Abstract
Sarcoidosis is a granulomatous disorder of unknown etiology, associated with an accumulation of CD4+ T cells and a TH1 immune response. Since previous studies of HLA associations with sarcoidosis were limited by serologic or low-resolution molecular identification, we performed high-resolution typing for the HLA-DPB1, HLA-DQB1, HLA-DRB1, and HLA-DRB3 loci and the presence of the DRB4 or DRB5 locus, to define HLA class II associations with sarcoidosis. A Case Control Etiologic Study of Sarcoidosis (ACCESS) enrolled biopsy-confirmed cases (736 total) from 10 centers in the United States. Seven hundred six (706) controls were case matched for age, race, sex, and geographic area. We studied the first 474 ACCESS patients and case-matched controls. The HLA-DRB1 alleles were differentially distributed between cases and controls (P<.0001). The HLA-DRB1*1101 allele was associated (P<.01) with sarcoidosis in blacks and whites and had a population attributable risk of 16% in blacks and 9% in whites. HLA-DRB1-F47 was the amino acid residue most associated with sarcoidosis and independently associated with sarcoidosis in whites. The HLA-DPB1 locus also contributed to susceptibility for sarcoidosis and, in contrast to chronic beryllium disease, a non–E69-containing allele, HLA-DPB1*0101, conveyed most of the risk. Although significant differences were observed in the distribution of HLA class II alleles between blacks and whites, only HLA-DRB1*1501 was differentially associated with sarcoidosis (P<.003). In addition to being susceptibility markers, HLA class II alleles may be markers for different phenotypes of sarcoidosis (DRB1*0401 for eye in blacks and whites, DRB3 for bone marrow in blacks, and DPB1*0101 for hypercalcemia in whites). These studies confirm a genetic predisposition for sarcoidosis and present evidence for the allelic variation at the HLA-DRB1 locus as a major contributor.
Introduction
Sarcoidosis (MIM 181000) is a systemic granulomatous disease that was originally described over 100 years ago (Newman et al. 1997). Pulmonary involvement occurs in about 90% of cases, but any organ system can be involved. Skin, eye, cardiac, liver and neurologic manifestations are not uncommon. The disorder has been described in all races and in every continent except Antarctica (Siltzbach et al. 1974). While many cases may undergo spontaneous regression, one-third of cases may be persistent and require therapy. An infectious agent has been suspected as the cause of this disorder, but no specific etiology has been identified (Newman et al. 1997).
The inflammatory response in sarcoidosis has been characterized by the accumulation of CD4+ T cells at the sites of disease activity (Hunninghake and Crystal 1981). A TH1 type response has been suspected because of the granulomatous response and has been supported by the findings of increased levels of IL-2 and INF-gamma (Pinkston et al. 1983). The T cells expressed oligoclonal T cell receptors consistent with a response to an environmental antigen (Moller 1998). These findings strongly supported an excessive inflammatory reaction or hypersensitivity to some innocuous agent. The utility of corticosteroids and other anti-inflammatory drugs in the treatment of this disorder supported this idea.
A genetic susceptibility to sarcoidosis has been supported by studies demonstrating familial clustering (Rybicki et al. 2001b). First-degree relatives have a higher prevalence of disease, and familial clustering has been observed in different populations but most noticeably in African Americans (Rybicki et al. 2001a). Polymorphisms of a number of candidate genes have been associated with sarcoidosis (Arbustini et al. 1996; Ishihara et al. 1996; Rybicki et al. 1999; Pandey and Frederick 2002; Planck et al. 2002; Zorzetto et al. 2002). Serologic and molecular analyses of HLA molecules have also been investigated in sarcoidosis. Unfortunately, because of the limitations of study design and technique of identification of HLA molecules (serological and low-resolution molecular typing), a consensus of the importance of HLA molecules in sarcoidosis has not been achieved (reviewed by Martinetti et al. [2002]).
HLA genes have long been recognized for their importance in transplantation, but only in the last decade has their role in the immune response been elucidated (Klein and Sato 2000). The HLA region is located on the short arm of chromosome 6 and is one of the most gene-dense regions of the genome (MHC Sequencing Consortium 1999). Within this region, the highly polymorphic HLA class I and class II genes are located. HLA class II molecules consist of heterodimers of α and β chains which contain a groove that anchors a 10–25–amino acid peptide. However, this groove is occupied by the invariant chain, which prevents it from binding peptides until this HLA molecule is transferred to the endosomal-lysosomal compartment. In this compartment, 10–25–amino acid peptides of extracellular origin bind to the groove of the class II molecule. The amino acids that line this groove determine the affinity of the groove for these peptides. This bimolecular complex is then expressed on the cell surface where CD4+ T cells can recognize it. The polymorphism of the HLA class I and II molecules is predominately accounted for by differences in the amino acids that line the peptide-binding groove. This groove can be grouped into specific areas or pockets that interact with the amino acids of the peptide. Certain amino acid substitutions are critical for determining peptide affinity, and alleles can be grouped according to these amino acid substitutions. Thus, HLA gene products play a direct role in the immune response, and disease associations with the HLA genes have been associated not only with specific alleles but also with specific amino acids that define a specific class of alleles.
Previous studies of HLA associations with sarcoidosis were limited by including patients without histological confirmation of disease and by methods that relied on the inexact serologic or low-resolution molecular identification of the HLA molecules (Pasturenzi et al. 1993; Ishihara et al. 1994; Martinetti et al. 1995). Since the ability to develop a CD4+ T cell response in sarcoidosis probably depends on specific immunogenic peptides binding to HLA class II molecules, the aim of this study was to perform high-resolution molecular HLA class II typing in cases with histologically confirmed sarcoidosis and well-matched controls to determine whether susceptibility to sarcoidosis could be associated with specific HLA class II alleles or groups of HLA class II alleles with common amino acid residues involved in peptide binding.
Materials and Methods
Study Design
Blood specimens were collected prospectively from patients and matched controls who were entered into a case-control etiologic study of sarcoidosis (ACCESS) (Group 1999). Ten U.S. academic medical centers, one coordinating center, a DNA core laboratory, and a designated HLA laboratory participated in the design, enrollment, data entry, and analysis. The National Heart, Lung, and Blood Institute (NHLBI), which appointed a Data and Safety Monitoring Board and study chair, provided oversight and funding. Between November 1996 and June 1999, 736 cases with 706 matched controls were entered into the study. The first 474 cases and matched controls make up the study sample for which HLA studies were performed. All cases and controls completed informed consent forms, in accordance with a protocol approved by the institutional review boards of all participating centers.
Cases
Cases of sarcoidosis were recruited prospectively within geographic regions surrounding the 10 participating clinical centers between 1996 and 1999 (Group 1999). Cases met the following inclusion criteria: (1) first tissue confirmation of noncaseating granulomas on biopsy within 6 mo of enrollment, (2) clinical signs or symptoms consistent with sarcoidosis, and (3) age ⩾18 years. Specific phenotypes of sarcoidosis were determined with an instrument developed by the ACCESS group (Judson et al. 1999). Excluded were individuals with fungal disease, with active tuberculosis, or who were taking antituberculosis therapy. Patients with a history of beryllium exposure were excluded, unless they had a negative blood beryllium lymphocyte proliferation test. Pathology slides were reviewed by a study pathologist at each clinical center, and the medical records, chest radiographs, and study tests were reviewed by the principal investigator at each clinical center. An interviewer-administered questionnaire was given to each participant. The clinical characteristics of the study patients were reported elsewhere (Baughman et al. 2001).
Controls
Controls were recruited by random digit dialing (RDD) methods (Waksberg 1978; Group 1999; Baumgartner et al. 2000) from within the same geographic region as cases. Controls were matched to cases on the basis of age (within 5 years), gender, and self-reported race and ethnicity. Controls were excluded if they reported a history of sarcoidosis or medical conditions that made the determination of sarcoidosis uncertain (e.g., granulomatous hepatitis, idiopathic uveitis).
DNA Preparation
Heparinized blood was collected from each case and control at the time of the interview and was sent by overnight courier to the DNA core laboratory for purification of the DNA. High–molecular-weight DNA was isolated from anticoagulated blood by detergent lysis and organic extraction (Zoghbi et al. 1989). Purified DNA samples were diluted to a standard concentration in 10 mM Tris, 5 mM EDTA buffer, and were frozen at −70°C until used. DNA integrity and concentration were monitored using agarose gel electrophoresis and ethidium bromide staining.
HLA Identification
Masked samples of DNA were sent to the HLA typing laboratory for analysis of HLA class II alleles. Low- to intermediate-resolution typing was performed for DRB1, DRB3, DQB1, and DPB1 by use of the SSOP method and the NMDP set of probes by Orchid Diagnostics (formerly “Lifecodes”).
PCR with sequence-specific primers
PCR with sequence-specific primers, performed using ARMS technology, was utilized for high-resolution typing of HLA-DRB1, HLA-DRB3, and HLA-DPB1. Depending on the low-resolution results, appropriate kits (Olerup SSP, by Genovision) were chosen for subtyping of DRB1, DRB3, and DPB1 loci (high-resolution typing). The amplification product was then electrophoresed on a 1.5% agarose gel. The pattern of PCR products and product size was analyzed so that specific alleles could be identified. The DR4- and DR5- loci were identified only as being present or absent, and no allelic assignments were made.
Quality Control
Repeat typing of HLA-DRB1, HLA-DQB1, and HLA-DPB1 alleles was performed on 5% of the study sample. Unblinding of the quality-control samples was performed only after both samples had been completely analyzed.
Statistical Evaluation
ACCESS data were analyzed using matched case-control analysis methods. Analyses of categorical variables used McNemar’s test (McNemar 1947). Matched logistic regression analyses were performed using the methods described by Breslow and Day (1980). The PHREG procedure in SAS was used to analyze these data.
A large number of statistical tests were performed in the analysis of these data. Two analytical techniques were used to address the potential for spurious associations. Our first approach to reduction of the number of spurious associations involved a global χ2 test, which was performed within each of the DRB1, DQB1, and DPB1 allele groups. Each test addressed the null hypothesis that all of the odds ratios (ORs) for the alleles in the grouping were equal to 1 versus the alternative that at least one OR was different from 1. Alleles and epitopes that can be tracked back to a significant allele grouping are more likely not to be spurious associations, since a single test has determined that it is more likely that the grouping is related to sarcoidosis. Alleles and amino acid epitopes that were identified as being related to case-control status but were not members of a significant allele grouping are downplayed in this article.
