Abstract
Kindler syndrome is an autosomal recessive disorder characterized by neonatal blistering, sun sensitivity, atrophy, abnormal pigmentation, and fragility of the skin. Linkage and homozygosity analysis in an isolated Panamanian cohort and in additional inbred families mapped the gene to 20p12.3. Loss-of-function mutations were identified in the FLJ20116 gene (renamed “KIND1” [encoding kindlin-1]). Kindlin-1 is a human homolog of the Caenorhabditis elegans protein UNC-112, a membrane-associated structural/signaling protein that has been implicated in linking the actin cytoskeleton to the extracellular matrix (ECM). Thus, Kindler syndrome is, to our knowledge, the first skin fragility disorder caused by a defect in actin-ECM linkage, rather than keratin-ECM linkage.
Introduction
In 1954, Theresa Kindler described a 14-year-old English girl with unusual congenital blistering of her hands and feet (Kindler 1954). Later in childhood, the patient developed reticulate erythema and diffuse cutaneous atrophy, beginning in sun-exposed areas. Her gums bled readily, and the skin of the dorsa on her hands and feet had a thin, wrinkled appearance. She also had webbing between the second and third toes on both feet. By 10 years of age, the blistering and sun sensitivity had resolved, but the skin remained thin and fragile.
Thus far, the genetic basis of Kindler syndrome (MIM 173650) has not been delineated. Clinically, it resembles both inherited blistering skin disorders such as dystrophic epidermolysis bullosa (MIM 226600) and congenital poikilodermas such as Rothmund-Thomson syndrome (MIM 268400). Although Kindler syndrome is rare, with <100 cases reported in the world literature, we have identified a group of 26 Native American patients with Kindler syndrome who are members of the Ngöbe-Buglé tribe in the Bocas del Toro province, on the northwestern Caribbean coast of Panama. This corresponds to an incidence of ∼21/100,000 in this province, which has a population mostly of indigenous people living in small, isolated villages. In the present study, we have localized the gene responsible for Kindler syndrome to 20p12.3, by genomewide linkage analysis in the affected Panamanian subjects. We then confirmed the locus in individuals with Kindler syndrome from diverse geographic backgrounds, some of whom have been described previously (Wiebe et al. 1996; Shimizu et al. 1997; Senturk et al. 1999; Suga et al. 2000; Al Aboud et al. 2002). We have identified loss-of-function mutations in the causative gene, KIND1. Kindlin-1 protein is expressed in the epidermis, and in vitro it colocalizes with actin and vinculin in focal contacts. Here, we report an actin–extracellular-matrix (ECM) linkage protein deficiency causing an inherited skin fragility and photosensitivity syndrome.
Subjects and Methods
Affected Individuals
All affected individuals were referred to the present study by their primary dermatologists. Blood samples were obtained after informed consent from members of 24 families from Panama, the United States, Britain, Italy, Oman, Jordan, Turkey, Saudi Arabia, Afghanistan, Pakistan, and Japan, with the approval of the institutional review board at the University of California, San Francisco, and the Ethics Committee at St Thomas’ Hospital, London.
Genomewide Screen and Genetic Linkage Analysis
DNA was isolated from whole blood by using a commercial kit (Gentra). Genotypes were generated for a panel of 811 microsatellite dinucleotide markers (Weber and May 1989) by using fluorescently labeled PCR primers, under conditions recommended by the manufacturer (HD5, version 2.0; Applied Biosystems). The sizes of the PCR products for the markers were determined from electropherograms produced with an ABI 3700 DNA sequencer. The sizes of marker amplimers were determined blind to pedigree structure and diagnosis, using Genotyper (ABI). The sex-averaged marker map order was obtained from ABI, from Généthon, or by interpolation of the genomic sequence (see the UCSC Genome Bioinformatics Web site), assuming a uniform marker to physical distance map.
The program PedCheck was used to detect non-Mendelian inheritance (O’Connell and Weeks 1998). Two-point LOD scores were calculated using Vitesse (O’Connell and Weeks 1995). The subjects were genotyped for additional simple-sequence repeat markers from the linked region that were either from Research Genetics or were developed from genomic sequence data. These were evaluated for conserved haplotypes. Multipoint LOD scores were calculated for families in which there were at least two individuals affected or in which there was a known history of consanguinity, using Simwalk2 (Sobel and Lange 1996).
Cell Culture and RNA Extraction
Skin-biopsy samples for keratinocyte culture were subjected to dispase (Sigma-Aldrich) digestion in Dulbecco’s PBS (Gibco BRL), to separate the epidermis from the dermis, as described elsewhere (Bleck et al. 1999). Keratinocytes were isolated from the epidermis by incubation in 0.5% trypsin (Invitrogen) at 37°C for 10 min. These cells were plated with a mitomycin C (Sigma-Aldrich)–treated 3T3 feeder layer in keratinocyte growth medium, as described elsewhere (Navsaria et al. 1994). Total RNA was extracted from cultured keratinocytes by using the Qiagen RNeasy Kit (Qiagen). Total RNA quality was checked on a 1% agarose gel in 1 × morphopropane sulfonic acid buffer, to view the ribosomal subunits, 60s and 40s.
cDNA Synthesis
RT-PCR of control RNA was performed in two identical reactions each comprising 2.5 μg total RNA, 4 μl of 10 mM dNTPs, and 2 μl of 50 μM first-strand random primers with the addition of nuclease-free water to make a total of 16 μl, followed by denaturation at 85°C for 3 min in a thermal cycler. Samples were placed on ice, and 2 μl of 10 × RT-PCR buffer and 1 μl of RNasin (Promega) were added. To one sample, 100 U of M-MuLV reverse transcriptase (Promega) was added, and, to the other, 0.5 μl of water was added as a negative control. Samples were incubated in a thermal cycler at 42°C for 1 h, and cDNA was then stored at −20°C.
Multiple-Tissue Northern Blot
A 583-bp cDNA probe of the last 457 bp of the KIND1 ORF and the first 126 bp of the 3′ UTR was generated from control keratinocyte cDNA. The primers cDNA2F, 5′-ACACAAATCCAAACAGCTGGCC-3′, and cDNA2R, 5′-GTGACCAGCGGTGAATGTAT-3′, were used under standard PCR conditions (described below, in the “Multiple-Tissue cDNA Panels” subsection), at an annealing temperature of 62°C. The forward primer cDNA2F crosses the splice site between exons 12 and 13 of KIND1 and therefore is cDNA specific. Fifty microliters of probe was loaded on a 2% agarose gel against a 1-kb DNA ladder of known concentration (New England Biolabs UK) and was gel purified with Prep-A-Gene (Bio-Rad), ready for radiolabeling. Twenty-five nanograms of probe was radiolabeled by random priming (Invitrogen) with [32P]-labeled α-dCTP and was purified using the QIAquick Nucleotide Removal Kit (Qiagen). A multiple-tissue northern blot (Clontech) was hybridized in ExpressHyb solution (Clontech) with 1–2 × 106 cpm/ml of labeled probe, followed by washing, according to manufacturer’s instructions. In addition, a northern blot was made using total RNA extracted from control keratinocytes, blotted overnight onto a nitrocellulose membrane, and then hybridized in Church solution with 1–2 × 106 cpm/ml of labeled probe, followed by washing in Church wash, as described elsewhere (Church et al. 1994). Both blots were subsequently stripped and hybridized with a control human β-actin probe (Clontech), to ensure that the RNA signal intensity was the same in each lane.
