL6488, a strain of S. pombe that was a stable diploid. Another important feature of this strain was that it contained no endogenous copies of Tf1. The strategy used to generate a collection of target sites relied on screening pools of DNA from cells induced for the transposition of Tf1-ori/neo. The transcription of Tf1-ori/neo was induced in five independent patches of S. pombe. The patches were replica printed onto agar containing 5-FOA, and cells from these plates were grown in liquid medium for two to four divisions. The cells were subsequently extracted to isolate genomic DNA. The pools of DNA were digested with either BamHI/BglII, SpeI/NheI, or BglII alone. Different sets of restriction enzymes were chosen to avoid favoring the isolation of target sites that were fortuitously located near a specific restriction site. The restricted DNAs were subjected to two rounds of agarose gel electrophoresis, and all fragments within a range of 7 to 30 kb were excised and circularized with T4 DNA ligase. The batches of DNA were separately transformed into DH10B, a strain of Escherichia coli that was genetically altered to take up large fragments of DNA. The transformants were selected on medium containing kanamycin to enrich for circularized copies of Tf1-ori/neo containing target sequences. Kanamycin-resistant transformants were screened by colony hybridization by using a probe specific for Tf1 Gag. The initial step in the characterization of each transformant identified by colony hybridization was to determine the restriction patterns produced by SnaBI and EcoRI digestion. The appearance of 3.0- and 3.2-kb SnaBI fragments indicated the integration of an intact copy of Tf1.
The sequences flanking each end of Tf1-ori/neo in the plasmids isolated from the bacteria were determined, and these data were used to identify the positions of the insertions within the genome of S. pombe. The sequences flanking the insertions were also used to evaluate whether the Tf1-ori/neo elements were introduced into the DNA of S. pombe as the result of true transposition events. The sequences flanking Tf1-ori/neo in the plasmids were compared to their counterpart sequences from the Sanger Centre genome database. In all, we found 51 plasmids that resulted from the simple insertion of Tf1-ori/neo and the duplication of five nucleotides of the target site. The five nucleotide duplications are the result of IN-mediated cleavages and demonstrated that the introduction of Tf1-ori /neo into the S. pombe sequence was the result of true transposition events (27).
Integration sites for Tf1-ori/neo.
The 51 insertions in Table 3 are listed with their coordinates relative to those of cosmids sequenced for the S. pombe genome project. The coordinates were translated into the chromosome coordinates assigned by the Sanger Centre for the “publication-freeze” version of the genome data. Figure 2 is a plot of the positions of the insertions throughout the genome. Each of the three chromosomes received substantial numbers of insertions, and the positions of the target sites were distributed throughout their lengths. The one exception is that no insertions occurred in the 1.2 Mb of ribosomal DNA (rDNA) repeats located on either end of chromosome 3 (not shown in Fig. 2).
TABLE 3.
Insertions
| Chromosome | Insertion | Duplicate insertion | Cosmid | Coordinate | Target site duplication | Orientation of flanking genes |
|---|---|---|---|---|---|---|
| 1 | pTS178 | No | SPAC24B11 | 31,440 | CTTAT | Divergent |
| pTS169 | No | SPAC1751 | 5,968 | ATAAC | Tandem | |
| pTS1257 | No | SPAC1D4 | 11,054 | TTATA | Divergent | |
| pTS1232b | No | SPAC10F6 | 30,945 | ACAAT | Divergent | |
| pTS372 | Yes | SPAC3A12 | 24,899 | TAACA | Divergent | |
| pTS522 | Yes | SPAC3A12 | 24,899 | TAACA | Divergent | |
| pTS206 | No | SPAC2E1P3 | 3,668 | GTTTT | Tandem | |
| pTS747 | Yes | pB24D3 | 5,389 | AAGCA | Tandem | |
| pTS814 | Yes | pB24D3 | 5,389 | AAGCA | Tandem | |
| pTS104 | No | SPAC2F3 | 17,556 | ACCTT | Tandem | |
| pTS1237 | No | SPAC27E2 | 13,549 | TTTAA | Divergent | |
| pTS512 | Yes | SPAC25B8 | 36,479 | ATAAC | Tandem | |
| pTS944 | Yes | SPAC25B8 | 36,479 | ATAAC | Tandem | |
| pTS1156 | Yes | SPAC25B8 | 36,479 | ATAAC | Tandem | |
| pTS1217 | Yes | SPAC25B8 | 36,479 | ATAAC | Tandem | |
| pTS1226 | Yes | SPAC25B8 | 36,479 | ATAAC | Tandem | |
| pTS1263 | No | SPAC1F7 | 2,937 | TTGTT | Divergent | |
| pTS362 | No | SPAC12B10 | 28,557 | TCGTA | Tandem | |
| 2 | pTS192 | No | SPBC947 | 15,380 | TAACG | Tandem |
| pTS114 | No | SPBC2F12 | 35,189 | CTAGG | Tandem | |