In our second approach, a stepwise logistic regression was used to obtain the most parsimonious model that relates alleles and amino acid epitopes to sarcoidosis. Some of the variables mentioned above were present in very low frequencies. The inclusion of these variables resulted in singular information matrices that destabilized the model that was being generated. The variables resulting in singularities that destabilized the model were excluded from the logistic regression models but were part of the univariate analyses. In our logistic regression analyses, P values >.05 were deemed to be statistically significant, and P values between .05 and .1 were deemed to indicate trends.
To assess whether the influence of alleles and amino acid epitopes was the same across different age, race, or sex groups, we performed interaction analyses with gender, race, or age serving as an effect modifier. The effect modifier was introduced as an interaction in the matched logistic regression model. The null hypothesis associated with this test was that the OR relating the impact of the allele/epitope was the same in the two groups (e.g., white vs. black) versus the alternative that the ORs were different. In model terms, we tested the null hypothesis that the interaction coefficient was equal to 0 versus the alternative that the coefficient was different from 0. A P value of .05 was used to determine whether the interaction term was statistically different from 0. Since not all individuals expressed a DRB4 or DRB5 allele, the association of the presence of any of these alleles with sarcoidosis was determined by χ2 analysis.
To address whether alleles and amino acid epitopes were associated with different clinical phenotypic expressions of sarcoidosis, a χ2 test was performed to test whether the clinical phenotype was equally prevalent in sarcoidosis patients who had the allele/amino acid epitope compared to those who did not have the allele/amino acid epitope. The significance level for the P value was set at .05. Statistical tests and logistic regression models were performed for the ACCESS group as a whole and within each age, race, and gender group.
To determine the proportion of the excess disease rate that might be attributable to a specific allele, the population attributable risk (PAR%) (Schildkraut 1998) was calculated from the following formula:
 |
where Pe is the prevalence of the allele in the general population, and RR is the relative risk.
Although controls represented a population age-/race-/sex-matched to the cases, we approximated the prevalence of an allele in the general population by using the prevalence of the allele in the control population and the RR by using the OR (rare disease assumption).
Results
Overview of the Study Population
The HLA typing laboratory received 1,016 DNA samples. All samples, except for one, were successfully typed. The one sample that could not be typed was contaminated. Final results were tabulated on 474 matched cases and controls (948 total samples). In addition, the presence and absence of a DR3, DR4, or DR5 were also determined. There were 52 quality-control samples (5.5%). The remaining 16 samples were from cases and their matched controls and, on administrative review, were withdrawn from the study, either because the cases did not meet study criteria for sarcoidosis or because the samples were from cases without a matched control. Among the 948 samples, there were 53 different HLA-DRB1 alleles, 4 different DRB3 alleles, 19 different DQB1 alleles, and 41 different DPB1 alleles identified.
The demographics of this subpopulation of the ACCESS study sample for which HLA typing was performed were similar to those of the population as a whole. There were 311 (66%) that were female pairs in the HLA study compared with 64% female pairs for the ACCESS study sample of 706 pairs. For the HLA study, there were 268 (56%) pairs that self reported as white, 193 (41%) that self reported as black, and 13 (3%) that were grouped together as “other.” This is similar to the entire study sample, in which 53% were white, 44% black, and 3% other. Finally, 206 (44%) of the cases were <40 years of age in the HLA study compared to 45% <40 years of age in the entire study sample.
Quality control of the HLA typing was performed by performing duplicate analyses on 52 masked samples. For each sample, the DRB1, DQB1, and DPB1 alleles were identified. Thus, the repeatable typing of 312 alleles was utilized to assess quality control. Only six alleles (2%) were not typed similarly. This satisfies the requirements set by ASHI (American Society for Histocompatibility and Immunogenetics) for HLA typing laboratories.
Analysis of the HLA-DPB1, HLA-DQB1, and HLA-DRB1 Alleles in the Study Population
To determine whether there were different allelic distributions in sarcoidosis patients and their matched controls, a global test was performed for each allele group of the 53 DRB1 alleles, the 19 DQB1 alleles, or the 41 DPB1 alleles detected. Allelic frequencies are given in tables 1, 2, and 3. The distributions of the HLA-DRB1 alleles were significantly different between sarcoidosis patients and their matched controls (P<.0001), whereas the distributions of the alleles for HLA-DQB1 and HLA-DPB1 were not significantly different (P=.40 and P=.27 for HLA-DQB1 and HLA-DPB1, respectively).
Table 1.
HLA-DRB1 Allele Frequency by Case and Control
|
Frequency in |
||||||
| Entire Cohort |
Whites |
Blacks |
||||
| DRB1Allele | Cases | Controls | Cases | Controls | Cases | Controls |
| 0101 | .039 | .0527 | .0560 | .0728 | .0181 | .0233 |
| 0102 | .0222 | .0253 | .0168 | .0187 | .0311 | .0363 |
| 0103 | .0011 | .0042 | .0019 | .0056 | … | .0026 |
| 0301 | .1023 | .1002 | .1381 | .1231 | .0492 | .0674 |
| 0302 | .0401 | .0264 | … | … | .0959 | .0622 |
| 0303 | .0011 | .0011 | … | .0019 | .0026 | … |
| 0305 | .0021 | .0011 | .0019 | .0019 | .0026 | … |
| 0306 | … | .0011 | … | .0019 | … | … |
| 0401 | .0390 | .0675 | .0616 | .1101 | .0104 | .0130 |
| 0402 | .0222 | .0084 | .0392 | .0131 | … | .0026 |
| 0403 | .0074 | .0074 | .0112 | .0075 | .0026 | .0026 |
| 0404 | .0137 | .0285 | .0205 | .0429 | .0026 | .0104 |
| 0405 | .0084 | .0074 | .0056 | .0019 | .0078 | .0130 |
| 0407 | .0021 | .0116 | .0037 | .0168 | … | .0052 |
| 0408 | .0042 | .0042 | … | … | … | … |
| 0411 | … | .0021 | .0075 | .0075 | … | .0026 |
| 0701 | .0897 | .1086 | .0951 | .1213 | .0881 | .0959 |
| 0801 | .0084 | .0137 | .0131 | .0243 | … | … |
| 0802 | … | .0042 | … | … | … | .0026 |
| 0803 | … | .0032 | … | .0037 | … | … |
| 0804 | .0243 | .0169 | … | … | .0596 | .0415 |
| 0806 | .0021 | .0032 | … | .0019 | .0052 | .0052 |
| 0901 | .0137 | .0200 | .0131 | .0131 | .0130 | .0311 |
| 1001 | .0158 | .0105 | .0093 | .0056 | .0259 | .0155 |
| 1101 | .1150 | .0686 | .0840 | .0466 | .1632 | .0984 |
| 1102 | .0084 | .0148 | … | .0019 | .0181 | .0337 |
| 1103 | .0032 | .0053 | .0037 | .0075 | .0026 | .0026 |
| 1104 | .0179 | .0274 | .0280 | .0392 | .0052 | .0078 |
| 1110 | … | .0011 | … | … | … | .0026 |
| 1114 | .0011 | … | … | … | .0026 | … |
| 1119 | … | .0011 | … | … | … | .0026 |
| 1201 | .0411 | .0222 | .0187 | .0112 | .0751 | .0363 |
| 1202 | .0021 | .0032 | … | … | .0052 | .0052 |
| 1301 | .0633 | .0665 | .0578 | .0597 | .0699 | .0777 |
| 1302 | .0464 | .0496 | .0373 | .0410 | .0596 | .0648 |
| 1303 | .0190 | .0148 | .0093 | .0112 | .0285 | .0207 |
| 1304 | .0042 | .0084 | … | .0019 | .0104 | .0181 |
| 1305 | .0011 | .0021 | .0019 | .0037 | … | … |
| 1306 | … | .0011 | … | … | … | .0026 |
| 1310 | .0011 | .0011 | … | .0019 | .0026 | … |
| 1315 | .0011 | … | .0019 | … | … | … |
| 1316 | .0042 | .0063 | .0037 | .0093 | .0052 | .0026 |
| 1401 | .0390 | .0232 | .0448 | .0243 | .0259 | .0207 |
| 1404 | .0011 | .0042 | … | .0019 | … | .0052 |
| 1407 | .0021 | … | … | … | .0052 | … |
| 1408 | .0011 | … | .0019 | … | … | … |
| 1421 | .0011 | … | .0019 | … | … | … |
| 1501 | .1160 | .0770 | .1940 | .1119 | .0104 | .0311 |
| 1502 | .0074 | .0053 | .0093 | .0093 | .0026 | … |
| 1503 | .0338 | .0506 | … | .0019 | .0803 | .1218 |
| 1601 | .0053 | .0116 | .0075 | .0187 | … | .0026 |
| 1602 | .0053 | .0042 | … | … | .0130 | .0104 |
| 1607 | … | .0011 | … | .0019 | … | … |
Table 2.