Multiple-Tissue cDNA Panels
Primer pairs were designed to amplify cDNA fragments for KIND1, KIND2, and KIND3. Clontech human normalized multiple-tissue cDNA panels MTCI and -II, in addition to cDNA from control keratinocyte and fibroblast cultures, were amplified by RT-PCR. RT-PCR was performed in duplicate for the housekeeping gene, G3PDH (Clontech), to ensure consistent normalization. Negative cDNA controls from keratinocytes and fibroblasts were included and were synthesized as described above (see the “Multiple-Tissue Northern Blot” subsection). PCRs consisting of 5 ng of cDNA as template in a 50-μl reaction containing 0.4 μM of each primer, 1.5 mM of MgCl2, 20 μM of each nucleotide, and 2.5 U of AmpliTaq Gold polymerase (PE Biosystems) were cycled at 95°C for 5 min, followed by 95°C for 30 s, 60°C for 30 s, and 72°C for 75 s, for a total of 32 cycles. Aliquots of 5 μl were removed and analyzed after 26, 28, 30, and 32 cycles, enabling the evaluation of tissue-expression levels prior to the PCR plateau phase. The time point for the assessment of expression was set at 30 cycles.
Mutation Detection
Intronic primer pairs were designed to amplify individual exons and flanking splice sites of the KIND1 gene. Forward and reverse primer sequences are given in table 1. PCR was performed using 250 ng of patient and control genomic DNA as template in a 25-μl reaction that contained 1 μM of each primer, 1.5 mM of MgCl2, 50 μM of each nucleotide, and 1.25 U of Taq polymerase (PE Biosystems). Amplification conditions were 95°C for 5 min, followed by 95°C for 45 s, annealing temperature (58°C–60°C) for 45 s, and 72°C for 45 s for 35 cycles in a PE Biosystems 9700 thermal cycler. Five microliters of PCR product was checked on a 2% agarose gel prior to heteroduplex analysis by conformation-sensitive gel electrophoresis, as described elsewhere (Ganguly et al. 1993). PCR products from patients with known consanguinity were mixed with the PCR product amplified from control DNA, to enable heteroduplex formation. Products displaying aberrant band shifts were purified and sequenced using Big Dye Terminator Cycle Sequencing chemistry on an ABI 310 genetic analyzer (PE Biosystems).
Table 1.
Genomic Primers for PCR Amplification of KIND1
| Primer | Sequence | Position Before/After Exon | Annealing Temperature(°C) | Product Size(bp) |
| KIND1F | AAATCTGCAGACTGCGCCTC | −372a | ||
| KIND1R | GAGGCTGCAGAAAGAAAGGG | +101 | 60 | 473 |
| KIND2F | ATATCTGGAGCACCTGGAAC | −150 | ||
| KIND2R | ATTGCTCTCCAGGGCATTAC | +116 | 58 | 401 |
| KIND3F | TGAGGAGCTGGAGATCAGTT | −123 | ||
| KIND3R | GAAGTAGGCAGAATGCACAC | +83 | 58 | 416 |
| KIND4F | GACCCTGAGTCTTAGAAGGA | −67 | ||
| KIND4R | GCCTTTCCTCATCACATCAG | +129 | 58 | 343 |
| KIND5F | CAGTGCCCAGCTTGACTTAT | −144 | ||
| KIND5R | ATCCCTAGGCCTACCAACTT | +97 | 58 | 455 |
| KIND6F | CAGTGCTCAGAAAGTGTCAG | −186 | ||
| KIND6R | GCTAAACAGGCGATCACACA | +118 | 58 | 407 |
| KIND7F | CTGAGCTGAAGTTTGCTGCA | −117 | ||
| KIND7R | GTGTGTGGATTATGAGGAGC | +209 | 58 | 434 |
| KIND8F | AAGGAGACCTCTGTTTAGGA | −133 | ||
| KIND8R | CTTGTTAGGTGAAGAGCATC | +139 | 56 | 404 |
| KIND9F | GTAGCGAGTGTAAACTGAAG | −168 | ||
| KIND9R | ACCTTTGAACCATGAACCTG | +109 | 56 | 315 |
| KIND10F | TGCAGCGTGTTCCACATTTC | −60 | ||
| KIND10R | GGATTACAGGTTTGAGCCAC | +72 | 58 | 257 |
| KIND11F | ACAGATGCCTCAGAACTCAG | −46 | ||
| KIND11R | TGCTCTTAGGCTTAGTGGAG | +55 | 58 | 208 |
| KIND12F | GCTTTGCACTTGAGCTTGCT | −43 | ||
| KIND12R | GTGCTGGAATTACAGGTGTG | +110 | 58 | 375 |
| KIND13F | CTAACAGGGTGATCACAGAG | −66 | ||
| KIND13R | CTAAATGAGAAAACTGGGGCT | +78 | 56 | 269 |
| KIND14F | CTTCATTGTCCATTCCTCTG | −123 | ||
| KIND14R | CAATTCTGAGGGACACACAT | +60 | 58 | 328 |
| KIND15F | CCAGTCCAGCAAAGCACTTT | −74 | ||
| KIND15R | GTCCAGAATCTACATGCTGG | +95b | 58 | 343 |
Before end of exon.
After TGA.
Anti-Kindlin-1 Antibody Generation
Anti-rabbit polyclonal antibody was made using the 15 carboxyl-terminal amino acids of kindlin-1, which were cross-linked to keyhole limpet hemocyanin as immunogen (Moravian Biotech). The antiserum was affinity purified against the peptide by using the HiTrap Protein G HP kit (Amersham Biosciences UK), according to the manufacturer’s instructions, and was then concentrated using a 10-kDa Microcon-10 concentrator (Amicon).
Immunohistochemistry
Skin was washed in PBS for 30 min before being embedded in OCT compound (Agar Scientific) and was snap frozen in isopentane cooled by liquid nitrogen. Five-micrometer cryosections were fixed in 1:1 acetone and methanol at −20°C for 20 min and were then rehydrated in PBS for 2 × 15 min and incubated in 0.1% Triton-X-100 in PBS prior to blocking with 10% normal goat serum in PBS for 20 min. Sections were then incubated in the same well with primary kindlin-1 rabbit polyclonal antibody at 37°C for 1 h, followed by goat anti-rabbit Alexa Fluor 488 conjugate (Cambridge BioScience) for 1 h. The samples were thoroughly washed in PBS and were subsequently mounted on cover slips. Microscopy was performed using a Nikon Optiphot Microscope with Kodak Microscopy Document System 290.
Kindlin-1 Transfections
A sequence-verified RT-PCR clone in pCR2.1, encompassing the entire kindlin-1 cDNA, was subcloned into pEGFP-C2 (Clontech) for expression of a fusion protein consisting of kindlin-1 fused to the C-terminus of enhanced green fluorescent protein (hereafter, “EGFP-kindlin”). PtK2 cells (potoroo kidney epithelial cell line) were transfected with kindlin constructs using FuGene 6 transfection reagent (Roche Diagnostics). The cells were fixed 24 h posttransfection with 3% paraformaldehyde and were permeabilized using 0.2% Triton X-100 (BDH Chemicals). Fixed cells were stained with Alexa Fluor 594-phalloidin conjugate (1 U/ml) (Molecular Probes) or vinculin mouse monoclonal antibody (Sigma Chemicals) at 1:100 dilution and Alexa Fluor 594 secondary antibody (Cambridge BioScience). Images were taken on a Zeiss LSM510 META laser-scanning confocal microscope, using an α-Plan Fluar × 100 objective and an optical section of 0.7 μm.