| pTS1283 | No | SPBC9B6 | 15,870 | TTAAC | Divergent | |
| pTS1307 | No | SPBP23A10 | 10,313 | TTTAC | Inside open reading frame | |
| pTS112 | No | SPBC21H7 | 6,566 | TACGG | Divergent | |
| pTS116 | Yes | SPBC12C2 | 25,671 | CTTAT | Tandem | |
| pTS333 | Yes | SPBC12C2 | 25,671 | CTTAT | Tandem | |
| pTS441 | Yes | SPBC12C2 | 25,671 | CTTAT | Tandem | |
| pTS365 | No | SPBC365 | 21,569 | ATTAA | Tandem | |
| pTS118 | Yes | SPBC365 | 25,788 | CTAAA | Divergent | |
| pTS189 | Yes | SPBC365 | 25,788 | CTAAA | Divergent | |
| pTS226 | Yes | SPBC365 | 25,788 | CTAAA | Divergent | |
| pTS389 | Yes | SPBC365 | 25,788 | CTAAA | Divergent | |
| pTS366 | No | SPBC3E7 | 28,605 | CTCTG | Tandem | |
| pTS1245 | No | SPBC1105 | 5,688 | TACGT | Tandem | |
| pTS1250 | No | SPBC1105 | 8,041 | TTAAG | Divergent | |
| pTS103 | No | SPBC23E6 | 25,420 | GTATT | Tandem | |
| 3 | pTS1243 | No | SPCC970 | 13,657 | TTATT | Divergent |
| pTS211 | No | SPCC521 | 7,284 | ATATT | Divergent | |
| pTS117 | Yes | SPCC4B3 | 7,624 | TACTC | Tandem | |
| pTS202 | Yes | SPCC4B3 | 7,624 | TACTC | Tandem | |
| pTS310 | Yes | SPCC4B3 | 7,624 | TACTC | Tandem | |
| pTS439 | Yes | SPCC4B3 | 7,624 | TACTC | Tandem | |
| pTS1253 | No | SPCC191 | 18,558 | ATAAT | Tandem | |
| pTS363 | No | SPCC1450 | 11,562 | ATATC | Tandem | |
| pTS311 | Yes | SPCC1450 | 21,288 | CTATT | Tandem | |
| pTS407 | Yes | SPCC1450 | 21,288 | CTATT | Tandem | |
| pTS318 | Yes | SPCC1450 | 21,288 | CTATT | Tandem | |
| pTS1251 | No | SPCC74 | 15,870 | GTTTT | Divergent | |
| pTS910 | Yes | SPCC4F11 | 6,673 | AATAG | Tandem | |
| pTS786 | Yes | SPCC4F11 | 6,673 | AATAG | Tandem | |
| pTS330 | Yes | SPCC4F11 | 6,673 | AATAG | Tandem | |
| pTS5 | No | SPCP1E11 | 13,992 | ATAGC | Divergent |
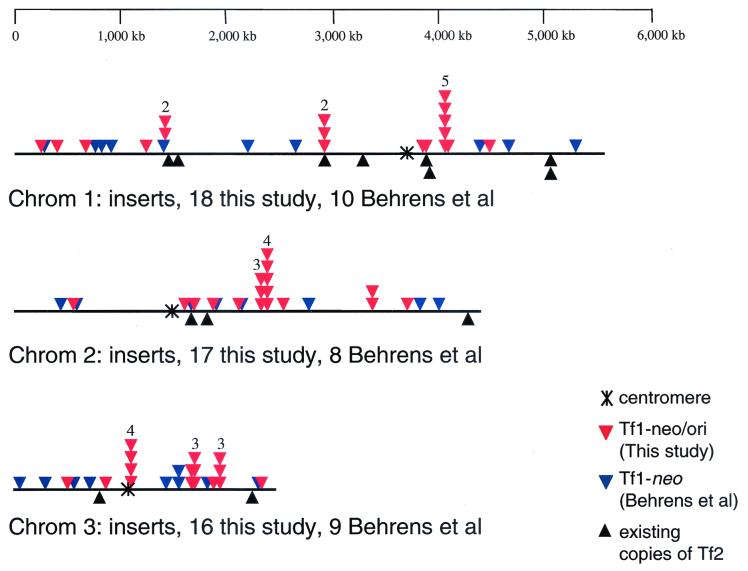
FIG. 2.
Map of the positions of integration of Tf1 in the genome of S. pombe. Fifty-one sites of integration from this study (red triangles) were found to contain intact copies of Tf1-ori/neo flanked by target site duplications. In a recent study (6), Behrens et al. identified 27 sites of integration for Tf1-neo (blue triangles). All but one of the 78 insertions occurred in intergenic regions. The 1.2 Mb of rDNA repeats in chromosome (Chrom) 3 are not shown in this map. The number of times that identical insertions were isolated from genetically independent cultures ranged from two to five and is indicated by a number above multiple triangles.
The map of insertions in Fig. 2 reveals the surprising finding that insertions at identical sites were identified at eight different positions in the genome. In each case, the number of times that identical insertions were isolated from genetically independent cultures ranged from two to five (Fig. 2). The sequences flanking each of the eight positions were examined for any exceptional features that correlated with the sites of multiple insertions. No single feature could be associated with these sites to distinguish them from sites that received single insertions.
We examined whether the integration sites that we isolated multiple times were in regions more likely to serve as insertion sites than other locations of the genome. One way in which we did this was to compare the positions of the endogenous Tf2 LTR retrotransposons to the sites that we isolated multiple times (Fig. 2). Since Tf2 is closely related to Tf1 and encodes the same IN, the positions of Tf2 elements in the genome are likely to represent sites that are active for Tf1 insertion. Although the integration sites were not immediately adjacent to copies of Tf2, we found that of the eight sites, one was 21 kb (pTS372) and another was 10.6 kb (pTS747) from endogenous copies of Tf2. Another indication that the sites with multiple insertions represent regions with high transposition potential was that seven of the eight were within 76 kb of another site of insertion that was described in this study and in another report (6) (Fig. 2). The average distance from the duplicate sites to the nearest neighboring sites was 54 kb. For these seven sites, the distances from adjacent insertions were 25.3 kb (pTS372), 10.6 kb (pTS747), 29.2 kb (pTS512), 47.9 kb (pTS116), 4.2 kb (pTS118), 9.7 kb (pTS311), and 75.9 kb (pTS910). Most of these distances were significantly shorter than 98 kb, the average of the distances between all the insertions and their nearest neighbors. This analysis suggests that the integration sites that we isolated multiple times were in domains of the genome that were more likely to serve as insertion sites than other regions.
