Abstract
The yeast C-type cyclin Ume3p/Srb11p and its cyclin-dependent kinase partner Ume5p/Srb10p repress the transcription of several genes required for meiotic recombination or meiosis I nuclear division. To relieve this repression, Srb11p is destroyed early in meiosis, prior to the first meiotic division. This report identifies two roles for Srb11p in regulating meiotic development. First, SRB11 is required for the normal exit from the mitotic cell cycle prior to meiotic induction. Specifically, mutants lacking SRB11 (srb11Δ) uncouple bud growth from chromosome segregation, producing small buds with nuclei. The bud growth defect is most likely due to the failure of srb11Δ mutants to reestablish polarized actin fibers at the bud tip following exposure to sporulation medium. Second, Srb11p is required for the efficient execution of meiosis I. srb11Δ mutants either exhibited a delay in performing meiosis I and meiosis II or skipped meiosis I entirely. This meiotic defect is not due to the activation of the recombination or spindle assembly checkpoint pathways. However, the expression of several meiotic genes is delayed and reduced in the mutant strains. These results suggest a positive role for Srb10-Srb11p in regulating the transcription program. This model is supported by the finding that overexpression of the meiotic inducer IME2 partially restored the ability of srb11 mutants to perform meiosis I. In conclusion, these findings indicate that Srb11p is required for both entry into and execution of the meiotic program, thus describing multiple roles for a C-type cyclin in the regulation of a developmental pathway.
Meiosis is the process by which diploid organisms produce haploid gametes capable of sexual reproduction. During meiosis, the cell undergoes one round of DNA synthesis (premeiotic S phase) followed by homolog pairing and extensive genetic recombination. Haploidization is achieved through two sequential chromosome divisions (meiosis I and meiosis II) followed by gamete differentiation. In budding yeast cells, entry into meiosis is controlled by both genetic and nutritional pathways (15). Yeast cells that are heterozygous at the mating type alleles enter the meiotic program from the G1 phase of the cell cycle when starved for nitrogen and a fermentable carbon source (20). Mutations that inappropriately drive the cell through the cell cycle, such as activated RAS2, or the overexpression of G1 cyclins prevents meiotic induction (9, 50). Entry into meiosis is initiated by the transcriptional induction of the IME1 gene (24). Ime1p, in turn, activates the transcriptional cascade of meiotic genes, including the protein kinase IME2 (45, 58). Ime2p is required for many aspects of meiosis, including premeiotic S phase (14) and early meiotic gene transcription (37), and has a later, poorly understood role in spore maturation (16, 37). Therefore, the concerted activity of Ime1p and Ime2p is necessary for normal entry and execution of meiosis and spore formation.
As in mitotic cell division, checkpoint pathways are present that monitor the completion of landmark meiotic events. The DNA damage checkpoint pathway involves several proteins that are activated by unreplicated DNA and/or the persistence of DNA breaks (e.g., Rad17p, Rad24p, and Mec1p) (32, 41). Another pathway arrests meiotic progression in pachytene until the final resolution of recombination intermediates (27, 40, 51). Finally, spindle status and chromosome attachment are monitored by a Mad2p-dependent pathway (43). Interestingly, loss of Mad2p activity causes a significant reduction in normal meiosis I disjunction but has little effect on meiosis II segregation, suggesting that the regulation of these two divisions is not identical. Activation of the DNA damage or spindle checkpoint pathway arrests cells in pachytene or metaphase of meiosis I, respectively. The underlying mechanism by which the checkpoint pathway halts meiotic progression is not completely understood. However, the DNA damage checkpoint negatively regulates the Ndt80p transcription factor, which is required for middle meiotic gene expression (8, 18).
Many genes that are required for the execution of landmark meiotic events in yeasts are expressed in both transient and temporal fashions (36). Different classes of meiotic genes (early, middle, and late) have been established based on the timing of their expression (7, 34). As expected, genes expressed early are required for premeiotic S phase and progression through meiotic prophase I. Middle genes are involved in both meiotic divisions and spore wall assembly, while late genes are required for spore wall maturation. The repression of early meiotic genes (e.g., SPO13 and HOP1) in vegetative cells requires the concerted action of several genes, including those for the histone deacetylation complex Sin3p-Ume6p-Rpd3p (23, 48, 53, 54), which functions through the URS1 promoter element (4, 52). In addition to Sin3p-Ume6p-Rpd3p, another set of transcriptional repressors, encoded by UME2, UME3, and UME5 (10, 47), functions through a different mechanism. UME2, UME3, and UME5 (also called SRB9, SRB11, and SRB10, respectively) copurify with the RNA polymerase II holoenzyme (17, 30). Ume3/Srb11p is a C-type cyclin that is required for activation of the cyclin-dependent kinase Ume5p/Srb10p (12, 30, 49). Unlike those of cyclins that regulate mitotic cell division, Srb11p levels do not vary throughout the cell cycle (10). Rather, Srb11p is rapidly destroyed early in development, prior to meiosis I. This destruction is important, as a degradation-resistant allele of SRB11 reduces SPO13 transcript accumulation (10). These findings suggest that Srb11p may play a role in regulating the early stages of meiotic development.
To further investigate the role of Srb11p in controlling meiosis, the effect of a null allele on meiotic induction and progression was analyzed. These studies identified two functions for Srb11p in normal meiotic development. First, Srb11p is required for the efficient execution of meiosis I. Mutants lacking this cyclin either performed the first division late or skipped the event entirely, forming two-spore asci. This delay was also observed at the level of gene expression, as mRNA levels were reduced and expression from several genes was delayed, providing a possible explanation for the meiotic defect. This hypothesis is supported by the finding that overexpression of the IME2 meiotic inducer partially restored the ability of an srb11 mutant to undergo meiosis I. In addition, Srb11p couples bud growth with nuclear division in the last cell cycle prior to meiotic entry, as mutants produce small buds with a nucleus. Taken together, these results define new roles for the Srb11-Srb10p kinase in both the cellular response to sporulation signals and the execution of meiotic development itself.
MATERIALS AND METHODS
Strains, media, and plasmids.
The strains used in this study are listed in Table 1. These strains are derived from crosses between rapidly sporulating SK1 and W303, which provided efficient sporulation without premature meiotic induction. Yeast strains were grown and induced to undergo meiosis as described previously (10). The IME2 overexpression construct pMR1 was a gift from E. Winter (Thomas Jefferson University, Philadelphia, Pa.); this construct contains the IME2 gene from pHS400 (44) inserted into pRS426 (6). Disruption of RAD17 was accomplished by using pAAA19 (a gift from T. Weinert, University of Arizona). The MAD2 disruption was generated by using oligonucleotide-directed recombination as described previously (31). The srb11::TRP1 allele was constructed by replacing the internal SphI-BglII fragment of SRB11 with an SphI-BglII TRP1 DNA fragment derived from pJJ280 (22).
