Abstract
We previously reported a genomewide scan to identify autism-susceptibility loci in 110 multiplex families, showing suggestive evidence (P < .01) for linkage to autism-spectrum disorders (ASD) on chromosomes 5, 8, 16, 19, and X and showing nominal evidence (P < .05) on several additional chromosomes (2, 3, 4, 10, 11, 12, 15, 18, and 20). In this follow-up analysis we have increased the sample size threefold, while holding the study design constant, so that we now report 345 multiplex families, each with at least two siblings affected with autism or ASD phenotype. Along with 235 new multiplex families, 73 new microsatellite markers were also added in 10 regions, thereby increasing the marker density at these strategic locations from 10 cM to ∼2 cM and bringing the total number of markers to 408 over the entire genome. Multipoint maximum LOD scores (MLS) obtained from affected–sib-pair analysis of all 345 families yielded suggestive evidence for linkage on chromosomes 17, 5, 11, 4, and 8 (listed in order by MLS) (P < .01). The most significant findings were an MLS of 2.83 (P = .00029) on chromosome 17q, near the serotonin transporter (5-hydroxytryptamine transporter [5-HTT]), and an MLS of 2.54 (P = .00059) on 5p. The present follow-up genome scan, which used a consistent research design across studies and examined the largest ASD sample collection reported to date, gave either equivalent or marginally increased evidence for linkage at several chromosomal regions implicated in our previous scan but eliminated evidence for linkage at other regions.
Introduction
Autism [MIM 209850] is a neuropsychiatric developmental disorder with a prevalence of 4–10 per 10,000 individuals (Smalley et al. 1988; Gillberg and Wing 1999; Fombonne et al. 2002) and a three- to fourfold higher incidence in boys than in girls (Folstein and Rosen-Sheidley 2001). Essential diagnostic features of autism include severe impairment in development of social interactions, a marked and sustained impairment of both verbal and nonverbal communication, and restricted, repetitive, or stereotyped behaviors and interests (American Psychiatric Association 2000), occurring within the first 3 years of life. Autism describes the most severe manifestation of a broad spectrum of disorders that include Asperger syndrome (AS [MIM 209850]) and are collectively categorized as “pervasive developmental disorders” (PDDs [MIM 209850]). It is recognized that autism and these autism spectrum disorders (ASD), which have a much higher prevalence of 10–60 individuals per 10,000 (Chakrabarti and Fombonne 2001; Charman 2002; Yeargin-Allsopp et al. 2003), share essential clinical and behavioral features although they differ in severity and age at onset.
Evidence from twin and family studies clearly establishes the importance of genetic factors in the development of autism and ASD (Folstein and Rutter 1977; Bailey et al. 1995). MZ twins show 40%–60% and 70%–90% concordance for autism and ASD, respectively, compared with rates of 0%–25% concordance, depending on diagnosis, among DZ twins (Folstein and Rutter 1977; Steffenburg et al. 1989; Bailey et al. 1995; Lauritsen et al. 2001). Although an accurate estimation of the population prevalence is complicated by an apparent increase in autism over recent decades (Gillberg and Wing 1999), it is clear that the estimated sibling recurrence risk of 2%–6% is significantly greater than risk of .04%–.1% in the general population (Smalley et al. 1988; Smalley 1997; Szatmari et al. 1998; Gillberg and Wing 1999). The estimated sibling risk for autism is therefore 50–100 times greater than the population risk (Lamb et al. 2000). The studies cited above indicate that autism and ASD are highly heritable, with some studies of ASD reporting heritability >90% (Bailey et al. 1995). However, these studies also make it clear that ASDs are complex genetic disorders that most likely result from the combinatorial effects of multiple genetic and environmental factors.
Nine independent genomewide scans with DNA markers have been performed to try to detect genetic variation related to autism or ASD (International Molecular Genetic Study of Autism Consortium [IMGSAC] 1998, 2001b; Barrett et al. 1999; Risch et al. 1999; Philippe et al. 1999; Buxbaum et al. 2001; Liu et al. 2001; Auranen et al. 2002; Shao et al. 2002). Although nominal suggestive evidence for linkage between DNA markers and given ASD phenotypes has been reported on 17 of the 22 autosomes and at various locations on the X chromosome, only a few regions appear to be supported by independent studies (Folstein and Rosen-Sheidley 2001). The most consistent finding to date, which is supported by meta-analysis, has been on chromosome 7q (Badner and Gershon 2002). A study from the IMGSAC provided initial evidence of genomewide linkage to 7q, in a study of 99 families, and subsequently strengthened the findings with the addition of 83 sib pairs and high-density markers across the implicated region (IMGSAC 1998, 2001b, 2001a). Several independent studies have produced nominal-to-suggestive evidence of linkage to ASD phenotypes on 7q as well (Barrett et al. 1999; Buxbaum et al. 2001; Liu et al. 2001; Alarcón et al. 2002; Auranen et al. 2002; Shao et al. 2002).