An analysis of all the insertion sites that we isolated revealed a strong bias for insertion into intergenic regions. Although the coding sequence of S. pombe represents 60.2% of the genome (European Consortium and Sanger Centre, unpublished data), only 1 of the 51 insertions disrupted a coding sequence, as indicated by the data from the Sanger Centre.
An analysis of the intergenic regions disrupted by the insertions revealed a strong preference for sequences between genes that are transcribed in either divergent or tandem directions. While 18 insertions occurred between divergent genes and 32 occurred between tandem genes, none of the insertions occurred between genes transcribed in convergent directions. Given that in the genome of S. pombe 1,299 gene pairs are divergent, 1,302 are convergent, and 2,289 are tandem (European Consortium and Sanger Centre, unpublished), we predicted that based on just these ratios, 13.3 of our inserts should be in divergent regions, 13.8 should be in convergent regions, and 24 should be in tandem regions. The lack of any insertion between a convergent pair of genes was not expected. However, these calculations do not reflect the fact that the average regions between divergent and tandem genes are larger than the average space between convergent genes. If we assume that the average size of the region between divergent genes is 1.34 kb and that the average sizes between tandem and convergent genes are 0.97 and 0.56 kb, respectively, then unbiased insertion into intergenic regions would be expected to produce 18.9 insertions between divergent pairs, 24.0 between tandem pairs, and 7.6 between convergent pairs for the 51 events. Since no insertions were found between convergent genes, the insertion of Tf1-ori/neo demonstrated a strong bias against this type of intergenic region.
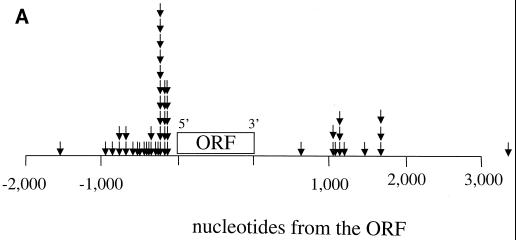
We examined whether the insertion of Tf1-ori/neo occurred at any specific positions within the intergenic regions. To test whether insertions occurred near the 5" or 3" ends of genes, the distance from the integration site to the closest end of a gene was determined. In Fig. 3A these events were plotted upstream of the open reading frame if the insertions were closest to the 5" end of a gene and downstream of the open reading frame if they were closest to the 3" end. In this analysis, 74% of the insertions were associated with the 5" ends of genes. Although the association with the 5" ends of genes occurred with distances of up to 1.54 kb, there was significant clustering within 300 nucleotides of the start of translation.
FIG. 3.
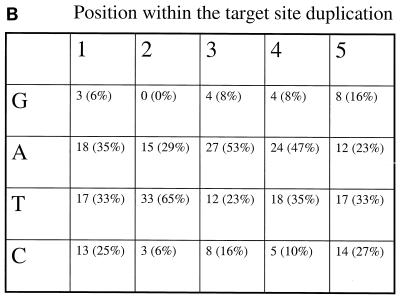
Integration sites for Tf1-ori/neo. (A) Positions of the insertion sites relative to the nearest end of an open reading frame (ORF) (arrows). If an insertion was closer to a 5" end of an ORF than to a 3" end, it was mapped upstream of the ORF in this diagram. The insertions closer to the 3" end of a gene were mapped downstream of the ORF in this diagram. (B) Frequency of each nucleotide in the target site duplications. The target site duplications for each of the 51 insertions were analyzed to determine the frequency of a nucleotide at each position of the duplication. The numbers in parentheses are the percentages of all the target sites that have a specific nucleotide at the position shown.
The sizes of the intergenic regions that were disrupted by Tf1-ori/neo were compared to the average sizes to examine whether transposition required specific classes of intergenic spaces. For the transposition events that occurred between tandem gene pairs, the average size of the intergenic space was 1.44 kb. This size was larger than the average intergenic space of 0.97 kb between tandem genes of the genome (European Consortium and Sanger Centre, unpublished). The average size of the divergent intergenic spaces that received inserts was 1.55 kb. This size was somewhat larger than the 1.3-kb average size of the divergent spaces in the genome (European Consortium and Sanger Centre, unpublished).
The nucleotide compositions at the sites of insertion were analyzed to determine whether Tf1 recognized specific sequence patterns. Figure 3B is a compilation of the five nucleotides that were duplicated during each of the transposition events. Although no consensus sequence existed, the target site duplications were particularly rich in AT. Positions 2, 3, and 4 were 94, 76, and 82% AT, respectively. These levels were higher than the 65% AT content of the genome of S. pombe and suggested that the IN of Tf1 had a bias for insertion sites that were AT rich. In addition, the lack of any G's at position 2 indicated a specific bias against this nucleotide at this position.
The integration of Tf1-ori/neo exhibited a significant preference for chromosome 3.
