Abstract
We report the results of a genomewide scan for age-related macular degeneration (AMD) in 158 multiplex families. AMD classification was based on fundus photography and was assigned a grade ranging from 1 (no disease) to 5 (exudative disease). Genotyping was performed by the National Heart, Lung, and Blood Institute Mammalian Genotyping Service at Marshfield (404 short tandem repeat markers). The sample included 158 families with two or more siblings with AMD, 490 affected individuals, 101 unaffected individuals, and 38 whose affection status was unknown. Relative pairs included 511 affected sibling, 28 avuncular, 53 cousin, 7 grandparent-grandchild, and 9 grand-avuncular pairs. Two-point parametric and multipoint parametric and nonparametric analyses were performed. Maximum two-point LOD scores of 1.0–2.0 were found for markers on chromosomes 1, 2, 8, 10, 14, 15, and 22. Multipoint analyses were consistent with the two-point results for chromosomes 1, 2, 8, 10, and 22 and provided evidence for additional linkage regions on chromosomes 3, 6, 8, 12, 16, and X. Our signals on chromosomes 1q, 6p, and 10q are consistent with some other previously published results. Significant linkage to AMD was found for one marker on chromosome 2, two adjacent markers on chromosome 3, two adjacent markers on chromosome 6, and seven contiguous markers on chromosome 8, with empirical P values of .00001. The consistency of many of the other signals across both two-point and multipoint, as well as parametric and nonparametric, analyses indicate several other regions worthy of follow-up.
Introduction
Age-related macular degeneration (ARMD1 [MIM 603075]; hereafter referred to as “AMD”) is the leading cause of visual impairment and blindness among older adults in the United States and all developed countries around the world. It affects the central regions of the retina and choroid and therefore can lead to central visual loss and inability to read, write, and drive, which results in loss of independence. Approximately 30% of individuals aged ⩾75 years have some sign of the disease, and ∼6%–8% of those in this age group have the advanced stages that cause visual loss (National Advisory Eye Council 2001). The prevalence of this important eye disease will continue to rise as the population ages. Since treatment options are limited to a small subset of patients and have short-term effects, it is imperative to identify the causes of this disease so that it can be prevented.
Familial aggregation has been demonstrated for AMD (Seddon et al. 1997; Klaver et al. 1998b; Klein et al. 2001). It is likely that both genetic susceptibility (Heiba et al. 1994; Allikmets et al. 1997; De Jong et al. 1997; Seddon et al. 1997; Klaver et al. 1998b; Klein et al. 1998, 2001; Allikmets 2000; Weeks et al. 2001; Hammond et al. 2002) and environmental risk factors (The Eye Disease Case-Control Study Group 1992; Seddon et al. 1996; Seddon 2001) play a role. The relative paucity of information about the specific genetic origins of AMD can be attributed to several factors. Unlike juvenile-onset retinal diseases with clear Mendelian inheritance for which genetic causes have been identified, AMD occurs late in life, typically only one generation has the disease phenotype, parents and some of the siblings are often deceased, and the offspring are too young to manifest the disease. Furthermore, considerable evidence indicates that the pathogenesis of AMD is multifactorial, involving a combination of genetic, environmental, and biologic factors. At this point, the evidence for environmental risk factors appears to be more clearly delineated than evidence for any particular gene.
Cigarette smoking (Seddon et al. 1996; Smith et al. 1996; Vingerling et al. 1996; Delcourt et al. 1998; Seddon 2001) and increasing age have been consistently shown to be related to the onset of this disease. Several other lifestyle and medical factors may possibly play a role, such as systemic hypertension (Sperduto et al. 1986; Friedman 1997; Hyman et al. 2000) and elevated cholesterol levels (The Eye Disease Case-Control Study Group 1992). In addition, nutritional factors are related to AMD (Seddon et al. 1994, 2001b; Mares-Perlman et al. 1995; VandenLangenberg et al. 1998). Foods rich in antioxidants may be protective, whereas fat consumption is associated with an elevated risk for AMD (Seddon et al. 2001b). Omega-3 fatty acids and fish intake have been shown to have a beneficial effect (Cho et al. 2001; Seddon et al. 2001b). A multicenter randomized clinical trial demonstrated that a supplement with vitamins C, E, beta-carotene, and zinc reduces the risk of progression to the advanced forms of AMD and subsequent visual loss among individuals with intermediate stages of AMD (Age-Related Eye Disease Study [AREDS] Research Group 2001b).
Genetic linkage to chromosome 1q has been reported in one family with AMD (Klein et al. 1998). Weeks et al. (2001) reported potential loci on chromosomes 1q31, 9p13, 10q26, and 17q25. This study corroborates the finding of Klein et al. (1998), in which linkage to chromosome 1q25-q31 and loci on chromosomes 2 and 12 were also shown to have LOD scores >1.5. A recent report by Schick et al. (2003) shows evidence for suggestive linkage to early AMD on chromosomes 3, 5, 6, 12, 15, and 16, with the strongest evidence for linkage on chromosome 12 near D12S46.
We hypothesize that underlying susceptibility gene(s) are critical to the development of AMD and that they most likely interact with environmental factors and possibly other genes to trigger the development and progression of this disease. We are therefore conducting both candidate-gene searches and genomic screening to identify these genes, in an effort to elucidate the etiology of AMD. In this article, we report the results of a genomewide scan of multiplex families with affected sibling pairs.
Material and Methods
Recruitment of Families and Clinical Procedures
The methods employed in this study conformed to the tenets of the Declaration of Helsinki and received approval from the institutional review board at the Massachusetts Eye and Ear Infirmary, Boston. Informed consent statements were signed by probands and family members who participated in this study.
Families with two or more siblings reported to have AMD were eligible for recruitment into the study. Potentially eligible probands were recruited from several sources, including the AMD database of the principal investigator (J.M.S.), from other ophthalmologists throughout the country, from some of the clinical centers participating in a multicenter randomized trial of vitamins and minerals (AREDS), and through various media such as newsletters, brochures, and newspapers. After obtaining written consent, all records and photographs were obtained from eye doctors and reviewed by J.M.S. Families with two or more living siblings with diagnoses of AMD (large drusen, geographic atrophy, or exudative disease) were then asked to participate.
Data collection procedures included an ocular examination, ocular fundus photography, blood drawing, and questionnaires about risk factors for AMD. We developed protocols and standardized forms for data collection, which were sent to a participating ophthalmologist near the subject’s home for use during the examination. Prior to the examination of a participant, J.M.S. called the ophthalmologist to review these procedures and answer any questions, to ensure uniformity and accuracy in data collection techniques and procedures.