We previously reported genomewide screens for autism- and ASD-related loci in 110 families and for a language-development quantitative-trait locus (QTL) in 152 multiplex families, along with a dense-marker screen covering much of chromosome 7 in a total of 160 multiplex families from the Autism Genetic Resource Exchange (AGRE) (Liu et al. 2001; Alarcón et al. 2002). Analysis of these initial studies yielded suggestive evidence for linkage on chromosomes 5q, 19p, and X (Liu et al. 2001), as well as suggestive evidence of a speech- and language-related locus on 7q (Alarcón et al. 2002). In the present study, we have added 235 new multiplex families affected by autism. The ascertainment scheme, diagnostic methods, microsatellite markers, and genetic analysis were all maintained from the initial study, although the pedigree and phenotype information for some of the original families has been updated since our last analyses. Here, we present the results of a genomewide scan with 408 microsatellite markers and 345 multiplex families, each with a minimum of two individuals who met criteria for a diagnosis of autism or ASD.
Families and Methods
Family Recruitment and Diagnosis
The sample comprises 345 multiplex families with ASD from the AGRE, a publicly available database consisting of biomaterials and genotype and phenotype data that was founded by the Cure Autism Now Foundation and is now supported by the National Institute of Mental Health (NIMH) as part of the NIMH genetics initiative. Families are recruited through a variety of methods (e.g., physician referral, Web site contact, and family meetings and seminars). They are ascertained on the basis that at least two family members met criteria for a diagnosis of an ASD (autism, AS, or PDD). Of these families, 110 have been analyzed elsewhere by our group (Liu et al. 2001; Alarcón et al. 2002); the present study adds 235 new families. Family recruitment and phenotypic assessment of the AGRE sample were conducted as described elsewhere (Geschwind et al. 2001). Diagnosis is established by the Autism Diagnostic Interview–Revised (ADI-R) (Lord et al. 1994). The ADI-R is currently the gold standard for research diagnosis and is based on the classifications of both the International Statistical Classification of Diseases and Related Health Problems, 10th revision (World Health Organization 1992) as well as the Diagnostic and Statistical Manual of Mental Disorders, 4th edition (American Psychiatric Association 2000). To be scored as affected, individuals must meet criteria in all three content areas of the ADI-R: (1) quality of social interaction, communication, and language; (2) repetitive, restricted, and stereotyped interests and behavior; and (3) age at onset <3 years (Lord et al. 1994). Collection of additional clinical and neurodevelopmental information is ongoing and has been obtained from 160 families; additional information was obtained through a medical examination and use of the Autism Diagnostic Observation Scale (Lord et al. 2001). Possible nonidiopathic autism cases are flagged in the AGRE data set (e.g., fragile X syndrome or perinatal insult) and are excluded from any genotyping and analysis.
In accordance with methods described elsewhere (Liu et al. 2001), the present study used two diagnostic categories for genetic linkage analysis: narrow and broad. The narrow category includes individuals with a diagnosis of autism based on the ADI-R (664 individuals of a total of 1,795), as well as individuals who were categorized as having a disorder that was “not quite autism” (NQA) (52 individuals of a total of 1,795). The inclusion of NQA individuals in the narrow category is consistent with results of the genome scan we reported elsewhere (Liu et al. 2001). The categorization of NQA represents individuals who are no more than 1 point away from meeting criteria for autism in any or all of the three content domains or individuals who meet criteria in all domains but do not meet age-at-onset criteria. Thus, they are individuals who would be identified by most clinicians as autistic but who narrowly miss meeting the ADI-R criteria. The broad category includes all individuals in the narrow category, as well as those who show patterns of impairment along the spectrum of PDDs (117 individuals designated as “broad spectrum” plus 664 as “autism” plus 52 as “NQA”). This broad diagnostic category encompasses individuals ranging from mildly to severely impaired and includes such PDDs as PDD–not otherwise specified and Asperger syndrome (full diagnostic protocol is available on the Autism Genetic Resource Exchange Web page). Multipoint affected–sib-pair (ASP) analysis based on only the strictest definition of autism was also performed, thereby excluding the 52 NQA individuals as affected siblings, although this analysis was not used to determine linkage results. Our sample contains 345 families that have two or more individuals that meet the diagnostic criteria for the broad diagnosis. Of those families, 7 have no members with the narrow diagnosis, 53 have only 1 member with the narrow diagnosis, and 285 have ⩾2 members with the narrow diagnosis. Thus, the analyses for the narrow diagnosis are based on 285 families, whereas the analyses for the broad diagnosis utilize all 345 families. The majority of the families reported here have two affected siblings (307 of 345 families), although 34 families have three affected siblings, and 4 families have four affected siblings (table 1).