HLA-DPB1 Allele Frequency by Case and Control
|
Frequency in |
||||||
| Entire Cohort |
Whites |
Blacks |
||||
| DPB1Allele | Cases | Controls | Cases | Controls | Cases | Controls |
| 0101 | .1804 | .1521 | .0690 | .0616 | .3394 | .2727 |
| 0201 | .1023 | .1309 | .1045 | .1306 | .0933 | .1299 |
| 0202 | .0032 | .0042 | .0019 | .0075 | .0052 | … |
| 0301 | .0897 | .0739 | .1063 | .0896 | .0699 | .0571 |
| 0401 | .2795 | .2883 | .4310 | .4254 | .0674 | .1039 |
| 0402 | .1065 | .1193 | .0970 | .1213 | .1166 | .1117 |
| 0501 | .0137 | .0116 | .0224 | .0149 | … | .0052 |
| 0601 | .0063 | .0127 | .0112 | .0205 | … | .0026 |
| 0801 | .0011 | … | .0019 | … | … | … |
| 0901 | .0063 | .0042 | .0093 | .0037 | .0026 | .0052 |
| 1001 | .0105 | .0084 | .0168 | .0131 | … | .0026 |
| 1101 | .0306 | .0359 | .0205 | .0354 | .0466 | .0390 |
| 1301 | .0243 | .038 | .0112 | .0243 | .0415 | .0597 |
| 1401 | .0137 | .0084 | .0205 | .0056 | .0026 | .0026 |
| 1501 | .0063 | .0084 | .0093 | .0056 | .0026 | .0130 |
| 1601 | .0053 | .0053 | .0093 | .0075 | … | .0026 |
| 1701 | .0380 | .0348 | .0168 | .0093 | .0674 | .0701 |
| 1801 | .0295 | .0296 | … | … | .0725 | .0727 |
| 1901 | .0053 | .0063 | .0075 | .0075 | .0026 | .0052 |
| 2001 | .0011 | .0011 | .0019 | .0019 | … | … |
| 2301 | .0084 | .0021 | .0149 | .0037 | … | … |
| 2501 | .0011 | … | .0019 | … | … | … |
| 2601 | .0011 | … | … | … | .0026 | … |
| 2701 | .0095 | .0084 | … | … | .0233 | .0208 |
| 2901 | .0021 | .0011 | .0037 | … | … | .0026 |
| 3001 | .0032 | .0011 | … | … | .0078 | .0026 |
| 3201 | .0011 | … | .0019 | … | … | … |
| 3401 | .0011 | … | … | … | .0026 | … |
| 3601 | … | .0011 | … | .0019 | … | … |
| 3901 | .0042 | .0042 | … | .0037 | .0104 | .0052 |
| 4001 | .0063 | .0032 | … | … | .0155 | .0078 |
| 4101 | .0011 | … | .0019 | … | … | … |
| 4901 | .0011 | .0011 | … | … | .0026 | .0026 |
| 5001 | .0011 | … | .0019 | … | … | … |
| 5101 | … | .0011 | … | .0019 | … | … |
| 5201 | .0011 | .0011 | .0019 | .0019 | … | … |
| 5501 | .0011 | .0011 | .0019 | … | … | .0026 |
| 6501 | … | .0011 | … | .0019 | … | … |
| 7201 | .0011 | … | .0019 | … | … | … |
| 7601 | .0011 | … | … | … | .0026 | … |
| 8001 | .0011 | … | … | … | .0026 | … |
Table 3.
HLA-DQB1 Allele Frequency by Case and Control
|
Frequency in |
||||||
| Entire Cohort |
Whites |
Blacks |
||||
| DQB1Allele | Cases | Controls | Cases | Controls | Cases | Controls |
| 0201 | .1055 | .1023 | .1362 | .1250 | .0596 | .0699 |
| 0202 | .1023 | .1150 | .0877 | .1007 | .1269 | .1399 |
| 0203 | .0032 | .0011 | … | … | .0078 | .0026 |
| 0301 | .1751 | .1888 | .1828 | .1978 | .1658 | .1736 |
| 0302 | .0622 | .0833 | .0951 | .1082 | .0181 | .0440 |
| 0303 | .0200 | .0253 | .0224 | .0354 | .0155 | .0130 |
| 0304 | .0042 | .0032 | .0075 | .0056 | … | … |
| 0305 | .0053 | .0011 | .0093 | .0019 | … | … |
| 0401 | .0032 | .0011 | .0037 | .0019 | … | … |
| 0402 | .0454 | .0454 | .0093 | .0261 | .0933 | .0674 |
| 0501 | .1255 | .1297 | .0858 | .1026 | .1865 | .1684 |
| 0502 | .0327 | .0179 | .0224 | .0205 | .0466 | .0155 |
| 0503 | .0422 | .0264 | .0466 | .0261 | .0285 | .0207 |
| 0601 | .0074 | .0053 | .0112 | .0093 | .0026 | … |
| 0602 | .1688 | .1519 | .1754 | .1194 | .1632 | .2021 |
| 0603 | .0538 | .0549 | .0672 | .0765 | .0337 | .0259 |
| 0604 | .0253 | .0285 | .0317 | .0336 | .0155 | .0233 |
| 0608 | .0011 | .0021 | … | … | .0026 | .0052 |
| 0609 | .0169 | .0169 | .0056 | .0093 | .0337 | .0285 |
To determine which of the HLA-DRB1 alleles accounted for the differences between cases and their matched controls, univariate analyses were performed. Seven HLA-DRB1 alleles were significant at P<.05 (table 4). Three alleles were identified as risk factors for sarcoidosis (HLA-DRB1*1101, HLA-DRB1*1501, and HLA-DRB1*1201), and four alleles were identified as protective for the development of sarcoidosis (HLA-DRB1*0401, HLA-DRB1*0404, HLA-DRB1*0407, and HLA-DRB1*1503).
Table 4.
HLA-DRB1 Alleles Associated with Sarcoidosis or Controls with Univariate P<.05
| HLA-DRB1 Allele | Frequencyin Cases(%) | Frequencyin Controls(%) | P | OR (95% CI) |
| Associated with Sarcoidosis: | ||||
| 1101 | 22 | 13 | <.001 | 1.98 (1.37–2.90) |
| 1501 | 22 | 14 | .003 | 1.70 (1.18–2.46) |
| 1201 | 8 | 4 | .015 | 2.13 (1.14–4.12) |
| Associated with Controls: | ||||
| 0401 | 8 | 13 | .003 | .48 (.28–.80) |
| 0407 | 0 | 2 | .022 | .18 (.02–.83) |
| 0404 | 3 | 6 | .034 | .46 (.21–.95) |
| 1503 | 6 | 10 | .048 | .57 (.32–1.00) |
Analysis of the DRB3, DRB4, and DRB5 Loci in the Study Population
The presence of the DRB3 locus was identified as a significant risk factor for sarcoidosis (P<.006; OR 1.54), while the presence of the DRB4 locus was significantly associated with a protective effect (P<.007; OR 0.69). The presence of the DRB5 locus was not associated with sarcoidosis (P=.193; OR 1.22). Since the presence of the DRB3 locus was associated with sarcoidosis, high-resolution typing of the DRB3 locus was performed. Four alleles were detected, DRB3*0101, DRB3*0201, DRB3*0202, and DRB3*0301. Only the presence of DRB3*0101 was identified as a risk factor for sarcoidosis with an OR of 1.34 (P=.044). This allele was present in 33% of cases and 27% of controls.
Analysis of Amino Acid Residues of the HLA Class II Molecules in the Study Population
Thirty-three (33) HLA-DPB1 residues, 51 HLA-DQB1 residues, and 60 HLA-DRB1 residues were tested for their involvement as risk factors for the development of sarcoidosis. By univariate analyses (table 5), 12 residues were associated with either cases or controls at the P⩽.05 level, and 13 had trends indicating a possible association (.05<P<.1).
Table 5.
HLA-DRB1-, DQB1-, and DPB1- Amino Acid Residues Associated with Sarcoidosis
| Amino Acid Residue | P | OR (95% CI) |
| Associated with Sarcoidosis: | ||
| DRB1-F47 | <.00 | 1.63 (1.18–2.27) |
| DPB1-V76 | <.00 | 1.49 (1.13–1.98) |
| DRB1-S13 | .02 | 1.43 (1.07–1.92) |
| DRB1-E58 | .03 | 1.41 (1.03–1.95) |
| DQB1-A86 | .03 | 1.35 (1.02–1.78) |
| DQB1-E84 | .05 | 1.33 (1.01–1.77) |
| DRB1-Y10 | .06 | 1.38 (.99–1.94) |
| DRB1-S11 | .06 | 1.38 (.99–1.94) |
| DQB1-R55 | .06 | 1.32 (.99–1.78) |
| DQB1-G89 | .06 | 1.31 (.99–1.74) |
| DQB1-Q53 | .06 | 1.31 (.99–1.74) |
| DQB1-G70 | .06 | 1.30 (.99–1.72) |
| DQB1-S57 | .07 | 1.87 (.96–3.76) |
| DPB1-D84 | .08 | 1.30 (.97–1.75) |
| DRB1-A74 | .09 | 1.45 (.95–2.23) |
| DRB1-H32 | .09 | 1.27 (.97–1.66) |
| Associated with Controls: | ||
| DRB1-H13 | <.00 | .59 (.41–.83) |
| DRB1-Q70R71 | .01 | .64 (.47–.88) |
| DRB1-Q10 | .01 | .66 (.49–.89) |
| DRB1-V11 | .01 | .67 (.48–.93) |
| DRB1-K12 | .02 | .69 (.51–.94) |
| DRB1-I31 | .03 | .68 (.47–.97) |
| DPB1-E69 | .06 | .77 (.59–1.01) |
| DPB1-I65 | .06 | .36 (.10–1.05) |
| DRB1-F13 | .09 | .75 (.53–1.04) |
Independent Determinants for the Risk/Protection of Sarcoidosis in the Study Population
To determine multivariate independence of HLA alleles and amino acid residues with sarcoidosis, a forward logistic regression analysis was performed using all alleles and residues that were significant at least at the 0.1 alpha level. Six alleles and two amino acid residues were multivariately associated with cases or controls (table 6). Five DRB1 alleles (DRB1*1101, DRB1*0402, DRB1*1201, DRB1*1501, and DRB1*1401), one DRB3 allele (DRB3*0101), and one DPB1 amino acid residue (DPB1-V76) were identified as risk factors for sarcoidosis, whereas only the DRB1 amino acid residue (DRB1-H13) was identified as protective for sarcoidosis.
Table 6.
