Abstract
Mutations in TBX5, a T-box–containing transcription factor, cause cardiac and limb malformations in individuals with Holt-Oram syndrome (HOS). Mutations that result in haploinsufficiency of TBX5 are purported to cause cardiac and limb defects of similar severity, whereas missense mutations, depending on their location in the T box, are thought to cause either more severe heart or more severe limb abnormalities. These inferences are, however, based on the analysis of a relatively small number of independent cases of HOS. To better understand the relationship between mutations in TBX5 and the variable expressivity of HOS, we screened the coding and noncoding regions of TBX5 and SALL4 for mutations in 55 probands with HOS. Seventeen mutations, including six missense mutations in TBX5 and two mutations in SALL4, were found in 19 kindreds with HOS. Fewer than 50% of individuals with nonsense or frameshift mutations in TBX5 had heart and limb defects of similar severity, and only 2 of 20 individuals had heart or limb malformations of the severity predicted by the location of their mutations in the T box. These results suggest that neither the type of mutation in TBX5 nor the location of a mutation in the T box is predictive of the expressivity of malformations in individuals with HOS.
Introduction
Congenital heart defects are the most frequent birth defect observed in humans and the most common cause of death in infancy (Guyer et al. 1998). Although the etiology of most heart defects is unknown, many of them are thought to have a genetic basis (Burn et al. 1998; Loffredo 2000). Consequently, understanding the molecular basis of heart malformations has been a major focus of biomedical research over the past decade. To this end, investigators have sought to gain insights about the genetic etiology of cardiac malformations by identifying genes underlying rare, Mendelian syndromes in which heart defects are a major feature. A prototypical example of such a condition is Holt-Oram syndrome (HOS [MIM 142900]), a disorder characterized by limb as well as heart malformations. Most individuals with HOS have either a secundum atrial septal defect (ASD) or a ventricular septal defect (VSD), although many other cardiac malformations have been reported, ranging from mitral valve prolapse to hypoplastic left heart (Newbury-Ecob et al. 1996; Sletten and Pierpont 1996; Bruneau et al. 1999).
In 1997, HOS was discovered to be caused by mutations in TBX5 (Basson et al. 1997; Li et al. 1997), one of a family of transcription factors, called “T-box” genes, that have been associated with a variety of malformations (Bamshad et al. 1997; Braybrook et al. 2001; Merscher et al. 2001). Most of the mutations in TBX5 were predicted to encode, if translated, a truncated protein that lacked a functional DNA-binding domain. These mutations often were found in individuals in whom the heart and limbs were affected with similar severity. This led to the inference that most cases of HOS were caused by haploinsufficiency of TBX5, which, in turn, disrupted both limb and heart development to a similar extent.
In several kindreds with HOS, individuals who had heart and limb defects of disparate severity were found to have missense mutations, suggesting that, in some cases, HOS might be caused by a dominant-negative mechanism (Basson et al. 1994, 1997). This observation led to the proposal that missense mutations predicted to disturb different regions of the T box might be associated with organ-specific defects (Basson et al. 1999). Specifically, an amino acid substitution in the N-terminal end of the T box (i.e., Gly80Arg), predicted to affect TBX5 binding to the major groove of target DNA, was found to cause severe heart malformations but relatively mild limb defects in a single, large kindred. Alternatively, two amino acid substitutions in the C-terminal end of the T box (Arg237Gln and Arg237Trp), which were predicted to perturb TBX5 binding to the minor groove of target DNA, were found to cause mild cardiac abnormalities but severe limb defects.
These findings suggested that TBX5 genotypes might be predictive of the expressivity of HOS, a compelling interpretation for several reasons. First, it implied that different regions of the T box might impart functional specificity to TBX5 (Schneider and Schwartz 1999). This inference was reinforced by transactivation studies that demonstrated that Tbx5 mutants with a Gly80Arg substitution associated with severe heart malformations failed to activate atrial natriuretic factor (ANF), whereas Tbx5 mutants with an Arg237Gln substitution activated ANF to a level similar to wild-type Tbx5 (Hiroi et al. 2001). Second, it suggested that genetic counseling of individuals with HOS might be facilitated by making inferences about anticipatory guidance, based on TBX5 genotypes. Third, it increased the plausibility that some mutations in the T box of TBX5 might cause isolated heart malformations, particularly ASDs or VSDs.
For several reasons, it is not clear that TBX5 genotypes are predictive of the expressivity or the extent of pleiotropy in HOS. First, all of the study subjects with HOS caused by a Gly80Arg substitution reported by Basson et al. (1999) were members of the same kindred, and some of the individuals with Arg237Gln or Arg237Trp were related to one another. Thus, the suggested genotype-phenotype correlation was based on only a few independent cases. Second, only several other missense mutations in the T box of TBX5 have been reported to date, and no study has compared TBX5 genotypes to HOS phenotypes in a large cohort of cases. Third, more recent studies of the DNA-binding characteristics of mutant Tbx5 proteins have demonstrated that Tbx5 mutants with either the Gly80Arg or the Arg237Gln substitution failed to bind to a preferred target DNA sequence identified via an in vitro selection procedure (Ghosh et al. 2001; Fan et al. 2003). Therefore, if neither mutant binds the target DNA of TBX5, it is unclear how missense mutations in different regions of the T box of TBX5 might cause heart and limb defects of disparate severity.
To further explore the relationship between TBX5 genotypes and HOS phenotypes, we screened the coding and noncoding regions of TBX5 for mutations in 55 probands with HOS—to our knowledge the largest collection of patients with HOS studied to date. Fourteen different mutations, including six missense mutations, were found in 17 kindreds in which affected individuals spanned the spectrum of HOS phenotypes. We found no evidence that either the type of mutation or the location of a mutation in TBX5 was predictive of the severity of limb or heart malformations in a patient with HOS. Only two mutations in TBX5 were found in probands with phenotypes atypical of HOS (i.e., individuals with heart and limb defects accompanied by physical findings—e.g., situs inversus—not commonly found in patients with HOS), and novel mutations in SALL4 were found in an additional two of these atypical cases.