Table 1.
Summary of AGRE Families[Note]
|
No. in Families with |
||||||||
|
Total No. of |
Two AffectedSiblings |
Three AffectedSiblings |
Four AffectedSiblings |
|||||
| PhenotypeDefinition | Families | Probands | Families | Probands | Families | Probands | Families | Probands |
| Narrow | 285 | 716 | 265 | 530 | 17 | 51 | 3 | 12 |
| Broad | 345 | 833 | 307 | 614 | 34 | 102 | 4 | 16 |
Note.— Data summarize structure of families in the narrowly and broadly defined phenotype groups (as described in “Families and Methods” section).
A total of 362 families were phenotyped and genotyped, including 12 families whose only affected siblings were MZ twins. These 12 families were purposely included as an internal control, to estimate detectable genotyping errors, but were excluded from linkage analysis. After removal of an additional 5 families in which genotyping was unsuccessful, we were left with 345 families that included ⩾2 affected siblings, providing a total of 381 sib pairs that fall into the broad disease classification. The narrow disease classification, comprising autism and NQA, consists of 285 families and a total of 321 sib pairs.
Since our previous report (Liu et al. 2001), AGRE has updated family relationships and diagnostic information, leading to corrections in six pedigrees and 35 phenotypes among the original 110 families. Reanalysis of the 110 families with the new pedigree and phenotype information reflects these changes in relation to the linkage results reported elsewhere (data indicated as appropriate in the “Results” section). In addition, given that MZ twins are identical at every locus, both twins cannot be used in genetic analysis, because they will inflate linkage values at every point in the genome. Therefore, as in our previous study, one of each MZ twin pair was randomly removed. In those families in which the MZ twins were the only affected individuals, the entire family was removed, because it was no longer informative for ASP analysis. We also previously reported a fine-map analysis of chromosome 7 that included an additional 50 families (Liu et al. 2001). Among those 50 families, 46 have now been genotyped over the entire genome and represent a fraction of the 235 new families added in this study. However, 2 of the 50 families were eliminated when purported DZ twins were determined to be MZ, and 2 other families were removed because they were found to have only one affected child, on the basis of the updated phenotypic information.
Genotyping
Laboratory and genotyping procedures have been described in detail elsewhere (Liu et al. 2001). Blood was drawn from affected individuals, as well as from parents and unaffected siblings (when available). DNA was extracted from whole blood or immortalized lymphoblast cell lines after standard proteinase digestion and salting-out protocols (Medrano et al. 1990). The DNA marker panel consisted of a set of 408 microsatellite markers, which includes the entire 335 microsatellite marker panel used in our previous genomewide scan (Liu et al. 2001) plus an additional 73 microsatellite markers specifically selected to augment information from peak MLS regions determined after an intermediate analysis (110 plus 46 additional families), increasing marker density to an average of 2 cM in these regions. Thirty of these 73 additional markers span a region of ∼60 cM on chromosome 7q. The majority of all microsatellite markers were selected from version 8.0 of the Marshfield genome screening set, as described elsewhere (Liu et al. 2001). The average heterozygosity of the markers used in this study is 0.77, with an average density of 10 cM and a few regions of denser coverage. Linkage map distances, in Kosambi centimorgans, were obtained from the Marshfield Center for Medical Genetics (Broman et al. 1998).