The ocular examination consisted of assessment of refractive error, best-corrected visual acuity, intraocular pressure, and notation of iris color compared with standard photographs (Seddon et al. 1990). After dilation of the pupils, cataract status was recorded by comparing the slit-lamp examination to standard photographs (LOCS II) (Chylack et al. 1993). Signs of macular degeneration were evaluated with the aid of example photographs that were chosen from the clinical files of J.M.S. The photographs depicted small hard drusen and retinal pigment epithelial hyperpigmentation and hypopigmentation, as well as geographic atrophy, retinal pigment epithelial detachment, and signs of choroidal neovascularization, including disciform scarring. Retinal photography was performed according to the standard protocol we provided, which required stereo pair 30° fundus photographs centered on the optic disc, fovea, and temporal to the fovea of each eye. Blood specimens were obtained for genetic analyses, as well as for biochemical analyses to assess the effect of various potential biomarkers on the risk of AMD. DNA was extracted using standard techniques.
Classification/Phenotype Assessment
Classification of AMD was based on fundus photography in combination with the study examination data. Macular characteristics seen on the photographs were graded within an area with a 3,000-micron radius centered on the foveal center and included drusen, retinal pigment epithelial irregularities, geographic atrophy, retinal pigment epithelial detachment, and evidence for choroidal neovascularization associated with AMD, including disciform scarring. AMD status was assigned by J.M.S. according to our modification of the grading system used in the AREDS (AREDS Research Group 2001a) as follows: grade 1, no evidence of AMD; grade 2, minimal maculopathy; grade 3, intermediate dry stage of AMD; grade 4, geographic atrophy, the advanced form of dry AMD; and grade 5, the advanced, exudative form of AMD (table 1). Past ocular records, photographs, and fluorescein angiograms were also reviewed, when available, to confirm the diagnosis. Maculopathies other than AMD, including those that could be associated with choroidal neovascularization, were noted and excluded from the case group. To evaluate reliability of the AMD grading, 222 photographs were sent to the Wisconsin Fundus Photographic Reading Center for detailed grading as used in the AREDS (AREDS Research Group 2001a). The level of agreement between J.M.S. and the Reading Center grade was determined using the kappa statistic. The kappa statistic was 0.77 and the weighted kappa was 0.84. Inconsistencies were adjudicated by J.M.S., using all available clinical and photographic data.
Table 1.
Fundus Characteristics Corresponding to AMD Grade
| AMDGrade | Fundus Characteristics |
| 1 | No drusen or small nonextensive drusen without retinal pigment epithelial abnormalities |
| 2 | Extensive hard drusen or nonextensive intermediate drusen, and/or retinal pigment epithelial changes |
| 3 | Extensive intermediate drusen or any large drusen with or without drusenoid retinal pigment epithelial detachment |
| 4 | Geographic atrophy |
| 5 | Exudative AMD including serous retinal pigment epithelial detachment and choroidal neovascular membrane |
| 6 | Other maculopathy |
Final AMD status for the genetic analysis was assigned, according to grade of AMD and age, as follows: “AMD present” (grade 3, 4, or 5), “AMD not present” (grade 1 or 2 if aged ⩾60 years), or “unknown” (grade 1 or 2 if aged <60 years). Grade 3 represents a maculopathy that is more likely to progress to the advanced stages of AMD (Klein et al. 1997; AREDS Research Group 2001b) and was included in the definition of AMD. Grade 2, however, can occur in a large proportion of individuals aged >60 years, and very few develop advanced AMD (Klein et al. 1992, 1997; AREDS Research Group 2001b); these individuals were therefore not included as affected. Because individuals aged <60 years with grades 1 or 2 may develop maculopathy in the future, they were designated as having unknown affection status.
Genotyping
Genotyping was performed by the National Heart, Lung, and Blood Institute (NHLBI) Mammalian Genotyping Service at Marshfield (Center for Medical Genetics Web site), under the direction of J. Weber. A total of 404 STR markers from Weber Screening Set 10 were used, 397 of which were either autosomal or on the X chromosome. Marker heterozygosity, excluding sex chromosome markers, ranged from 0.50 to 0.89 and averaged 0.756±0.066. The average marker spacing across the genome was 9.26 cM.
Error Checking
Genotyping data were entered into a locally developed data management system in the laboratory of S.L.S. Because there were very few parents or founders in the data set, marker allele frequencies were recalculated from the data via the USERM13 module of the program Mendel (Lange et al. 1988; Mendel Web site), which employs a maximum-likelihood method to provide unbiased estimates of the allele frequencies based on data from related individuals. Prior to statistical analysis, the data were checked for Mendelian and sex inconsistencies through use of the subroutine UNKNOWN from the LINKAGE software package. We found 2,103 Mendelian and sex inconsistencies in the data. If it is assumed that each inconsistency was due to one genotype or binning error, then the error rate is 0.0086, or 0.86%. For cases in which the removal of one individual resolved the inconsistency over several markers, that individual was excluded from further analysis. For all other cases, the inconsistent genotypes, which were generally specific to one marker, were excluded. Sometimes this necessitated the exclusion of entire families for certain markers. Finally, we used the relationship estimation program RELPAIR version 0.90 (Duren et al. 1997; Epstein et al. 2000; Center for Statistical Genetics Web site) to check the accuracy of the stated relationships between individuals in a given pedigree against the actual level of allele sharing across the genome. For instance, we used RELPAIR to determine whether siblings who claim to be full sibs are actually full or half sibs. The RELPAIR software program infers the relationships of pairs of individuals on the basis of genetic marker data, either within families or across an entire sample. It is based on the pairwise likelihood ratio proposed by Boehnke and Cox (Boehnke et al. 1997) that calculates the multipoint probability of the observed marker genotypes, conditioned on each relationship considered. Unlike some other relationship checking programs, RELPAIR is able to detect undiagnosed MZ twins. We found no errors in relationships in any of the pedigrees.
Statistical Analyses
Two-point parametric analyses were run using the MLINK module of FASTLINK version 4.1p (Cottingham et al. 1993; Schaffer et al. 1994; Fastlink Home Page) and were followed by multipoint parametric analyses with the Genehunter version 2.1 (Kruglyak et al. 1996; Kruglyak Labs Web site) software package. It is highly likely that AMD is oligogenic and may result from gene-by-environment as well as epistatic interactions. Therefore, the correct model is unknown, and a single-gene model for the disorder is almost certainly incorrect. Nevertheless, it is generally accepted that, in the absence of the correct model, it is possible to obtain increased power to detect linkage by calculating LOD scores under both a dominant and a recessive parametric model (Vieland et al. 1992; Durner et al. 1999) and allowing for locus heterogeneity. Thus, for the two-point and multipoint parametric analyses, we calculated two LOD scores, one dominant and one recessive, and report the maximum of the two, the maximum LOD score (MLS). For the multipoint analyses, we calculated the heterogeneity LOD score (HLOD) via the admixture model of Smith (1963), as implemented in Genehunter, under both a dominant and a recessive model. We report the maximum of the two (dominant and recessive) and henceforth refer to the maximum multipoint heterogeneity LOD score as the MMHLS. The parameter estimates used in the parametric models were as follows: for the recessive model, the disease allele frequency q=0.10, and penetrance values are f1=f2=0 and f3=0.5. For the dominant model, the disease allele frequency q=0.01 and penetrance values are f1=0 and f2=f3=0.5.