TABLE 1.
Yeast strains
| Strain | Characteristics | Source or reference |
|---|---|---|
| JX212 | MATa/MATα ade2/ade2 can1-100/CAN1 CYH2/cyh2r-z his3-11,15/his3-11,15 leu1-12/leu1-c lys2-1/LYS2 MET13/met13-B trp1-1/trp1-1 tyr1-1/tyr1-2 ura3/ura3 | This study |
| JX213 | MATa/MATα ade2/ade2 can1-100/CAN1 CYH2/cyh2r-z his3-11, 15/his3-11, 15 leu1-12/leu1-c lys2-1/LYS2 MET13/met13-B trp1-1/trp1-1 tyr1-1/tyr1-2 ura3/ura3 srb11::TRP1/srb11::TRP1 | This study |
| RSY335 | MATa/MATα cyh2r-z/cyh2r-z ho::LYS2/ho::LYS2 leu2::hisG/leu2::hisG lys2/lys2 trp1::hisG/trp1::hisG ura3/ura3 | 10 |
| RSY389 | MATa/MATα cyh2r-z/cyh2r-z ho::LYS2/ho::LYS2 leu2::hisG/leu2::hisG lys2/lys2 trp1::hisG/trp1::hisG ura3/ura3 srb11::LEU2/srb11::LEU2 | 10 |
| RSY503 | MATa/MATα leu2::hisG/leu2::hisG ho::LYS/ho::LYS2 lys2/lys2 trp1::hisG/trp1::hisG ura3-1/ura3-1 rad17::LEU2/rad17::LEU2 | This study |
| RSY504 | MATa/MATα leu2::hisG/leu2::hisG ho::LYS/ho::LYS2 lys2/lys2 trp1::hisG/trp1::hisG ura3-1/ura3-1 rad17::LEU2/rad17::LEU2 srb11::TRP1/srb11::TRP1 | This study |
| RSY738 | MATa/MATα leu2::hisG/leu2::hisG ho::LYS/ho::LYS2 lys2/lys2 trp1::hisG/trp1::hisG ura3-1/ura3-1 mad2::TRP1/mad2::TRP1 | This study |
| RSY770 | MATa/MATα leu2::hisG/leu2::hisG ho::LYS/ho::LYS2 lys2/lys2 trp1::hisG/trp1::hisG ura3-1/ura3-1 mad2::TRP1/mad2::TRP1 srb11::LEU2/srb11::LEU2 | This study |
Northern analysis.
Northern blot analyses were performed as described previously (35) with 25 μg of total RNA. Probes were obtained from PCR amplification of the gene in question and labeled with 32P-UTP by using a PrimeI+II random priming kit (Stratagene). Signals were quantitated by phosphorimaging (Fuji Inc.). The same blot was stripped and reprobed for various transcripts as previously described (10).
Meiotic cell synchrony.
Synchronous meiotic cultures were generated as described previously (25). To monitor the execution of meiosis I and meiosis II, samples were taken at various times, fixed in 70% ethanol, and then stained with the nucleic acid-specific stain 4",6"-diamidino-2-phenylindole (DAPI). For each time point, at least 200 cells were assayed for the appearance of bi- and tetranucleated cells to indicate the execution of meiosis I and meiosis II, respectively. Flow cytometic analyses of sporulating cultures were conducted as described previously (42). Briefly, yeast cells were harvested and fixed in 70% ethanol. The cells were washed briefly with water, sequentially treated with RNase A and pepsin, and then stained with propidium iodide (10 μg/ml). Cells were washed, lightly sonicated to disrupt clumps, and sorted by using a Becton Dickinson FACStarPLUS. At least 50,000 cells were analyzed per sample. All meiotic time courses were conducted at least twice.
Recombination assays.
JX212 and JX213 were induced to enter meiosis by using standard protocols. At various times, samples were taken and lightly sonicated, and the cell numbers were determined by direct counting. The cells were serially diluted and then plated on solid rich medium to determine total viable cell numbers and minimal medium lacking leucine to monitor prototrophy due to intergenic recombination or gene conversion. The plates were incubated for 3 days at 30°C, and the numbers of colonies were determined.
Cell biological techniques.
Nuclear divisions were monitored by using the DNA-specific stain DAPI. Cells were fixed in 70% ethanol for at least 12 h at 4°C, washed twice with water, and then stained for 15 min with DAPI (1 μg/ml). The cells were washed with water twice and visualized by fluorescence microscopy. Actin staining was performed essentially as described previously (1). A 0.5-ml volume of formaldehyde (37%) was added to 4.5 ml of sporulating culture and incubated for 18 h at 4°C. Cells were washed twice with phosphate-buffered saline and stained with 3 U of rhodamine-phalloidin (Molecular Probes Inc.) for 2 h at room temperature. Cells were washed five times in phosphate-buffered saline and resuspended in mounting medium (2) containing 50 ng of DAPI/ml to determine whether large-budded cells had undergone nuclear division. Budded cells were first identified and categorized (small or large) under the DAPI channel, and then the filter was switched to visualize phalloidin. To score polarized actin growth, cells containing greater than 50% total actin staining in the bud were considered polarized (13).
RESULTS
Srb11p is required for the efficient execution of meiosis I.
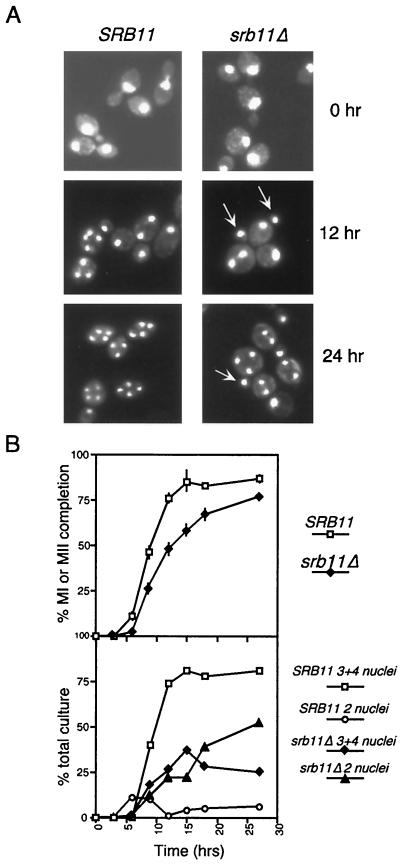
To assess the role that Srb11p plays in regulating meiotic development, a homozygous diploid harboring an SRB11 null allele (srb11Δ) was constructed (RSY389). This strain was induced to enter meiosis, and samples were taken at various times. First, we monitored the progression through meiosis I and meiosis II by monitoring nuclear divisions by using DAPI staining. The mutant strain exhibited two significant phenotypes in this assay. First, there was an approximate 3-h delay in the appearance of bi- and tetranucleated cells indicative of cells undergoing meiosis I and meiosis II, respectively (Fig. 1A; quantitated in Fig. 1B). In addition, the maximum sporulation level was achieved 9 h earlier in the wild type than in the mutant (15 and 24 h, respectively). These results indicate that Srb11p is required for the timely execution of the first and second meiotic divisions.