Results
Patients with Kindler Syndrome from Bocas del Toro, Panama
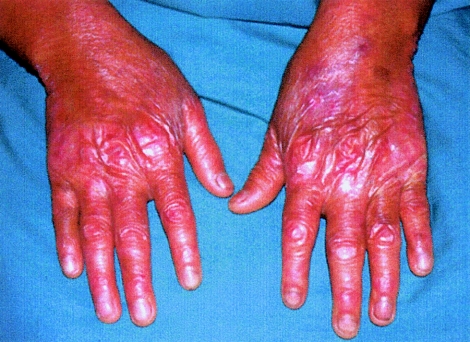
Patients in the Panamanian cohort that we studied have clinical findings quite similar to those of the patient whom Kindler (1954) originally studied. These include congenital acral blisters, blistering after trauma or sun exposure, erythema and itching after sun exposure, and patchy hyper- and hypopigmentation with atrophy and telangiectases (poikiloderma) developing in early childhood in both sun-exposed and nonexposed skin. Other features include hyperkeratosis of the palms and soles and diffuse cutaneous atrophy and wrinkling, particularly on the dorsa of the hands and feet (fig. 1). Other mucocutaneous features include periodontal disease, dental caries, and phimosis. Typically, the blistering and photosensitivity improve markedly in adulthood, but the poikiloderma persists. Some variability in phenotypic severity existed, particularly in the degree of photosensitivity, the age at onset of poikiloderma, and the degree of hyperkeratosis. All patients examined were <40 years of age.
Figure 1.
Clinical features. Atrophy, wrinkling, and abnormal pigmentation are shown on the hands of a Panamanian young adult with Kindler syndrome.
Linkage of Kindler Syndrome to 20p12.3
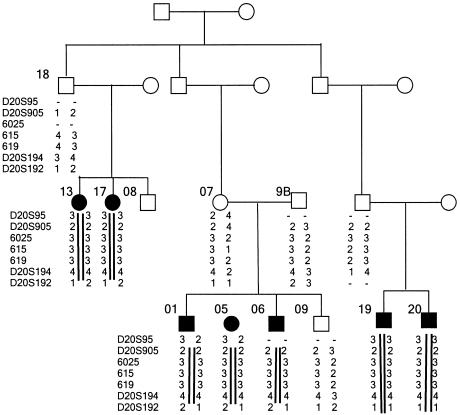
After excluding linkage to the RecQ1 and RecQ5 genes, candidates selected because of the phenotypic similarity of Kindler syndrome to Rothmund-Thomson syndrome and Bloom syndrome (MIM 210900), we performed a genomewide scan on DNA from 24 individuals from the Panamanian cohort—16 with Kindler syndrome and 8 unaffected family members. A cluster of markers (D20S846, D20S115, and D20S851) on 20p12.3 cosegregated with the disease, giving a maximum two-point LOD score of 2.48 at recombination fraction (θ) 0 with D20S846. False linkage was detected initially with D20S107 on 20q12 (Zmax = 2.59 at θ = 0) but subsequently was excluded by analysis of additional Panamanian families. We then genotyped further Panamanian individuals over a 3.6-Mb area surrounding D20S846. Observed recombinants and the shared haplotype narrowed the critical interval to a 0.6-cM area of homozygosity between D20S95 and D20S192 (Généthon genetic map), corresponding to 280 kb of genomic DNA (fig. 2). Patients with Kindler syndrome from consanguineous families from Italy, Oman, Jordan, Turkey, Afghanistan, and Saudi Arabia showed homozygosity across the same region, leading to a maximum multipoint LOD score of 11.7 at θ = 0 with D20S905.
Figure 2.
Haplotype analysis of a Panamanian family with Kindler syndrome. Genotype data are shown below the symbol for each affected individual. Haplotypes that delineate the Kindler syndrome critical interval are indicated by black bars. Markers 6025, 615, and 619 were derived from analysis of the sequence of the Human Genome Browser (UCSC Genome Bioinformatics).
Identification of the Kindler Syndrome Gene, KIND1
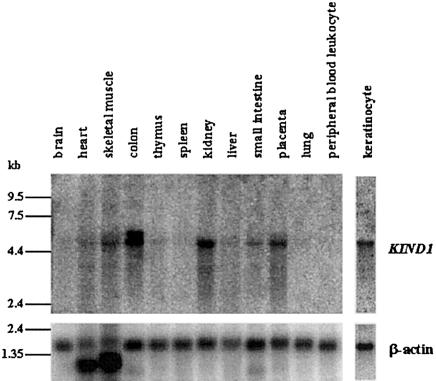
Six experimentally verified and/or strongly predicted genes were identified within the critical interval between D20S95 and D20S192, by study of the Human Genome Browser (UCSC Genome Bioinformatics). These were CHGB, LOC51605, MGC4816, LOC54675, FLJ20116, and BMP2. We sequenced the predicted exons for all these genes and identified mutations in only one of the six: FLJ20116, subsequently renamed “KIND1” (encoding the protein kindlin-1). The UCSC annotation for FLJ20116 predicted a gene of seven exons that encodes a predicted protein of 230 amino acids; however, RT-PCR of keratinocyte mRNA identified a 4,931-bp transcript encoded by a 15-exon gene, with the initiating methionine in exon 2 and a stop codon in exon 15. Northern blotting confirmed the latter, 4.9-kb mRNA as the major transcript (see the “Tissue Expression of KIND1” subsection, below; also see fig. 3). The gene spans 48.5 kb of genomic DNA, and exon sizes range from 47 to 234 bp. The predicted ORF is 2,034 bp, encoding a protein of 677 amino acids with a calculated molecular weight of 77.3 kDa. The predicted exon sizes and acceptor/donor splice sites (table 2) all were confirmed by RT-PCR.
Figure 3.
Multiple-tissue and keratinocyte northern blots hybridized with KIND1 and β-actin probes. Expression of a 4.9-kb KIND1 transcript is present predominantly in keratinocyte, colon, kidney, and placenta and at lower levels in heart, skeletal muscle, liver, and small intestine. An additional transcript, of ∼5.8-kb, is present in colon.
Table 2.