In addition, we used nonparametric or model-free methods to assess evidence for linkage, in which specifying the mode of inheritance (which is unknown for AMD) is not required. We assessed the extent to which the sib and other relative pairs shared ancestral alleles identical by descent (IBD) by calculating nonparametric Z scores (NPL scores), first using Genehunter version 2.1 and then using MERLIN version 0.8.8 (Abecasis et al. 2002; MERLIN Web site), combined with SimWalk2 version 2.83 (Weeks et al. 1995; Sobel and Lange 1996; Statgen Software Web site). Since some pedigrees in the data set were too large to be analyzed with MERLIN, we used SimWalk2 in combination with MERLIN to handle the larger pedigrees. MERLIN calculates exact results for the sib pairs and other relative pairs, and SimWalk2 uses Markov Chain Monte Carlo methods to estimate results for the combined sample of relative pairs and larger pedigrees, which couldn’t be analyzed with MERLIN. In Genehunter, we chose the Spairs option to calculate the NPL scores (labeled “NPLpairs” in table 3 below). The Spairs option assesses pairwise allele sharing among all affected relatives by counting the number of pairs of alleles from the affected members of a family that are shared IBD (Whittemore et al. 1994; Kruglyak et al. 1996). The MERLIN/SimWalk2 output reports clustering statistic D, which is very similar to the Whittemore and Halpern Spairs statistic as implemented in Genehunter. Statistic D, like Spairs, is a pairwise statistic. It measures the extent of IBD gene sharing among affected relative pairs by summing over their kinship coefficients (Sobel and Lange 1996). The power of such clustering statistics is increased in SimWalk2 by use of the information in the unaffecteds as well as the affecteds to sample all the IBD configurations, proportional to their true likelihood. In tables 3 and 4 below, we use the term “NPLpairs” to indicate both the Genehunter Spairs statistic (table 3), and the P value of the SimWalk2 statistic D (table 4), since the two statistics are so similar.
Table 3.
Results of Multipoint Parametric and Nonparametric Analyses Using Genehunter
| Chromosomeand Locationa(in cM) | Marker | MMHLS(Model)b | αc | NPLpairsScore | P Valued |
| X: | |||||
| 92.9 | DXS8084 | 1.19 (r) | .13 | 2.53 | .006 |
| 96.9 | DXS998 | 1.53 (r) | .17 | 2.54 | .006 |
| 2: | |||||
| 64.3 | D2S1356 | .53 (d) | .11 | 1.82 | .03 |
| 77.6 | D2S1352e | 1.80 (d) | .24 | 1.58 | .06 |
| 90.8 | D2S1394 | .55 (d) | .55 | 1.94 | .03 |
| 186.2 | D2S1391 | 1.94 (d) | .24 | 2.32 | .01 |
| 190.5 | D2S1384f | 2.37 (d) | .29 | 2.03 | .02 |
| 22: | |||||
| 36.2 | D22S683 | .94 (d) | .17 | 2.20 | .01 |
| 3: | |||||
| 117.1 | D3S4529g | 1.50 (r) | .16 | 1.67 | .05 |
| 124.2 | D3S3045 | 1.19 (r) | .12 | 2.03 | .02 |
| 129.4 | D3S2460h | 1.39 (r) | .14 | 1.86 | .03 |
| 16: | |||||
| 81.2 | GATA138C05 | 1.32 (d) | .16 | 2.01 | .02 |
| 1: | |||||
| 151.9 | D1S534 | .52 (d) | .12 | 1.73 | .04 |
| 170.9 | D1S1679 | .55 (d) | .14 | 1.94 | .03 |
| 192.1 | D1S1589 | 2.41 (d) | .25 | 1.95 | .03 |
| 10: | |||||
| 156.3 | D10S1222 | 1.90 (d) | .27 | 1.73 | .04 |
| 12: | |||||
| 83.2 | D12S1052 | 1.56 (r) | .16 | 1.71 | .04 |
| 8: | |||||
| 4.3 | D8S262 | 1.23 (r) | .15 | 1.43 | .08 |
Location is given in Kosambi centimorgans from the telomere.
Models are the same as for the two-point analyses. A “d” indicates the MMHLS was maximized under the dominant model, and an “r” indicates the MMHLS was maximized under the recessive model.
Proportion of families with linkage to a given locus.
P values indicate nominal (not genomewide) significance of NPL scores and are known to be conservative.
This signal, at 77.6 cM, falls between markers GATA27D04 (D2s1352), at 73.6 cM, and GATA8F03 (D2s1779), at 86.8 cM.
This signal, at 190.5 cM, falls between markers GATA65C03 (D2S1391), at 186.2 cM, and GATA52A04 (D2S1384), at 200.4 cM.
This signal, at 117.1 cM, falls between markers GATA128C0 (D3s4529), at 112.42 cM, and GATA84B12 (D3S3045), at 124.2 cM.
This signal, at 129.4 cM, falls between markers GATA84B12 (D3s3045), at 124.2 cM, and GATA68F07 (D3s2460), at 134.6 cM.
Table 4.
Results of Nonparametric Multipoint Analysis Using MERLIN and SimWalk2
| Chromosomeand Marker | DNamea | Locationb(cM) | NPLpairsP Valuec |
| 2: | |||
| GATA11H10 | D2S1360 | 38.3 | .00001 |
| GATA69E12 | D2S1394 | 90.8 | .06 |
| GATA65C03 | D2S1391 | 186.2 | .01 |
| GATA52A04 | D2S1384 | 200.4 | .04 |
| GATA30E06 | D2S2944 | 210.4 | .02 |
| 22: | |||
| GATA11B12 | D22S683 | 36.2 | .01 |
| ATA37D06 | D22S1045 | 42.8 | .05 |
| 3: | |||
| GATA84B12 | D3S3045 | 124.2 | .04 |
| GATA68F07 | D3S2460 | 134.6 | .00001 |
| ATA34G06 | D3S4523 | 138.0 | .00001 |
| 16: | |||
| GATA138C05 | Unknown | 81.2 | .04 |
| 1: | |||
| ATA4E02 | D1S1589 | 192.1 | .03 |
| GATA48B01 | D1S1660 | 212.4 | .10 |
| 10: | |||
| ATA22D02 | D10S1222 | 156.3 | .05 |
| 12: | |||
| GATA26D02 | D12S1052 | 83.2 | .06 |
| 8: | |||
| GATA151F02 | Unknown | 27.4 | .00001 |
| AFM123xg5 | D8S261 | 37.0 | .00001 |
| AFMa127ye5 | D8S560 | 43.4 | .00001 |
| UT7129 | D8S1048 | 54.3 | .00001 |
| GGAA20C10 | D8S1477 | 60.3 | .00001 |
| GATA8G10 | D8S1110 | 67.3 | .00001 |
| CGAA8G07 | D8S113 | 77.9 | .00001 |
| 6: | |||
| GATA64D02 | D6S1053 | 80.5 | .00001 |
| ATA28B11 | D6S1031 | 88.6 | .00001 |
Alternate marker name.