FIG. 1.
Phenotypic analysis of srb11Δ diploids during meiosis. (A) Fluorescence microscopy (magnification, ×1,000) of DAPI-stained RSY335 (SRB11) and RSY389 (srb11Δ) at 0, 12, and 24 h after transfer to sporulation medium. Arrows indicate the presence of small buds containing nucleus-sized DAPI-stained material. (B) Rate of appearance of bi- and tetranucleated cells in SRB11 and srb11Δ strains during meiosis. (Top) Percentage of cells in the culture executing at least one meiotic division, presented as a function of time following transfer to sporulation medium. MI, meiosis I; MII, meiosis II. Error bars indicate standard deviation. (Bottom) Breakdown between binucleated (2 nuclei) and tri- and tetranucleated (3+4 nuclei) cells in the total population for each culture.
The second phenotype observed for the mutant strain was the dramatic increase in the final percentage of two-spore asci or dyads compared to the percentage in the wild-type strain (52 versus 6%) (Table 2 and Fig. 1A, 24-h time point). Dyads are formed when one of the two meiotic divisions fails to occur, producing two diploid spores instead of the normal four haploid products. To analyze the srb11Δ dyads further, they were dissected onto rich medium, and spore clones were obtained. No significant loss in viability was observed (data not shown), indicating that chromosome segregation was not significantly affected. To investigate which of the two divisions occurred in the dyads, a mating test was performed with the resulting spore colonies. If the reductional, meiosis I division occurred, then a majority of the cells would be genetically MATa/a or MATα/α at the mating type locus due to linkage of the MAT locus to the centromere on chromosome III. These cell types would be competent for mating. Conversely, a single equational, meiosis II division would produce MATa/α cells that would be unable to mate. These mating studies revealed that 83% of the spore colonies (22 dyads tested) were unable to mate and indicated that the srb11Δ mutant strain bypassed meiosis I and completed equational (meiosis II) division. The genetic background used in these initial experiments was derived from SK1, a strain known to exhibit rapid sporulation kinetics. To determine if elevated dyad formation was dependent on the strain background, these experiments were repeated with isogeneic strains derived from W303. Identical results were obtained (data not shown), indicating that the failure to fully execute the first meiotic division was strain independent. Taken together, these data indicate that SRB11 is required for the timely and efficient execution of the first meiotic division.
TABLE 2.
Phenotypic analysis of an srb11Δ/srb11Δ homozygous diploid during meiosis
| Strain | Genotype | Budding index (%)a at the following h after shift:
|
Bi- and tetra-nucleated cells (% of total) | % Binu-cleated cellsb | |||
|---|---|---|---|---|---|---|---|
| 0 | 3 | 15 | 36 | ||||
| RSY335 | SRB11/SRB11 | 31 | 6 | 0 | 0 | 78 | 6 |
| RSY389 | srb11Δ/srb11Δ | 28 | 22 | 22 | 20 | 73 | 52 |
The budding index was calculated as the fraction of cells containing buds in the total population during vegetative growth (0 h) and at 3, 15, and 36 h after a shift to sporulation medium.
(Total number of binucleated cells/total number of bi- and tetranucleated cells) × 100.
Srb11p is required for normal recombination kinetics.
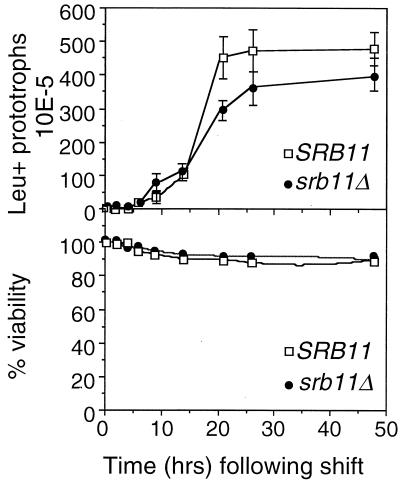
The studies presented above indicate that mutants lacking Srb11p exhibit either a delay in executing meiosis I or skip the division entirely. To more precisely define the stage of meiosis that requires Srb11p, two early landmark events were examined. First, premeiotic S phase was monitored by using flow cytometry (see Materials and Methods for details). No significant differences were detected in the timing or efficiency of premeiotic S phase (data not shown), indicating that Srb11p is dispensable for bulk DNA synthesis during meiosis. We next asked whether Srb11p was required for meiotic recombination. To address this question, two diploid strains were constructed either wild type or mutant for SRB11 and containing heteroallelic markers at the leu1 locus (JX212 or JX213, respectively) (Table 1). Three independent isolates of each strain were induced to enter meiosis. Aliquots were taken at various times and plated on rich medium to determine overall viability and defined minimal medium to detect leucine prototrophs due to intergenic recombination at the leu1 locus (see Materials and Methods for details). The timing of appearance of Leu+ prototrophs was similar in both strains (Fig. 2, top panel) although the wild type produced recombinants with a slightly higher frequency and completed the process 4 h before the mutant. The reduction in colonies on leucine-lacking medium observed with the mutant was not due to a general loss in viability, as determined by plating on rich medium (Fig. 2, bottom panel). Therefore, these data suggest a delay and perhaps a reduction in the efficiency of recombination events.
FIG. 2.
Srb11p is required for normal recombination during meiosis. Wild-type JX212 (SRB11) and mutant JX213 (srb11Δ) strains containing heteroalleles at leu1 were induced to enter meiosis. (Top) Three independent samples were taken at the indicated times following the shift and plated on medium to select for recombinants. Error bars indicate standard deviations. (Bottom) Overall cell viability was determined by plating the appropriate dilutions of the same samples on rich medium without selection.
The meiosis I defect in srb11 mutants is independent of RAD17 DNA damage or recombination or MAD2 spindle checkpoint pathways.