Material and Methods
Clinical Status
All studies were performed with the approval of the institutional review board of the University of Utah and the general counsel of the Shriners Hospitals for Children. After obtaining informed consent, most living members of each kindred were evaluated by review of their medical history, physical examination, radiographs of the upper limbs, electrocardiogram, and echocardiogram. Heart and limb malformations were classified as mild or severe according to criteria established elsewhere (Basson et al. 1999). In brief, individuals with unilateral or bilateral hypoplasia or aplasia of both the humerus and radius were categorized as having severe limb defects; otherwise, affected individuals were considered to have mild limb defects. Individuals with multiple septation defects (e.g., an ASD and a VSD), a single septal defect plus another cardiac anomaly, or complex congenital heart disease were categorized as having severe heart defects. Individuals with a single septal defect or only an electrocardiographic abnormality were categorized as having a mild heart defect. All subjects included in the analysis had normal karyotypes.
Mutation Analysis
Genomic DNA was extracted from peripheral lymphocytes through use of standard techniques. DNA sequences were amplified using 25 ng genomic DNA as a template in 1× buffer (1.5 mM MgCl2; 10 mM Tris.Cl, pH 8.3; 20% Q solution [Qiagen]), 0.5 U QIAGEN hotstar Taq polymerase, and 10 pmol of each primer. Samples were cycled 30 times in an MJ Research DNA Engine Tetrad, using a standard three-step PCR profile with an initial denaturing step at 94°C for 15 min and a final extension step at 72°C for 10 min. Annealing temperatures and primer sequences can be found in table A (online only). PCR products were purified by size exclusion or gel extraction using a QIAquick column. Purified PCR products were sequenced using ABI BigDye terminator cycle sequencing version 2.0 reagent. Sequenced products were loaded on an ABI 377 automated sequencer and were analyzed by Sequencing Analysis 3.4.1 and SEQUENCHER 4.1 software (Genecodes). The forward and reverse strand of exons 1–9 of TBX5, including the flanking splice-recognition sequences, were analyzed. Complete sequence data were available for 49 of 55 probands.
Table A.
Primers Used for Amplification and Sequencing of TBX5 Exons 1–9 and SALL4 Exons 1–4
| Gene, Exon,and Primer | Sequence | PCRProductSize(bp) | AnnealingTemperature(°C) |
| TBX5: | |||
| 1: | |||
| ab-TBX5-1FPa,b | 5′ AAGAGGGCACTGAGTTATCGCATC 3′ | 832 | 55 |
| ab-TBX5-1F1b | 5′ CTGATAGGCGAAGACGGAGAGAAA 3′ | ||
| ab-TBX5-1RPa,b | 5′ GAGACGTCACGAGTCACGCAACCG 3′ | ||
| ab-TBX5-1R1b | 5′ GACTAAGACGGGGTGAAAAGCCAA 3′ | ||
| ab-TBX5-1R2 | 5′ GGGAATAAATAAAGACATAAACCA 3′ | ||
| 2: | |||
| TBX5-exon2Fa,b | 5′ GTGCTCTCCAACCTTTCTCTCGT 3′ | 333 | 62 |
| 2GRa,b,d | 5′ CAAGAGAAGCCGAGCAGGAAAGCCA 3′ | ||
| 3: | |||
| 3GFa,b,d | 5′ AGTTTGGGGAAGGAATGCCCACTAC 3′ | 200 | 62 |
| 3GRa,b,d | 5′ TTCTCCTCGTCCCTCTCTCTACACA 3′ | ||
| 4: | |||
| 4FPa,b,d | 5′ AACGGGGCTAGTTTCCGCTTCCACG 3′ | 315 | 62 |
| 4GRa,b,d | 5′ CTTTTCAACTTTTTGGGAGAAGGTTCCACTTTTC 3′ | ||
| TB5-K5302-AseI-Fc | 5′ GCCGACGATCACAGATACAAATTCGCAGATAATTA 3′ | 107 | 59 |
| TB5-K5302-AseI-Rc | 5′ CAACTTTTTGGGAGAAGGTTCCAC 3′ | ||
| 5: | |||
| TBX5-exon5Fa,b | 5′ AGATACCTAAGGGAGACGGGA 3′ | 450 | 62 |
| TBX5-exon5Ra | 5′ TAGAGGCAGAAAGCGACGAAAGTG 3′ | ||
| TBX5-exon5R2b | 5′ AGAGAGGACAAGAGGGAGACAAGGC 3′ | ||
| 6: | |||
| TBX5-exon6Fa | 5′ GAGCCGATATAACAAGGCGAAT 3′ | 377 | 66 |
| TBX5-exon6F2b | 5′ GCGGGGAGCAGGGTTTTA 3′ | ||
| TBX5-exon6Ra,b | 5′ GTCGAAGTTGGTGACTGCTGC 3′ | ||
| Ser196Ter-AlwI-F2c | 5′ CATCGTGAAAGCGGATGAAAATAATGGATTTGGAT 3′ | 125 | |
| Ser196Ter-AlwI-Rc | 5′ GCTTCAGGCTTACCTTGTGGTTCT 3′ | ||
| 7: | |||
| TBX5-exon7Fa,b | 5′ TTAATTTGCTTCTTTTGGTTGCCA 3′ | 203 | 66 |
| TBX5-exon7Ra,b | 5′ GGGTATGTGGGGAGGAGAAAGTT 3′ | ||
| Thr223Met-NsiI-Fc | 5′ GATATTTATTATTAGCTCATGTCC 3′ | 126 | 54 |
| Thr223Met-NsiI-Rc | 5′ CCTTTGGCAAAGGGATTATTCTCAATCTTTAAATG 3′ | ||
| TB5-K3598-EcoRI-Fc | 5′ TTAAAGATTGAGAATAATCCCTTTGCCAAAGGAAT 3′ | 116 | 59 |
| TB5-K3598-EcoRI-Rc | 5′ GGGAGGAGAAAGTTGAGGAATCCA 3′ | ||
| 8: | |||
| 8GFa,d | 5′ GTATCAGGGCACTGATAGGCGTT 3′ | 515 | 62 |
| TBX58F3a,b | 5′ CTGGTGGATTCTCTCACACCTGG 3′ | ||
| 8GRa,d | 5′ GGGTAGGAACATGTCAACGGAACT 3′ | ||
| TBX58R3a,b | 5′ ACTGGGGGTAGGAACATGTCAAGG 3′ | ||