Location is given in Kosambi centimorgans from the telomere.
P values are computed empirically, indicate nominal (not genomewide) significance of NPL scores, and may be conservative.
Results
Of the 7,107 families screened to date, 6,208 did not meet the eligibility criterion requiring two or more living affected siblings. Of the 899 families reported to be potentially eligible, 75% did not have two or more living siblings with AMD or had relatives with other maculopathies or retinal disease (diabetic retinopathy, vein occlusion, epiretinal membrane, macular hole, etc.), and a few eligible families refused to participate.
Of the 224 families that were enrolled, 198 completed the study procedures. A sample of 158 of these families (645 individuals) was genotyped with 404 microsatellite markers. After exclusions for Mendelian inconsistencies, the sample used for the analyses included 629 individuals, 401 women and 228 men. There were 490 affected individuals, with a mean age of 76.6 years and a mean age at diagnosis of 69.9 years (grade 3 or higher); 101 unaffected individuals, with a mean age of 70.3 years (grades 1 or 2, and over age 60 years); and 38 whose affection status was unknown. The sample included 511 affected sibling pairs, as well as 13 parent-child, 28 avuncular, 53 cousin, 7 grandparent-grandchild, and 9 grand-avuncular pairs.
Table 2 lists the results of the two-point parametric analyses; all LOD scores >1.0 are listed in descending order. In every case, the two-point MLSs were maximized under the dominant model. Marker locations are given in Kosambi centimorgans from the telomere. Markers on chromosomes 1, 2, 8, 10, 14, 15, and 22 had two-point MLSs between 1.0 and 2.0; the highest was a MLS of 2.00 on chromosome 22 (ATA37D06; 42.8 cM). Although there were no significant or suggestive linkage findings from the two-point analyses (Lander et al. 1995), there were five markers with MLSs between 1.5 and 2.0.
Table 2.
Results of Two-Point Parametric Analyses
| Chromosome | Marker | DNamea | Locationb(cM) | θc | LOD | P Value |
| 22 | ATA37D06 | D22S1045 | 42.8 | .19 | 2.00 | .001 |
| 2 | GATA65C03 | D2S1391 | 186.2 | .23 | 1.81 | .002 |
| 8 | AFM127xh2 | D8S262 | 4.3 | .22 | 1.67 | .003 |
| 10 | ATA22D02 | D10S1222 | 156.3 | .23 | 1.61 | .003 |
| 14 | ATA29G03Z | D14S599 | 40.7 | .24 | 1.54 | .004 |
| 1 | ATA4E02 | D1S1589 | 192.1 | .24 | 1.33 | .007 |
| 15 | GATA151F03 | D1S1507 | 60.2 | .23 | 1.06 | .014 |
Alternate marker name.
Location is given in Kosambi centimorgans from the telomere.
Recombination fraction, or distance from the marker.
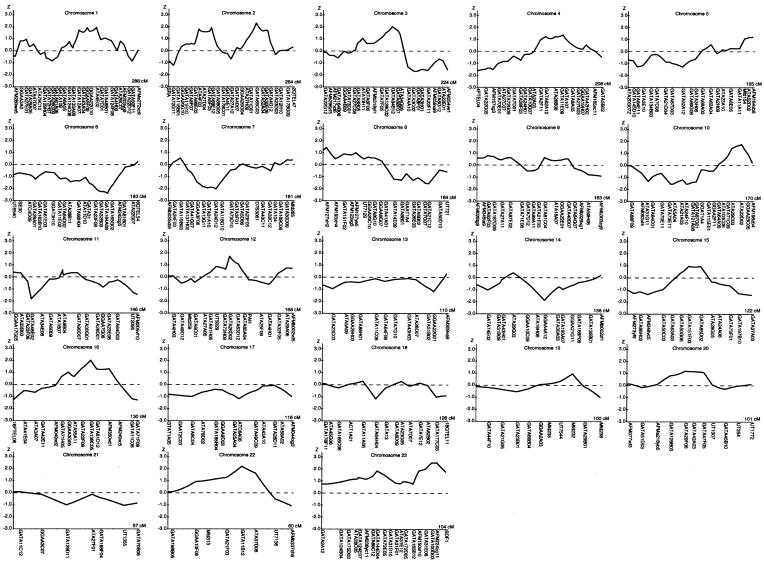
Tables 3 and 4 list the most interesting results from the multipoint analyses. Figure 1 graphically displays the plots of the NPLpairs scores from the Genehunter analyses for each of the chromosomes.
Figure 1.
Multipoint NPL analysis results for the AMD genome scan. Multipoint NPL scores were calculated with the Genehunter version 2.1 software package. In each graph, the chromosome number represented is indicated at the top of the chart, the NPL score is indicated on the left vertical axis and is traced on the curve. The positions of the microsatellite markers and the length of each chromosome are indicated on the horizontal axis.
From table 3, which summarizes the results of the Genehunter parametric and nonparametric analyses, it is clear that the signals on chromosomes 22 and 2 were again among the highest. The Spairs option in Genehunter yielded NPL scores of 2.32 at 186 cM and 2.03 at 191 cM on chromosome 2 (P=.01 and .02, respectively) and an NPL score of 2.20 at 36 cM on chromosome 22 (P=.01). The MMHLSs associated with these regions were 1.94 and 2.37 (both under the dominant model) on chromosome 2 and 0.94 (dominant) on chromosome 22. However, these analyses also implicated four new regions, on chromosomes X, 3, 16, and 12, that were not seen in the two-point results. Adjacent markers on the X chromosome at 93 cM and 97 cM produced NPL scores of 2.53 and 2.54, respectively (P=.006), with MMHLSs of 1.2 and 1.5 (both recessive); on chromosome 3, adjacent markers at 117 cM, 124 cM, and 129 cM yielded NPL scores of 1.67, 2.03, and 1.86 (P=.05, .02, and .03, respectively) and MMHLSs of 1.50, 1.19, and 1.39 (all recessive). On chromosome 16, there was an NPL score of 2.01 (P=.02) and an MMHLS of 1.32 (dominant) at a marker located at 81 cM, while a marker on chromosome 12 at 83 cM produced an NPL score of 1.71 (P=.04) and MMHLS of 1.56 (recessive). Altogether, these analyses identified 10 regions on nine chromosomes (2 regions on chromosome 2) with either MMHLS >1.0, NPL nominal P values <.05, or both.