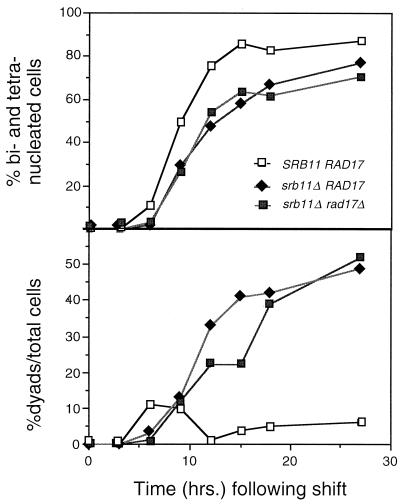
The findings described above suggest that srb11 mutants exhibit a small delay in finishing recombination. Persistent recombination intermediates trigger a RAD17-dependent checkpoint pathway that leads to pachytene arrest (32), providing a possible explanation for the srb11 mutant meiotic delay. To test this possibility, the ability of srb11 mutants to undergo meiosis in a rad17 strain defective for the DNA damage checkpoint was examined. Isogenic strains were constructed with a deletion of rad17 and with or without the wild-type SRB11 allele. These strains were induced to enter meiosis, and samples were taken as described above. Deleting RAD17 alone (RSY503) had no effect on sporulation kinetics, as determined by the appearance of bi- and tetranucleated cells (data not shown). The srb11 rad17 double mutant (RSY504) continued to display the delayed kinetics (Fig. 3, top panel) and the aberrant production of binucleated cells (Fig. 3, bottom panel) observed for the srb11Δ single mutant. These results suggest that the meiotic defect observed in srb11 mutants is not due to the accumulation of recombination intermediates. In addition to the recombination checkpoint, Rad17p is involved in the general DNA damage response (57), suggesting that the srb11 mutant does not generate this type of damage. These experiments were repeated with the srb11 mad2 strain (RSY770) to determine whether srb11 mutants possess a problem with chromosome capture by the spindle. Like previous results, the mad2 mutation did not rescue the srb11Δ-induced delay, indicating that spindle checkpoint activation was also not involved (data not shown). These findings argue against the observed meiotic defect being due to accumulated cellular damage and suggest a more direct role for Srb11p in the regulation of meiotic development.
FIG. 3.
The srb11Δ meiotic defect is independent of checkpoint pathways. Wild-type strain RSY335 (SRB11), mutant strain RSY389 (srb11Δ), or double-mutant strain RSY504 (srb11Δ rad17Δ) was induced to enter meiosis, and samples were taken at the indicated times. The samples were fixed, stained with DAPI, and examined for the production of binucleated and tetranucleated cells. A total of 200 cells were counted for each time point.(Top) Total percentage of the population completing either meiotic division plotted against time in sporulation medium. (Bottom) Percentage of the population containing only binucleated cells.
Srb11p is required for the normal meiotic transcription program.
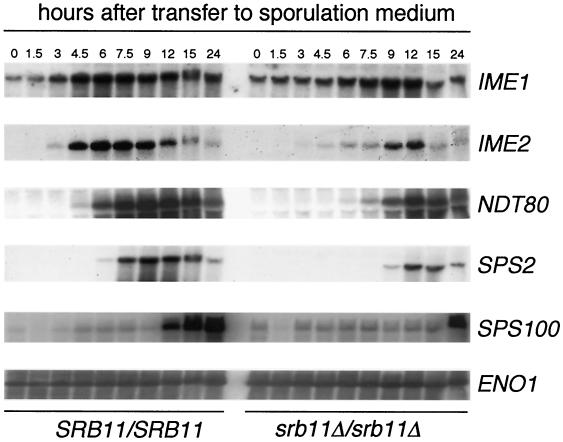
The results described above indicate that Srb11p is required for normal progression through meiosis I. Since Srb11p is involved in transcriptional regulation, we next examined the fate of the transcription program in wild-type and mutant cells. The mRNA accumulation profiles of several genes representing the various expression classes were examined. Specifically, IME1, the master regulatory switch for meiosis, is induced as the cell enters the program (24). Genes representative of the early (IME2) (45), early-middle (NDT80) (8), middle (SPS2) (39), and late (SPS100) (39) classes were also examined. RSY335 and RSY389 were induced to enter meiosis, and samples were taken at various times. Total RNA samples prepared from each time point were blotted and subjected to Northern analysis (see Materials and Methods for details). IME1 mRNA induction peaked later in the mutant strain (9 h) than in the wild-type strain (6 h) (Fig. 4). Similar results were obtained with an srb10 mutant (38). Although the timing was slower, the peak levels of accumulation were similar in both strains, as determined by phosphorimager quantitation. Similarly, the timing of IME2 induction was also delayed in the srb11 mutant strain. However, total IME2 mRNA accumulation was only 47% that of the wild-type control, indicating that the loss of Srb11p activity affected both the timing and the levels of IME2 transcription. Comparable results were obtained for SPS2 and perhaps SPS100, although the time course for the latter was not extended far enough to see the entire profile. NDT80 expression was also delayed and reduced; the expression level was 60% that of the wild type. The finding that NDT80 mRNA was detected at relatively robust levels also argues against the activation of the pachytene checkpoint. We conclude that Srb11p is required for the normal transcriptional timing of meiotic genes from several classes. Moreover, total mRNA levels, especially those of IME2, are also affected.
FIG. 4.
Meiotic gene expression kinetics in the wild type and an srb11Δ mutant. Wild-type RSY335 (SRB11) and mutant RSY389 (srb11Δ) cultures were induced to enter meiosis, and samples were taken at the indicated times. Total RNA was prepared, and transcript levels for IME1, IME2, NDT80, SPS2, and SPS100 were determined (see Materials and Methods for details). ENO1 mRNA levels were used to standardize loadings.
Overexpression of IME2 partially relieves the srb11 meiotic defect.
Ime2p plays an important role in promoting early meiotic gene transcription, including its own (37). Therefore, the reduction of IME2 mRNA levels may provide an explanation for the srb11Δ mutant phenotype. To determine whether increased IME2 mRNA levels can compensate for the loss of SRB11, the IME2 gene was placed in a high-copy-number plasmid (pMR1; see Materials and Methods for details). This plasmid and a vector control were introduced into the diploid srb11Δ mutant (RSY389), and the transformants were induced to enter meiosis. Northern blot analysis revealed that IME2 mRNA was indeed elevated to levels equal to or greater than those observed when the gene is fully induced (data not shown). DAPI analysis revealed that the final percentages of cells undergoing at least one meiotic division were similar in the strains overexpressing IME2 (83%) and the vector control (79%) (Table 3). Importantly, the percentages of srb11Δ cells that overexpressed IME2 and that were able to undergo both divisions (percentages of tri- and tetranucleated cells) increased significantly compared to the results obtained with the vector control (62 versus 37%). These findings suggest that the srb11Δ meiotic defect may be due at least in part to reduced Ime2p activity. However, since the mutant phenotype was not completely rescued by IME2 overexpression, Srb11p may also regulate meiosis through an IME2-independent pathway.
TABLE 3.