Forward Logistic Regression Analysis of the Association of HLA-DRB1, HLA-DRB3, HLA-DQB1, and HLA-DPB1 Alleles and Amino Acid Residues and Sarcoidosis
| Allele orAmino Acid Residue | OR (95% CI) | P |
| Associated with Sarcoidosis: | ||
| DRB1*1101 | 2.68 (1.78–4.03) | <.001 |
| DRB1*0402 | 4.59 (1.76–12.0) | .002 |
| DRB1*1201 | 2.62 (1.38–4.95) | .003 |
| DRB1*1501 | 1.84 (1.22–2.77) | .004 |
| DRB3*0101 | 1.60 (1.16–2.20) | .004 |
| DPB1-V76 | 1.56 (1.14–2.12) | .005 |
| DRB1*1401 | 2.29 (1.21–4.34) | .011 |
| Associated with Controls: | ||
| DRB1-H13 | .32 (.12–.87) | .026 |
The Influences of Sex and of Age at Diagnosis on the Relationship Between HLA Alleles and the Development of Sarcoidosis
Because prior studies had suggested that there was an interaction between sex and age at diagnosis of sarcoidosis, we performed analyses to determine whether there were significant interactions of these variables with the HLA alleles and sarcoidosis. Only four alleles were identified as having possible interactions (P<.10) with sex or age at diagnosis. Interestingly, two of these alleles were also independently associated with sarcoidosis. HLA-DRB1*1101, which was a risk factor for sarcoidosis in the study population, had a higher OR in males than in females (OR 3.6 for males and 1.51 for females; P=.053). HLA-DRB1*0401, which was identified as a protective factor, had a lower OR in males than in females (OR 0.25 for males and 0.69 for females; P=.078). DQB1*0603 was not originally identified as associated with sarcoidosis in the study population (P=1.000; OR 0.98). The interaction analysis identified DQB1*0603 as a risk factor for females and a protective factor for males (OR 1.32 for females and 0.50 for males; P=.062). Similarly, HLA-DRB1*1302 was not associated with sarcoidosis in the study population (P=1.000; OR 1.00) but was identified as a risk factor for early-onset sarcoidosis and a protective factor for late-onset sarcoidosis (OR 1.65 for sarcoidosis at age <40 years vs. OR 0.56 for sarcoidosis at age ⩾40 years; P=.028 for interaction).
Analyses Based Within Race Groups
Using analytical methods analogous to those used for the entire study population, we investigated the interrelationships of race with HLA alleles and amino acid residues in sarcoidosis. The global test identified that at least one of the HLA-DRB1 alleles was associated with sarcoidosis in whites and blacks (P<.001 for whites and P=.0084 for blacks). There were no global effects identified for the HLA-DQB1 loci in either whites or blacks (P=.31 for whites and P=.17 for blacks). However, the global test identified that at least one of the HLA-DPB1 alleles was associated with sarcoidosis in blacks (P=.03).
Table 7 shows the results of univariate analyses for whites and blacks (tables 1, 2, and 3 give the allele frequencies). We identified three alleles that were risk factors for blacks with sarcoidosis (HLA-DRB1*1101, HLA-DRB1*1201, and HLA-DPB1*0101) and one allele that was protective (HLA-DRB1*1503). Three alleles were identified as risk factors for sarcoidosis in whites (HLA-DRB1*1101, HLA-DRB1*0402, and HLA-DRB1*1501), and one allele was identified as a protective factor (HLA-DRB1*0401). Only a single HLA-DPB1 allele (HLA-DPB1*0101) was identified in blacks as a risk factor for sarcoidosis.
Table 7.
Race-Specific HLA-DRB1 Allele Associations with Sarcoidosis
| HLA Allele | Frequencyin Cases(%) | Frequencyin Controls(%) | P | OR | PopulationAttributableRisk |
| Associated with Sarcoidosis among Blacks: | |||||
| DRB1*1101 | 31 | 19 | .006 | 2.04 (1.22–3.53) | 16.5 |
| DRB1*1201 | 15 | 7 | .014 | 2.67 (1.20–6.52) | 10.5 |
| DPB1*0101 | 58 | 44 | .008 | 1.72 (1.14–2.62) | 24.1 |
| Associated with Controls among Blacks: | |||||
| DRB1*1503 | 15 | 23 | .044 | .56 (.30–.99) | |
| Associated with Sarcoidosis among Whites: | 0 | 2 | .022 | .18 (.02–.83) | |
| DRB1*1501 | 36 | 21 | <.001 | 2.08 (1.39–3.15) | 18.5 |
| DRB1*1101 | 17 | 9 | .010 | 2.05 (1.17–3.69) | 8.6 |
| DRB1*0402 | 7 | 3 | .043 | 2.57 (1.02–7.28) | 4.5 |
| Associated with Controls among Whites: | |||||
| DRB1*0401 | 12 | 21 | .003 | .44 (.25–.77) |
Population attributable risk estimates in blacks were as follows: for HLA-DRB1*1101, 16%; HLA-DRB1*1201, 11%; and HLA-DPB1*0101 was 24%. For whites, the population attributable risk estimates were as follows: for HLA-DRB1*1101, 9%; for DRB1*0402, 5%; and for DRB1*1501, 18%. Thus, if the prevalence of these alleles in our black and white control populations is similar to the prevalence of these alleles in the black and white populations of the United States, these alleles represent substantial risk factors for the development of sarcoidosis.
Interaction tests were performed to determine if alleles were differentially associated with sarcoidosis in whites and blacks. Only one allele, HLA-DRB1*1501, had a significant interaction between blacks and whites (OR 0.36 for blacks and 2.08 for whites; P=.003 for interaction).
By univariate analysis, 22 amino acid residues in blacks and 16 amino acid residues in whites were associated with sarcoidosis (P<.05), and 11 amino acid residues in blacks and 13 amino acid residues in whites were identified as having interesting trends (.05<P<.10 [data not shown]).
The presence of the DRB3 locus was identified as a risk factor for blacks but not for whites (for blacks, OR 2.11, P=.010; for whites, OR 1.30, P=.191). The presence of the DRB4 locus was identified with protection from sarcoidosis for blacks but not for whites (for blacks, OR 0.61, P=.042; for whites, OR 0.72, P=.071). Since the test for interaction between blacks and whites for the presence of the DRB3 or DRB4 locus was not significant, these differences only indicate trends rather than significant findings. The interaction test identified the presence of the DRB5 locus as being differentially associated with sarcoidosis between blacks and whites, despite the lack of association with the entire study population (OR 0.57 for blacks and 1.23 for whites; P=.002 for interaction). High-resolution typing identified three DRB3 alleles in blacks (DRB3*0101, DRB3*0202, and DRB3*0301). Whereas none of these alleles was significantly associated with sarcoidosis, by univariate analyses, DRB3*0303 was trending as a risk factor for sarcoidosis (OR 1.64; P=.064).
To determine multivariate independence of alleles and amino acid residues in whites and blacks with sarcoidosis, the amino acid residues and the alleles of HLA-DRB1, HLA-DRB3, HLA-DPB1, and HLA-DQB1 were entered into a forward logistic regression (table 8). In blacks, four alleles (DPB1*0101, DRB1*1201, DRB1*1101, and DQB1*0502) were identified as risk factors for sarcoidosis, and two amino acid residues (DRB1-Y26 and DRB1-Q10) were identified as protective factors (P<.05). In whites, four alleles (DRB1*1101, DRB1*1501, DRB1*1401, and DRB1*0402) and three amino acid residues (DPB1-F35, DPB1-H9, and DRB1-F47) were identified as risk factors for sarcoidosis, and two amino acid residues (DPB1-I65 and DRB1-F26) and one allele (DQB1*0602) were identified as protective factors (P<.05).
Table 8.
Forward Logistic Regression Analysis of the Association of HLA-DRB1, HLA-DRB3, HLA-DQB1, and HLA-DPB1 Alleles and Amino Acid Residues in Blacks and Whites with Sarcoidosis
| Marker | OR (95% CI) | P |
| Associated with Sarcoidosis among Blacks: | ||
| DPB1*0101 | 2.20 (1.38–3.53) | .001 |
| DRB1*1201 | 3.23 (1.36–7.67) | .008 |
| DRB1*1101 | 2.24 (1.22–4.09) | .009 |
| DQB1*0502 | 3.19 (1.07–9.50) | .037 |
| Associated with Sarcoidosis among Whites: | ||
| DRB1*1101 | 3.31 (1.70–6.43) | <.001 |
| DRB1*1501 | 16.6 (3.24–85.0) | .001 |
| DRB1*1401 | 4.66 (1.85–11.8) | .001 |
| DRB1*0402 | 5.25 (1.90–14.5) | .001 |
| DPB1-F35 | 12.6 (2.03–77.5) | .006 |
| DPB1-H9 | 2.73 (1.26–5.93) | .011 |
| DRB1-F47 | 2.04 (1.15–3.64) | .015 |
| Associated with Controls among Blacks: | ||
| DRB1-Y26 | .38 (.19–.77) | .008 |
| DRB1-Q10 | .53 (.33–.87) | .012 |
| Associated with Controls among Whites: | ||
| DPB1-I65 | .04 (.01–.31) | .002 |
| DRB1-F26 | .31 (.14–.69) | .004 |
| DQB1*0602 | .11 (.02–.56) | .008 |
Sarcoidosis Phenotypes Associated with HLA Alleles
The alleles associated with sarcoidosis were evaluated to determine whether they correlated with different phenotypes of sarcoidosis at presentation. On the basis of the ACCESS questionnaire (Judson et al. 1999), involvement at presentation was assessed in 16 different systems as either definite, probable, possible, or not involved. If a subject had definite or probable involvement with sarcoidosis for a particular system, this system was considered involved with sarcoidosis for this analysis. In three organ systems (bone and joint, muscle, and renal), involvement was detected in <2% of cases. Ten different alleles were tested for their association with specific systems in all patients and in blacks and whites. Univariate (P<.05) associations are shown in table 9. The most significant finding was the relationship between DRB1*0401 and eye involvement (P<.0008; OR 3.49) in the study population. Although only 2% of blacks had a DRB1*0401 allele, a similar OR was present in both blacks and whites. The association of DRB1*1503 with extrathoracic lymph node and neurological involvement in the study population was confounded, since DRB1*1503 was only present in blacks and extrathoracic lymph node and neurological involvement were also associated with blacks. When these associations were examined in just the black population, the association was less striking, with a decreased OR and a decreased level of significance. The presence of the DRB3 locus was strongly associated with bone marrow involvement in blacks (OR 6.71; P=.004), but, because of the rarity of bone marrow involvement in whites (∼0.4%), no association could be demonstrated. DPB1*0101 was not associated with any particular organ involvement in blacks but was strongly associated with hypercalcemia in whites (P<.005; OR 4.28). The other significant association (P<.01) present in this analysis was the presence of DRB1*0401 and parotid and salivary gland involvement in blacks (P<.001; OR 78.0).
Table 9.