The transposon sequences throughout the genome sequence of S. pombe were examined by using the genome data from the Sanger Centre (European Consortium and Sanger Centre, unpublished); it was found that the density of endogenous Tf LTRs was twofold higher in chromosome 3 than in the other two chromosomes (N. J. Bowen and H. L. Levin, unpublished data). We therefore examined whether the enrichment of endogenous Tf sequences in chromosome 3 could be due to a bias in integration. Each insertion reported here was tabulated by the chromosome that was disrupted. These numbers were then normalized to the portions of the chromosomes that are nonrepetitive and associated with pol II transcription. To do this, the 1.2 Mb of rDNA repeats on chromosome 3 and the telomeres were excluded from the calculation. One justification for excluding the rDNA repeats is that this region is not a target for Tf1 integration. In a recent study of Tf1 transposition, 27 insertions were generated (6). When these data were pooled with ours, a total of 78 insertions of Tf1 were characterized. Even though the 1.2 Mb of rDNA repeats constitute 35% of chromosome 3, none of the 25 insertions in chromosome 3 occurred in rDNA repeats. Thus, by considering the insertions reported in our study and only the region of the genome associated with pol II transcription and not the telomeres or rDNA repeats, we found the surprising result that, per unit length, chromosome 3 was significantly more likely to be the target of transposition than either of the other two chromosomes (Table 4). When all the insertions were tabulated, chromosomes 1 and 2 received 3.3 and 3.9 inserts of Tf1-ori/neo per Mb, respectively, while chromosome 3 had 6.7 per Mb. This value represents a frequency of transposition into the DNA of chromosome 3 that was about twofold higher than expected based on its length.
TABLE 4.
Chromosome preference
| Chromosome | Length (Mb)
|
Source of data
|
||||||
|---|---|---|---|---|---|---|---|---|
| This study
|
Behrens et al. (6)
|
|||||||
| Total | Excluding rDNA and telomeres | No. (%) of insertions of Tf1-ori/neo | No. of inserts/Mb, excluding rDNA and telomeres | No. (%) of insertions | No. of inserts/Mb, excluding rDNA and telomeres | |||
| 1 | 5.7 | 5.5 | 18 (35) | 3.3 | 10 (37) | 1.8 | ||
| 2 | 4.6 | 4.4 | 17 (33) | 3.9 | 8 (30) | 1.8 | ||
| 3 | 3.5 | 2.4 | 16 (31) | 6.7 | 9 (33) | 3.7 | ||
We used chi-square analysis to test whether this high number of insertions in chromosome 3 was statistically significant. Table 5 includes a pairwise test of the hypothesis that the bias for chromosome 3 verses the other two chromosomes could be due to chance. The P value is less than 0.025, so the preference for chromosome 3 is significant. We also tested whether the transposition events reported by Behrens et al. (6) showed a similar preference for chromosome 3 sequences. Table 4 shows that the proportion of inserts per chromosome was very similar to what we observed. This finding is significant in that the transposition events generated by Behrens et al. (6) were isolated by the direct selection of G418 resistance in haploid cells. Thus, although different procedures were used to identify sites of Tf1 insertions, very similar results were obtained. Although the percentages of insertions in chromosome 3 isolated by Behrens et al. (6) were very similar to ours, their total number of events was only large enough to provide a marginal level of statistical significance when tested for the bias for chromosome 3 (chi-square calculation not shown). When we pooled our data with theirs and used the same chi-square analysis, the statistical significance of the bias for chromosome 3 was greatly increased. The P value dropped to 0.0034. By pooling both data sets, we found that the total numbers of inserts per megabase of nonrepetitive DNA were 5.1, 5.7, and 10.4 for chromosomes 1, 2, and 3, respectively. These values were very similar to the twofold preference for chromosome 3 seen with our data alone.
TABLE 5.
Analysis of chromosome 3 bias
| Chromosome | Fraction of genome, excluding rDNA and telomeres | Source of data
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| This study alone
|
Behrens et al. (6) plus this study
|
||||||||
| No. of insertions
|
(Obs − Exp)2/Exp | No. of insertions
|
(Obs − Exp)2/Exp | ||||||
| Observed | Expected, excluding rDNA and telomeres | Observed | Expected | ||||||
| 1 and 2 | 0.81 | 35 | 41.3 | 0.96 | 53 | 63.2 | 1.65 | ||
| 3 | 0.19 | 16 | 9.7 | 4.09 | 25 | 14.8 | 7.03 | ||
| Total | 51 | 5.05 (P < 0.025) | 78 | 8.68 (P = 0.0034) | |||||
The unusual preference for transposition into sequences of a specific chromosome led us to examine whether chromosome 3 had a greater proportion of intergenic sequences than the other two chromosomes. The proportions of chromosomes 1, 2, and 3 that are intergenic are 0.41, 0.42, and 0.45, respectively (European Consortium and Sanger Centre, unpublished). Although the fraction of intergenic sequences in chromosome 3 is slightly higher than those in the other two chromosomes, it alone is unlikely to account for the higher fraction of transposition events in chromosome 3. This is because the larger amount of intergenic sequence in chromosome 3 is due to a slight increase in the average size of the space between divergent genes (V. Wood, personal communication). The inserts that we isolated from chromosome 3 were not found between divergent genes any more often than the inserts isolated from the other chromosomes. In addition, the average size of the intergenic sequence disrupted by Tf1 insertions into chromosome 3 was no larger than the sizes of the intergenic regions disrupted in chromosomes 1 and 2.
We examined whether the preference for insertion into chromosome 3 could be due to its having a higher proportion of tandem and divergent gene pairs. This was not the case. The fractions of the intergenic sequences in chromosomes 1, 2, and 3 that were located between divergent gene pairs were 26.5, 27, and 27%, respectively. For intergenic sequences located between tandem gene pairs, the fractions in chromosomes 1, 2, and 3 were 47.5, 46.0, and 48.5%, respectively (Wood, personal communication). The results of these tabulations indicate that the gene densities and polarities are nearly equivalent for all three chromosomes.