IME2 overexpression partially suppresses the srb11 meiotic defecta
| Overexpression plasmid | % of cells that were
|
|||
|---|---|---|---|---|
| Mononucleated | Binucleated | Tri- and tetranucleated | Totally sporulated | |
| SRB11 | 17 | 4 | 79 | 83 |
| Vector | 21 | 42 | 37 | 79 |
| IME2 | 17 ± 6 | 21 ± 1 | 62 ± 5 | 83 ± 4 |
RSY389 (srb11Δ) was transformed with the indicated plasmids: SRB11, pKC342 (10); vector, pRS425; pMR1, IME2 inserted into a 2μm high-copy-number vector (see Materials and Methods for details). Transformants were subjected to a standard sporulation regimen in liquid medium. Cells were fixed and stained with DAPI 24 h after a shift to sporulation medium. Three independent cultures harboring pMR1 were assayed; data are means and standard deviations.
Srb11p is required for coupling bud growth with nuclear division in cells entering the meiotic program.
During the studies described above, we observed that a significant percentage of srb11Δ cells maintained their buds throughout the sporulation time course. When transferred to sporulation medium, wild-type cells complete their current cell cycle and enter the meiotic program early in G1 (unbudded) (20). Accordingly, we found that the percentage of wild-type cells with buds decreased rapidly following transfer to sporulation medium (Fig. 1A; quantitated in Table 2). However, even after 36 h in sporulation medium, 20% of mutant cells still retained buds. Since the budding frequencies were similar in both strains during vegetative culturing, these results suggest that Srb11p is required for the normal completion of the last cell cycle prior to entry into the meiotic program.
Normally, bud development and nuclear division are coupled through the activity of the cyclin-Cdc28 protein kinase (reviewed in reference 28). Bud emergence occurs early in S phase, and bud growth continues throughout the remainder of the cell cycle. Nuclear division commences only when the bud reaches an appropriate size (reviewed in reference 19). Surprisingly, we found that many small buds which persisted in sporulation medium were equivalent in size to those indicative of S phase and contained a nucleus-sized DAPI-stained body (Fig. 1A). As indicated in the 24-h micrograph, the mother of the abnormal bud was still able to undergo meiosis, indicating that the cell was not significantly crippled by this characteristic. These findings suggest that nuclear division was not altered in the srb11 mutant but rather that bud growth was defective. Taken together, these findings indicate that nuclear division and bud growth have become uncoupled in the srb11 mutant. Moreover, these cells (i.e., small buds with a nucleus) were not observed in vegetative cultures, suggesting that this phenotype is restricted to cells induced to enter meiosis.
Srb11p is required for actin repolarization following exposure to sporulation medium.
The above findings indicated that bud growth was defective in srb11 mutants only entering meiosis. Bud emergence and growth require actin polarization to efficiently deliver necessary cell wall components to the growing bud tip (1). To determine whether polarization was affected in srb11Δ strains, a fluorochrome-labeled phallotoxin was used to monitor actin patch localization (see Materials and Methods). Patches represent actin and actin binding protein foci that associate with the membrane. They are dispersed in unbudded cells but congregate at the newly selected bud site and in the growing bud itself (reviewed in reference 33). For this analysis, the cells were divided into those containing small buds (≤10% the mother size) and large buds that had not undergone nuclear division (as determined by DAPI staining; data not shown). This distinction was made due to the observation that the actin cytoskeleton behaved in different ways in large and small buds in response to heat shock (13).
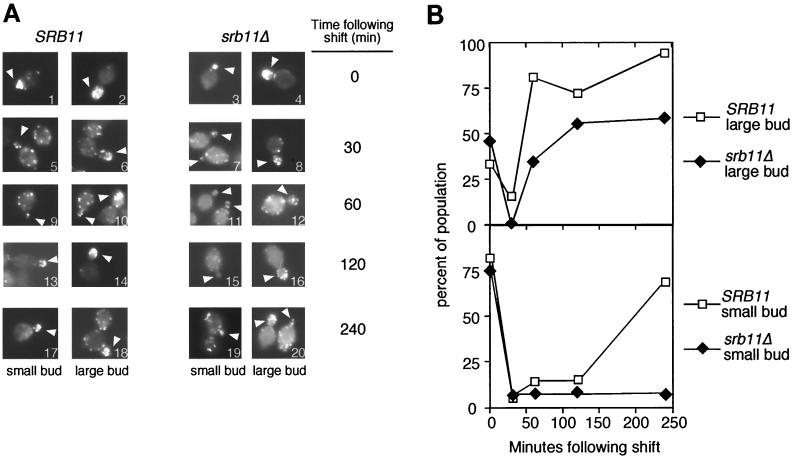
During vegetative growth, the wild type displayed actin patches localized to both small and large growing buds (0 h) (Fig. 5A, panels 1 and 2; quantitated in Fig. 5B). However, within 30 min following transfer to sporulation medium, significant actin depolarization was observed in both bud types (Fig. 5A, panels 5 and 6). Within 60 min, the wild type relocalized actin patches to the large buds (Fig. 5A, panel 10) but not the small buds (panel 9) (compare the small-budded cell on the left in panel 10 with the large-budded cell on the right). Small-budded cells began to regain oriented actin patches by 120 min (Fig. 5A, panel 13) and were back to nearly vegetative levels by 240 min (panel 17). It should be noted that the presence of small-budded cells in wild-type sporulation cultures after 4 h was very rare. These findings indicate that yeast cells depolarize their actin cytoskeleton in response to sporulation medium. Moreover, a difference in repolarization kinetics was observed between cells with large buds and those with small buds.
FIG. 5.
Srb11p is required for reestablishing polarized actin microtubules following transfer to sporulation medium. (A) Wild-type RSY335 (SRB11) or mutant RSY389 (srb11Δ) cells were harvested at the indicated times and stained with phalloidin (see Materials and Methods for details). Cells were scored as small buds (sizes, <10% those of the mothers) or large buds (arrowheads). Image 6 contains a cell with small (left) and large (right) buds. Image 20 contains a large-budded cell (left) and a small-budded cell (right). Final magnification, ×860. (B) Quantitation of actin studies. Wild-type and mutant cells were identified as cells with large and small buds as defined in panel A. Polarized growth was scored as ≥50% the total actin localized to the bud and neck region in the cell (13). At least 100 cells were counted for each time point, except for the wild-type 240-min sample; for this sample, only 25 cells were counted due to the infrequency of finding budded cells in wild-type sporulating cultures after 4 h.