Genomic Organization of KIND1
| Exon | 3′ Acceptor Sequencea | Exon Size(bp) | 5′ Donor Sequencea | Intron Size(bp) |
| 1 | … | 771 | AAGGAGgtgggtgctc | 3,201 |
| 2 | tgttcagcagACACCA | 169 | AGATCAgtaagttact | 3,359 |
| 3 | atttttgcagATATAT | 234 | TCCTGAgtaagtaccc | 3,187 |
| 4 | ttatttctagATATTA | 147 | CATCAGgtaagacttg | 1,965 |
| 5 | tgccttgcagTAAGTC | 214 | TGCAGGgtaaggacac | 2,663 |
| 6 | cattttctagTTGGCT | 103 | CCTAAAgtaagcaact | 9,900 |
| 7 | ttgtctctagTATGAT | 108 | CTACAGgtatgggaac | 490 |
| 8 | ttcattttagTACCAC | 132 | CTTTTGgtatgaactt | 1,913 |
| 9 | tttttcttagGAGGAC | 50 | ATTTAGgtaagtaaac | 5,899 |
| 10 | gaatttgcagGCCCAA | 125 | TTAGAGgtaagagtac | 1,081 |
| 11 | tgtccttcagGCTGCG | 107 | GACCATgtgagtaaaa | 2,489 |
| 12 | tttgctgcagGAGAAT | 222 | AAACAGgtactgttaa | 901 |
| 13 | ttctgttcagCTGGCC | 125 | TGTCAGgtgattacaa | 4,462 |
| 14 | ttcttttcagATTTAA | 142 | CGGCAGgtaaagtgaa | 2,089 |
| 15 | cctcttccagGTGGTC | 2,282b | … |
Lowercase letters represent the intronic sequence, and uppercase letters represent the exonic sequence. The intronic acceptor/donor splice sites are shown in boldface italic.
Up to and including polyadenylation signal AATAAA.
Kindlin-1 Protein Structure
Analysis of the primary protein structure of the kindlin-1 polypeptide revealed a number of features predicting that the function of this molecule relates to anchorage of the actin cytoskeleton to the plasma membrane:
First, BLAST analysis (NCBI) revealed that kindlin-1 has C-terminal homology to talin and N-terminal homology to the Dictyostelium talin homolog, filopodin (fig. 4A). Both of these proteins are involved in anchorage of the actin cytoskeleton in focal contacts (Critchley 2000), which are membrane-substrate attachment structures observable in cultured cells (Zamir and Geiger 2001a, 2001b).
Second, Conserved Domain BLAST analysis showed that kindlin-1 possesses a centrally located pleckstrin homology (PH) domain (fig. 4A). PH domains mediate associations with specific phosphatidylinositol phospholipid species in the plasma membrane and are a feature of a number of cytoskeletal-associated and/or cell-signaling molecules (Maffucci and Falasca 2001; Itoh and Takenawa 2002; Lemmon et al. 2002).
Third, kindlin-1 also possesses two regions of homology with the FERM (filopodin and ezrin/radixin/moesin) domain, which is shared by erythrocyte protein 4.1, ERM (ezrin, radixin, and moesin) proteins, and a number of proteins that again mediate anchorage of the cytoskeleton to the plasma membrane (Chishti et al. 1998). In kindlin-1, the first FERM-domain homology region aligns to residues 49–133 of the 207-amino-acid FERM-domain consensus sequence (NCBI Conserved Domain Database); the second region aligns to residues 126–207 of the consensus sequence. Thus, kindlin-1 possesses an unusual, bipartite FERM domain that is interrupted by a PH domain. Both of these structural features imply that kindlin-1 is involved in membrane and/or cytoskeleton association.
Figure 4.
Kindlin-1 protein-domain organization. A, Schematic representation of the FERM, PH, and talin homology domains of kindlin. B, Amino acid sequence alignment between members of the human kindlin family and their C. elegans ortholog UNC-112. The overall amino acid sequence identity is high (41%) between the two species. C, Evolutionary tree analysis by UPGMA, showing that kindlin-1 and kindlin-2 are more related and more recently diverged from kindlin-3. Protein alignments and UPGMA analysis were performed by use of the Geneworks 2.5 analysis package (Oxford Molecular Group).
A Family of Kindlin-Related Proteins
Kindlin-1 shows closest homology (30% amino acid identity) to the Caenorhabditis elegans protein UNC-112 (fig. 4B). By a translated BLAST search of the human genome sequence, we identified two further proteins with significant overall homology to UNC-112. These were MIG-2 (with 42% amino acid identity) and the predicted protein MGC10966 (with 38% amino acid identity) (fig. 4B). The three human proteins and UNC-112 are very similar in size and share the same protein-domain organization and talin/filopodin homology regions (fig. 4A). This domain organization appears to be unique to the kindlin family of proteins (NCBI Conserved Domain Database and Pfam). Evolutionary analysis by the unweighted pair group method with arithmetic mean (UPGMA) (Nei 1987, pp. 293–298) revealed that kindlin-1 and MIG-2 are more closely related and more recently diverged from MGC10966 (fig. 4C). Overall, MIG-2 shows the closest amino acid similarity to UNC-112.
MIG-2 (mitogen-induced gene 2 [GenBank accession number Z24725]) has 62% amino acid identity with kindlin-1 and is located on 14q22.1. (Note that a C. elegans gene named “mig-2” [Zipkin et al. 1997] is neither structurally nor functionally related to the human MIG-2 gene [Wick et al. 1994]; therefore, to avoid confusion, we suggest that human MIG-2 be renamed “kindlin-2” [gene name “KIND2”] because of its close homology to kindlin-1.) MIG-2 was originally isolated by differential cDNA library screening as a serum-inducible gene expressed in human fibroblasts (Wick et al. 1994). The gene was thought to be encoded partly by an antisense transcript of the transcription factor gene TCF12 (also known as “HTF4”) (Wick et al. 1994); however, in agreement with published mapping data (Zhang et al. 1995), the near-complete human genome data localize TCF12 to 15q21.3, not 14q22.1. We found that the antisense strand of the 3′ UTR of MIG-2 indeed possesses regions of similarity to the TCF12 transcript; however, the most homologous area is only 42% identical. Furthermore, this homology is shared by the 3′ UTRs of the KIND1 and KIND3 transcripts. The functional significance, if any, of this antisense homology remains to be elucidated.
The other human homolog of KIND1 is the predicted gene MGC10966 (GenBank accession number NM_031471), located in a gene-dense region on 11q13.1. We propose that this gene, encoding the protein kindlin-3, be renamed “KIND3.” The predicted polypeptide is 49% identical to kindlin-1 and 53% identical to kindlin-2. Interestingly, this transcript has an additional region of antisense homology within its 3′ UTR. Part of the 3′ UTR is shared with the 3′ UTR of a gene located upstream and expressed from the opposite strand—the human ortholog of Tpt1h, the murine tRNA splicing 2′ phosphotransferase gene. The entire KIND3 and TPT1H genes exist within intronic sequences of the LRP16 gene (see the UCSC Genome Bioinformatics Web site).
Tissue Expression of KIND1
Hybridization of a KIND1 cDNA probe to a multiple-tissue northern blot showed tissue-specific expression. The KIND1 transcript is predicted to be 4.9 kb, and a transcript of this size was detected in cultured epidermal keratinocytes, colon, kidney, and placenta and at lower expression levels in heart, skeletal muscle, liver, and small intestine (fig. 3). Colon contains an additional transcript that is ∼5.8 kb. The June 2002 UCSC Human Genome Working Draft includes an intragenic EST found in colon (FLJ21712 [clone COL10231] [GenBank accession number AK025365]), which could represent a further translated exon between exons 8 and 9. Furthermore, the November 2002 UCSC Human Genome Working Draft also provides evidence for alternative exons 11 and 15.