DISCUSSION
The preferences of target site selection have been observed in vivo for the integration of Tf1 into the genome of its host, S. pombe. Fifty-one insertion events that spanned all three chromosomes were isolated. The target sites revealed a strong association with intergenic sequences. Further examination revealed that the integration sites were more likely to occur in regions between divergent or tandem gene pairs than in regions between convergent pairs of genes. The two most surprising observations were that duplicate insertions occurred at eight sites in the genome and that per unit length, chromosome 3 was preferred for integration over the other two chromosomes.
The techniques used in this study were designed to detect sites of insertion whether or not they disrupted essential genes or contained silent chromatin structures. Nevertheless, our collection of 51 insertions showed similarities to a set of 27 insertions of Tf1-neo that were recently reported (6). Behrens et al. (6) identified sites of integration by direct screening of haploid cells of S. pombe for the G418 resistance that resulted from the transposition of Tf1-neo. The collection of Behrens et al. (6) showed a preference for integration into intergenic regions, just as we observed. In addition, they also reported the preference for integration into spaces between divergent or tandem gene pairs. In addition, they found that insertions occurred in intergenic regions that were larger than average. The positions of their insertions were also associated with the 5" ends of pol II genes, suggesting an interaction with transcription factors. Despite these similarities, there were important differences between the two studies. These include our surprising isolation of duplicate insertions and the unique observation that Tf1 has a preference for integration into chromosome 3.
The association of Ty1 and Ty3 insertions with specific transcription units has been observed. In each instance, the target site specificity protects host genes from being disrupted by integration events (7). The Ty3 element integrates just a few base pairs upstream of pol III genes (11), and this positioning is due to an interaction with the pol III transcription factors in TF-IIIB (22, 41). Ty1 also inserts upstream of pol III genes, but the integration window extends 75 to 700 bp upstream of the transcription start site (14). The examples of Ty1 and Ty3 indicate that for host genomes with a dense coding sequence, targeting strategies are required to prevent the disruption of genes. The genome of S. pombe has a dense coding sequence, and the preference for integration into intergenic regions indicates that this mechanism is also designed to avoid the disruption of host genes. Since Tf1 integration is associated with pol II promoters, it is possible that insertion alters the expression of pol II genes. However, eight different insertions of Tf1-neo were tested, and none was found to change significantly the expression of the adjacent genes (6). This result suggests that the insertions occurred upstream of the sequences critical for transcription.
A dramatic bias was represented by the duplicate insertions that occurred at eight positions in the genome. Each of the duplicate events was obtained from a separate culture of S. pombe and was therefore genetically independent. The possibility that the duplicate events were the result of cross-contaminated pools of DNA was unlikely, since at least one of the pools of transposition events was generated and characterized 6 months later at a different research institution. The close association of the duplicate sites with other sites of insertion provided additional evidence of high frequencies of integration. The lack of any obvious similarities between the duplicate sites makes it difficult to identify a mechanism for the high frequencies of repetition. The lack of sequence similarity between the eight multiply isolated sites suggests that they could represent landmarks in the superstructure of the chromosomes. Possible examples include sites of unique chromosomal structures, such as folds or regions of contact with other nuclear structures, such as the nuclear matrix. It it likely that the eight repeated sites do not represent the complete set of such hot spots. Their isolation was likely subject to biases, such as the efficiency of transformation in bacteria of the circularized DNA. Nevertheless, the repeated and independent isolation of the same insertion sites indicates that the integration of Tf1 cDNA was mediated by factors with surprising specificity.
An important question is why did not we identify insertions closer than 2 kb to the duplicate sites. This class of events would presumably retain their selective advantages in bacteria as well as their relationship to the restriction sites used in gel purification and circularization. The answer may be that for any given intergenic region, there is one dominant site with the potential for Tf1 integration. This notion would imply that integration is precisely controlled by DNA binding proteins, as is the case for Ty3.
Perhaps the most significant bias that we observed was that when normalized for size, the nonrepetitive portion of chromosome 3 was more likely to be targeted for integration than the sequences of chromosomes 1 and 2. In fact, we observed about twice the number of insertions per kilobase of chromosome 3 than we observed for chromosomes 1 and 2. It was also interesting to observe that chromosomes 1 and 2 received equal frequencies of insertions per kilobase. Although Behrens et al. (6) did not report a preference for chromosome 3, their data exhibited the same pattern as ours. When we pooled both data sets and normalized the data for chromosome size, we observed the same twofold preference for insertion into chromosome 3. This similarity in the data sets demonstrates that the bias for integration into chromosome 3 is not likely due to an artifact of either isolation procedure. Additional documentation of this bias was obtained when the genome sequence of S. pombe was analyzed for the locations of the 230 Tf LTRs. The nonrepetitive sequences of chromosome 3 had twice the density of insertions as the other two chromosomes (Bowen and Levin, unpublished).
The association with a specific chromosome is unusual for a transposon, but some examples have been observed. The endogenous retrovirus gypsy of Drosophila has potentially active elements that are predominantly located on the Y chromosome (12, 20, 34). However, the high numbers of gypsy elements on the Y chromosome are not due to preferences of integration but instead result from the loss of gypsy elements from the other chromosomes. The presence of gypsy elements on chromosomes other than Y is selected against because damaging levels of transposition occur only in females of the permissive strains. Since the Y chromosome is not present in females, active copies of gypsy elements can reside on the Y chromosome without resulting in high levels of transposition. In the human genome, similar enrichments of the endogenous retroviruses HERV K and HERV L have been observed on the Y chromosome, and these may also be due to selective pressure against their presence in other chromosomes (24).