Like the wild-type strain, the srb11Δ mutant displayed localized actin patch patterns in both large- and small-budded vegetative cells (Fig. 5A, panels 3 and 4) that were rapidly lost upon exposure to sporulation medium (panels 7 and 8). Polarization returned to large-budded mutant cells with kinetics somewhat slower than those for the wild-type control (Fig. 5A, panels 12, 16, and 20). However, small buds failed to reestablish a substantial degree of polarized staining, even by 4 h (Fig. 5A, panels 11, 15, and 19) (see a direct comparison in panel 20). Further incubation (24 h) did not significantly alter these results, suggesting that this phenotype was terminal and not the result of slowed kinetics (data not shown). These findings indicate that Srb11p is required for the efficient recovery of polarized actin, at least in cells harboring small buds. Moreover, this observation, combined with the DAPI analysis, suggests that Srb11p couples bud growth to nuclear division, but only in cells switching from mitotic cell growth to meiotic development. These results define a new function for the C-type cyclin-Cdk kinase in regulating cell fate decisions (see Discussion).
DISCUSSION
The Ume3p/Srb11p-Ume5p/Srb10p kinase represses the transcription of several diverse gene sets, including those for sucrose utilization (SUC2) (26), the stress response (SSA1) (10), and meiosis (SPO13) (47). This study demonstrates a new requirement for Srb11p in normal entry into the meiotic program and the execution of meiosis I itself. Following exposure to sporulation medium, Srb11p is required for the completion of the last cell cycle prior to entry into meiosis. Specifically, Srb11p is important for reestablishing polarized actin necessary for the continued growth of new buds. Following entry into the meiotic program, Srb11p is also necessary for the efficient execution of meiosis I. A mutant lacking SRB11 either exhibits a delay in the execution of meiosis I or skips the event entirely. These phenotypes are independent of both the recombination and the spindle damage checkpoint pathways, arguing against the cause of the meiotic defect being cellular damage. Rather, a delay and a reduction in the mRNA levels of several meiotic genes were observed, suggesting that the underlying defect lies in the transcription program. This model is supported by the result that the overexpression of IME2, a critical inducer of meiotic gene expression, can partially suppress the srb11 meiotic defect. Taken together, these findings reveal new roles for C-type cyclins in controlling entry into and execution of a developmental process.
This study presents evidence that srb11Δ mutants uncouple nuclear division from bud growth. Specifically, small buds stop growing, while nuclear division does not stop, resulting in a small bud containing a nucleus. Since small buds containing DAPI masses are not observed in vegetative cells, the requirement of Srb11p for this process appears to be restricted to cells that are switching from mitotic to meiotic cell division. Moreover, small buds fail to reassemble their actin cytoskeleton following transfer to sporulation medium, providing a likely mechanism for this phenotype. Why are small buds more likely to stop growing than large buds during the final cell cycle prior to meiosis? During vegetative growth, bud site selection occurs in G1, with emergence initiating as the cell enters S phase (reviewed in reference 28). Bud site selection and initial bud emergence require the G1 cyclin-Cdc28 kinase (29). Specifically, actin localization to the growing bud tip is dependent on Cln-Cdc28 activity. As the bud continues to grow, it shifts from apical to isotropic growth, which requires the B-type cyclins Clb1p and Clb2p. However, G1 cyclins antagonize meiotic induction by inhibiting Ime1p function (9). Therefore, the G1 cyclins are rapidly downregulated in cells exposed to sporulation conditions to promote meiotic induction. This system effectively excludes continuation of the mitotic cell cycle and funnels cells toward meiosis. However, the downregulation of the G1 cyclins may have an adverse effect on cells harboring recently emerged buds, since actin disorganization accompanies the shift to sporulation medium. One model consistent with our data is that the Srb11-Srb10p kinase fills this gap by providing Cdk activity, which allows the continuation of bud growth until B-type cyclin-Cdc28 activity takes over. It is interesting that the large-budded cells, which no longer required Cln-Cdc28 activity, although delayed, were able to reorganize their actin in the srb11 mutant strain. This strategy would allow efficient entry into meiosis without interference from Cln-Cdc28 or sacrifice of newly born buds.
This report demonstrates that exposure to sporulation medium results in a transient depolarization of actin fibers. Similarly, vegetative cells exposed to heat shock undergo a transient depolymerization of actin that halts bud growth (13). As the cells adapt to the higher temperature, polarized actin fibers return and cell division resumes. These findings suggest that the cell may view exposure to sporulation medium as a “stress response.” Does Srb11-Srb10p also control actin polarization in vegetative cells exposed to stress? Delley and Hall showed that actin polarization returned to cells approximately 120 min following heat shock (13). Previous studies from our laboratory found that Srb11p is destroyed in response to several types of stress, including heat shock (10, 11). With heat shock, the reduction in Srb11p levels is also transient. Within 10 min following heat shock at 37°C, Srb11p levels are reduced to below the limits of detection, but they return to pre-heat shock concentrations within 120 min. Therefore, the kinetics of Srb11p destruction and accumulation mirror those reported for depolarizing-repolarizing actin filaments. These findings are consistent with Srb11p also playing a role in actin reorganization during vegetative growth. Preliminary experiments do suggest a role for Srb11p in this process, as mutant strains displayed a 20% delay in actin repolarization kinetics compared to the results for the wild type following exposure to heat shock (L. H. Rutkowski and R. Strich, unpublished results). However, if Srb11p-Srb10p does indeed control actin repolarization following heat shock, its role is modest, and additional regulatory factors must be involved.
The most pronounced phenotype associated with the loss of Srb11p activity is the 10-fold increase in dyad formation, with these cells predominantly skipping meiosis I. How does Srb11p-Srb10p regulate meiosis I? A possible answer to this question was provided by the findings that the mRNA levels of many meiotic genes were reduced and that their peak accumulation was delayed in the srb11 mutant. The reduction or delay in the expression of key meiotic genes required for meiosis I may lead to the exclusive execution of meiosis II by default. These results formally suggest that the Srb11-Srb10p kinase plays a positive role in meiotic gene expression. A caveat to this interpretation is that Srb11-Srb10p function may be indirect. For example, Srb11-Srb10p may repress a repressor, therefore indirectly playing a positive role in meiotic gene expression. Indeed, genetic analyses have indicated that the Srb11-Srb10p kinase functions as a transcriptional repressor for several meiotic and nonmeiotic genes (5, 10, 26, 47, 56). Alternatively, Srb11-Srb10p may play a more direct role in transcriptional activation. Recent studies reported that this cyclin-Cdk enhances transcription through phosphorylation of the transcriptional activator Gal4p or Sip4p (21, 55). This result may suggest that Srb11-Srb10p can switch from being a repressor in mitotic cells to performing positive activity during meiosis, similar to a current model for Ume6p function (3, 46). However, this model for Srb11-Srb10p function cannot be so simple. Previous studies found that Srb11p is destroyed as cells enter meiotic prophase and prepare for meiosis I (10). This destruction is important for the full induction of SPO13, an early meiotic gene repressed by Srb11-Srb10p during vegetative growth (47). This finding appears at odds with the positive role that Srb11p-Srb10p plays in early meiotic events. However, a model satisfying both observations is suggested by the timing of IME2 and SPO13 expression and previous genetic studies. Ime2p is required for the full induction of several early meiotic genes, including SPO13 and itself (37). IME2 mRNA induction occurs rapidly, due to this positive feedback, before the full induction of SPO13 (7). This step in IME2 mRNA accumulation is one possible entry point for Srb11p-dependent control. A requirement for Srb11-Srb10p in the positive-feedback loop would account for the significant decrease in IME2 mRNA levels in the srb11 mutant and the ability of IME2 overexpression to partially suppress the meiotic defect. This possibility is currently under investigation. Since Srb11p degradation begins at approximately the time when Ime2p accumulation peaks, it is possible that the combination of these two events leads to the full induction of SPO13 and the rest of the early genes. This model would also predict that the downregulation of Srb11p provides a mechanism to attenuate early meiotic gene expression by reducing Ime2p-dependent induction of its own expression. This mechanism could be important to allow the normal transition from the first meiotic prophase to the later stages of meiotic development.