Differential Expression of Human Kindlin Genes
Multiple-tissue cDNA panels showed significant differences in expression of the three kindlin genes (table 3). In particular, KIND1 is highly expressed in keratinocytes, with lower expression in prostate, ovary, colon, kidney, and pancreas and weak expression in spleen, thymus, testis, heart, brain, placenta, lung, liver, and fibroblasts. The high expression in cultured keratinocytes and the low expression in cultured fibroblasts were confirmed by quantitative PCR (data not shown). No expression is seen in small intestine, peripheral blood leukocytes, or skeletal muscle. KIND2 shows only moderate expression in spleen, prostate, testis, ovary, small intestine, colon, heart, placenta, lung, liver, kidney, pancreas, and fibroblasts and weak expression in thymus, brain, skeletal muscle, and keratinocytes. In contrast, KIND3 shows high expression in spleen, thymus, and peripheral blood leukocytes and moderate-to-low expression in prostate, testis, ovary, small intestine, colon, heart, brain, placenta, lung, liver, skeletal muscle, kidney, and pancreas. No expression of KIND3 was observed in cultured keratinocytes or fibroblasts.
Table 3.
RT-PCR Tissue-Expression Profiles of the KIND-Family Genes
|
Expressiona of |
|||
| KIND1 | KIND2 | KIND3 | |
| Spleen | + | ++ | +++ |
| Thymus | + | + | +++ |
| Prostate | ++ | ++ | + |
| Testis | + | ++ | + |
| Ovary | ++ | ++ | ++ |
| Small intestine | − | ++ | ++ |
| Colon | ++ | ++ | ++ |
| Leukocyte | − | − | +++ |
| Heart | + | ++ | + |
| Brain | + | + | + |
| Placenta | + | ++ | ++ |
| Lung | + | ++ | ++ |
| Liver | + | ++ | ++ |
| Skeletal muscle | − | + | + |
| Kidney | ++ | ++ | + |
| Pancreas | ++ | ++ | ++ |
| Keratinocyte | +++ | + | − |
| Fibroblast | + | ++ | − |
| Control cDNA | ++ | ++ | ++ |
− = No expression; + = weak expression; ++ = moderate expression; +++ = strong expression.
Detection of Loss-of-Function Mutations in KIND1
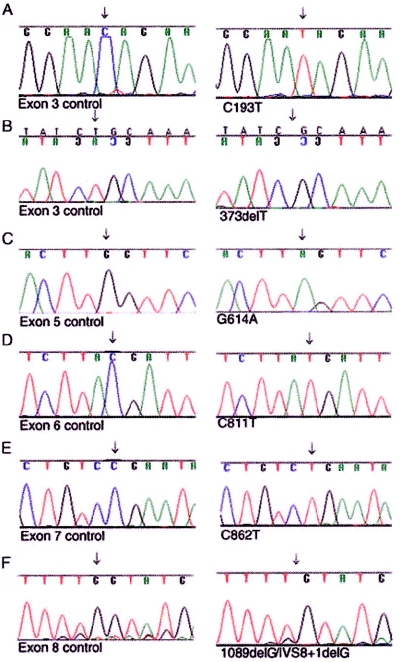
By direct nucleotide sequencing, we identified homozygous nonsense or frameshift mutations in 17 families with Kindler syndrome (table 4). All 26 patients from Bocas del Toro shared the same homozygous nonsense mutation, R271X (fig. 5D). This mutation also was identified in patients from three additional families (two white families from the United States and one Middle Eastern Omani family). Haplotype analysis of common KIND1 polymorphisms in these families indicated that the mutation likely arose on different genetic backgrounds in geographically diverse families (data not shown). A second recurrent nonsense mutation, R288X (fig. 5E), was identified in British and Turkish families, again embedded within different haplotypes. Details of the other pathogenic mutations are shown in table 4 and figures 5A–5F. All sequence variants were excluded in 100 ethnically matched control chromosomes by direct sequencing or restriction-endonuclease digestion.
Table 4.
Mutations Identified in Individuals with Kindler Syndrome[Note]
| Geographic Origin (No. of Patients) | Nucleotide Change | Predicted Consequencea | Exon |
| Jordan (2) | C193T | Q65X | 3 |
| Italy (1) | 373delT | I124fs | 3 |
| Pakistan (1) | G614A | W205X | 5 |
| Panama (26) | C811T | R271X | 6 |
| United States (2) | C811T | R271X | 6 |
| Oman (2) | C811T | R271X | 6 |
| Britain (1) | C862T | R288X | 7 |
| Turkey (1) | C862T | R288X | 7 |
| Japan (2) | 1089delG or IVS8+1delG (splice donor) | L363fs or IVS8+1delG (splice donor) | 8 |
Figure 5.
Nucleotide sequencing of DNA from patients with Kindler syndrome and unaffected control individuals, demonstrating pathogenic homozygous nonsense or frameshift mutations. A, Nucleotide transition 193C→T, predicting amino acid change Q65X, in two Jordanian siblings. B, Deletion mutation 373delT in an Italian patient. C, 614G→A, predicting W205X, in a Pakistani patient. D, 811C→T, predicting amino acid change R271X, in the Panamanian families, the Omani sibship, and two unrelated Americans. E, Transition mutation 862C→T, predicting R288X, in a British patient and a Turkish patient. F, Deletion mutation in two unrelated Japanese patients, representing either 1089delG or IVS8+1delG.
In five additional inbred families, homozygosity by descent was observed at the KIND1 locus; however, we were unable to detect mutations by our genomic PCR strategy. It is possible that these families carry promoter defects or other cryptic mutations. Two additional families (one from Canada [Haber and Hanna 1996] and the other from the United States, with one parent of European descent and the other of African descent) did not show linkage to the KIND1 locus; however, both of these families showed clinical differences from the Panamanian kindred (Haber and Hanna 1996). Thus, there may be a disorder that is clinically similar to but genetically different from Kindler syndrome.
Localization of Kindlin-1 in Skin
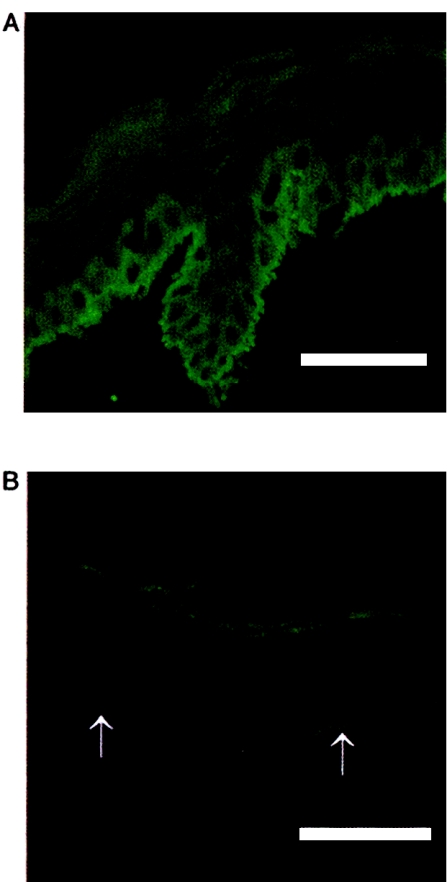
An affinity-purified polyclonal antibody was produced against a C-terminal synthetic peptide of kindlin-1. This stained normal control skin throughout the epidermis and, in particular, gave strong, cytoplasmic labeling of basal keratinocytes with a linear pattern at the dermal-epidermal junction (fig. 6A). In contrast, there was markedly reduced staining in skin from patients with Kindler syndrome (fig. 6B), and, specifically, staining of basal keratinocytes was virtually absent. There was no visible staining in either control or patient skin in the dermis. Thus, the kindlin-1 protein, as recognized by this antibody, appears to be localized solely within the epidermis and particularly in basal keratinocytes.