PCR amplification of microsatellite markers was performed as described elsewhere (Aita et al. 1999; Liu et al. 2001). PCR products were resolved using the PRISM 377XL data collection software, and base pair size was called using GENESCAN, v. 2.1, and GENOTYPER, v. 1.1.1, software packages (Applied Biosystems). Computer-generated genotypes were checked by an independent researcher who was blind to each individual’s identity, pedigree, sex, and disease diagnosis. The genotypes were then imported to the LABMAN database (Adams 1994). LABMAN was used to identify allele-binning errors; LABMAN and PedCheck (O’Connell and Weeks 1998) were both used to identify Mendelization errors. Biological relationships were reconfirmed using genotypic information with the program RelCheck (Broman and Weber 1998).
Statistical and Genetic Analysis
Marker allele frequencies were estimated with GCONVERT (Liu et al. 2002), using all genotyped individuals; this conservative estimation of frequencies diminishes the risk of type I error due to lack of parental genotypes in a few families (Göring and Terwilliger 2000a). Across the entire sample, 322 families include both parents, 14 families are missing one parent, and 9 families are missing both parents. Multipoint ASP analysis of genotype data was performed using the Mapmaker/Sibs program (v. 2.1) that is part of the GENEHUNTER 2.1 software package (Kruglyak and Lander 1995; Kruglyak et al. 1996). Specifically, we used the weighted “all pairs” option and set the increment function to scan at a distance of 1.0 cM throughout the genetic map. ASP analysis calculates the probabilities of sharing zero, one, or two alleles (z0, z1, or z2) identical by descent (IBD) between sib pairs, since loci that are involved in susceptibility to a trait will have probabilities (z0, z1, and z2) that differ from the expected Mendelian proportions (Risch 1990a, 1990b). Under the assumption of dominance variation, we limited the sharing probabilities to the “possible triangle,” as described elsewhere (Faraway 1993; Holmans 1993), to ensure biological consistency; this is defined by:
Two-point parametric analysis was performed on the entire data set, using Fastlink (Lathrop et al. 1984; Cottingham et al. 1993). We chose to use the pseudomarker parameters for our two-point analysis (Göring and Terwilliger 2000b), which approximates a “model-free” affected–relative-pair analysis but maintains the important property of LOD-score analysis that pedigree correlations between all relatives are considered jointly rather than breaking the pedigree into a set of all possible relative pairs. Since the actual mode of inheritance for ASD is unknown, both dominant and recessive models were applied to our analyses.
Analysis on the X chromosome was performed using Mapmaker/Sibs (v 2.0) (Kruglyak and Lander 1995). This algorithm follows Cordell’s extension of Holmans’s method to analyze X-linked data by considering brother-brother (bb), sister-brother (sb), and sister-sister (ss) pairs separately (Cordell et al. 1995). To consider the three groups independently, the maximization is restricted to the following genetically valid values:
where z1 represents IBD sharing of maternal alleles (Nyholt 2000). The sum of these three MLS statistics, known as “X-MLS,” is a mixture of χ2 distributions with 1, 2, and 3 df (Cordell et al. 1995; Nyholt 2000). Therefore, to obtain a point-wise P value <.05, an X-MLS >1.18 is required, whereas evidence for suggestive and significant linkage is reached at X-MLS values of 3.06 and 4.62, respectively (Nyholt 2000). P values reported for all chromosomes were determined according to the guidelines presented by Nyholt (2000).
The broad and narrow data sets are not independent; in fact, the narrow data set makes up >82% (285/345) of the broad data set. No corrections were made to account for overlap between these analyses or for the overlap of the 110 families from our previous analysis and the combined sample of 345 families in the complete data set; instead, we present all information from the analysis and allow readers to make their own interpretations as to statistical significance.
To clarify the interpretation of our linkage results, we used simulated data sets to calculate empirical genomewide significance values. Under the assumption of no linkage, 100 replicates were simulated for both the narrow and broad data sets by use of the SIMULATE program (Terwillger and Ott 1994). Each replicate was then analyzed using GENEHUNTER 2.1. Genomewide significance was then determined on the basis of the number of times an MLS was reached in the 100 replications.