A comprehensive analysis of all the transposons in S. cerevisiae revealed that Ty1, Ty2, Ty3, and Ty4 are tightly linked to tRNA genes (21). Further analysis revealed that the density of insertions per kilobase of DNA is higher for the smaller chromosomes. The three smallest chromosomes, I, III, and VI, have an average of one insertion per 25.2 kb. In contrast, the three largest chromosomes, VII, XV, and IV, have an average of one insertion per 39.4 kb. Interestingly, when we normalized the data for the chromosomes of S. cerevisiae for number of tRNAs, chromosomes I, III, and VI had densities of Ty1 elements per tRNA at least sixfold higher than those of chromosomes VII, XV, and IV. Since chromosome 3 of S. pombe is the smallest chromosome, it is possible that chromosome size may in some way contribute to the selection of insertion sites. However, it is also possible that the chromosome bias observed in the genome of S. cerevisiae is not due to selective integration but instead is similar to the situation for gypsy, in that it may result from selective pressure that favors the loss of Ty1 from the larger chromosomes. To our knowledge, the preference of Tf1 for insertion into chromosome 3 sequences represents the first example of an LTR retroelement that has an integration mechanism with a chromosome-specific bias.
One important observation was that the existing copies of Tf2 identified by the S. pombe genome project do not show a bias for chromosome 3. Despite the high concentration of Tf2 LTRs on chromosome 3, the full-length elements are more numerous on chromosomes 1 and 2. In fact, eight are located on chromosome 1, and two are located on chromosome 3 (Fig. 2). The explanation may be found in the mechanism used by the current copies of Tf2 to mobilize (18). Tf2 mobilizes primarily through homologous recombination with preexisting sequences. Despite the high number of Tf2 LTRs on chromosome 3, this process of homologous recombination appears to favor the other two chromosomes.
Just how Tf1 favors integration into chromosome 3 remains unknown. One interesting issue is that the 1.2 Mb of rDNA repeats are located on chromosome 3. Perhaps all sequences of chromosome 3 are more tightly associated with the nucleolus than the sequences of the other chromosomes and this localization somehow favors the interaction with the preintegration complexes of Tf1. Another possibility is that the composition of the chromatin on chromosome 3 is distinct from that on the other chromosomes. For example, an alternative histone might be found in higher concentrations on chromosome 3 and could be recognized by the Tf1 IN. We suggest this possibility because the IN of Tf1 contains a chromodomain (30) and because chromodomains were recently found to interact with modified versions of histone proteins (5, 23, 33). Another explanation for the preference of chromosome 3 accounts for the observation that this bias is almost exactly twofold. Perhaps integration is linked to replication and chromosome 3 is replicated later than the other two chromosomes. If transposition were to occur after the replication of chromosomes 1 and 2 but before that of chromosome 3, then the number of Tf1 elements inserted into chromosome 3 would be effectively amplified by a factor of 2. The higher density of insertions in chromosome 3 also could be due to the distribution of hot spots instead of being a global feature of the entire chromosome. If the duplicate insertions that we observed were the result of local chromatin environments, then these conditions might be more prevalent on chromosome 3.
Another important question about the integration of Tf1 into chromosome 3 is what the function of this bias is. Is it the result of a process that favors the transmission of the transposon or the viability of the host? One possibility is that the overall levels of transcription of genes in chromosome 3 are low so that higher densities of Tf1 in chromosome 3 are required to offset this difference. This scenario could be similar to the dosage compensations of Drosophila melanogaster or Caenorhabditis elegans, where the transcription of genes in the X chromosomes of females is altered by a factor of 2 (16, 31).
A different explanation that is more compelling centers on the possibility that the integration mechanism targets all the chromosomes with equal probabilities. Because the nonrepetitive sequence on chromosome 3 is half the size of the sequences on chromosomes 1 and 2, transposition events would be twice as likely to insert into a kilobase of sequence in chromosome 3 than into a kilobase in the other two chromosomes. This model would also explain the sixfold-higher density of Ty1 on the smaller chromosomes of S. cerevisiae than on the larger chromosomes. Chromosomes I, III, and VI are three- to sixfold smaller than chromosomes VII, XV, and IV.
The explanation for why chromosomes might be targeted with equal probabilities may lie in the method likely used by LTR retrotransposons in the wild to populate the genomes of strains that lack them. Since LTR retrotransposons are not infectious in the classical sense, the only way in which they can integrate into genomes that lack copies of an element is through mating and subsequent transposition into the vacant chromosomes. To avoid damage to the element that could occur during recombination or perhaps to avoid host mechanisms that would inhibit the recombination of transposons, integration may take place after meiosis is complete. If this is the case, the most effective way of dispersing the transposon into naive genomes is by mediating integration into chromosomes with equal probabilities. Thus, with a random assortment of chromosomes, the chance is the greatest that each product of meiosis will carry the transposon.
Because the propagation of transposons ultimately depends on the fitness of the host, it is critical for the elements to develop mechanisms of integration that avoid the disruption of coding sequences. The ability to study the transposition of LTR retrotransposons in S. pombe provides the unique opportunity to compare the strategies that transposons use to select insertion sites in two yeasts that diverged from each other 109 years ago (38). In addition, the analysis of both of these yeasts is particularly informative because their coding sequences represent the majority of their genomes. Our analysis of the sites chosen by Tf1 for integration revealed interactions with the host genome that are entirely different from the strategies used by the Ty elements in the genome of S. cerevisiae. Ty1, Ty2, Ty3, and Ty4 avoid damaging host genes by inserting into gene-poor regions upstream of pol III genes (7, 11, 14, 21, 37). Ty5 does not disrupt essential coding sequences because it specifically inserts into regions of silent chromatin (42, 43). In contrast to the strategies used by the Ty elements in S. cerevisiae, the integration of Tf1 shows no preference for tRNA genes or any other pol III transcribed unit. Nor does Tf1 insert into regions of silent chromatin. Instead, we found that Tf1 specifically inserts in regions between genes that are likely transcribed by pol II. Although this strategy is significantly different from that used by the Ty elements, the result is the same. The disruption of coding sequences is avoided.