Figure 6.
Immunohistochemical changes in Kindler syndrome epidermis. A, Immunofluorescence labeling with affinity-purified anti-kindlin-1 polyclonal antibody in normal control skin reveals bright intracellular and cell-surface labeling within the epidermis, especially in the basal-keratinocyte layer. Linear labeling is seen at the dermal-epidermal junction. B, Skin from a patient with Kindler syndrome, showing almost-complete absence of immunostaining. The dermal-epidermal junction is indicated with arrows. Scale bar = 50 μm.
Localization of Kindlin-1 in Epithelial Cells
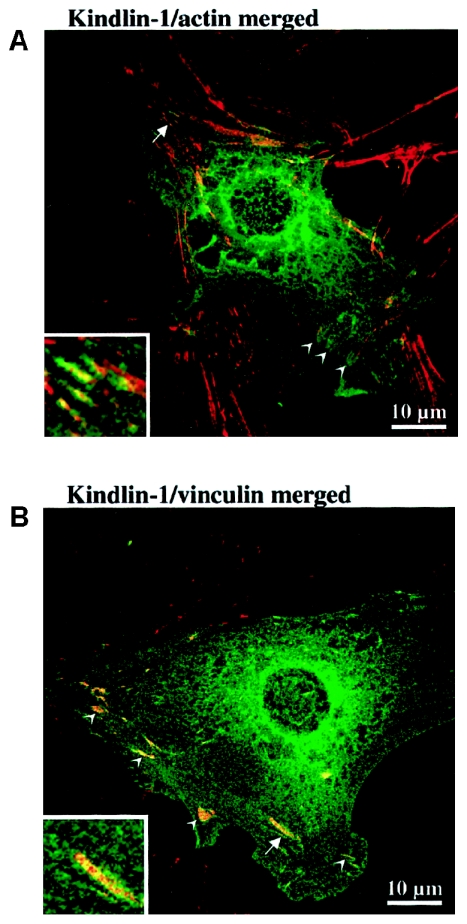
We transiently expressed full-length EGFP-kindlin-1 (i.e., EGFP fused to the N-terminus of kindlin-1) in epithelial cell line PtK2 and examined the resultant expression pattern by confocal laser-scanning microscopy. Cells expressing high levels of the fusion protein gave only diffuse staining patterns (data not shown). In some cells that transiently expressed moderate levels of EGFP-kindlin-1, we observed filamentous staining that colocalized with filamentous actin labeled with fluorescent phalloidin (fig. 7A). These actin filament bundles were seen to terminate in putative attachment structures reminiscent of focal contacts. In cells that expressed EGFP-kindlin-1 at very low levels, in which the EGFP background haze was much lower, these attachment structures could be seen more clearly, and EGFP-kindlin-1 colocalized with vinculin, a marker for focal contacts (fig. 7B). Localization to focal contacts was very reproducible but was not observed in all EGFP-expressing cells. These data indicate that kindlin-1 is likely a hitherto-unrecognized component of focal contacts directly or indirectly associated with filamentous actin within focal contacts. Identical colocalization data were obtained when EGFP-kindlin-1 was expressed in normal human primary keratinocytes (data not shown); however, because of their flat morphology, colocalization was seen more readily in PtK2 cells.
Figure 7.
Intracellular localization of kindlin-1 in epithelial cell line PtK2. A, Merged confocal laser-scanning micrograph of PtK2 cells, transiently transfected with EGFP-kindlin-1 (green) and stained with Alexa Fluor 594 phalloidin to reveal filamentous actin (red). The ends of actin stress fiber bundles exhibit coalignment with EGFP-kindlin-1 to produce the orange coloration in this merged image (arrowheads). These structures were reminiscent of focal contacts. The area marked by the arrow is enlarged in the inset, where the colocalization can be seen more easily. B, Merged confocal laser-scanning micrograph of PtK2 cells transiently transfected with EGFP-kindlin (green) and stained for vinculin by indirect immunofluorescence using Alexa Fluor 594 (red) to reveal focal contacts. Focal contacts (arrowheads) are seen to coalign with EGFP-kindlin to produce the orange coloration in this merged image. Some faint colocalization with cytoskeletal structures also can be seen. The area marked by the arrow is enlarged in the inset, where the colocalization can be seen more easily. These data confirmed that kindlin-1 is a component of focal contacts.
Discussion
We have showed that loss of epidermal kindlin-1 expression is the molecular basis of Kindler syndrome, an autosomal recessive genodermatosis characterized by skin fragility, poikiloderma, photosensitivity, and premature skin aging. Kindlin-1 is a protein of predicted molecular mass 77.3 kDa that contains regions of homology to talin and filopodin and has both a bipartite FERM domain and a PH domain, all of which are indicative of association with plasma-membrane adhesion structures and the actin cytoskeleton (Maffucci and Falasca 2001; Itoh and Takenawa 2002; Lemmon et al. 2002) (fig. 4A). Here, we have experimentally showed that kindlin-1 colocalizes with vinculin and therefore is a component of focal contacts (fig. 7B), which are structures involved in membrane-substratum attachment in cultured cells, and also can associate, to some extent, with filamentous actin (fig. 7A). We have identified two additional kindlinlike proteins from the human genome: kindlin-2 (MIG-2) and kindlin-3 (MGC10966). All three kindlins are closely related, in amino acid sequence and protein-domain organization, to the C. elegans focal-contact protein UNC-112 (Rogalski et al. 2000). This protein-domain configuration appears to be unique to the kindlin family.
In nematodes, loss-of-function mutations in UNC-112 give rise to the pat (paralyzed, arrested elongation at twofold) phenotype (Rogalski et al. 2000). This phenotype can result from mutations in molecules that anchor the actin-myosin cytoskeleton in the muscle via transmembrane complexes (especially α- and β-integrins) to the ECM—in particular, to the major heparin sulfate proteoglycan, perlecan. UNC-112 binds to a molecule with potential adapter and signaling functions—namely PAT-4, the worm homolog of integrin-linked kinase (ILK) (Mackinnon et al. 2002). During assembly of junctions of muscle cells to the ECM, PAT-4/ILK and UNC-112 function after integrins have marked the sites of the nascent junctions and before deposition of vinculin and attachment of actin filaments (Rogalski et al. 2000; Mackinnon et al. 2002). ILK has been shown to function primarily as a structural molecule by direct binding to the first 32 N-terminal residues of UNC-112, and kinase-deficient forms of ILK can rescue the pat-4 phenotype in worms (Mackinnon et al. 2002). However, the N-termini of all three human kindlin proteins lack homology to the ILK binding site of UNC-112 (fig. 4B). Therefore, this function may have been modified or lost in mammalian homologs. Ultrastructurally, in Kindler syndrome, there is marked disorganization of skin basement membrane, with reduplication of lamina densa and cleavage at or close to the dermal-epidermal junction (Shimizu et al. 1997). We too have found extensive basement-membrane reduplication, in several of the patients described here. Significantly, therefore, the loss of cytoskeletal-ECM adhesion noted in nematodes carrying mutations in UNC-112 would seem to be analogous to the adhesive defect observed in the skin of patients with Kindler syndrome. These morphological features underscore the significance of kindlin-1 in the construction/regulation of a normally functioning epidermal basement membrane.