| 9: | |||
| 9FPa,b,e | 5′ TACTTTGGCCAAATAACTGTCTCC 3′ | 707 | 48 |
| ab-TBX5-9F3b | 5′ AGCCTGTGCCCAGCCTAGAGGACA 3′ | ||
| ab-TBX5-9R2b | 5′ CCAGCCAGCCGAGGGACCAGGGGC 3′ | ||
| ab-TBX5-x9tera,b | 5′ TCTCTCTCTCTCTTTCTCTAGGAAA 3′ | ||
| SALL4: | |||
| 1: | |||
| SALL4-1Fa,b | 5′ ATTACTGGGACATGCGCGTTC 3′ | 361 | 65 |
| SALL4-1Ra,b | 5′ AAATCTCGGCTCCTGAATTTGCG 3′ | ||
| 2: | |||
| SALL4-2(1)Fa,b | 5′ GTGCTAGGATTATAGATGTGAGCG 3′ | 683 | 65 |
| SALL4-2(1)Ra,b | 5′ ATGTTCACCTGGATGCGGATCTGC 3′ | ||
| SALL4-2(2)a,b | 5′ GTGCCTGGTGCCAACAGCATCC 3′ | 731 | 65 |
| SALL4-2(2)Ra,b | 5′ TTGTCCTGGAACTCGGCAAACAG 3′ | ||
| SALL4-2(3)Fa,b | 5′ CCGCTCCCACACTGGAGAGAGA 3′ | 681 | 68 |
| SALL4-2(3)Ra,b | 5′ CACGAATGCTGCGTCTTAATGGAT 3′ | ||
| SALL4-2(4)Fa,b | 5′ GGTAACCTGAAGACACACCTTG 3′ | 719 | 65 |
| SALL4-2(4)Ra,b | 5′ GTAAAGTTCAACCCAGGCTCCTT 3′ | ||
| SALL4-2(1)del-Fa,b | 5′ CGGATGCGGAGTCTGTGGTGTAC 3′ | 507 | 57 |
| SALL4-2(1)del-Ra,b | 5′ TCAGAGTGAAGGGTGCCAGCC 3′ | ||
| 3: | |||
| SALL4-3Fa,b | 5′ GCTTTGAAGAGAAAGAATGAAG 3′ | 379 | 60 |
| SALL4-3Ra,b | 5′ AAGACACCTGGTGCCTAGCCC 3′ | ||
| 4: | |||
| SALL4-4Fa,b | 5′ GGCTTGCCAGTGAGCTTCAAATCT 3′ | 539 | 60 |
| SALL4-4Ra,b | 5′ AAAGCAGATTCTAGAGATTTCAC 3′ |
The presence of each missense and nonsense mutation was confirmed in each affected individual by either restriction digestion, performed according to manufacturers’ instructions, or hybridization to an allele-specific oligonucleotide (see table A [online only]). For each mutation, 150 chromosomes from unrelated, unaffected individuals from a matched ethnic background were also screened. Small deletions and insertions were assessed in at least one affected individual in each kindred by transforming DH5a cells (Invitrogen) with a ligation mixture containing the PCR-amplified product and pCR2.1 plasmid (Invitrogen TA cloning kit). Plasmid DNA was extracted from 10 transformed clones through use of QIAGEN miniprep columns and was subjected to direct sequencing as described above.
Results
Fourteen different mutations in TBX5 were found in 17 (35%) of 49 probands with putative diagnoses of HOS, and 12 of these mutations were novel (fig. 1). Either of two mutations (Thr223Met and Arg237Trp) was found in five unrelated kindreds with HOS. Mutations were detected in 9 of 17 familial cases (53%) of HOS, whereas we were able to identify a mutation in TBX5 in only 8 of 32 sporadic cases (25%). Thus, the rate of detection in familial cases was approximately twice that in sporadic cases.
Table 1.
Phenotype, Severity Score, and TBX5/SALL4 Genotype in Individuals Diagnosed with HOS
|
Phenotypea |
|||||||
| Limb |
Severity of Phenotypeb |
||||||
| Gene Mutated, Kindred, and Patient | Heart | Hand | Arm | Other | Mutation | Heart | Limb |
| TBX5: | |||||||
| K1: | |||||||
| II-2 | ASD | Absent digit (B1) | Absent radius (B) | 456delC | M | S | |
| K2: | |||||||
| I-1 | Normal echo | Absent digit (B1), hypoplastic digit (B2-5) | Hypoplastic radius, ulna, humerus (L) | Hypoplastic chest wall, abnormal shoulder girdle | 100delG | M | S |
| II-1 | Sec-ASD, mus-VSD | Hypoplastic digit (B1) | Normal | Hypoplastic scapula (B) | S | M | |
| II-2 | Complete AV canal, multiple mus-VSDs | Absent digit (L1), hypoplastic digit (R1) | Radial deviation | Hypoplastic chest wall, abnormal shoulder girdle | S | M | |
| K3: | |||||||
| II-1 | ASD | Ulnar deviation of distal phalanx (B1) | Normal | Abnormal shoulder girdle | 100-101insG | M | M |
| K4: | |||||||
| I-2 | ASD | Normal | Hypoplastic radius (L) | Hypoplastic chest wall | Trp121Gly | M | M |
| II-1 | ASD | Normal | Hypoplastic radius (L) | Hypoplastic chest wall, narrow shoulders | M | M | |
| III-2 | ASD | Hypoplastic digit (L1) | Hypoplastic radius (L), fused radial-ulnar joint (L) | Narrow shoulders | M | M | |
| III-3 | ND | Normal | Hypoplastic radius (L) | Hypoplastic chest wall | M | M | |
| IV-1 | ASD, 4 mus-VSDs | Clinodactyly (B5) | Normal | S | M | ||
| K5: | |||||||
| II-1 | Normal echo and ECG | Triphalangeal thumb (L), hypoplastic digit (R1) | Limited supination of forearm (B) | Thr223Met | M | M | |
| III-1 | Sec-ASD, multiple mus-VSDs | Triphalangeal thumb (L) | Normal | S | M | ||
| III-2 | ASD, multiple mus-VSDs | Triphalangeal thumb (B) | Hypoplastic radius (B) | S | M | ||
| K6: | |||||||
| II-3 | ASD, memb-VSD | Absent digit (B1) | Hypoplastic radius (B) | 798delA | S | M | |