The detailed information about the interaction of transposons with two greatly divergent genomes may contribute to the understanding of how these types of elements propagate in the genomes of more complex eukaryotes. Now that the human genome project is complete, it may be particularly interesting to examine the patterns of integration throughout the genome and compare these results to those for the two yeasts.
Acknowledgments
We thank Nathan Bowen for reading the manuscript and for thoughtful discussions. Susan Vitale provided invaluable guidance with the statistical analysis of our data.
REFERENCES
- 1.Asante-Appiah, E., and A. M. Skalka. 1997. Molecular mechanisms in retrovirus DNA integration. Antiviral Res. 36:139-156. [DOI] [PubMed] [Google Scholar]
- 2.Atwood, A., J. Choi, and H. L. Levin. 1998. The application of a homologous recombination assay revealed amino acid residues in a long terminal repeat retrotransposon that were critical for integration. J. Virol. 72:1324-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atwood, A., J. Lin, and H. Levin. 1996. The retrotransposon Tf1 assembles virus-like particles with excess Gag relative to integrase because of a regulated degradation process. Mol. Cell. Biol. 16:338-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balasundaram, D., M. J. Benedik, M. Morphew, V. D. Dang, and H. L. Levin. 1999. Nup124p is a nuclear pore factor of Schizosaccharomyces pombe that is important for nuclear import and activity of retrotransposon Tf1. Mol. Cell. Biol. 19:5768-5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 6.Behrens, R., J. Hayles, and P. Nurse. 2000. Fission yeast retrotransposon Tf1 integration is targeted to 5" ends of open reading frames. Nucleic Acids Res. 28:4709-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boeke, J. D., and S. E. Devine. 1998. Yeast retrotransposons: finding a nice quiet neighborhood. Cell 93:1087-1089. [DOI] [PubMed] [Google Scholar]
- 8.Boeke, J. D., J. Trueheart, G. Natsoulis, and G. R. Fink. 1987. 5-Fluoro-orotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154:164-175. [DOI] [PubMed] [Google Scholar]
- 9.Boeke, J. D., and J. P. Stoye. 1998. Retrotransposons, endogenous retroviruses, and the evolution of retroelements, p. 343-435. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 10.Brown, P. O. 1997. Integration. In J. M. Coffin et al. (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 11.Chalker, D. L., and S. B. Sandmeyer. 1992. Ty3 integrates within the region of RNA polymerase III transcription initiation. Genes Dev. 6:117-128. [DOI] [PubMed] [Google Scholar]
- 12.Chalvet, F., C. di Franco, A. Terrinoni, A. Pelisson, N. Junakovic, and A. Bucheton. 1998. Potentially active copies of the gypsy retroelement are confined to the Y chromosome of some strains of Drosophila melanogaster possibly as the result of the female-specific effect of the flamenco gene. J. Mol. E vol. 46:437-441. [DOI] [PubMed] [Google Scholar]
- 13.Dang, V. D., M. J. Benedik, K. Ekwall, J. Choi, R. C. Allshire, and H. L. Levin. 1999. A new member of the sin3 family of corepressors is essential for cell viability and required for retroelement propagation in fission yeast. Mol. Cell. Biol. 19:2351-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Devine, S. E., and J. D. Boeke. 1996. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev. 10:620-633. [DOI] [PubMed] [Google Scholar]
- 15.Eickbush, T. H. (ed.). 1994. The evolutionary biology of viruses. Raven Press, New York, N.Y.
- 16.Franke, A., and B. S. Baker. 2000. Dosage compensation rox! Curr. Opin. Cell Biol. 12:351-354. [DOI] [PubMed] [Google Scholar]
- 17.Hanahan, D., J. Jessee, and F. R. Bloom. 1991. Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol. 204:63-113. [DOI] [PubMed] [Google Scholar]
- 18.Hoff, E. F., H. L. Levin, and J. D. Boeke. 1998. Schizosaccharomyces pombe retrotransposon Tf2 mobilizes primarily through homologous cDNA recombination. Mol. Cell. Biol. 18:6839-6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji, H., D. P. Moore, M. A. Blomberg, L. T. Braiterman, D. F. Voytas, G. Natsoulis, and J. D. Boeke. 1993. Hotspots for unselected Ty1 transposition events on yeast chromosome III are near tRNA genes and LTR sequences. Cell 73:1007-1018. [DOI] [PubMed] [Google Scholar]
- 20.Junakovic, N., A. Terrinoni, C. Di Franco, C. Vieira, and C. Loevenbruck. 1998. Accumulation of transposable elements in the heterochromatin and on the Y chromosome of Drosophila simulans and Drosophila melanogaster. J. Mol. E vol. 46:661-668. [DOI] [PubMed] [Google Scholar]
- 21.Kim, J. M., S. Vanguri, J. D. Boeke, A. Gabriel, and D. F. Voytas. 1998. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 8:464-478. [DOI] [PubMed] [Google Scholar]
- 22.Kirchner, J., C. M. Connolly, and S. B. Sandmeyer. 1995. Requirement of RNA polymerase III transcription factors for in vitro position-specific integration of a retroviruslike element. Science 267:1488-1491. [DOI] [PubMed] [Google Scholar]