Clinical Involvement in Sarcoidosis Associated with HLA Class II Alleles
|
Value in |
||||||
| Entire Population |
Blacks |
Whites |
||||
| Allele andSarcoidosis Phenotypea | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P |
| DRB1*1101: | ||||||
| Calcium metabolism | 1.66 (.66–4.19) | .28 | .55 (.06–5.00) | .59 | 3.28 (1.13–9.54) | .02 |
| Extra-thoracic lymph nodes | 1.75 (1.00–3.05) | .05 | 1.59 (.76–3.33) | .21 | 1.53 (.61–3.80) | .36 |
| DRB1*1201: | ||||||
| Ear, nose and throat | 4.03 (1.23–13.1) | .01 | 4.62 (.98–21.8) | .04 | 3.47 (.31–30.8) | .24 |
| Neurological | .97 (.21–4.27) | .96 | .00b | .15 | 5.13 (.98–26.8) | .03 |
| DRB3: | ||||||
| Bone marrow | 2.33 (.81–6.71) | .11 | 6.71 (2.09–21.5) | .004 | .00b | .50 |
| DRB1*0401: | ||||||
| Eye | 3.49 (1.62–7.54) | .0008 | 4.73 (.64–34.8) | .10 | 5.77 (2.27–14.6) | .0001 |
| Neurological | 3.37 (1.18–9.59) | .02 | 5.97 (.57–62.6) | .09 | 3.23 (.95–11.0) | .05 |
| Parotid and salivary glands | 2.61 (.84–8.10) | .09 | 78.0 (7.18–848) | <.001 | .55 (.07–4.38) | .57 |
| DRB1*0402: | ||||||
| Erythema nodosum | 3.05 (.96–9.73) | .048 | …c | 3.92 (1.16–13.2) | .02 | |
| DRB1*1503: | ||||||
| Bone marrow | 6.06 (1.80–20.3) | .001 | 2.46 (.72–8.46) | .14 | …c | |
| Extra-thoracic lymph nodes | 3.20 (1.43–7.17) | .003 | 2.56 (1.07–6.08) | .03 | …c | |
| Neurological | 3.10 (.99–9.69) | .04 | 3.59 (.98–13.2) | .04 | …c | |
| DRB5: | ||||||
| Neurological | 1.28 (.55–2.97) | .56 | 3.91 (1.12–13.6) | .02 | .62 (.19–2.02) | .42 |
| Parotid and salivary glands | 2.36 (1.03–5.38) | .04 | 1.88 (.46–7.64) | .37 | 3.01 (.98–9.26) | .04 |
| DPB1*0101: | ||||||
| Calcium metabolism | 1.22 (.50–2.98) | .66 | .47 (.08–2.90) | .41 | 4.28 (1.45–12.6) | .005 |
| Eye | 1.75 (1.01–3.04) | .05 | 1.49 (.69–3.19) | .31 | .54 (.12–2.41) | .42 |
Specific definitions are given by Judson et al. (1999).
The CI of the OR could not be determined, since no cases of neurological involvement occurred in blacks with HLA-DRB1*1201.
No alleles were detected in the subpopulation.
Discussion
Although prior studies have suggested that there was an association between HLA alleles and sarcoidosis, sarcoidosis has not been generally recognized as a disease with significant HLA mediation in its development (Hansen et al. 1993; Klein and Sato 2000). A genetic predisposition to sarcoidosis has been suspected (reviewed by Rybicki et al. [2001a]), and a recent study of a genomewide search for predisposing genes in sarcoidosis suggested that not only was there a locus heterogeneity of susceptibility but the most prominent peak was noted in the HLA region (Schurmann et al. 2001). Our study extends this observation by showing that the DRB1 locus appears to be an important class II locus associated with susceptibility to sarcoidosis.
In this, the largest case-control study of high-resolution identification of HLA polymorphisms in sarcoidosis, our global test to evaluate allelic heterogeneity clearly showed a very strong association between sarcoidosis and the HLA class II DRB1 locus (P<.0001) and confirmed the initial observations of a DR association with sarcoidosis (Krause and Goebel 1987; Kunikane et al. 1987). Using logistic regression, we demonstrated that both specific alleles and amino acid residues appeared to independently confer risk for sarcoidosis. This could mean that single and multiple amino acid changes might be instrumental in defining the population of individuals who are at risk for developing sarcoidosis. In addition, the logistic regression identified two additional HLA-DRB1 alleles not identified by univariable analysis and an amino acid residue on HLA-DPB1 that appeared to be associated with sarcoidosis. Although univariate associations were noted for DRB3, DRB4, and DRB5 loci, only the DRB3*0101 allele was associated with sarcoidosis when considered with the other candidate alleles and amino acid residues in the logistic regression. This suggests that the other univariate associations identified in this analysis could be due to linkage disequilibrium or multiple comparisons
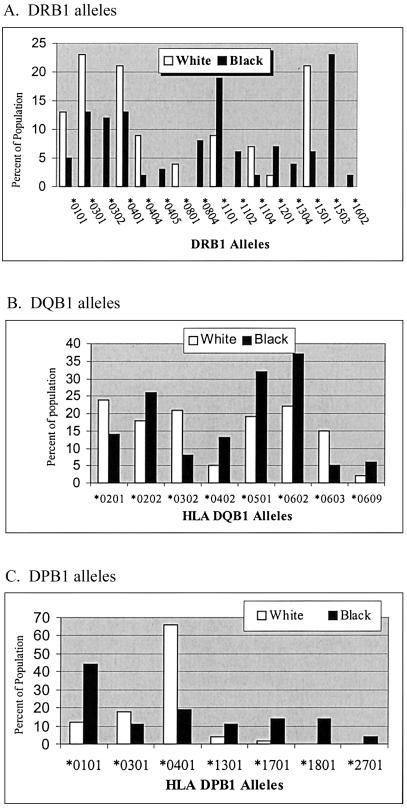
Because HLA class II allele distributions differ among different populations, we also investigated whether there were differences in the distribution of HLA alleles between whites and blacks in our control population and whether our ACCESS control population was representative of the general population. The high-resolution HLA-DRB1, HLA-DPB1, and HLA-DQB1 typing in the control populations may represent the largest population to date in which high-resolution typing of HLA-DRB1, HLA-DQB1, and HLA-DPB1 was performed to compare the HLA class II differences between blacks and whites in the United States. Our findings (fig. 1) confirm previous differences noted for HLA-DRB1*01 (Collins et al. 2000), HLA-DRB1*03 (Tang et al. 2002), HLA-DRB1*04 (Chen et al. 2002), HLA-DRB1*11 (Tang et al. 2000), and HLA-DRB1*13 (Sintasath et al. 1999), and were similar to findings for blacks in New York City (Just et al. 1997) except for the HLA-DRB1*1503 allele. In addition, differences between blacks and whites were noted in the frequencies of the HLA-DRB1*08, HLA-DRB1*12, and HLA-DRB1*16 alleles. Differences in the expression of DQB1 locus alleles were also noted, especially for DQB1*0302, DQB1*0402, DQB1*0501, DQB1*0602, and DQB1*0603. Similar differences have also previously been published (Dorman and Bunker 2000). Major racial differences for the DPB1 locus alleles were noted for DPB1*0101 and DPB1*0401 and, to a lesser degree, for DPB1*1301, DPB1*1701, and DPB1*1801. These findings suggest that the distribution of HLA alleles in our control populations is similar to the distribution of HLA alleles in the black and white populations in the United States and illustrate the need to analyze black and white populations separately and with their own matched control groups. It has been known for years that U.S. blacks, on average, have 30% white admixture (Steinberg 1969). Although this could be an important possible weakness of this study, it is important to realize that this was a matched case-control study and was matched on self-designated race.
Figure 1.
HLA class II alleles differentially expressed in blacks and whites. The alleles that were significantly different (χ2 P<.05) are illustrated. The proportion of the population expressing a specific allele is shown on the abscissa (white bars for whites, black bars for blacks). A, HLA-DRB1 alleles. B, HLA-DQB1 alleles. C, HLA-DPB1 alleles.
HLA-DRB1*1101 appears to be the most important susceptibility factor among the HLA class II alleles for sarcoidosis as this allele was significantly associated with sarcoidosis in both blacks and whites. Since the distribution of alleles in our black and white control populations appears to be similar to the distribution of alleles in the black and white populations in the United States, we were able to calculate the population attributable risk for sarcoidosis due to this allele. The population attributable risk was 16.5% for blacks and 8.5% for whites. Similarly, in a study of Japanese patients (Ishihara et al. 1994) with sarcoidosis predominantly affecting the eye (58 of 63 subjects had eye involvement), HLA-DRB1*1101 was significantly increased compared with controls. This finding is of added interest because of the recent observation of a decreased prevalence of HLA-DRB1*11 alleles in patients with hepatitis C (Yenigun and Durupinar 2002), a disease that worsens with corticosteroid treatment of sarcoidosis.
In the evaluation of the study sample, HLA-DPB1 alleles did not appear to be associated with sarcoidosis. However, in blacks, a significant association was observed for HLA-DPB1 that appeared to be accounted for entirely by HLA-DPB1*0101. Because of the high prevalence of this allele in the black control population, this allele had a population attributable risk of 24.1%. Importantly, only one allele, HLA-DRB1*1501, appeared to be differentially associated with sarcoidosis in blacks and whites (associated with controls in blacks [OR 0.36] and with sarcoidosis in whites [OR 2.08; P=.003 for interaction]). However, this study was underpowered to detect significant differential associations in alleles of low frequency. Nevertheless, the majority of HLA class II alleles that are associated with sarcoidosis may be similar in both blacks and whites, and the increased frequency of sarcoidosis in blacks could be due to the different distribution of HLA alleles in blacks compared to that in whites. Similarly, Grunewald and Eklund (Grunewald and Eklund 2001) recently reported a case of sarcoidosis in an African that had an HLA type (DR 17) that has been associated with a specific immunological profile (i.e., increased lung T cells that express the AV2S3 T cell receptor) and clinical presentation, Lofgren syndrome, that is frequently seen in Scandinavians and is unusual in Africans.
Because linkage disequilibrium complicates the interpretation of the HLA associations, controversy exists whether several HLA factors or a single HLA allele or genotype can explain sarcoidosis associations within the MHC region. For instance, in white populations, the DQB1 and DRB1 loci are in very tight linkage disequilibrium, with the allele at one locus generally predicting the allele at the other locus (Begovich et al. 1992). The degree of linkage disequilibrium between the DQB1 and DRB1 loci in blacks may not be as strong (Zachary et al. 2001). This might explain the recent findings suggesting that the DQB1 locus is the primary HLA class II susceptibility gene in blacks (Rybicki et al. 2003). Although we chose to focus on the DRB1 and DPB1 sarcoidosis associations that were most prominent in our data, on the basis of our statistical approach (logistic regressions), we did observe an association of HLA-DQB1*0502 in blacks that is consistent with a role for HLA-DQB1 in susceptibility to sarcoidosis.