Table 4 displays the results of multipoint nonparametric analyses, through use of the combination of MERLIN and SimWalk2. We report all P values <.05 for the NPLpairs statistic (SimWalk2 statistic D), as defined above.
Results of the MERLIN/SimWalk2 analyses are quite consistent with the Genehunter results (tables 3 and 4). Both analyses identified the same regions on chromosomes 2, 22, 3, 16, 1, 10, 12, and 8. However, the X chromosome signals identified by Genehunter were not as strong in the MERLIN/SimWalk2 results, and the MERLIN/SimWalk2 analyses identified a region on chromosome 6 that was new. For several of the signals that were identified by both multipoint analyses, the level of significance assessed by the MERLIN/SimWalk2 analyses was much greater than that indicated by Genehunter; there were 12 markers in four regions with P values of .00001 with MERLIN/SimWalk2. However, this apparent discrepancy in significance levels is not so surprising, since it is well known that, although the empirical P values calculated by SimWalk2 are somewhat conservative, the NPL score P values computed by Genehunter are extremely conservative whenever the IBD information is incomplete, as it almost always is (Kong and Cox 1997). The conservative P values computed by Genehunter are the result of the program’s overestimation of the variance of the NPL statistic, and this overestimation can be quite severe when information on IBD status is very incomplete. In this situation, the Genehunter P values can be unacceptably conservative (Kong and Cox 1997)
Significant linkage to AMD was found for one marker on chromosome 2, two adjacent markers on chromosome 3, two adjacent markers on chromosome 6, and seven contiguous markers on chromosome 8 (table 4). These findings await replication in a larger sample. In addition, the consistency of many other signals across the two-point and multipoint—as well as parametric and nonparametric—analyses indicate several other regions worthy of follow-up.
Discussion
This genomewide scan for AMD in 158 multiplex families with 511 affected sib pairs indicated several regions with internally consistent evidence (from two-point, multipoint, parametric, and nonparametric analyses) for linkage to regions on chromosomes 1, 2, 3, 8, 10, 12, 16, and 22. In addition, regions on chromosomes 6 and 23 (X) were identified by one of the multipoint analysis methods. Significant linkage was found for regions on chromosomes 2, 3, 6, and 8.
Our results on chromosome 1q are consistent with those of Klein et al. (1998), who found significant linkage in a large family, mapping to 1q25-31 between markers D1S466 and D1S413 (at 198.3 and 212.4 cM, respectively) and with those of Weeks et al. (2001), who report HLODs >2.0 for three markers in this location, D1S1660, D1S1647, and D1S1678 (212.44–218.46 cM) in an extended genome scan for AMD. Our results for chromosome 1 markers ATA4E02 and GATA48B01 (at 192.1 and 212.4 cM, respectively) are also internally consistent across the two-point and multipoint analyses, although they fall short of the criterion for suggestive linkage. Taken together, the internal consistency of our signal in this region of 1q, as well as the signals of Klein et al. (1998) and Weeks et al. (2001), provide support for a susceptibility locus on chromosome 1q.
Our finding of significant linkage (MERLIN/SimWalk 2; NPLpairs P value .00001) to two adjacent markers on chromosome 6, at positions 80.5 and 88.6 cM, agrees with the finding of possible linkage at locations at 84.0, 86.0, and 88.0 cM (P values .044, .042, and .044, respectively) reported by Schick et al. (2003). In addition, our linkage signal on chromosome 10 (D10S1222 at 156.3 cM) is within 20 cM of marker D10S1230, where Weeks et al. found some evidence for linkage in both the initial (Weeks et al. 2000) and extended (Weeks et al. 2001) genome scans for AMD. In a separate study of two large pedigrees (J.M.S., unpublished data), we also observed some evidence for linkage at marker D10S1693 (at 137.4 cM), within 5 cM of D10S1230.
With the exception of the possible role of mutations in the ABCA4 gene (MIM 60169) in a small percentage of cases of AMD (Allikmets et al. 1997; Allikmets 2000), no gene has yet been identified as playing a major role in the development of AMD. Candidate genes that have been evaluated and found not to be associated with AMD include TIMP3 (22q12.1-q13.2; MIM 188826) (De La Paz et al. 1997), ELOVL4 (6q14.1; MIM 605512) (Ayyagari et al. 2001), VMD2 (11q13; MIM 153700) (White et al. 2000; Seddon et al. 2001a), RDS (6p21.1-cen; MIM 179605) (Shastry et al. 1999), and EFEMP1 (2p16; MIM 601548) (Stone et al. 1999). Other genes, such as PON1 (7q21.3; MIM 168820) (Ikeda et al. 2001), ACE (17q23; MIM 106180) (Hamdi et al. 2002), SOD2 (6q25.3; MIM 147460) (Kimura et al. 2000), APOE (19q13.2; MIM 107741) (Klaver et al. 1998a; Souied et al. 1998), and CST3 (20p11.21; MIM 604312) (Zurdel et al. 2002) have shown a positive association in at least one study.
We found no evidence for linkage on chromosomes 11 (VMD2), 7 (PON1), 17 (ACE), or 19 (APOE) in this study. However, CST3 is within 30 cM of GATA47F05, where we calculated a two-point MLS of 1.25 on chromosome 20.
Although the SOD2 gene is located on chromosome 6, it is >80 cM away from our significant linkage peak on that chromosome. However, chromosome 6 contains a number of retinal genes—MCDR1 (6q15-q16.2; MIM 136550), RP25 (6q14-q21; MIM 602772), LCA5 (6q11-q16; MIM 604537), and RDS (6p21.1-cen; MIM 179605)—that are mapped to within 20 cM of our linkage peak, including a newly identified gene located at 6q14 (Lagali et al. 2002).
Four genes, MERTK (2q14.1; MIM 604705) (Gal et al. 2000), RP1 (8q11-q13; MIM 603937) (Liu et al. 2002), CORD9 (8p12-8q11; MIM unreviewed) (Danciger et al. 2001), and TTPA (8q13.1-q13.3; MIM 600415) (Yokota et al. 1996), might also be of potential interest as candidate genes, because of their locations on chromosomes 2 and 8.
Evaluation of the genetic contribution to late-onset diseases such as AMD poses several challenges. Many individuals in these families are young and have not yet reached the age of risk, and many of the older family members, particularly siblings and parents, have died. Therefore, several generations with informative individuals are rarely available for genetic studies. Even when large multiplex families are identified, there are other notable challenges, including the relatively high disease prevalence; ∼30% of individuals aged ⩾75 years have some form of the disease, with 6%–8% having the advanced stage causing visual loss (National Advisory Eye Council 2001). This can lead to the ubiquitous problems of genetic heterogeneity, wherein a disease may result from mutations in two or more different and unrelated genes, and pleiotropy or phenotypic heterogeneity, in which a single gene may be expressed in a variety of manifestations in different individuals, even within the same families.