Results
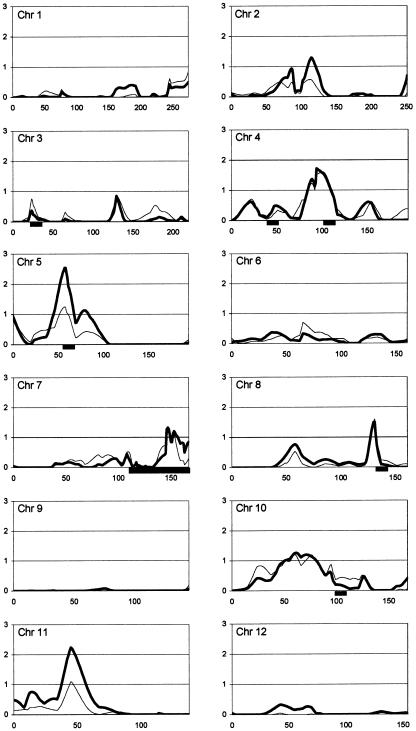
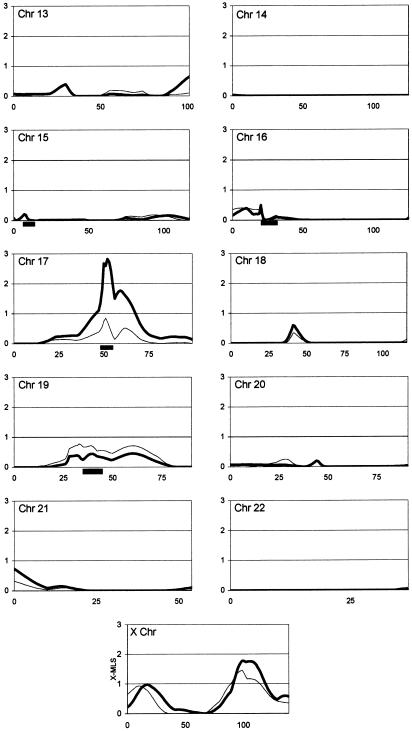
Results from the genomewide multipoint ASP analysis are shown in figure 1. Linkage analysis produced MLS peaks with a significance level less than a pointwise threshold of P<.05 on 12 chromosomes (1, 2, 3, 4, 5, 7, 8, 10, 11, 17, 19, and X). The single largest peak from the total set of 345 families is located on chromosome 17q near marker D17S1800, with an MLS of 2.83 (P=.00029), using the broad diagnostic classification (MLS of 0.85 with the narrow classification). The second highest MLS found in the entire genome was 2.54 (P=.00059) on chromosome 5p near marker D5S2494, again using the broad classification (MLS of 1.25 with the narrow classification). Other regions of interest include chromosome 11p between markers D11S1392 and D11S1993, which produced an MLS of 2.24 (P=.0012) with the broad classification (MLS of 1.10 with the narrow classification). The region on chromosome 4q between markers D4S2361 and D4S2909 gave MLS values of 1.6 (P=.0058) and 1.72 (P=.0043) with the narrow and broad classifications, respectively. Also, the region on chromosome 8q near marker D8S1832 gave an MLS of 1.6 (P=.0058) with the narrow classification and 1.5 (P=.0074) with the broad classification. All other chromosomal regions produced MLS values <1.4 (P>.01) or an X-MLS < 1.9 (P>.01) (Nyholt 2000). Multipoint ASP analysis was also preformed for the strictest definition of autism, as opposed to our narrow category, which includes NQA; however, the results were almost identical to those for the narrow category (data not shown). Genomewide two-point analysis (data not shown) identified peaks in the same chromosomal regions as multipoint ASP analysis, although with consistently lower LOD scores, as shown for several peaks in table 2.
Figure 1.
Multipoint MLS results from Mapmaker/Sibs ASP analysis, shown by chromosome. The X-axis depicts genetic distance in Kosambi centimorgans from pter (zero coordinate) to qter; the Y-axis represents the MLS for all autosomes and the X-MLS for the X chromosome. Thin lines represent the narrow disease classification; thick lines represent the broad disease classification (see the “Families and Methods” section). Black bars below the X-axis show regions with dense microsatellite marker coverage.
Table 2.
Results of Analyses in 345 Families[Note]
|
Multipoint MLS |
Two-Point LOD |
|||||
| Chromosome | Score | Pa | Peak(cM)b | Score | Pa | Peak(cM)b |
| 17 | 2.83 | .000298 | 52 | 1.236c | .0085 | 48 |
| 5 | 2.54 | .000595 | 58 | 1.43c | .0051 | 57 |
| 11 | 2.24 | .0012 | 45 | .496c | .065 | 54 |
| 4 | 1.72 | .0043 | 94 | 1.69c | .0026 | 101 |
| 8 | 1.60 | .0058 | 131 | .812d | .0266 | 135 |
Note.— Only the five chromosomal regions with MLS > 1.4 (P<.01) in the analysis of all 345 families are shown.