| K7: | |||||||
| II-1 | PFO, asymmetrical aortic valve | Hypoplastic digit 1 (B), absent scaphoid | Normal | Abdominal situs inversus, vertebral defects (C2C3 and C6C7 fusion) | Gly195Ala | M | M |
| K8: | |||||||
| II-1 | Sec-ASD, multiple mus-VSDs | Hypoplastic, triphalangeal thumb (B) | Hypoplastic radius (L) | Arg279Ter | S | M | |
| K9: | |||||||
| II-3 | VSD | Triphalangeal thumb (R) | Absent radius (L), hypoplastic ulna (L) | Ser196Ter | M | S | |
| K10: | |||||||
| II-4 | AV canal, memb-VSD | Hypoplastic digit (L1), absent digit (R1) | Hypoplastic radius (B) | Arg237Trp | S | M | |
| K11: | |||||||
| II-2 | PFO, normal ECG | Hypoplastic thenar eminence (L) | Normal | Hypoplastic chest wall | Arg237Gln | M | M |
| III-1 | Sec-ASD | Hypoplastic thenar eminence (L) triphalangeal thumb (B) | Normal | Hypoplastic chest wall | M | M | |
| III-2 | Sec-ASD | Absent digit (L1, L2, R1) | Hypoplastic humerus, radius, ulna (B) | M | S | ||
| K12: | |||||||
| I-2 | Normal echo | Normal | Normal | Thr223Met | M | M | |
| II-2 | Sec-ASD, multiple mus-VSDs | Syndactyly (B1 and 2) | Hypoplastic radius (B) | Abnormal shoulder girdle musculature | S | M | |
| II-3 | Multiple mus-VSDs | Syndactyly (B1 and 2) | Hypoplastic radius (B) | S | M | ||
| II-5 | VSD | Normal | Normal | M | M | ||
| K13: | |||||||
| I-1 | Normal echo | Hypoplastic digit (L1) | Normal | Arg237Trp | M | M | |
| II-2 | Sec-ASD | Hypoplastic digit (L1) | Limited supination of forearm (L) | M | M | ||
| K14: | |||||||
| II-1 | Normal | Hypoplastic distal phalanges | Normal | Ser261Cys | M | M | |
| III-3 | Double outlet right ventricle, AV canal | Absent digit (L2) | Hypoplastic radius (L) | Cleft palate, facial asymmetry, micrognathia, hypoplastic nails | S | M | |
| III-4 | Normal echo | Hypoplastic distal phalanges (B1-5) | Normal | Hypoplastic nails | M | M | |
| K15: | |||||||
| II-2 | VSD | Unknown | Unknown | 400-401insC | M | ND | |
| III-1 | VSD | Absent digit (B1) | Hypoplastic radius (R) | M | M | ||
| III-2 | ASD or VSD | Digitalized thumb (B) | Hypoplastic radius (R) | M | M | ||
| IV-1 | ASD | Digitalized thumb (B) | Normal | M | M | ||
| IV-2 | Multiple VSDs | Hypoplastic digit (L1), triphalangeal thumb (R) | Normal | S | M | ||
| K16: | |||||||
| II-1 | ASD or VSD | Hypoplastic digit (1) | Radio-ulnar synostosis | 426-427insC | M | M | |
| III-3 | VSD | Absent digit (B1) | Absent ulna and radius (L), hypoplastic humerus (L), hypoplastic ulna and radius (R) | M | S | ||
| IV-1 | Sec-ASD, 2 mus-VSDs | Absent digit (L1), hypoplastic digit (R1) | Hypoplastic radius (L) | S | M | ||
| IV-2 | Mus-VSD | Extra digit (R1), triphalangeal thumb (L) | Normal | M | M | ||
| K17: | |||||||
| II-5 | Sec-ASD, memb-VSD, mus-VSDs | Hypoplastic metacarpal (B1), hypoplastic digit (L1), triphalangeal thumb (R) | Hypoplastic radius (B) | Thr223Met | S | M | |
| SALL4: | |||||||
| K18: | |||||||
| I-2 | Normal | Normal | Normal | V752M | M | M | |
| II-1 | Truncus arteriosus | Absent digit (L1), hypoplastic digit (R1) | Absent radius (L), hypoplasic radius (R) | S | S | ||
| K19: | |||||||
| I-2 | Normal, murmur in childhood | Absent digit (L1), digitalized thumb (R), syndactyly (L2 and 3) | Absent radius (B) | Pelvic kidneys | 614-615insCCGT | M | S |
| II-2 | Mus-VSD | Absent digit (L1), digitalized thumb (R) | Absent radius (L) | Hydronephrosis (B), imperforate anus, Duane’s anomaly | M | S | |
ND = no data; echo = echocardiogram; ECG = electrocardiogram; B = bilateral; L = left; R = right; PFO = patent foramen ovale; mus-VSD = muscular VSD; memb-VSD = membranous VSD; sec-ASD = secundum ASD; normal = normal physical examination. Digits are numbered 1–5; from anterior (thumb) to posterior.
M = mild; S = severe; ND = no data.
Figure 1.
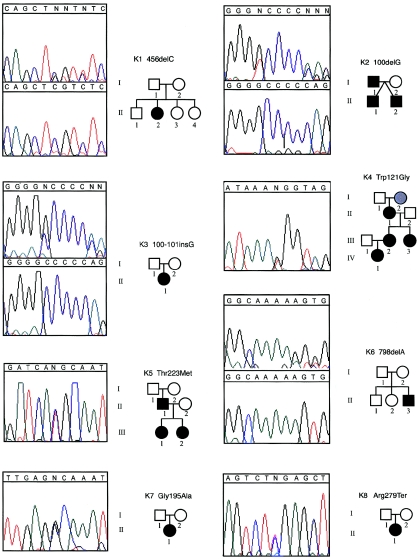
Pedigrees and electropherograms of 17 families with HOS in which mutations in TBX5 were found. Affected individuals are denoted by a blackened symbol, unaffected individuals by an unblackened symbol, and unknown phenotypes by a symbol filled with gray. Numbers below each symbol correspond to the identification numbers in table 1, which is a detailed summary of the clinical findings of each affected individual. For frameshift mutations, the mutant and wild-type DNA sequences are shown in the top and bottom boxes, respectively.