AMD is heterogeneous in its phenotypic appearance, which also adds to its complexity. The broad spectrum of clinical presentation of disease raises the question of whether AMD is one disorder or perhaps several diseases with different etiologies. This variable presentation and progression of AMD leads to difficulty and imprecision in the classification of individuals as affected. A quantitative-trait analysis based on a continuous or ordinal scale of maculopathy could potentially provide more power and precision in the mapping of susceptibility genes for this relatively prevalent disorder. A few such scales are under development at the present time and may be useful tools for further genetic investigations.
Furthermore, the interaction of behavioral and environmental factors shown to be associated with AMD, such as smoking (Seddon et al. 1996) and nutritional factors (Seddon et al. 1994; Mares-Perlman et al. 1995; AREDS Research Group 2001b), may complicate genetic studies. It is possible that even if an individual inherits a putative susceptibility allele, that individual may not manifest the disease if he or she has practiced a protective lifestyle. Once one or more positional or functional candidate genes are identified, it may be informative to investigate gene-by-environment interactions, through use of some of the known environmental risk factors. It is also possible that there are several susceptibility genes that act additively (genetic heterogeneity) or multiplicatively (epistasis), as well as gene-by-environment interactions contributing to various forms of AMD.
In summary, the present study identifies significant linkage of AMD to regions on chromosomes 2, 3, 6, and 8, as well as regions of potential interest, on chromosomes 1, 10, 12, 16, 22, and X, that warrant further study. In addition, our results provide some support for the existence of potential candidate genes on chromosomes 1q, 10q, and 16q. AMD is most likely a complex disorder with both genetic and environmental components contributing to its development. Family-based association studies and linkage disequilibrium analyses will be useful tools as next steps for localizing AMD susceptibility genes.
Acknowledgments
This research was supported by National Eye Institute grant EY11309 (to J.M.S.), as well as by grants from The National Institutes of Health, Bethesda, MD; The Foundation Fighting Blindness, Owings Mills, MD; Research to Prevent Blindness, New York, NY; and Massachusetts Lions Research Fund, Inc. We wish to gratefully acknowledge all of the patients, their families, and the numerous ophthalmologists throughout the country who participated in this study. We wish also to acknowledge the technical assistance of the NHLBI Marshfield Mammalian Genotyping Service, Madison, Wisconsin; Chen-Hsing Yen, M.S., and Michael Dowd, Ph.D., each of whom assisted with the statistical analyses; Jane Armstrong at the University of Wisconsin Reading Center, for grading a subset of the fundus photographs; and, finally, the Advisory Committee Members of this project, who have provided their valuable time and expertise.
Electronic-Database Information
The URLs for data presented herein are as follows:
- Center for Medical Genetics, Marshfield Clinic, http://research.marshfieldclinic.org/genetics/
- Center for Statistical Genetics, University of Michigan, http://www.sph.umich.edu/statgen/boehnke/relpair.html (for RELPAIR)
- Fastlink Home Page, http://www.ncbi.nlm.nih.gov/CBBresearch/Schaffer/fastlink.html
- Kruglyak Labs, Fred Hutchinson Cancer Research Center, http://www.fhcrc.org/labs/kruglyak/Downloads/index.html (for Genehunter)
- Mendel, UCLA Human Genetics Software, http://www.genetics.ucla.edu/software/mendel.html
- MERLIN, http://www.sph.umich.edu/csg/abecasis/Merlin/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for ABCA4, ACE, ARMD1, APOE, CST3, EFEMP1, ELOVL4, LCA5, MCDR1, MERTK, PON1, RDS, RP25, SOD2, TIMP3, TTPA, and VMD2)
- Statgen Software, http://watson.hgen.pitt.edu/register (for SimWalk2)
References
- Abecasis GR, Cherny SS, Cookson WO,Cardon LR (2002) Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 [DOI] [PubMed] [Google Scholar]
- Allikmets R (2000) Further evidence for an association of ABCR alleles with age-related macular degeneration. The International ABCR Screening Consortium. Am J Hum Genet 67:487–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allikmets R, Shroyer NF, Singh N, Seddon JM, Lewis RA, Bernstein PS, Peiffer A, Zabriskie NA, Li Y, Hutchinson A, Dean M, Lupski JR, Leppert M (1997) Mutation of the Stargardt disease gene (ABCR) in age-related macular degeneration. Science 277:1805–1807 [DOI] [PubMed] [Google Scholar]
- AREDS Research Group (2001a) The Age-Related Eye Disease Study system for classifying age-related macular degeneration from stereoscopic color fundus photographs: the Age-Related Eye Disease Study report number 6. Am J Ophthalmol 132:668–681 [DOI] [PubMed] [Google Scholar]
- ——— (2001b) A randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins C and E, beta carotene, and zinc for age-related macular degeneration and vision loss: AREDS report number 8. Arch Ophthalmol 119:1417–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyagari R, Zhang K, Hutchinson A, Yu Z, Swaroop A, Kakuk LE, Seddon JM, Bernstein PS, Lewis RA, Tammur J, Yang Z, Li Y, Zhang H, Yashar BM, Liu J, Petrukhin K, Sieving PA, Allikmets R (2001) Evaluation of the ELOVL4 gene in patients with age-related macular degeneration. Ophthalmic Genet 22:233–239 [DOI] [PubMed] [Google Scholar]
- Boehnke M, Cox NJ (1997) Accurate inference of relationships in sib-pair linkage studies. Am J Hum Genet 61:423–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E, Hung S, Willett WC, Spiegelman D, Rimm EB, Seddon JM, Colditz GA, Hankinson SE (2001) Prospective study of dietary fat and the risk of age-related macular degeneration. Am J Clin Nutr 73:209–218 [DOI] [PubMed] [Google Scholar]