Two-tailed P values were calculated according to methods described by Nyholt (2000).
Position of the highest point/marker expressed as distance from pter = 0
Highest two-point LOD score obtained with a recessive model.
Highest two-point LOD score obtained with a dominant model.
On the basis of the linkage findings from the initial set of 110 families (Liu et al. 2001), as well as several independent studies, additional microsatellite markers were added to chromosome 7q to generate dense coverage (fine map) across this important region. On the basis of the linkage findings from an intermediate stage of analysis (156 families; P<.05), 10 additional chromosomal regions (3p, 4p, 4q, 5p, 8q, 10q, 15p, 16p, 17q, and 19q) were selected for dense-marker coverage. In only 4 of the 11 regions observed, the addition of fine-map markers diminished evidence of linkage, whereas in the remaining regions, evidence for linkage was either maintained or increased, as illustrated by the examples shown in table 3.
Table 3.
Multipoint MLSs and Peak Positions in Three Separate Analyses[Note]
|
Updated 110 Familiesa |
345 Familiesb |
345 Familiesand Fine-Map Markersc |
|||||||
| Chromosome | Peak(cM)d | MLS | Pe | Peak(cM)d | MLS | Pe | Peak(cM)d | MLS | Pe |
| 4 | 94 | .713 | .054 | 92 |
1.72 |
.0043 |
94 |
1.72 |
.0043 |
| 5 | 0 |
1.44 |
.0085 |
0 | .839 | .0388 | 0 | .869 | .0359 |
| 5 | 59 |
2.01 |
.0021 |
60 |
1.89 |
.00285 |
59 |
2.54 |
.00059 |
| 8 | 130 |
1.71 |
.0044 |
132 | .84 | .0388 | 132 |
1.6 |
.0058 |
| 11 | 53 | 1.08 | .021 | 46 |
2.24 |
.0012 |
… | … | … |
| 17 | 49 | .633 | .0667 | 52 |
2.04 |
.00198 |
52 |
2.83 |
.00029 |
| 19 | 35 |
3.36 |
.000085 |
39 | .686 | .058 | 33 | .778 | .045 |
| X | 139 |
2.27 |
.0044 |
99 | 1.78 | .003 | … | … | … |
Note.— Chromosomal regions with MLS > 1.4 (P < .01) in each analysis are underlined. For clarification and comparison, we show the corrected results of the original 110 families for these regions with the highest MLSs. Chromosomes 11 and X did not have any fine map markers added, so the two analyses of the 345 families are identical.
Original 110 families reanalyzed with the updated pedigree and phenotype information from AGRE.
Total data set, without the additional high-density fine-map markers; therefore, 335 microsatellite markers covering the genome at 10-cM density.
Total data set and fine-map markers, with all 408 microsatellite markers.
Position of the highest point expressed as distance from pter = 0.
Two-tailed P values were calculated according to methods described by Nyholt (2000).
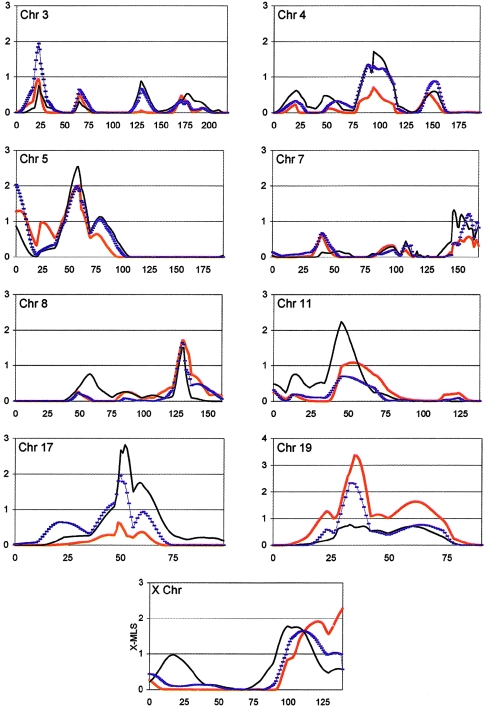
Figure 2 depicts several of the most interesting chromosomal regions at three different stages of analysis (stage 1 [110 families], stage 2 [156 families], and stage 3 [345 families]). Stage 2 represents a relatively small addition of families; however, it is important, primarily because the selection of dense-marker maps was based on this analysis. None of the regions initially identified in the stage 1 analysis showed substantial increase in statistical significance in the stage 3 analysis, although suggestive evidence was maintained in support of chromosomal regions 5p, 8q, and X. The significances of several regions implicated by the stage 2 analysis were either maintained or increased in stage 3, including regions on chromosomes 4, 11, and 17.