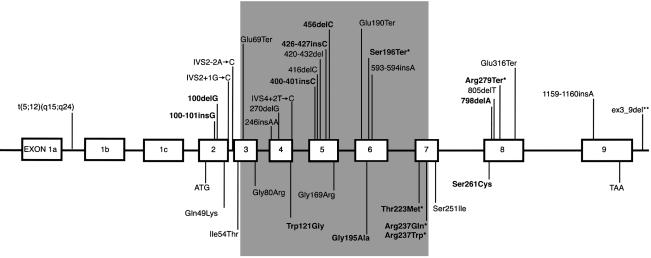
Six of the mutations found in TBX5 are predicted to encode a product that, if translated, would be prematurely truncated prior to or within the T box (fig. 2). This includes one nonsense mutation (Ser196Ter) and five frameshift mutations (100-101insG, 100delG, 400-401insC, 426-427insC, and 456delC). Since deletion of even the most C-terminal amino acid residues of the T box results in loss of TBX5 binding to its consensus binding sequence in vitro (Ghosh et al. 2001), these mutations likely represent null alleles. A partial TBX5 protein that includes the T box could be produced in the presence of either of two mutations, 798delA and Arg279Ter. However, both proteins would still lack much of the C-terminal domain of TBX5. The mechanism(s) by which 798delA and Arg279Ter disrupt the function of TBX5 is unclear, because deletion of the entire C-terminal region (amino acids 238–518 inclusive) enhances DNA binding affinity, whereas deletion of amino acids 242 onwards results in loss of binding to the T-binding element (Ghosh et al. 2001).
Figure 2.
Schematic of genomic structure of TBX5 (not drawn to scale), indicating the locations of mutations identified to date. The exons encoding the T box are shaded, and each of the mutations reported in this study is in boldface type. Asterisks (*) denote mutations that have been found in more than one proband.
Six of the mutations found in TBX5 were missense mutations, four of which have not been reported previously. Each of these mutations is predicted to result in substitution of an amino acid residue of TBX5 that is highly conserved among species (fig. 3). Five of these amino acid residues are located within the DNA-binding domain of TBX5. One predicted amino acid substitution (Ser261Cys) is in the C-terminal domain of TBX5, farther downstream of the T box than any missense mutation reported previously. One mutation—Thr223Met, caused by a missense mutation of a CG doublet—was found in three unrelated kindreds, suggesting that this may be a mutational hotspot. Overall, missense mutations were found in ∼50% of the probands in whom mutations were identified.
Figure 3.
Deduced amino acid sequence of partial TBX5 protein in the human, mouse, chicken, and zebrafish. The conserved T-box region is shaded. Missense mutations are indicated in boldface type.
Most individuals in whom a mutation in TBX5 was discovered had a phenotype typical of HOS, although a wide spectrum of limb and heart malformations was found both within and between families. A total of 16 individuals had mutations predicted to cause truncation of TBX5 and, in turn, heart and limb malformations of similar severity. Although 6 of these individuals had both mild heart and mild limb defects, 10 individuals had either more severe heart (n=6) or more severe limb (n=4) defects. Moreover, the categorization of phenotypes frequently varied, even within the same kindred. For example, individual I-1 in kindred K2 (fig. 1) has hypoplasia of the left humerus, radius, and ulna and absence of the thumb bilaterally (i.e., severe limb defects) without an accompanying heart defect. In contrast, each of his two sons (II-1 and II-2) have multiple septal defects (i.e., severe heart defects) and unilateral or bilateral hypoplasia/aplasia of the thumb (i.e., mild limb defects).
Twenty-three individuals had missense mutations in TBX5. A Trp121Gly substitution in the N-terminal region of the DNA-binding domain of TBX5 was found in five members of a single kindred. This mutation is predicted to cause severe heart and mild limb defects, and, although each individual had mild limb malformations, only one had a severe heart defect (an ASD and multiple VSDs in individual IV-1). The other four affected individuals had either a single ASD or no detectable structural defect. Fourteen individuals had missense mutations in the C-terminal region of the DNA-binding domain of TBX5 that were predicted to cause severe limb anomalies and mild cardiac defects. Eight of these individuals, from three kindreds, had a Thr223Met substitution. In contrast to the anticipated phenotype, all eight individuals had mild limb defects, and five of them had severe heart malformations. Six individuals had substitutions of amino acid residue 237, only one of whom had a severe limb anomaly, and two of these individuals with an Arg237Trp substitution had severe heart defects. Overall, 8 of 20 individuals with a missense mutation causing HOS had heart and limb defects of disparate severity. Only 2 of the 20 affected individuals had a pattern of limb and heart malformations consistent with the prediction that mutations in the carboxy- and amino-terminal regions of the T box cause more severe limb and heart malformations, respectively.
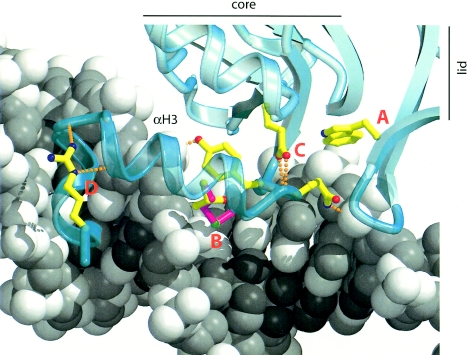
The mechanism by which missense mutations in TBX5 cause HOS remains unclear. We found five different missense mutations in TBX5 that are predicted to cause an amino acid substitution in the T box (Trp121Gly, Gly195Ala, Thr223Met, Arg237Gln, and Arg237Trp). To gain further insight into the effect of these mutations on the function of TBX5, we compared the predicted structure of TBX5 mutants to wild-type TBX5 by creating a model of the DNA-binding domain of TBX5 based on the crystal structure of TBX3 (Coll et al. 2002). The justification for using TBX3 as a starting point is based, in part, on the observation that the T-box domains of human TBX3 and TBX5 share 62% amino acid identity.
The core of the T box consists of a seven-stranded β barrel, which is closed by a lid structure comprising a two-stranded β sheet (fig. 4). The protein contacts the DNA through amino acids located in loops and strands in the lid and from residues in two perpendicular C-terminal helices, α helix 3 and helix 310C. TBX5 residue Trp121 corresponds to TBX3 residue Trp170 in the lid structure where it packs against the core β barrel (“A” in fig. 4). The Trp121Gly substitution in TBX5 is predicted to destabilize the lid structure, which would likely alter the folding or stability of TBX5 and disrupt protein-DNA interactions.
Figure 4.