The many forms of epidermolysis bullosa and related skin fragility diseases can be considered as analogous to the muscular dystrophies. Thus, genetic diseases of both organ systems can be caused by defects in any one component of a chain of molecules that connect the cytoskeleton to the ECM. In human inherited muscle diseases, the underlying genetic defects more commonly occur in a series of molecules that connect the actin-myosin cytoskeleton via actin-binding proteins (dystrophin) through a transmembrane complex (dystroglycans) to the basement membrane (laminin 2) and, subsequently, to the ECM (collagens such as collagen VI). Alternatively, defects in the muscle intermediate-filament protein desmin can cause myopathy (Munoz-Marmol et al. 1998). Similarly, in the skin, a series of proteins connect the basal-keratinocyte intermediate-filament cytoskeleton (keratins K5 and K14) through the hemidesmosome transmembrane complex (plectin, BPAG1, α6β4 integrin, and collagen XVII) to the basement membrane (laminin 5) and, subsequently, to the ECM (collagen VII). Interestingly, loss of plectin causes both muscle disease and skin blistering (McLean et al. 1996), a phenotype explicable by plectin’s participation in linking keratins to the inner plaque of hemidesmosomes in skin (Smith et al. 1996) and also in linking desmin within the Z-lines of striated muscle (Reipert et al. 1999). Kindler syndrome appears to complete the tetrad of actin-ECM and intermediate-filament–ECM linkage diseases of skin and muscle. The apparent lack of muscle disease in patients with Kindler syndrome suggests that the function performed by UNC-112 in C. elegans muscle presumably has been replaced in mammals by the dystrophin-dystroglycan system, in which a more specialized and robust linkage system may be needed to withstand the more demanding structural requirements of vertebrate muscle. Since kindlin-1 is very strongly expressed in keratinocytes but is absent from skeletal muscle (table 3) and since the other two kindlins are more widely expressed, it is possible that the latter make a more important contribution to actin-ECM linkage in muscle and other tissues. In particular, kindlin-3 is very strongly expressed in leukocytes, spleen, and thymus, so the main function of this protein may be immune system related (table 3).
More than 50 proteins have been shown to associate with focal contacts, where they perform structural and/or signaling functions (Zamir and Geiger 2001a, 2001b). It remains to be seen whether the main function of kindlin-1 is primarily that of structural tethering or whether it is that of regulation and/or recruitment of other molecules mediating actin-ECM adhesion. Here, we have observed that not all cells expressing EGFP-kindlin-1 localize this protein to focal contacts. One possible explanation is that kindlin-1 may have to associate with another factor(s) to enter focal contacts. In cells expressing very high levels of EGFP-kindlin-1, the cytoplasmic pool of this recruitment factor may become exhausted, preventing most of the putatively overexpressed EGP-kindlin-1 from associating with focal contacts. In view of this observation and the immensely complex molecular architecture of focal contacts (Zamir and Geiger 2001a, 2001b), the identification of protein and phospholipid binding partners of kindlin-1 and its relatives by yeast two-hybrid analysis, coimmunoprecipitation, and blot overlays is a logical next step in elucidating the function of this protein family.
In view of the skin blistering observed in younger patients with Kindler syndrome, it is tempting to speculate that kindlin-1 is a structural molecule, although this mechanical defect also could be secondary to a signaling or organizational/recruitment role. The extensive reduplication of basement membrane seen in the skin of these patients suggests that loss of kindlin-1 from membrane-bound attachment structures at the dermal-epidermal junction induces basal keratinocytes to oversecrete basement-membrane components. Since many proteins containing PH domains are involved in signal transduction (Maffucci and Falasca 2001), kindlin-1 could play a regulatory role in the inhibition of basement-membrane synthesis.
One unexplained clinical phenomenon is the waning of blistering in older children, although such changes are well recognized in other inherited skin fragility disorders, especially autosomal dominant cases of epidermolysis bullosa simplex resulting from mutations in keratin 14 or keratin 5 (Smith 2003). However, spontaneous improvement is rare in autosomal recessive blistering skin disorders. Mechanistically, in Kindler syndrome, it is possible that the reliance of newborn epidermis on the actin-ECM and intermediate-filament–ECM associations is more balanced but that, by adulthood, the contribution of the former lessens. Genotype-phenotype correlations are difficult to assess, because of the variability of clinical severity both between patients at different ages and among patients within the same family with the same mutation. More puzzling is the nature of the link between loss of kindlin-1 protein and the pathogenesis of other clinical findings—in particular, the photosensitivity and skin atrophy, resembling findings in helicase-deficient diseases, such as Rothmund-Thomson syndrome and Bloom syndrome. Future functional studies will help elucidate the mechanisms causing these interesting phenotypic abnormalities.