Acknowledgments
This work was supported by grants from the National Science Foundation (MCB-9513479), the U.S. Army Breast Cancer Research Program (DAMD 17-1-7296), and the National Institutes of Health (GM 57842) to R.S. and a postdoctoral award by the National Institutes of Health (CA-09035) to K.F.C. Institutional support was provided by the National Cancer Center (Comprehensive Cancer Center core grant CA 06927) and by an appropriation from the Commonwealth of Pennsylvania.
REFERENCES
- 1.Adams, A. E., and J. R. Pringle. 1984. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J. Cell Biol. 98:934-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams, A. E. M., and D. Botstein. 1989. Dominant suppressors of yeast actin mutations that are reciprocally suppressed. Genetics 121:675-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowdish, K. S., H. E. Yuan, and A. P. Mitchell. 1995. Positive control of yeast meiotic genes by the negative regulator UME6. Mol. Cell. Biol. 15:2955-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckingham, L. E., H.-T. Wang, R. T. Elder, R. M. McCarroll, M. R. Slater, and R. E. Esposito. 1990. Nucleotide sequence and promoter analysis of SPO13, a meiosis-specific gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 87:9406-9410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlson, M., B. C. Osmond, L. Neigeborn, and D. Botstein. 1984. A suppressor of snf1 mutations causes constitutive high-level invertase synthesis in yeast. Genetics 107:19-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119-122. [DOI] [PubMed] [Google Scholar]
- 7.Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein, P. O. Brown, and I. Herskowitz. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699-705. [DOI] [PubMed] [Google Scholar]
- 8.Chu, S., and I. Herskowitz. 1998. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol. Cell 1:685-696. [DOI] [PubMed] [Google Scholar]
- 9.Colomina, N., E. Gari, C. Gallego, E. Herrero, and M. Aldea. 1999. G1 cyclins block the Ime1 pathway to make mitosis and meiosis incompatible in budding yeast. EMBO J. 18:320-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper, K. F., M. J. Mallory, J. S. Smith, and R. Strich. 1997. Stress and developmental regulation of the yeast C-type cyclin UME3 (SRB11/SSN8). EMBO J. 16:4665-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper, K. F., M. J. Mallory, and R. Strich. 1999. Oxidative stress-induced destruction of the yeast C-type cyclin Ume3p requires the phosphatidylinositol-specific phospholipase C and the 26S proteasome. Mol. Cell. Biol. 19:3338-3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper, K. F., and R. Strich. 1999. Functional analysis of the yeast C-type cyclin Ume3p/Srb11p-RNA polymerase II holoenzyme interaction. Gene Expr. 8:43-57. [PMC free article] [PubMed] [Google Scholar]
- 13.Delley, P. A., and M. N. Hall. 1999. Cell wall stress depolarizes cell growth via hyperactivation of RHO1. J. Cell Biol. 147:163-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dirick, L., L. Goetsch, G. Ammerer, and B. Byers. 1998. Regulation of meiotic S phase by IME2 and a Clb5,6-associated kinase in Saccharomyces cerevisiae. Science 281:1854-1857. [DOI] [PubMed] [Google Scholar]
- 15.Esposito, R. E., and S. Klapholz. 1981. Meiosis and ascospore development, p. 211-287. In J. N. Strathern, E. W. Jones, and J. R. Broach (ed.), The molecular biology of the yeast Saccharomyces: life cycle and inheritance. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 16.Foiani, M., E. Nadjar-Boger, R. Capone, S. Sagee, T. Hashimshoni, and Y. Kassir. 1996. A meiosis-specific protein kinase, Ime2, is required for the correct timing of DNA replication and for spore formation in yeast meiosis. Mol. Gen. Genet. 253:278-288. [DOI] [PubMed] [Google Scholar]
- 17.Hengartner, C. J., C. M. Thompson, J. Zhang, D. M. Chao, S.-M. Liao, A. J. Koleske, S. Okamura, and R. A. Young. 1995. Association of an activator with an RNA polymerase II holoenzyme. Genes Dev. 9:897-910. [DOI] [PubMed] [Google Scholar]
- 18.Hepworth, S. R., H. Friesen, and J. Segall. 1998. NDT80 and the meiotic recombination checkpoint regulate expression of middle sporulation-specific genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:5750-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herskowitz, I. 1988. Life style of the budding yeast Saccharomyces cerevisiae. Microbiol. Rev. 52:536-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirschberg, J., and G. Simchen. 1977. Commitment to the mitotic cell cycle in yeast in relation to meiosis. Exp. Cell Res. 105:245-252. [DOI] [PubMed] [Google Scholar]
- 21.Hirst, M., M. S. Kobor, N. Kuriakose, J. Greenblatt, and I. Sadowski. 1999. GAL4 is regulated by the RNA polymerase II holoenzyme-associated cyclin-dependent protein kinase SRB10/CDK8. Mol. Cell 3:673-678. [DOI] [PubMed] [Google Scholar]
- 22.Jones, J. S., and L. Prakash. 1990. Yeast Saccharoyces cerevisiae selectable markers in pUC18 polylinkers. Yeast 6:363-366. [DOI] [PubMed] [Google Scholar]
- 23.Kadosh, D., and K. Struhl. 1997. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89:365-371. [DOI] [PubMed] [Google Scholar]
- 24.Kassir, Y., D. Granot, and G. Simchen. 1988. IME1, a positive regulator of meiosis in S. cerevisiae. Cell 52:853-862. [DOI] [PubMed] [Google Scholar]
- 25.Klapholz, S., and R. E. Esposito. 1980. Recombination and chromosome segregation during the single division meiosis in spo12-1 and spo13-1 diploids. Genetics 96:567-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuchin, S., P. Yeghiayan, and M. Carlson. 1995. Cyclin-dependent protein kinase and cyclin homologs SSN3 and SSN8 contribute to transcriptional control in yeast. Proc. Natl. Acad. Sci. USA 92:4006-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leu, J. Y., and G. S. Roeder. 1999. The pachytene checkpoint in S. cerevisiae depends on Swe1-mediated phosphorylation of the cyclin-dependent kinase Cdc28. Mol. Cell 4:805-814. [DOI] [PubMed] [Google Scholar]
- 28.Lew, D. J., and S. I. Reed. 1995. Cell cycle control of morphogenesis in budding yeast. Curr. Opin. Genet. Dev. 5:17-23. [DOI] [PubMed] [Google Scholar]
- 29.Lew, D. J., and S. I. Reed. 1993. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J. Cell Biol. 120:1305-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao, S.-M., J. Zhang, D. A. Jeffery, A. J. Koleske, C. M. Thompson, D. M. Chao, M. Viljoen, H. J. J. van Vuuren, and R. A. Young. 1995. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature 374:193-196. [DOI] [PubMed] [Google Scholar]
- 31.Longtine, M. S., A. R. McKenzie, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953-961. [DOI] [PubMed] [Google Scholar]
- 32.Lydall, D., Y. Nikolsky, D. K. Bishop, and T. Weinert. 1996. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature 383:840-843. [DOI] [PubMed] [Google Scholar]
- 33.Madden, K., and M. Snyder. 1998. Cell polarity and morphogenesis in budding yeast. Annu. Rev. Microbiol. 52:687-744. [DOI] [PubMed] [Google Scholar]
- 34.Magee, P. T. 1987. Transcription during meiosis, p. 355-382. In P. Moens (ed.), Meiosis. Academic Press, Inc., New York, N.Y.