Ribbon diagram of TBX3/TBX5, shown in turquoise, interacting with DNA, shown as a space-filling model in shades of gray. Important side chains are shown as ball-and-stick models, colored by atom type. Hydrogen bond interactions are shown as dashed orange lines. Perturbations of TBX5 introduced by missense mutations are indicated by A, B, C, and D (see text for details). The TBX5 model is based on the crystal structure of TBX3 from Swiss Prot deep view (Coll et al. 2002). Atomic coordinates for the structure of a TBX3-DNA complex (Protein Database entry 1H6F) were used to create a homology model of TBX5 in complex with DNA. Model manipulations, including changes to the identity and positions of amino acid side chains and examination of hydrogen-bonding networks, were performed with the molecular graphics program O (Jones et al. 1991). Mutations were modeled by changing the relevant amino acid side chains and exploring standard side chain rotamer positions for steric clashes. Atomic figures were generated with the programs MOLSCRIPT (Kraulis 1991) and Raster3D (Merritt and Murphy 1994; Merritt and Bacon 1997).
TBX5 residue Thr223 corresponds to residue Thr270 in TBX3 (“B” in fig. 4). Thr270 is in α helix 3, where the side chain contacts DNA phosphate backbone atoms, and projects into a relatively open environment in the major groove. Helix 3 and the preceding loop (residues 264–268 in TBX3) make important contacts in the major groove. Their position is determined, in part, by an important hydrogen bonding network including Glu229 (Glu180 in TBX5), which bridges between the core β barrel and the loop preceding helix 3 (“C” in fig. 4). Thus, substitution of residue Thr270 or Trp170 is expected to influence DNA recognition residues that normally contact the major groove. Thr270 occupies only part of a small void between the protein and the major groove. Modeling predicts that this space can accommodate a methionine side chain, suggesting that alteration in the activity of TBX5 does not result from steric clashes between a bulky side chain and DNA.
Arg237Gln and Arg237Trp, corresponding to Arg284 in TBX3 (“D” in fig. 4), are predicted to disrupt the position and stability of the C-terminal α helix, (equivalent to α helix 310C in TBX3 and α helix 4 in Xbra), thereby affecting binding of TBX5 to the minor groove of the DNA target (Basson et al. 1999). Gly195 is analogous to Lys242 in TBX3. Lys242 is located within a short α helix that is not present in the Xbra crystal structure of the DNA-binding domain of TBX3 (Muller and Herrmann 1997). This helix is on the surface of the core region distal to the DNA-binding residues and is positioned next to a stretch of four amino acid residues predicted to make weak interactions with a second TBX3 monomer in complex with the palindromic binding site. Since the function of this part of the protein has not been established, we cannot predict the impact of the Gly195Ala substitution in TBX5. One missense mutation in TBX5 (Ser261Cys) is downstream of the T box. A predicted function of this region of TBX5 is to interact with modifier proteins (Ghosh et al. 2001). Consequently, this substitution might, if translated into a stable protein, alter the interaction of TBX5 with a cofactor.
Two individuals with mutations in TBX5 had findings atypical of HOS: abdominal situs inversus in one individual (II-1 in kindred K7), and cleft palate, micrognathia, and facial asymmetry in another (III-3 in kindred K14). These facial abnormalities are, in particular, uncommon in HOS (Allanson and Newbury-Ecob 2003). Interestingly, in the latter case, HOS was caused by a Ser261Cys substitution downstream of the DNA-binding domain of TBX5, in a region whose function is unknown. However, other affected individuals in kindred K14 also had atypical findings, such as hypoplastic distal phalanges and fingernails, and a sib without a mutation in TBX5 had thin hair and hypoplastic teeth. Thus, it is possible that two etiologically distinct disorders are segregating in this family.
Of the remaining 32 kindreds in which mutations in the noncoding and coding regions of TBX5 were not found, haplotype analysis could not exclude TBX5, and no pedigree provided enough statistical power to independently establish linkage to another locus. Some of the affected individuals in these kindreds had defects that are uncommon in HOS (e.g., imperforate anus and truncus arteriosus), suggesting that the etiology of heart and limb malformations in our cohort might be heterogeneous.
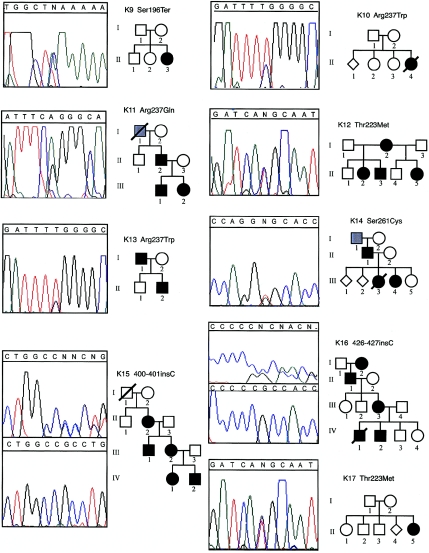
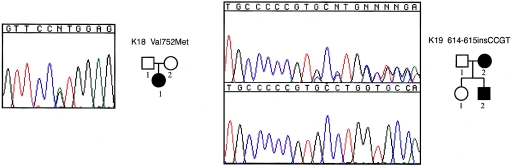
Fanconi anemia and Okihiro syndrome (MIM 607323) can be challenging to distinguish from HOS. Okihiro syndrome is thought to be distinguished from HOS by the presence of Duane anomaly, which includes an external ophthalmoplegia. However, Duane anomaly has a variable age of presentation and can be difficult to detect in the absence of a complete ophthalmological exam. Because Okihiro syndrome has recently been found to be caused by mutations in SALL4 (Kohlhase et al. 2002), we screened the four exons of SALL4 in 29 of the remaining kindreds with HOS and discovered two novel mutations (see fig. A [online only]). One mutation, in an individual with sporadic disease, was a G→A missense mutation that causes a Val752Met substitution. This mutation was not found on 230 control chromosomes. However, one parent had the same mutation, but no phenotypic information about this parent was available. Therefore, it is unclear whether this variant is the definitive cause of HOS in this individual. The other mutation identified was a 4-bp insertion that causes a frameshift in SALL4 in a mother and her son. Only one of these three individuals with a mutation in SALL4 had an ophthalmoplegia.
Figure A.
Pedigrees and electropherograms of two families with HOS in which mutations in SALL4 were found. Affected individuals are denoted by a blackened symbol. Numbers below each symbol correspond to the identification numbers in table 1. For the frameshift mutation, the mutant and wild-type DNA sequences are shown in the top and bottom boxes, respectively.