Acknowledgments
We are grateful to the patients for their participation in this study. The work was supported by an Individual National Research Service Award from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (AR 123456) and a fellowship from the Society for Pediatric Dermatology (to D.H.S.), Action Research, the Dystrophic Epidermolysis Bullosa Research Association (UK) and the British Skin Foundation. W.H.I.M. and F.J.D.S. are funded by a Wellcome Trust Senior Research Fellowship (to W.H.I.M.). The Centre for High-Resolution Imaging and Processing, Dundee, is supported by the Medical Research Council and the Wellcome Trust. We thank Galderma, for the generous donation of sunscreen for patients in Panama, and Christina Walker and Fintan Coleman, for their help in preparation of the manuscript.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for KIND1 [accession number AY137240], MIG-2 [accession number Z24725], MGC10966 [accession number NM_031471], and FLJ21712 [clone COL10231] [accession number AK025365])
- NCBI Conserved Domain Database, http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd/shtml
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for Kindler syndrome, dystrophic epidermolysis bullosa, Rothmund-Thomson syndrome, and Bloom syndrome)
- Pfam, http://www.sanger.ac.uk/Software/Pfam/search.shtml
- UCSC Genome Bioinformatics, http://genome.ucsc.edu/ (for Human Genome Browser)
References
- Al Aboud K, Al Hawsawi K, Al Aboud D, Al Githami A (2002) Kindler syndrome in a Saudi kindred. Clin Exp Dermatol 27:673–676 [DOI] [PubMed] [Google Scholar]
- Bleck O, Abeck D, Ring J, Hoppe U, Vietzke JP, Wolber R, Brandt O, Schreiner V (1999) Two ceramide subfractions detectable in Cer(AS) position by HPTLC in skin surface lipids of non-lesional skin of atopic eczema. J Invest Dermatol 113:894–900 [DOI] [PubMed] [Google Scholar]
- Chishti AH, Kim AC, Marfatia SM, Lutchman M, Hanspal M, Jindal H, Liu SC, et al (1998) The FERM domain: a unique module involved in the linkage of cytoplasmic proteins to the membrane. Trends Biochem Sci 23:281–282 [DOI] [PubMed] [Google Scholar]
- Church DM, Stotler CJ, Rutter JL, Murrell JR, Trofatter JA, Buckler AJ (1994) Isolation of genes from complex sources of mammalian genomic DNA using exon amplification. Nat Genet 6:98–105 [DOI] [PubMed] [Google Scholar]
- Critchley DR (2000) Focal adhesions—the cytoskeletal connection. Curr Opin Cell Biol 12:133–139 [DOI] [PubMed] [Google Scholar]
- Ganguly A, Rock MJ, Prockop DJ (1993) Conformation-sensitive gel electrophoresis for rapid detection of single-base differences in double-stranded PCR products and DNA fragments: evidence for solvent-induced bends in DNA heteroduplexes. Proc Natl Acad Sci USA 90:10325–103329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber RM, Hanna WM (1996) Kindler syndrome: clinical and ultrastructural findings. Arch Dermatol 132:1487–1490 [DOI] [PubMed] [Google Scholar]
- Itoh T, Takenawa T (2002) Phosphoinositide-binding domains: functional units for temporal and spatial regulation of intracellular signalling. Cell Signal 14:733–743 [DOI] [PubMed] [Google Scholar]
- Kindler T (1954) Congenital poikiloderma with traumatic bulla formation and progressive cutaneous atrophy. Br J Dermatol 66:104–111 [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM, Abrams CS (2002) Pleckstrin homology domains and the cytoskeleton. FEBS Lett 513:71–76 [DOI] [PubMed] [Google Scholar]
- Mackinnon AC, Qadota H, Norman KR, Moerman DG, Williams BD (2002) C. elegans PAT-4/ILK functions as an adaptor protein within integrin adhesion complexes. Curr Biol 12:787–797 [DOI] [PubMed] [Google Scholar]
- Maffucci T, Falasca M (2001) Specificity in pleckstrin homology (PH) domain membrane targeting: a role for a phosphoinositide-protein co-operative mechanism. FEBS Lett 506:173–179 [DOI] [PubMed] [Google Scholar]
- McLean WHI, Pulkkinen L, Smith FJD, Rugg EL, Lane EB, Bullrich F, Burgeson RE, Amano S, Hudson DL, Owaribe K, McGrath JA, McMillan JR, Eady RAJ, Leigh IM, Christiano AM, Uitto J (1996) Loss of plectin causes epidermolysis bullosa with muscular dystrophy: cDNA cloning and genomic organization. Genes Dev 10:1724–1735 [DOI] [PubMed] [Google Scholar]
- Munoz-Marmol AM, Strasser G, Isamat M, Coulombe PA, Yang Y, Roca X, Vela E, Mate JL, Coll J, Fernandez-Figueras MT, Navas-Palacios JJ, Ariza A, Fuchs E (1998) A dysfunctional desmin mutation in a patient with severe generalized myopathy. Proc Natl Acad Sci USA 95:11312–11317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navsaria H, Sexton C, Bouvard V, Leigh I (1994) Growth of keratinocytes with a 3T3 feeder layer: basic techniques. In: Leigh I, Watt F (eds) Keratinocyte methods. Cambridge University Press, Cambridge, UK, pp 5–12 [Google Scholar]
- Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York [Google Scholar]
- O’Connell JR, Weeks DE (1995) The VITESSE algorithm for rapid exact multilocus linkage analysis via genotype set-recoding and fuzzy inheritance. Nat Genet 11:402–408 [DOI] [PubMed] [Google Scholar]
- ——— (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reipert S, Steinbock F, Fischer I, Bittner RE, Zeold A, Wiche G (1999) Association of mitochondria with plectin and desmin intermediate filaments in striated muscle. Exp Cell Res 252:479–491 [DOI] [PubMed] [Google Scholar]
- Rogalski TM, Mullen GP, Gilbert MM, Williams BD, Moerman DG (2000) The UNC-112 gene in Caenorhabditis elegans encodes a novel component of cell–matrix adhesion structures required for integrin localization in the muscle cell membrane. J Cell Biol 150:253–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senturk N, Usubutun A, Sahin S, Bukulmez G, Erkek E, Topaloglu R, Akan T (1999) Kindler syndrome: absence of definite ultrastructural feature. J Am Acad Dermatol 40:335–337 [DOI] [PubMed] [Google Scholar]
- Shimizu H, Sato M, Ban M, Kitajima Y, Ishizaki S, Harada T, Bruckner-Tuderman L, Fine JD, Burgeson R, Kon A, McGrath JA, Christiano AM, Uitto J, Nishikawa T (1997) Immunohistochemical, ultrastructural, and molecular features of Kindler syndrome distinguish it from dystrophic epidermolysis bullosa. Arch Dermatol 133:1111–1117 [PubMed] [Google Scholar]
- Smith FJD (2003) The molecular genetics of keratin disorders. Am J Clin Dermatol 4:347–364 [DOI] [PubMed] [Google Scholar]
- Smith FJD, Eady RAJ, Leigh IM, McMillan JR, Rugg EL, Kelsell DP, Bryant SP, Spurr NK, Geddes JF, Kirtschig G, Milana G, de Bono AG, Owaribe K, Wiche G, Pulkkinen L, Uitto J, McLean WHI, Lane EB (1996) Plectin deficiency results in muscular dystrophy with epidermolysis bullosa. Nat Genet 13:450–457 [DOI] [PubMed] [Google Scholar]
- Sobel E, Lange K (1996) Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet 58:1323–1337 [PMC free article] [PubMed] [Google Scholar]
- Suga Y, Tsuboi R, Hashimoto Y, Yaguchi H, Ogawa H (2000) A Japanese case of Kindler syndrome. Int J Dermatol 39:284–286 [DOI] [PubMed] [Google Scholar]
- Weber JL, May PE (1989) Abundant class of human DNA polymorphisms which can be typed using the polymerase chain reaction. Am J Hum Genet 44:388–396 [PMC free article] [PubMed] [Google Scholar]
- Wick M, Bürger C, Brüsselbach S, Lucibello FC, Müller R (1994) Identification of serum-inducible genes: different patterns of gene regulation during G0→S and G1→S progression. J Cell Sci 107 Pt 1:227–239 (erratum 107[Pt 3]:precedi) [DOI] [PubMed] [Google Scholar]
- Wiebe CB, Silver JG, Larjava HS (1996) Early-onset periodontitis associated with Weary-Kindler syndrome: a case report. J Periodontol 67:1004–1010 [DOI] [PubMed] [Google Scholar]
- Zamir E, Geiger B (2001a) Components of cell-matrix adhesions. J Cell Sci 114:3577–3579 [DOI] [PubMed] [Google Scholar]
- ——— (2001b) Molecular complexity and dynamics of cell-matrix adhesions. J Cell Sci 114:3583–3590 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Flejter WL, Barcroft CL, Riviere M, Szpirer J, Szpirer C, Bina M (1995) Localization of the human HTF4 transcription factors 4 gene (TCF12) to chromosome 15q21. Cytogenet Cell Genet 68:235–238 [DOI] [PubMed] [Google Scholar]
- Zipkin ID, Kindt RM, Kenyon CJ (1997) Role of a new Rho family member in cell migration and axon guidance in C. elegans. Cell 90:883–894 [DOI] [PubMed] [Google Scholar]