- Chylack LT Jr, Wolfe JK, Singer DM, Leske MC, Bullimore MA, Bailey IL, Friend J, McCarthy D, Wu SY (1993) The lens opacities classification system III. The Longitudinal Study of Cataract Study Group. Arch Ophthalmol 111:831–836 [DOI] [PubMed] [Google Scholar]
- Cottingham RW Jr, Idury RM, Schaffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed] [Google Scholar]
- Danciger M, Hendrickson J, Lyon J, Toomes C, McHale JC, Fishman GA, Inglehearn CF, Jacobson SG, Farber DB (2001) CORD9 a new locus for arCRD: mapping to 8p11, estimation of frequency, evaluation of a candidate gene. Invest Ophthalmol Vis Sci 42:2458–2465 [PubMed] [Google Scholar]
- De Jong PT, Klaver CC, Wolfs RC, Assink JM, Hofman A (1997) Familial aggregation of age-related maculopathy. Am J Ophthalmol 124:862–863 [DOI] [PubMed] [Google Scholar]
- De La Paz MA, Pericak-Vance MA, Lennon F, Haines JL, Seddon JM (1997) Exclusion of TIMP3 as a candidate locus in age-related macular degeneration. Invest Ophthalmol Vis Sci 38:1060–1065 [PubMed] [Google Scholar]
- Delcourt C, Diaz J, Ponton-Sanchez A, Papoz L (1998) Smoking and age-related macular degeneration: the POLA study. Arch Ophthalmol 116:1031–1035 [DOI] [PubMed] [Google Scholar]
- Duren WL, Cox NJ, Hauser ER, Boenke M, FUSION Study Group (1997) Software for determining most likely relationships in relative pairs. Am J Hum Genet Suppl 61:A273 [Google Scholar]
- Durner M, Vieland VJ, Greenberg DA (1999) Further evidence for the increased power of LOD scores compared with nonparametric methods. Am J Hum Genet 64:281–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein MP, Duren WL, Boehnke M (2000) Improved inference of relationship for pairs of individuals. Am J Hum Genet 67:1219–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eye Disease Case-Control Study Group, The (1992) Risk factors for neovascular age-related macular degeneration. Arch Ophthalmol 110:1701–1708 [DOI] [PubMed] [Google Scholar]
- Friedman E (1997) A hemodynamic model of the pathogenesis of age-related macular degeneration. Am J Ophthalmol 124:677–682 [DOI] [PubMed] [Google Scholar]
- Gal A, Li Y, Thompson DA, Weir J, Orth U, Jacobson SG, Apfelstedt-Sylla E, Vollrath D (2000) Mutations in MERTK, the human orthologue of the RCS rat retinal dystrophy gene, cause retinitis pigmentosa. Nat Genet 26:270–271 [DOI] [PubMed] [Google Scholar]
- Hamdi HK, Reznik J, Castellon R, Atilano SR, Ong JM, Udar N, Tavis JH, Aoki AM, Nesburn AB, Boyer DS, Small KW, Brown DJ, Kenney MC (2002) Alu DNA polymorphism in ACE gene is protective for age-related macular degeneration. Biochem Biophys Res Commun 295:668–672 [DOI] [PubMed] [Google Scholar]
- Hammond CJ, Webster AR, Snieder H, Bird AC, Gilbert CE, Spector TD (2002) Genetic influence on early age-related maculopathy: a twin study. Ophthalmology 109:730–736 [DOI] [PubMed] [Google Scholar]
- Heiba IM, Elston RC, Klein BE, Klein R (1994) Sibling correlations and segregation analysis of age-related maculopathy: the Beaver Dam Eye Study. Genet Epidemiol 11:51–67 [DOI] [PubMed] [Google Scholar]
- Hyman L, Schachat AP, He Q, Leske MC (2000) Hypertension, cardiovascular disease, and age-related macular degeneration. Arch Ophthalmol 118:351–358 [DOI] [PubMed] [Google Scholar]
- Ikeda T, Obayashi H, Hasegawa G, Nakamura N, Yoshikawa T, Imamura Y, Koizumi K, Kinoshita S (2001) Paraoxonase gene polymorphisms and plasma oxidized low-density lipoprotein level as possible risk factors for exudative age-related macular degeneration. Am J Ophthalmol 132:191–195 [DOI] [PubMed] [Google Scholar]
- Kimura K, Isashiki Y, Sonoda S, Kakiuchi-Matsumoto T, Ohba N (2000) Genetic association of manganese superoxide dismutase with exudative age-related macular degeneration. Am J Ophthalmol 130:769–773 [DOI] [PubMed] [Google Scholar]
- Klaver CC, Kliffen M, van Duijn CM, Hofman A, Cruts M, Grobbee DE, van Broeckhoven C, de Jong PT (1998a) Genetic association of apolipoprotein E with age-related macular degeneration. Am J Hum Genet 63:200–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaver CC, Wolfs RC, Assink JJ, Van Duijn CM, Hofman A, de Jong PT (1998b) Genetic risk of age-related maculopathy. Population-based familial aggregation study. Arch Ophthalmol 116:1646–1651 [DOI] [PubMed] [Google Scholar]
- Klein BE, Klein R, Lee KE, Moore EL, Danforth L (2001) Risk of incident age-related eye diseases in people with an affected sibling: The Beaver Dam Eye Study. Am J Epidemiol 154:207–211 [DOI] [PubMed] [Google Scholar]
- Klein ML, Schultz DW, Edwards A, Matise TC, Rust K, Berselli CB, Trzupek K, Weleber RG, Ott J, Wirtz MK, Acott TS (1998) Age-related macular degeneration. Clinical features in a large family and linkage to chromosome 1q. Arch Ophthalmol 116:1082–1088 [DOI] [PubMed] [Google Scholar]
- Klein R, Klein BEK, Jensen SC, Meuer SM (1997) The five-year incidence and progression of age-related maculopathy. Ophthalmology 104:7–21 [DOI] [PubMed] [Google Scholar]
- Klein R, Klein BEK, Linton KLP (1992) Prevalence of age-related maculopathy. Ophthalmology 99:933–943 [DOI] [PubMed] [Google Scholar]
- Kong A, Cox NJ (1997) Allele sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Lagali PS, Kakuk LE, Griesinger IB, Wong PW, Ayyagari R (2002) Identification and characterization of C6orf37, a novel candidate human retinal disease gene on chromosome 6q14. Biochem Biophys Res Commun 293:356–365 [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Lange K, Weeks D, Boehnke M (1988) Programs for pedigree analysis: MENDEL, FISHER, and dGENE. Genet Epidemiol 5:471–472 [DOI] [PubMed] [Google Scholar]
- Liu Q, Zhou J, Daiger SP, Farber DB, Heckenlively JR, Smith JE, Sullivan LS, Zuo J, Milam AH, Pierce EA (2002) Identification and subcellular localization of the RP1 protein in human and mouse photoreceptors. Invest Ophthalmol Vis Sci 43:22–32 [PMC free article] [PubMed] [Google Scholar]