Discussion
When our results are included, a total of 35 different mutations in TBX5 have been found to cause HOS (Basson et al. 1997, 1999; Li et al. 1997; Cross et al. 2000; Yang et al. 2000; Akrami et al. 2001; Huang 2002). These include missense, nonsense, frameshift, and splice-site mutations; a deletion encompassing exons 3–9 (Akrami et al. 2001); and a translocation that putatively disrupts TBX5 (Basson et al. 1997). The Arg279Ter mutation, found in a total of six familial and sporadic cases of HOS, is caused by a C→T transition at bp 1500 that is part of a CG doublet. Accordingly, this site may represent a mutation hotspot in TBX5. Three missense mutations (Thr223Met, Arg237Gln, and Arg237Trp) and two nonsense mutations (Ser196Ter and Arg279Ter) account for ∼50% of all mutations found thus far in TBX5.
TBX5 mutations were found in only ∼30%–35% of sporadic and familial cases meeting the diagnostic criteria for HOS. This is similar to the summarized results of previous surveys of patients who have received diagnoses of HOS (Cross et al. 2000). There are several possible explanations for the low detection rate of mutations. First, some individuals with diagnoses of HOS might be phenocopies, particularly because sporadic heart and limb defects are common, and many of the putative HOS pedigrees were uninformative for linkage analysis. This is supported by the higher mutation detection rate in familial compared with sporadic cases in our cohort. Second, in most studies, only the coding regions of TBX5 have been screened for mutations. Thus, large deletions or mutations in noncoding regions that might disrupt the expression of TBX5 function would not be detected. However, a search for large deletions in 20 cases of HOS in which no mutations in TBX5 had been found yielded only one new mutation (Akrami et al. 2001). Similarly, screening the UTRs of TBX5 did not increase our detection rate. Third, HOS may be a genetically heterogeneous disorder. This is consistent with the observation that HOS does not map to 12q24 in some pedigrees (Terrett et al. 1994), although no additional loci have been identified. Finally, there may be regulatory regions important for normal TBX5 expression that have yet to be identified and that may harbor mutations causing HOS. None of these explanations are mutually exclusive, and each may, in part, be responsible for the low rate of detection of mutations in TBX5.
We found no evidence that either the type of mutation or the location of a mutation in TBX5 was predictive of the severity of limb or heart malformations in an individual with HOS. Most individuals with mutations predicted to truncate TBX5 did not exhibit heart and limb defects of similar severity. Moreover, the position of an amino acid substitution in the DNA-binding domain of TBX5 was not predictive of the severity of either the heart or the limb malformations. This result is, however, dependent on the definition of a mild versus a severe malformation. Although we used the classification of Basson et al. (1999) to score severity, the biological justification of this scheme is unclear. For example, it may not be meaningful, in either a clinical or developmental context, to consider absence of the hand and forearm as a mild limb defect while hypoplasia of the thumb, radius, and humerus is considered a severe malformation. Unfortunately, any such dichotomization of limb and heart defects is arbitrary, and, if we reanalyze the HOS phenotypes in our cohort by classifying each malformation separately (e.g., ASD, VSD, triphalangeal thumb, or hypoplastic thumb), TBX5 genotypes are still not predictive of the type of heart or limb malformation.
An accurate estimate of genotype-phenotype relationships in HOS would be a powerful resource for further exploring the molecular and developmental mechanisms underlying normal cardiac and limb development and for improving the clinical care for patients with HOS. However, this is a challenging objective for any rare malformation syndrome, including HOS. This is due, in part, to the small number of independent cases available for statistical analysis. The size of the sample often is limited further because rare malformation syndromes typically are caused by many different mutations. Inferences about genotype-phenotype relationships for such conditions could be corroborated by data from model organisms. Yet, even highly inbred strains of model organisms with disruptions of genes that cause human malformation syndromes (e.g., see Bruneau et al. 2001) often exhibit widely variable phenotypes. This underscores the substantial degree of environmental and/or stochastic variation that influences the expressivity of many malformation syndromes.
Acknowledgments
We would like to thank the families for their participation, generosity, and patience, and all of the clinicians who referred study subjects to us. We would like to thank C. T. Basson, J. C. Carey, J. Kohlhase, and R. Newbury-Ecob, for discussion, review, and comments, and L. Rasley and S. W. Watkins, for technical assistance. This project was completed with the support of the Shriners Hospitals for Children, the Primary Children’s Medical Center Foundation, the Clinical Genetics Research Program at the University of Utah, the Genomics Core Facility, and the General Clinical Research Center at the University of Utah (grant PHS MO1-00064).