Figure 2.
Comparison of the three stages of ASP analysis for selected chromosomes. The X-axis depicts genetic distance in Kosambi centimorgans from pter (zero coordinate) to qter; the Y-axis represents the MLS for all autosomes and the X-MLS for the X chromosome. Thick red lines represent analysis of updated stage 1 (110 families) (see the “Families and Methods” section); hatched blue lines represent analysis of stage 2 (156 families); and black lines represent analysis of stage 3 (345 families).
Our sample included a total of 30 pairs of MZ twins, the 12 whose families were removed from analysis, as well as 18 other pairs whose families were still usable because of the presence of other affected siblings. All 60 of these individuals were completely genotyped so that an estimate of the detectable genotyping error rate, which came to 0.58%, could be calculated for this study.
Discussion
ASP analysis of 345 multiplex families, using both a narrow and a broad diagnostic classification (for autism and ASD, respectively) revealed five chromosomal regions with pointwise significance values P<.01. In no single region, however, did evidence for linkage reach the LOD score threshold of 3.0 (P=10-4) or the more conservative threshold of 3.6 (P=2.2×10-5) recommended for genomewide significance with an infinitely dense map of markers (Lander and Kruglyak 1995). We conducted simulation studies to compute a genomewide occurrence expectation (OE), corresponding to our strongest MLS of 2.83. When the 95% CI is included, this finding is equivalent to an OE value of 0.09 (95% CI = 0.042–0.165). An OE value of 0.09 reflects the fact that one would expect to see an MLS of 2.83 once in every 11.1 (95% CI = 6–24) genome scans. These findings are of particular interest in view of the fact that this carefully controlled follow-up investigation represents the single largest linkage study of autism and related spectrum disorders reported to date. It is notable that the follow-up analysis of 235 new families with autism were carefully controlled to maintain consistent criteria and standards for ascertainment scheme, diagnostic methods, genotyping methods, and genetic analysis, to an extent that would be difficult or impossible to match when combining data from multiple independent studies.
The most significant evidence for linkage was detected on chromosome 17q, with an MLS of 2.83 (P=.00029). We had previously detected nominal evidence for linkage to 17q from the stage 2 analysis of 156 families, which yielded an MLS of 1.95 (P=.0024) (fig. 2). Although this region was not identified in the stage 1 scan, analysis of the next set of 189 families (families added between stage 2 and stage 3) also produced nominal evidence for linkage to this same region (MLS of 1.52) (P=.007). This same region of 17q was identified in an independent autism study, with an MLS of 2.34 (IMGSAC 2001a).
The next strongest linkage signal mapped to chromosome 5p, with an MLS of 2.54 (P=.00059) near marker D5S2494. Analysis of the original 110 families (stage 1), once updated for changes in pedigree structure and individual diagnoses (see the “Families and Methods” section), produced an MLS of 2.0 (P=.0022) at the same position near D5S2494 on 5p. Analysis of the new set of 189 families alone did not provide much evidence for linkage in this region (MLS of 0.82; P=.04), which helps explain why tripling the total number of multiplex families and including the addition of densely spaced DNA markers across this region produced only a slight increase in evidence for linkage to ASD (fig. 2). Although a previous study reported nominal evidence for linkage to chromosome 5p (MLS of .84) (Philippe et al. 1999), this region does not appear to overlap with that reported in the present study. Three other regions—on chromosomes 4q, 8q, and 11p—were identified in the present analysis, with MLSs >1.4, which appear to overlap with regions reported to have MLSs >1.0 in an independent genomewide scan with 75 families affected by autism (Barrett et al. 1999).