- 35.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 36.Mitchell, A. P. 1994. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol. Rev. 58:56-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitchell, A. P., S. E. Driscoll, and H. E. Smith. 1990. Positive control of sporulation-specific genes by the IME1 and IME2 products in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:2104-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohkuni, K., and I. Yamashita. 2000. A transcriptional autoregulatory loop for KIN28-CCL1 and SRB10-SRB11, each encoding RNA polymerase II CTD kinase-cyclin pair, stimulates the meiotic development of S. cerevisiae. Yeast 16:829-846. [DOI] [PubMed] [Google Scholar]
- 39.Percival-Smith, A., and J. Segall. 1986. Characterization and mutational analysis of a cluster of three genes expressed preferentially during sporulation of Saccharomyces cerevisiae. Mol. Cell. Biol. 6:2443-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roeder, G. S., and J. M. Bailis. 2000. The pachytene checkpoint. Trends Genet. 16:395-403. [DOI] [PubMed] [Google Scholar]
- 41.San-Segundo, P. A., and G. S. Roeder. 1999. Pch2 links chromatin silencing to meiotic checkpoint control. Cell 97:313-324. [DOI] [PubMed] [Google Scholar]
- 42.Sazer, S., and S. W. Sherwood. 1990. Mitochondrial growth and DNA synthesis occur in the absence of nuclear DNA replication in fission yeast. J. Cell Sci. 97:509-516. [DOI] [PubMed] [Google Scholar]
- 43.Shonn, M. A., R. McCarroll, and A. W. Murray. 2000. Requirement of the spindle checkpoint for proper chromosome segregation in budding yeast meiosis. Science 289:300-303. [DOI] [PubMed] [Google Scholar]
- 44.Smith, H. E., S. E. Driscoll, R. A. L. Sia, H. E. Yuan, and A. P. Mitchell. 1993. Genetic evidence for transcriptional activation by the yeast IME1 gene product. Genetics 133:775-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith, H. E., and A. P. Mitchell. 1989. A transcriptional cascade governs entry into meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 9:2142-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steber, C. M., and R. E. Esposito. 1995. UME6 is a central component of the developmental regulatory switch controlling meiosis-specific gene expression. Proc. Natl. Acad. Sci. USA 92:12490-12494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strich, R., M. R. Slater, and R. E. Esposito. 1989. Identification of negative regulatory genes that govern the expression of early meiotic genes in yeast. Proc. Natl. Acad. Sci. USA 86:10018-10022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strich, R., R. T. Surosky, C. Steber, E. Dubois, F. Messenguy, and R. E. Esposito. 1994. UME6 is a key regulator of nitrogen repression and meiotic development. Genes Dev. 8:796-810. [DOI] [PubMed] [Google Scholar]
- 49.Surosky, R. T., R. Strich, and R. E. Esposito. 1994. The yeast UME5 gene regulates the stability of meiotic mRNAs in response to glucose. Mol. Cell. Biol. 14:3446-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tatchell, K. 1986. RAS genes and growth control in Saccharomyces cerevisiae. J. Bacteriol. 166:364-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tung, K. S., E. J. Hong, and G. S. Roeder. 2000. The pachytene checkpoint prevents accumulation and phosphorylation of the meiosis-specific transcription factor Ndt80. Proc. Natl. Acad. Sci. USA 97:12187-12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vershon, A. K., N. M. Hollingsworth, and A. D. Johnson. 1992. Meiotic induction of the yeast HOP1 gene is controlled by positive and negative regulatory sites. Mol. Cell. Biol. 12:3706-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vidal, M., and R. F. Gaber. 1991. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:6317-6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vidal, M., R. Strich, R. E. Esposito, and R. F. Gaber. 1991. RPD1 (SIN3/UME4) is required for maximal activation and repression of diverse yeast genes. Mol. Cell. Biol. 11:6306-6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vincent, O., S. Kuchin, S. P. Hong, R. Townley, V. K. Vyas, and M. Carlson. 2001. Interaction of the Srb10 kinase with Sip4, a transcriptional activator of gluconeogenic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:5790-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wahi, M., and A. D. Johnson. 1995. Identification of genes required for alpha 2 repression in Saccharomyces cerevisiae. Genetics 140:79-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weinert, T. A., G. L. Kiser, and L. H. Hartwell. 1994. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 8:652-665. [DOI] [PubMed] [Google Scholar]
- 58.Yoshida, M., H. Kawaguchi, Y. Sakata, K. Kominami, M. Hirano, S. Harumasa, R. Akada, and I. Yamashita. 1990. Initiation of meiosis and sporulation in Saccharomyces cerevisiae requires a novel protein kinase homologue. Mol. Gen. Genet. 221:176-186. [DOI] [PubMed] [Google Scholar]