Electronic-Database Information
The URL for data presented herein is as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for HOS and Okihiro syndrome)
References
- Akrami SM, Winter RM, Brook JD, Armour JA (2001) Detection of a large TBX5 deletion in a family with Holt-Oram syndrome. J Med Genet 38:E44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allanson JE, Newbury-Ecob RA (2003) Holt-Oram syndrome: is there a “face?” Am J Med Genet 118:314–318 [DOI] [PubMed] [Google Scholar]
- Bamshad M, Lin RC, Law DJ, Watkins WC, Krakowiak PA, Moore ME, Franceschini P, Lala R, Holmes LB, Gebuhr TC, Bruneau BG, Schinzel A, Seidman JG, Seidman CE, Jorde LB (1997) Mutations in human TBX3 alter limb, apocrine and genital development in ulnar-mammary syndrome. Nat Genet 16:311–315 [DOI] [PubMed] [Google Scholar]
- Basson CT, Bachinsky DR, Lin RC, Levi, T, Elkins JA, Soults J, Grayzel D, Kroumpouzou E, Traill TA, Leblanc-Straceski J, Renault B, Kucherlapati R, Seidman JG, Seidman CE (1997) Mutations in human TBX5 cause limb and cardiac malformation in Holt-Oram Syndrome. Nat Genet 15:30–35 [DOI] [PubMed] [Google Scholar]
- Basson CT, Cowley GS, Traill TA, Soloman S, Seidman JG, Seidman CE (1994) The clinical and genetic spectrum of the Holt-Oram syndrome (heart-hand syndrome). N Engl J Med 330:885–891 [DOI] [PubMed] [Google Scholar]
- Basson CT, Huang T, Lin RC, Bachinsky DR, Weremowicz S, Vaglio A, Bruzzone R, Quadrelli R, Lerone M, Romeo G, Silengo M, Pereira A, Krieger J, Mesquita SF, Kamisago M, Morton CC, Pierpont ME, Muller CW, Seidman JG, Seidman CE (1999) Different TBX5 interactions in heart and limb defined by Holt-Oram syndrome mutations. Proc Natl Acad Sci USA 96:2919–2924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braybrook C, Doudney K, Marcano AC, Arnason A, Bjornsson A, Patton MA, Goodfellow PJ, Moore GE, Stanier P (2001) The T-box transcription factor gene TBX22 is mutated in X-linked cleft palate and ankyloglossia. Nat Genet 29:107–109 [DOI] [PubMed] [Google Scholar]
- Bruneau BG, Logan M, Davis N, Levi T, Tabin CJ, Seidman JG, Seidman CE (1999) Chamber-specific cardiac expression of tbx5 and heart defects in Holt-Oram syndrome. Dev Biol 211:100–108 [DOI] [PubMed] [Google Scholar]
- Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, Conner DA, Gessler M, Nemer M, Seidman CE, Seidman JG (2001) A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell 106:709–721 [DOI] [PubMed] [Google Scholar]
- Burn J, Brennan P, Little J, Holloway S, Coffey R, Somerville J, Dennis NR, Allan L, Arnold R, Deanfield JE, Godman M, Houston A, Keeton B, Oakley C, Scott O, Silove E, Wilkinson J, Pembrey M, Hunter AS (1998) Recurrence risks in offspring of adults with major heart defects: results from first cohort of British collaborative study. Lancet 351:311–316 [DOI] [PubMed] [Google Scholar]
- Coll M, Seidman JG, Müller CW (2002) Structure of the DNA-bound T-box domain of human TBX3, a transcription factor responsible for ulnar-mammary syndrome. Structure 10:343–356 [DOI] [PubMed] [Google Scholar]
- Cross SJ, Ching YH, Li QY, Armstrong-Buisseret L, Spranger S, Lyonnet S, Bonnet D, Penttinen M, Jonveaux P, Leheup B, Mortier G, Van Ravenswaaij C, Gardiner CA (2000) The mutation spectrum in Holt-Oram syndrome. J Med Genet 37:785–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Liu M, Want Q (2003) Functional analysis of TBX5 missense mutations associated with Holt-Oram syndrome. J Biol Chem 278:8780–8785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh TK, Packham EA, Bonser AJ, Robinson TE, Cross SJ, Brook JD (2001) Characterization of the TBX5 binding site and analysis of mutations that cause Holt-Oram syndrome. Hum Mol Genet 10:1983–1994 [DOI] [PubMed] [Google Scholar]
- Guyer B, MacDorman MF, Martin JA, Peters KD, Strobino DM (1998) Annual summary of vital statistics—1997. Pediatrics 102:1333–1349 [DOI] [PubMed] [Google Scholar]
- Hiroi Y, Kudoh S, Monzen K, Ikeda Y, Yazaki Y, Nagai R, Komuro I (2001) Tbx5 associates with Nkx2-5 and synergistically promotes cardiomyocyte differentiation. Nat Genet 28:276–280 [DOI] [PubMed] [Google Scholar]
- Huang T (2002) Current advances in Holt-Oram syndrome. Curr Opin Pediatr 14:691–695 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47:110–119 [DOI] [PubMed] [Google Scholar]
- Kohlhase J, Heinrich M, Schubert L, Liebers M, Kispert A, Laccone F, Turnpenny P, Winter RM, Reardon W (2002) Okihiro syndrome is caused by SALL4 mutations. Hum Mol Genet 11:2979–2987 [DOI] [PubMed] [Google Scholar]
- Kraulis PJ (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Cryst 24:946–950 [Google Scholar]
- Li QY, Newbury-Ecob RA, Terret JA, Wilson DI, Curtis ARJ, Yi CH, Gebuhr T, Bullen PJ, Robson SC, Strachan T, Bonner D, Lyonnet S, Young ID, Raeburn JA, Bucjler AJ, Law DJ, Brook DJ (1997) Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat Genet 15:21–29 [DOI] [PubMed] [Google Scholar]
- Loffredo CA (2000) Epidemiology of cardiovascular malformations: prevalence and risk factors. Am J Med Genet 97:319–325 [DOI] [PubMed] [Google Scholar]
- Merritt EA, Bacon DJ (1997) Raster3D photorealistic molecular graphics. Methods Enzymol 277:505–524 [DOI] [PubMed] [Google Scholar]
- Merritt EA, Murphy MEP (1994) Raster3D version 2.0—a program for photorealistic molecular graphics. Acta Crystallogr D 50:869–873 [DOI] [PubMed] [Google Scholar]
- Merscher S, Funke B, Epstein JA, Heyer J, Puech A, Lu MM, Xavier RJ, Demay MB, Russell RG, Factor S, Tokooya K, Jore BS, Lopez M, Pandita RK, Lia M, Carrion D, Xu H, Schorle H, Kobler JB, Scambler P, Wynshaw-Boris A, Skoultchi AI, Morrow BE, Kucherlapati R (2001) TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell 104:619–629 [DOI] [PubMed] [Google Scholar]
- Muller CW, Herrmann BG (1997) Crystallographic structure of the T domain-DNA complex of the Brachyury transcription factor. Nature 389:884–888 [DOI] [PubMed] [Google Scholar]
- Newbury-Ecob RA, Leanage R, Raeburn JA, Young, ID (1996) Holt-Oram syndrome: a clinical genetic study. J Med Genet 33:300–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider MD, Schwartz RJ (1999) Heart or hand? Unmasking the basis for specific Holt-Oram syndrome phenotypes. Proc Natl Acad Sci USA 96:2577–2578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sletten LJ, Pierpont ME (1996) Variation in severity of cardiac disease in Holt-Oram syndrome. Am J Med Genet 65:128–132 [DOI] [PubMed] [Google Scholar]
- Terrett JA, Newbury-Ecob R, Cross GS, Fenton I, Raeburn JA, Young ID, Brook JD (1994) Holt-Oram syndrome is a genetically heterogeneous disease with one locus mapping to human chromosome 12q. Nat Genet 6:401–404 [DOI] [PubMed] [Google Scholar]
- Yang J, Hu D, Xia J, Yang Y, Ying B, Hu J, Zhou X (2000) Three novel TBX5 mutations in Chinese patients with Holt-Oram syndrome. Am J Med Genet 92:237–240 [DOI] [PubMed] [Google Scholar]