- Mares-Perlman JA, Brady WE, Klein R, VandenLangenberg GM, Klein BEK, Palta M (1995) Dietary fat and age-related maculopathy. Arch Ophthalmol 113:743–748 [DOI] [PubMed] [Google Scholar]
- National Advisory Eye Council (2001) Vision research—a national plan: 1999–2003, vol 1. A report of the National Advisory Eye Council. National Institutes of Health, Bethesda, MD (NIH Publication Number 98–4120) [Google Scholar]
- Schaffer AA, Gupta SK, Shriram K, Cottingham RW Jr (1994) Avoiding recomputation in linkage analysis. Hum Hered 44:225–237 [DOI] [PubMed] [Google Scholar]
- Schick JH, Iyengar SK, Klein BE, Klein R, Reading K, Liptak R, Millard C, Lee KE, Tomany SC, Moore EL, Fijal BA, Elston RC (2003) A whole-genome screen of a quantitative trait of age-related maculopathy in sibships from the beaver dam eye study. Am J Hum Genet 72:1412–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seddon J (2001) Epidemiology of age-related macular degeneration. In: Ryan SJ (ed) Retina, vol 2, 3rd ed. Mosby, St. Louis, pp 1039–1050 [Google Scholar]
- Seddon JM, Afshari MA, Sharma S, Bernstein PS, Chong S, Hutchinson A, Petrukhin K, Allikmets R (2001a) Assessment of mutations in the Best macular dystrophy (VMD2) gene in patients with adult-onset foveomacular vitelliform dystrophy, age-related maculopathy, and bull’s-eye maculopathy. Ophthalmology 108:2060–2067 [DOI] [PubMed] [Google Scholar]
- Seddon JM, Ajani UA, Mitchell BD (1997) Familial aggregation of age-related maculopathy. Am J Ophthalmol 123:199–206 [DOI] [PubMed] [Google Scholar]
- Seddon JM, Ajani UA, Sperduto RD, Hiller R, Blair N, Burton TC, Farber MD, Gragoudas ES, Haller J, Miller DT (1994) Dietary carotenoids, vitamins A, C, and E, and advanced age-related macular degeneration. Eye disease case-control study group. JAMA 272:1413–1420 [PubMed] [Google Scholar]
- Seddon JM, Rosner B, Sperduto RD, Yannuzzi L, Haller JA, Blair NP, Willett W (2001b) Dietary fat and risk for advanced age-related macular degeneration. Arch Ophthalmol 119:1191–1199 [DOI] [PubMed] [Google Scholar]
- Seddon JM, Sahagian CR, Glynn RJ, Sperduto RD, Gragoudas ES (1990) Evaluation of an iris color classification system. The Eye Disorders Case-Control Study Group. Invest Ophthalmol Vis Sci 31:1592–1598 [PubMed] [Google Scholar]
- Seddon JM, Willett WC, Speizer FE, Hankinson SE (1996) A prospective study of cigarette smoking and age-related macular degeneration in women. JAMA 276:1141–1146 [PubMed] [Google Scholar]
- Shastry BS, Trese MT (1999) Evaluation of the peripherin/RDS gene as a candidate gene in families with age-related macular degeneration. Ophthalmologica 213:165–170 [DOI] [PubMed] [Google Scholar]
- Smith CAB (1963) Testing for heterogeneity of recombination fraction values in human genetics. Ann Hum Genet 27:175–182 [DOI] [PubMed] [Google Scholar]
- Smith W, Mitchell P, Leeder SR (1996) Smoking and age-related maculopathy. Arch Ophthalmol 114:1518–1523 [DOI] [PubMed] [Google Scholar]
- Sobel E, Lange K (1996) Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet 58:1323–1337 [PMC free article] [PubMed] [Google Scholar]
- Souied EH, Benlian P, Amouyel P, Feingold J, Lagarde JP, Munnich A, Kaplan J, Coscas G, Soubrane G (1998) The ε4 allele of the apolipoprotein E gene as a potential protective factor for exudative age-related macular degeneration. Am J Ophthalmol 125:353–359 [DOI] [PubMed] [Google Scholar]
- Sperduto RD, Hiller R (1986) Systemic hypertension and age-related maculopathy in the Framingham study. Arch Ophthalmol 104:216–219 [DOI] [PubMed] [Google Scholar]
- Stone EM, Lotery AJ, Munier FL, Heon E, Piguet B, Guymer RH, Vandenburgh K, Cousin P, Nishimura D, Swiderski RE, Silvestri G, Mackey DA, Hageman GS, Bird AC, Sheffield VC, Schorderet DF (1999) A single EFEMP1 mutation associated with both Malattia Leventinese and Doyne honeycomb retinal dystrophy. Nat Genet 22:199–202 [DOI] [PubMed] [Google Scholar]
- VandenLangenberg G, Mares-Perlman J, Klein R, Klein B, Brady W, Palta M (1998) Associations between antioxidant and zinc intake and the 5-year incidence of early age-related maculopathy in the Beaver Dam Eye Study. Am J Epidemiol 148:204–214 [DOI] [PubMed] [Google Scholar]
- Vieland VJ, Hodge SE, Greenberg DA (1992) Adequacy of single-locus approximations for linkage analyses of oligogenic traits. Genet Epidemiol 9:45–59 [DOI] [PubMed] [Google Scholar]
- Vingerling JR, Hofman A, Grobbee DE, deJong PT (1996) Age-related macular degeneration and smoking. Arch Ophthalmol 114:1193–1196 [DOI] [PubMed] [Google Scholar]
- Weeks DE, Conley YP, Mah TS, Paul TO, Morse L, Ngo-Chang J, Dailey JP, Ferrell RE, Gorin MB (2000) A full genome scan for age-related maculopathy. Hum Mol Genet 9:1329–1349 [DOI] [PubMed] [Google Scholar]
- Weeks DE, Conley YP, Tsai HJ, Mah TS, Rosenfeld PJ, Paul TO, Eller AW, Morse LS, Dailey JP, Ferrell RE, Gorin MB (2001) Age-related maculopathy: an expanded genome-wide scan with evidence of susceptibility loci within the 1q31 and 17q25 regions. Am J Ophthalmol 132:682–692 [DOI] [PubMed] [Google Scholar]
- Weeks DE, Sobel E, O’Connell JR, Lange K (1995) Computer programs for multilocus haplotyping of general pedigrees. Am J Hum Genet 56:1506–1507 [PMC free article] [PubMed] [Google Scholar]
- White K, Marquardt A, Weber BH (2000) VMD2 mutations in vitelliform macular dystrophy (Best disease) and other maculopathies. Hum Mutat 15:301–308 [DOI] [PubMed] [Google Scholar]
- Whittemore AS, Halpern J (1994) A class of tests for linkage using affected pedigree members. Biometrics 50:118–127 [PubMed] [Google Scholar]
- Yokota T, Shiojiri T, Gotoda T, Arai H (1996) Retinitis pigmentosa and ataxia caused by a mutation in the gene for the alpha-tocopherol-transfer protein. N Engl J Med 335:1770–1771 [DOI] [PubMed] [Google Scholar]
- Zurdel J, Finckh U, Menzer G, Nitsch RM, Richard G (2002) CST3 genotype associated with exudative age related macular degeneration. Br J Ophthalmol 86:214–219 [DOI] [PMC free article] [PubMed] [Google Scholar]