Because of linkage disequilibrium in the HLA region, specific HLA-A, HLA-B, and HLA-DR haplotypes have been identified (Schipper et al. 1997) that can extend from the HLA class II region through the class III region and into the class I region. Whether specific genes or cassettes of genes inherited as a haplotype account for the susceptibility to sarcoidosis is not known. Associations with sarcoidosis have been observed with polymorphisms of the class II gene DMB (Ishihara et al. 1996), the class III gene TNF-α (Grutters et al. 2002), and, in some studies, with specific phenotypes of sarcoidosis with TNF-α polymorphisms (Swider et al. 1999; Pandey and Frederick 2002) and with the serologically defined class I antigen B8 (Lenhart et al. 1990; Dubaniewicz and Szczerkowska 1999). Until there is identification of the genetic polymorphisms that comprise specific conserved haplotypes in the HLA region, the association of these haplotypes with sarcoidosis and the specific genes accounting for the susceptibility to sarcoidosis may be uncertain.
Peptides bind to HLA class II molecules via amino acid residues that project down into the HLA class II cleft into specificity pockets (Stern et al. 1994). The specific amino acids of the HLA class II specificity pockets have been shown to determine peptide binding (Androulakis et al. 1997; Chelvanayagam 1997). In addition, the presence or absence of specific amino acid residues has been associated with insulin-dependent diabetes, rheumatoid arthritis (Dorman and Bunker 2000; Zanelli et al. 2000), and chronic beryllium disease (a granulomatous disease similar to sarcoidosis) (Richeldi et al. 1993; Wang et al. 1999; Rossman et al. 2002b). We were also able to show here that specific HLA class II amino acid residues defined susceptibility to sarcoidosis. We were surprised to find that specific alleles appeared to be more important than amino acid residues for defining susceptibility to sarcoidosis. Nevertheless, certain amino acid residues appear to be important for the susceptibility to sarcoidosis. HLA-DRB1-F47, which was the amino acid residue most associated, by univariate analysis, with sarcoidosis (table 5), was present on the three alleles most associated with sarcoidosis—HLA-DRB1*1101, HLA-DRB1*1201, and HLA-DRB1*1501—but was also present on one of the alleles associated with the control population, HLA-DRB1*1503. Thus, in the logistic regression (table 6), this amino acid residue did not appear to independently add to the risk of sarcoidosis. However, in whites who did not have a HLA-DRB1*1503 allele, HLA-DRB1-F47 appeared to independently contribute to the risk of sarcoidosis (table 8) and may be the most important amino acid residue associated with peptide binding in sarcoidosis. Amino acid residues at position 47 would affect pocket 7 (Androulakis et al. 1997; Chelvanayagam 1997), and mutations of the amino acid residues of pocket 7 peptides have been shown to affect T cell recognition (Evavold et al. 1993; Spain et al. 1994; Hsu et al. 1995).
Foley et al., in a study of European patients with sarcoidosis that used low-resolution HLA typing, observed that HLA-DRB1-V11 and –L11 amino acid residues were associated with protection from sarcoidosis (Foley et al. 2001). In our study, HLA-DRB1-V11 was associated with the control population, “protection,” and HLA-DRB1-S11 was associated with sarcoidosis (table 5). However, in our study, the most significant association with protection was with HLA-DRB1-H13. This amino acid is always associated with HLA-DRB1-V11 with the HLA-DRB1*04 alleles but is not present on the HLA-DRB1*1001 allele, which does contain -V11. Thus, we feel that it is more likely that the association with HLA-DRB1-H13 explains their association with position 11 and suggests that pocket 4, which is associated with HLA-DRB1-H13, may be important in protection from sarcoidosis, rather than pocket 6, which is associated with position 11. This may have important implications for identifying immunogenic peptides that bind class II molecules in sarcoidosis.
Prior studies have demonstrated a strong association between HLA-DPB1-E69 and chronic beryllium disease (Richeldi et al. 1993; Wang et al. 1999; Rossman et al. 2002b). Because of the similarity of chronic beryllium disease to sarcoidosis, prior studies have looked for an association of sarcoidosis with HLA-DPB1-E69 (Maliarik et al. 1998; Foley et al. 1999). We also did not find any association of HLA-DPB1-E69 with sarcoidosis. However, we did find an association with the amino acid residue DPB1-V76 in the entire study sample. This amino acid residue is present in HLA-DPB1*0101, the frequency of which was significantly increased in blacks with sarcoidosis. In the European population that was typed for HLA-DPB1, all alleles containing DPB1-V76 (DPB1*0101, DPB1*0301, DPB1*0801, DPB1*1001, and DPB1*1401) except one (DPB1*0901) were more frequent in patients with sarcoidosis than in controls suggesting that DPB1-V76 might be an important marker for sarcoidosis. In a black population (Maliarik et al. 1998), DPB1-F35 was increased, and we also noted an increase in this amino acid residue in our white population. These findings suggest that the HLA-DPB1 locus alleles may also be markers for susceptibility to sarcoidosis.
In addition to being susceptibility markers for sarcoidosis, HLA class II genes may also be susceptibility markers for a particular form or presentation of sarcoidosis. Such findings have been reported for rheumatoid arthritis (Hellier et al. 2001; Vos et al. 2001), tuberculosis (Ravikumar et al. 1999; Dubaniewicz et al. 2000), and chronic beryllium disease (Rossman et al. 2002b). In sarcoidosis, Swedish (Berlin et al. 1997), Danish (Bogunia-Kubik et al. 2001), and German and Polish (Swider et al. 1999) patients who express DRB1*03 are more likely to have Lofgren syndrome and a nonchronic form of the disease, whereas cardiac sarcoidosis has been associated with Japanese patients with HLA-DQB1*0601 (Naruse et al. 2000). We also noted that the clinical presentation appeared to be associated with specific HLA class II alleles (table 6). Finding phenotypes of sarcoidosis associated with HLA class II alleles has several important implications. First, since sarcoidosis is a polygenic disorder, other genes must be associated with sarcoidosis. Thus, specific HLA class II alleles may determine the phenotype of sarcoidosis and not the predisposition to develop a sarcoidlike reaction. Candidate genes have been identified that could be linked to the HLA class II region as a specific haplotype (Ishihara et al. 1996; Rybicki et al. 1999; Pandey and Frederick 2002; Zorzetto et al. 2002) or be present on other chromosomes (Arbustini et al. 1996; Rybicki et al. 1999; Planck et al. 2002; Zorzetto et al. 2002). The report of other areas of the genome linked to sarcoidosis supports both possibilities (Schurmann et al. 2001). Second, because HLA class II alleles may select specific environmental agents, the phenotype of the disease may relate to the localization of the selected environmental agent. Preliminary studies suggest that environmental-HLA class II associations do exist in sarcoidosis (Rossman et al. 2002a), and HLA class II alleles may be a significant tool to determine the environmental causes/triggers of sarcoidosis.
Although it is attractive to speculate that specific environmental or infectious agents will be associated with one or more alleles or amino acid variants in the polymorphic positions of the HLA; unfortunately, this is not necessarily true. For small metals, such as beryllium, it appears to be true (Richeldi et al. 1993). However, for responses to bacteria or mycobacteria, it is not necessarily true, as there may be multiple immunogenic peptides and thus a predominant HLA allele or amino acid residue is not always found (Vejbaesya et al. 2002). Thus, our findings could support several theories about the cause of sarcoidosis. First, that it might be caused by multiple small molecules present in the environment like beryllium. Second, it might be caused by an organism like tuberculosis. Third, it might be caused by a combination of the two.
Acknowledgments
The authors would like to thank the patients with sarcoidosis and the control subjects who participated in this study and Mary McNichol for assistance with editing the manuscript. Support was provided by the National Heart, Lung, and Blood Institute (grants NO1-HR-56065, NO1-HR-56066, NO1-HR-56067, NO1-HR-56068, NO1-HR-56069, NO1-HR-56070, NO1-HR-56071, NO1-HR-56072, NO1-HR-56073, NO1-HR-56074, and NO1-HR-56075).
Appendix
Clinical Centers
Beth Israel Deaconess Medical Center: Steven E. Weinberger, M.D.; Patricia Finn, M.D.; Erik Garpestad, M.D.; and Allison Moran, R.N.
Georgetown University Medical Center:: Henry Yeager, Jr., M.D.; David L. Rabin, M.D.; and Susan Stein, M.A.
Case Western Reserve University, Henry Ford Health Sciences Center: Michael C. Iannuzzi, M.D.; Benjamin Rybicki, Ph.D.; Marcie Major, R.N.; Mary Maliarik, Ph.D.; and John Popovich, Jr., M.D.
Johns Hopkins University School of Medicine: David R. Moller, M.D.; Carol J. Johns, M.D. (deceased); Cynthia Rand, Ph.D.; and Joanne Steimel, R.N.
Medical University of South Carolina: Marc A. Judson, M.D.; Susan D’Alessandro, R.N.; Nancy Heister, R.N.; Theresa Johnson, R.N.; Daniel T. Lackland, Dr.P.H.; Janardan Pandey, Ph.D.; Steven Sahn, M.D.; and Charlie Strange, M.D.
Mount Sinai Medical Center: Alvin S. Teirstein, M.D.; Louis DePalo, M.D.; Sheldon Brown, M.D.; Marvin Lesser, M.D.; Maria L. Padilla, M.D.; and Marilyn Marshall
National Jewish Medical and Research Center: Lee S. Newman, M.D., M.A.; Cecile Rose, M.D., M.P.H.; and Juli Barnard, M.A.
University of Cincinnati Medical Center: Robert P. Baughman, M.D.; Elyse E. Lower, M.D.; and Donna B. Winget
University of Iowa College of Medicine: Geoffrey McLennan, M.D., Ph.D.; Gary Hunninghake, M.D.; Chuck Dayton, B.S.Pharm.; and Linda Powers, M.S.
University of Pennsylvania and MCP-Hahnemann University Medical Centers: Milton D. Rossman, M.D.; Eddy A. Bresnitz, M.D.; Ronald Daniele, M.D., Jackie Regovich, M.P.H.; and William Sexauer, M.D.