- 23.Lachner, M., N. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 24.Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody, J. Baldwin, K. Devon, K. Dewar, M. Doyle, W. FitzHugh, R. Funke, D. Gage, K. Harris, A. Heaford, J. Howland, L. Kann, J. Lehoczky, R. LeVine, P. McEwan, K. McKernan, J. Meldrim, J. P. Mesirov, C. Miranda, W. Morris, J. Naylor, C. Raymond, M. Rosetti, R. Santos, A. Sheridan, C. Sougnez, N. Stange-Thomann, N. Stojanovic, A. Subramanian, D. Wyman, J. Rogers, J. Sulston, R. Ainscough, S. Beck, D. Bentley, J. Burton, C. Clee, N. Carter, A. Coulson, R. Deadman, P. Deloukas, A. Dunham, I. Dunham, R. Durbin, L. French, D. Grafham, S. Gregory, T. Hubbard, S. Humphray, A. Hunt, M. Jones, C. Lloyd, A. McMurray, L. Matthews, S. Mercer, S. Milne, J. C. Mullikin, A. Mungall, R. Plumb, M. Ross, R. Shownkeen, S. Sims, R. H. Waterston, R. K. Wilson, L. W. Hillier, J. D. McPherson, M. A. Marra, E. R. Mardis, L. A. Fulton, A. T. Chinwalla, K. H. Pepin, W. R. Gish, S. L. Chissoe, M. C. Wendl, K. D. Delehaunty, T. L. Miner, A. Delehaunty, J. B. Kramer, L. L. Cook, R. S. Fulton, D. L. Johnson, P. J. Minx, S. W. Clifton, T. Hawkins, E. Branscomb, P. Predki, P. Richardson, S. Wenning, T. Slezak, N. Doggett, J. F. Cheng, A. Olsen, S. Lucas, C. Elkin, E. Uberbacher, M. Frazier, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 25.Levin, H. L. 1995. A novel mechanism of self-primed reverse transcription defines a new family of retroelements. Mol. Cell. Biol. 15:3310-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levin, H. L. 1996. An unusual mechanism of self-primed reverse transcription requires the RNase H domain of reverse transcriptase to cleave an RNA duplex. Mol. Cell. Biol. 16:5645-5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levin, H. L., and J. D. Boeke. 1992. Demonstration of retrotransposition of the Tf1 element in fission yeast. EMBO J. 11:1145-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levin, H. L., D. C. Weaver, and J. D. Boeke. 1993. Novel gene expression mechanism in a fission yeast retroelement: Tf1 proteins are derived from a single primary translation product. EMBO J. 12:4885-4895. (Erratum, 13:1494, 1994.) [DOI] [PMC free article] [PubMed]
- 29.Levin, H. L., D. C. Weaver, and J. D. Boeke. 1990. Two related families of retrotransposons from Schizosaccharomyces pombe. Mol. Cell. Biol. 10:6791-6798. (Erratum, 11:2334, 1991.) [DOI] [PMC free article] [PubMed]
- 30.Malik, H. S., and T. H. Eickbush. 1999. Modular evolution of the integrase domain in the Ty3/Gypsy class of long terminal repeat retrotransposons. J. Virol. 73:5186-5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meller, V. H. 2000. Dosage compensation: making 1× equal 2×. Trends Cell Biol. 10:54-59. [DOI] [PubMed] [Google Scholar]
- 32.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 33.Nakayam, J., J. C. Rice, B. D. Strahl, C. D. Allis, and S. I. S. Grewal. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292:110-113. [DOI] [PubMed] [Google Scholar]
- 34.Pimpinelli, S., M. Berloco, L. Fanti, P. Dimitri, S. Bonaccorsi, E. Marchetti, R. Caizzi, C. Caggese, and M. Gatti. 1995. Transposable elements are stable structural components of Drosophila melanogaster heterochromatin. Proc. Natl. Acad. Sci. USA 92:3804-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 36.Rose, R. E. 1988. The nucleotide sequence of Pacyc184. Nucleic Acids Res. 16:355.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sandmeyer, S. 1998. Targeting transposition: at home in the genome. Genome Res. 8:416-418. [DOI] [PubMed] [Google Scholar]
- 38.Sipiczki, M. 1989. Molecular biology of the fission yeast. Academic Press Ltd., London, England.
- 39.Xiong, Y., W. Burke, and T. Eickbush. 1993. Pao, a highly divergent retrotransposable element form Bombyx mori containing long terminal repeats with tandem copies of the putative R region. Nucleic Acids Res. 21:2117-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiong, Y., and T. H. Eickbush. 1990. Origin and evolution of retroelements based on their reverse transcriptase sequences. EMBO J. 9:3353-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yieh, L., G. Kassavetis, E. P. Geiduschek, and S. B. Sandmeyer. 2000. The Brf and TATA-binding protein subunits of the RNA polymerase III transcription factor IIIB mediate position-specific integration of the gypsy-like element, Ty3. J. Biol. Chem. 275:29800-29807. [DOI] [PubMed] [Google Scholar]
- 42.Zhu, Y. X., S. G. Zou, D. A. Wright, and D. F. Voytas. 1999. Tagging chromatin with retrotransposons: target specificity of the Saccharomyces Ty5 retrotransposon changes with the chromosomal localization of Sir3p and Sir4p. Genes Dev. 13:2738-2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zou, S., N. Ke, J. M. Kim, and D. F. Voytas. 1996. The Saccharomyces retrotransposon Ty5 integrates preferentially into regions of silent chromatin at the telomeres and mating loci. Genes Dev. 10:634-645. [DOI] [PubMed] [Google Scholar]
- 44.Zou, S., and D. F. Voytas. 1997. Silent chromatin determines target preference of the Saccharomyces retrotransposon Ty5. Proc. Natl. Acad. Sci. USA 94:7412-7416. [DOI] [PMC free article] [PubMed] [Google Scholar]