National Heart, Lung, and Blood Institute
Robert Musson, Ph.D.; Joanne Deshler; Paul Sorlie, Ph.D.; and Margaret Wu, Ph.D.
Study Chairman
Reuben Cherniack, M.D.
Study Cochairman
Lee Newman, M.D.
Clinical Coordinating Center
Clinical Trials & Surveys Corporation: Genell L. Knatterud, Ph.D.; Michael L. Terrin, M.D.; Bruce W. Thompson, Ph.D.; Kathleen Brown, Ph.D.; Margaret Frederick, Ph.D.; Frances LoPresti, M.S.; Patricia Wilkins, B.S.; Martha Canner, M.S.; and Judy Dotson
Central Repository
McKesson Bioservices (September 1996 to November 1998): Steve Lindenfelser
BBI-Biotech Research Laboratories (December 1998 to present): Mark Cosentino, Ph.D.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
References
- Androulakis IP, Nayak NN, Ierapetritou MG, Monos DS, Floudas CA (1997) A predictive method of the evaluation of peptide binding in pocket 1 of HLA-DRB1 via global minimization of energy interactions. Proteins Struct Funct Genet 29:87–102 [PubMed] [Google Scholar]
- Arbustini E, Grasso M, Leo G, Tinelli C, Fasani R, Diegoli M, Banchieri N, Cipriani A, Gorrini M, Semenzato G, Luisetti M (1996) Polymorphisms of angiotensin-converting enzyme gene in sarcoidosis. Am J Respir Crit Care Med 153:851–854 [DOI] [PubMed] [Google Scholar]
- Baughman R, Teirstein A, Judson M, Rossman MD, Yeager HJ, Bresnitz EA, DePalo L, Hunninghake G, Iannuzzi MC, Johns CJ, McLennan G, Moller DR, Newman LS, Rabin DL, Rose C, Rybicki B, Weinberger SE, Terrin ML, Knatterud GL, Cherniak R (2001) Clinical characteristics of patietns in a case control etiology study of sarcoidosis. Am J Respir Crit Care Med 164:1885–1889 [DOI] [PubMed] [Google Scholar]
- Baumgartner KB, Samet JM, Coultas DB, Stidley CA, Hunt WC, Colby TV, Waldron JA (2000) Occupational and environmental risk factors for idiopathic pulmonary fibrosis: a multicenter case-control study. Am J Epidemiol 152:307–315 [DOI] [PubMed] [Google Scholar]
- Begovich AB, McClure GR, Suraj VC, Helmuth RC, Fildes N, Bugawan TL, Erlich HA, Klitz W (1992) Polymorphism, recombination, and linkage disequilibrium within the HLA class II region. J Immunol 148:249–258 [PubMed] [Google Scholar]
- Berlin M, Fogdell-Hahn A, Olerup O, Eklund A, Grunewald J (1997) HLA-DR predicts the prognosis in Scandinavian patients with pulmonary sarcoidosis. Am J Respir Crit Care Med 156:1601–1605 [DOI] [PubMed] [Google Scholar]
- Bogunia-Kubik K, Tomeczko J, Suchnicki K, Lange A (2001) HLA-DRB1*03, DRB1*11 or DRB1*12 and their respective DRB3 specificities in clinical variants of sarcoidosis. Tissue Antigens 57:87–90 [DOI] [PubMed] [Google Scholar]
- Breslow NE, Day NE (1980) The analysis of case control studies. In: Davis W (ed) Statistical methods in cancer research. Vol 1: IARC Scientific Publication. No. 32. International Agency for Research on Cancer, Lyon, pp 84–119 [PubMed] [Google Scholar]
- Chelvanayagam G (1997) A roadmap for HLA-DR peptide binding specificities. Hum Immunol 58:61–69 [DOI] [PubMed] [Google Scholar]
- Chen DS, Tang TF, Pulyaeva H, Slack R, Tu B, Wagage D, Li L, Perlee L, Ng J, Hartzman RJ, Hurley CK (2002) Relative HLA-DRB1*04 allele frequencies in five United States populations found in a hematopeietic stem cell volunteer donor registry and seven new DRB1*04 alleles. Hum Immunol 63:665–672 [DOI] [PubMed] [Google Scholar]
- Collins MM, Tang T, Slack R, Sintasath D, Hartzman RJ, Ng J, Hurley CK (2000) The relative frequencies of HLA-DRB1*01 alleles in the major US populations. Tissue Antigens 55:48–52 [DOI] [PubMed] [Google Scholar]
- Dorman JS, Bunker CH (2000) HLA-DQ Locus of the human leukocyte antigen complex and type I diabetes mellitus: a HuGE review. Epidemiol Rev 22:218–227 [DOI] [PubMed] [Google Scholar]
- Dubaniewicz A, Lewko B, Moszkowska G, Zamorska B, Stepinski J (2000) Molecular subtypes of the HLA-DR antigens in pulmonary tuberculosis. Int J Infect Dis 4:129–33 [DOI] [PubMed] [Google Scholar]
- Dubaniewicz A, Szczerkowska Z (1999) HLA-A, B, C antigens in pulmonary sarcoidosis in Polish population. Arch Immunol Ther Exp (Warsz) 47:55–59 [PubMed] [Google Scholar]
- Evavold BD, Sloan-Lancaster J, Hsu BL, Allen PM (1993) Separation of T helper 1 close cytolysis from proliferation and lymphokine production using analog peptides. J Immunol 150:3131–3140 [PubMed] [Google Scholar]
- Foley PJ, Lympany PA, Puscinska E, Zielinski J, Welsh KI, du Bois RM (1999) Analysis of MHC encoded antigen-processing genes TAP1 and TAP2 polymorphisms in sarcoidosis. Am J Respir Crit Care Med 160:1009–1014 [DOI] [PubMed] [Google Scholar]
- Foley PJ, McGrath DS, Puscinska E, Petrek M, Kolek V, Drabek J, Lympany PA, Pantelidis P, Welsh KI, Zielinski J, du Bois RM (2001) Human leukocyte antigen-DRB1 position 11 residues are a common protective marker for sarcoidosis. Am J Respir Cell Mol Biol 25:272–277 [DOI] [PubMed] [Google Scholar]
- Group AR (1999) Design of a case control etiologic study of sarcoidosis (ACCESS). J Clin Epidemiol 52:1173–1186 [DOI] [PubMed] [Google Scholar]
- Grunewald J, Eklund A (2001) Human leukocyte antigen genes may outweigh racial backgroud when generating a specific immune response in sarcoidosis. Eur Respir J 17:1046–1048 [DOI] [PubMed] [Google Scholar]
- Grutters JC, Sato H, Pantelidis P, Lagan AL, McGrath DS, Lammers JW, van den Bosch JM, Wells AU, du Bois RM, Welsh KI (2002) Increased frequency of the uncommon tumor necrosis factor -857T allele in British and Dutch patients with sarcoidosis. Am J Respir Crit Care Med 165:1119–24 [DOI] [PubMed] [Google Scholar]
- Hansen TH, Carreno BM, Sachs DH (1993) The major histocompatibility complex. In: Paul WE (ed) Fundamental Immunology, Third Edition. Raven Press, Ltd., New York, pp 577–628 [Google Scholar]
- Hellier JP, Eliaou JF, Daures JP, Sany J, Combe B (2001) HLA-DRB1 genes and patients with late onset rheumatoid arthritis. Ann Rheum Dis 60:531–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu BL, Evavold BD, Allen PM (1995) Modulation of T cell development by an endogenous altered peptide ligand. J Exp Med 181:805–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunninghake GW, Crystal RG (1981) Pulmonary sarcoidosis: a disorder mediated by excess helper T-lymphocyte activity at sites of disease activity. New Engl J Med 305:429–434 [DOI] [PubMed] [Google Scholar]
- Ishihara M, Naruse T, Ohno S, Kawata H, Mizuki N, Yamagata N, Ishida T, Nose Y, Inoko H (1996) Analysis of HLA-DM polymorphisms in sarcoidosis. Hum Immunol 49:144–146 [DOI] [PubMed] [Google Scholar]
- Ishihara M, Ohno S, Ishida T, Ando H, Naruse T, Nose Y, Inoko H (1994) Molecular genetic studies of HLA class II alleles in sarcoidosis. Tissue Antigens 43:238–241 [DOI] [PubMed] [Google Scholar]
- Judson M, Baughman R, Teirstein A, Terrin M, Yeager HJ (1999) Defining organ involvement in sarcoidosis: the ACCESS proposed instrument. Sarcoidosis Vasc Diffuse Lung Dis 16:75–86 [PubMed] [Google Scholar]
- Just JJ, King M-C, Thomson G, Klitz W (1997) African-American HLA class II allele and haplotype diversity. Tissue Antigens 49:547–555 [DOI] [PubMed] [Google Scholar]
- Klein J, Sato A (2000) Advances in immunology: the HLA system—first of two parts. New Engl J Med 343:702–709 [DOI] [PubMed] [Google Scholar]
- Krause A, Goebel KM (1987) Class II MHC antigen (HLA-DR3) predisposed to sarcoid arthritis. J Clin Lab Immunol 24:25–27 [PubMed] [Google Scholar]
- Kunikane H, Abe S, Tsuneta Y, Nakayama T, Tajima Y, Misonou J, Wakisaka A, Aizawa M, Kawakami Y (1987) Role of HLA-DR antigens in Japanese patients with sarcoidosis. Am Rev Respir Dis 135:688–691 [DOI] [PubMed] [Google Scholar]
- Lenhart K, Kolek V, Bartova A (1990) HLA antigens associated with sarcoidosis. Dis Markers 8:23–29 [PubMed] [Google Scholar]
- Maliarik MJ, Chen KM, Major ML, Sheffer RG, Popovich JJ, Rybicki BA, Ianuzzi MC (1998) Analysis of HLA-DPB1 polymorphisms in African-Americans with sarcoidosis. Am J Respir Crit Care Med 158:111–114 [DOI] [PubMed] [Google Scholar]
- Martinetti M, Luisetti M, Cuccia M (2002) HLA and sarcoidosis: new pathogenetic insights. Sarcoidosis Vasc Diffuse Lung Dis 19:83–95 [PubMed] [Google Scholar]
- Martinetti M, Tinelli C, Kolek V, Cuccia M, Salvaneschi L, Pasturenzi L, Semenzato G, Cipriani A, Bartova A, Luisetti M (1995) “The sarcoidosis map”: a joint survey of clinical and immunogenetic findings in two European countries. Am J Respir Crit Care Med 152:557–564 [DOI] [PubMed] [Google Scholar]
- McNemar Q (1947) Note on the sampling error of the difference between correlated proportions or percentages. Psychometrika 12:153–157 [DOI] [PubMed] [Google Scholar]
- MHC Sequencing Consortium (1999) Complete sequence and gene map of a human major histocompatibility complex. Nature 401:829–938 [DOI] [PubMed] [Google Scholar]
- Moller DR (1998) Involvement of T cells and alterations in T cell receptors in sarcoidosis. Semin Respir Infect 13:174–183 [PubMed] [Google Scholar]
- Naruse TK, Matsuzawa Y, Ota M, Katsuyama Y, Matsumori A, Hara M, Nagai S, Morimoto S, Sasayama S, Inoko H (2000) HLA-DQB1*0601 is primarily associated with susceptibility to cardiac sarcoidosis. Tissue Antigens 56:52–57 [DOI] [PubMed] [Google Scholar]
- Newman LS, Rose CS, Maier LA (1997) Medical Progress: Sarcoidosis. New Engl J Med 336:1224–1234 [DOI] [PubMed] [Google Scholar]
- Pandey JP, Frederick M (2002) TNF-alpha, IL1-beta, and immunoglobulin (GM and KM) gene polymorphisms in sarcoidosis. Hum Immunol 63:485–491 [DOI] [PubMed] [Google Scholar]
- Pasturenzi L, Martinetti M, Cuccia M, Cipriani A, Semenzato G, Luisetti M (1993) HLA class I, II, and III polymorphism in Italian patients wtih sarcoidosis. The Pavia-Padova Sarcoidosis Study Group. Chest 104:1170–1175 [DOI] [PubMed] [Google Scholar]
- Pinkston P, Bitterman PB, Crystal RG (1983) Spontaneous release of interleukin-2 by lung T lymphocytes in active pulmonary sarcoidosis. New Engl J Med 308:793–800 [DOI] [PubMed] [Google Scholar]
- Planck A, Eklund A, Yamaguchi E, Grunewald J (2002) Angiotensin-converting enzyme gene polymorphism in relation to HLA-DR in sarcoidosis. J Intern Med 251:217–222 [DOI] [PubMed] [Google Scholar]
- Ravikumar M, Dheenadhayalan V, Rajaram K, Lakshmi SS, Kumaran PP, Paramasivan CN, Balakrishnan K, Pitchappan RM (1999) Associations of HLA-DRB1, DQB1 and DPB1 alleles with pulmonary tuberculosis in south India. Tuber Lung Dis 79:309–317 [DOI] [PubMed] [Google Scholar]
- Richeldi L, Sorrentino R, Saltini C (1993) HLA-DPB1 glutamate 69: a genetic marker of beryllium disease. Science 262:242–244 [DOI] [PubMed] [Google Scholar]
- Rossman M, Thompson B, Frederick M, Cizman B, Magira E, Monos D (2002a) Sarcoidosis: association with human leukocyte antigen class II amino acid epitopes and interaction with environmental exposures. Chest 121:14S [DOI] [PubMed] [Google Scholar]
- Rossman MD, Stubbs J, Lee CW, Argyris E, Magira E, Monos D (2002b) Human leukocyte antigen Class II amino acid epitopes: Susceptibility and progression markers for beryllium hypersensitivity. Am J Respir Crit Care Med 165:788–794 [DOI] [PubMed] [Google Scholar]
- Rybicki BA, Iannuzzi MC, Frederick MM, Thompson BW, Rossman MD, Bresnitz EA, Terrin ML, Moller DR, Barnard J, Baughman RP, DePalo L, Hunninghake GW, Johns C, Judson MA, Knatterud GL, McLennan G, Newman LS, Rabin DL, Rose C, Teirstein AS, Weinberger SE, Yeager H, Chrniack R, GROUP AR (2001a) Familial aggregation of sarcoidosis: A Case-Control Etiologic Study of Sarcoidosis (ACCESS). Am J Respir Crit Care Med 164:1885–1889 [DOI] [PubMed] [Google Scholar]
- Rybicki BA, Kirkey KL, Major M, Maliarik MJ, Popovich JJ, Chase GA, Iannuzzi MC (2001b) The familial risk-ratio of sarcoidosis in African-American sibs and parents. Am J Epidemiol 153:188–193 [DOI] [PubMed] [Google Scholar]
- Rybicki BA, Maliarik MJ, Malvitz E, Sheffer RG, Major M, Popovich JJ, Iannuzzi MC (1999) The influence of T cell receptor and cytokine genes on sarcoidosis susceptibility in African Americans. Hum Immunol 60:867–874 [DOI] [PubMed] [Google Scholar]
- Rybicki BA, Maliarik MJ, Poisson LM, Sheffer R, Chen KM, Major M, Chase GA, Iannuzzi MC (2003) The major histocompatibility complex gene region and sarcoidosis susceptibility in african americans. Am J Respir Crit Care Med 167:444–449 [DOI] [PubMed] [Google Scholar]
- Schildkraut JM (ed) (1998) Examining complex genetic interactions. Wiley-Liss, New York [Google Scholar]
- Schipper RF, D'Amaro J, Bakker JT, Bakker J, van Rood JJ, Oudshoorn M (1997) HLA gene and haplotype frequencies in bone marrow donors worldwide registries. Hum Immunol 52:54–71 [DOI] [PubMed] [Google Scholar]
- Schurmann M, Reichel P, Muller-Myhsok B, Schlaak M, Muller-Quernheim J, Schwinger E (2001) Results from a genome-wide search for predisposing genes in sarcoidosis. Am J Respir Crit Care Med 164:840–846 [DOI] [PubMed] [Google Scholar]
- Siltzbach LE, James DG, Neville E, Turiaf J, Battesti JP, Sharma OP, Hosoda Y, Mikami R, Odaka M (1974) Course and prognosis of sarcoidosis around the word. Am J Med 57:847–852 [DOI] [PubMed] [Google Scholar]
- Sintasath D, Tang T, Slack R, Tilley EE, Ng J, Hartzman RJ, Hurley CK (1999) Relative HLA-DRB1*13 allele frequencies and DRB3 associations of unrelated individuals from five US populations. Hum Immunol 60:1001–1010 [DOI] [PubMed] [Google Scholar]
- Spain LM, Jorgensen JL, Davis MM, Berg LJ (1994) A peptide antigen antagonist prevents the differentiation of T cell receptor transgenic thymocytes. J Immunol 152:1709–1717 [PubMed] [Google Scholar]
- Steinberg AG (1969) Globulin polymorphisms in man. Ann Rev Genet 3:25–52 [Google Scholar]
- Stern LJ, Brown JH, Jardetzky TS, Gorga JC, Urban RG, Strominger JL, Wiley DC (1994) Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature 368:457–462 [DOI] [PubMed] [Google Scholar]
- Swider C, Schnittger L, Bogunia-Kubik K, Gerdes J, Flad H, Lange A, Seitzer U (1999) TNF-a and HLA-DR genotyping as potential prognostic markers in pulmonary sarcoidosis. Eur Cytokine Netw 10:143–146 [PubMed] [Google Scholar]
- Tang T, Huang AY, Pappas A, Slack R, Ng J, Hartzman RJ, Hurley CK (2000) Relative frequencies of DRB1*11 alleles and thier DRB3 associations in five major population groups in a United States bone marrow registry. Hum Immunol 61:820–827 [DOI] [PubMed] [Google Scholar]
- Tang TF, Wang J, Slack R, Lin Y-S, Li L, Heine U, Ng J, Hartzman RJ, Hurley CK (2002) DRB1*03 diversity and DRB3 associations in five major population groups in the United States. Hum Immunol 63:221–228 [DOI] [PubMed] [Google Scholar]
- Vejbaesya S, Chierakul N, Luangtrakool K, Srinak D, Stephens H (2002) Associations of HLA class II alleles with pulmonary tuberculosis in Thais. Eur J Immunogenet 29:431–434 [DOI] [PubMed] [Google Scholar]
- Vos K, Visser H, Schreuder GMT, de Vries RRP, Zwinderman AH, Breedveld FC, Mieke J, Hazes MW, Zanelli EH (2001) Human leukocyte antigen-DQ and DR plymorphisms predict rheumatoid arthritis outcome better than DR alone. Hum Immunol 62:1217–1225 [DOI] [PubMed] [Google Scholar]
- Waksberg J (1978) Sampling methods for random digit dialing. J Am Stat Assoc 73:40–46 [Google Scholar]
- Wang Z, White PS, Petrovic M, Tatum OL, Newman LS, Maier LA, Marrone BL (1999) Differential susceptibilities to chronic beryllium disease contributed by different Glu69 HLA-DPB1 and -DPA1 alleles. J Immunol 163:1647–1653 [PubMed] [Google Scholar]
- Yenigun A, Durupinar B (2002) Decreased frequency of the HLA-DRB1*11 allele in patients with chronic hepatitis C virus infection. J Virol 76:1787–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachary AA, Bias WB, Johnson A, Rose SM, Leffell MS (2001) Antigen, allele, and haplotype frequencies report of the ASHI minority antigens workshops: part 1, African-Americans. Hum Immunol 62:1127–1136 [DOI] [PubMed] [Google Scholar]
- Zanelli E, Breedveld FC, de Vries RRP (2000) HLA Class II association with rheumatoid arthritis: facts and interpretations. Hum Immunol 61:1254–1261 [DOI] [PubMed] [Google Scholar]
- Zoghbi HY, Lodewyk S, Sandkuyl LA, Ott J, Daiger SP, Pollack M, O'Brien WE, Beaudet AL (1989) Extensive DNA polymorphism at the factor XIIIa (F13a) locus and linkage to HLA. Am J Hum Genet 44:255–263 [PMC free article] [PubMed] [Google Scholar]
- Zorzetto M, Bombieri C, Ferrarotti I, Medaglia S, Agostini C, Tinelli C, Malerba G, Carrabino N, Beretta A, Casali L, Pozzi E, Pignatti PF, Semenzato G, Cuccia MC, Luisetti M (2002) Complement receptor 1 gene polymorphisms in sarcoidosis. Am J Respir Cell Mol Biol 27:17–23 [DOI] [PubMed] [Google Scholar]