Figure 2 displays the comparative linkage patterns at each of the three stages of analysis (110, 156, and 345 families). As shown, evidence in support of a number of putative autism related regions is either maintained or increased in the follow-up analyses; specifically, chromosomes 4, 5, 7, 8, 11, 17, and X (most of which were also identified from stage 2 analysis). In contrast, evidence for linkage on chromosomes 16 and 19, which was reported in the first analysis of 110 families (Liu et al. 2001), decreased in the follow-up analysis. Reanalysis of chromosome 16 after incorporating the minor corrections in family relationship and diagnostic classification in the updated clinical database (see the “Families and Methods” section) eliminated evidence for linkage to autism in this region from the original 110 families. In contrast, these same changes had the effect of increasing statistical support for linkage to chromosome 19. However, in spite of suggestive-to-significant linkage support from the stage 1 analysis (MLS=3.36; P=.000085), we found no evidence of linkage to the chromosome 19 region in the next 56 families or the subsequent 189 families (table 3). ASP analysis of chromosome 3 at stage 2 (156 families) produced a relatively high LOD score (MLS=1.97; P=.002); however, evidence for linkage at this locus was not supported by either the stage 1 or stage 3 analyses (fig. 2). Thus, we suspect the chromosome 3 data represent false-positive results, especially because small sample sizes are more susceptible to false-positive findings due to reduced power. These findings may also reflect changes in the degree of genetic heterogeneity among sample subgroups that could influence the outcome of linkage analyses. Since we purposely adhered to the same sampling scheme at each stage of our analyses, we believe that biases arising from differences in sample heterogeneity will be greater in small sample subsets and that such effects will tend to be mitigated in the entire sample of 345 families.
Four of the five most-suggestive regions identified in this study are areas covered by high-density DNA markers. This led us to question whether the 10-cM scan might be suboptimal to detect autism-related genetic variation in the absence of the fine-map markers. Instead, analysis with the original 335 markers, which provide an average density of 10 cM over the entire genome, identified most of the same regions, albeit with lower levels of significance in most cases (table 3). Therefore, although the denser map is more informative, our data do not indicate that we are preferentially detecting linkage in the densely mapped regions.
The linkage region implicated on chromosome 17q is especially interesting, because it includes the serotonin transporter (5-HTT) gene locus (SLC6A4). The marker that gave the most significant linkage score over the entire genomewide scan, D17S1800, is ∼1 Mb distal of 5-HTT (UCSC Genome Bioinfomatics). The serotonin transporter has previously been implicated as a candidate gene for autism on the basis of some, but certainly not all, allelic association studies (Cook et al. 1997; Klauck et al. 1997; Yirmija et al. 2001; Kim et al. 2002). In addition, some reports show evidence of elevated blood serotonin levels both in patients with autism and in their unaffected first-degree relatives (Cook and Leventhal 1996), and other studies purport to show that drugs that selectively target 5-HTT can ameliorate some autism-related symptoms (Cook and Leventhal 1996). More recently, a functional magnetic resonance imaging study was coupled with a 5-HTT genotyping analysis, to monitor changes in blood flow to the amygdala in response to the subjects’ processing of changes in facial expression (Hariri et al. 2002). The study indicated that amygdala activation is highly correlated with a presumptive functional polymorphism in the 5-HTT promoter region. Although still quite speculative, these findings are interesting in view of the diminished capacity to discern facial expressions among patients with autism.
Linkage studies have been enormously successful for mapping disease loci corresponding to rare disorders with Mendelian inheritance patterns. However, when applied to common, multigenic disorders, the very connection that makes linkage studies possible (the high correlation between genotype and phenotype) is often lost (Weiss and Terwilliger 2000). The search for genotype-phenotype correlations between microsatellite DNA markers and highly reliable dichotomous disease classifications for autism and ASD has produced nominal-to-suggestive linkage evidence on varied chromosomes in many independent studies; however, only a few of these regions overlap between studies or even in follow-up studies performed by the same group. One possible explanation for these findings is that inadequate sample sizes not only increase the likelihood of false-positive findings but also lack the power to detect linkage when individual gene-effect sizes are small and/or when nonallelic heterogeneity is high. The present follow-up study extends the sample size significantly beyond other autism studies reported to date. The question remains whether this increase is significant in regard to the problems of false-positive and false-negative findings described above. In regard to false-positive findings, it is striking that the majority of putative linkage signals detected after stage 1 and stage 2 analyses were maintained after the largest stage 3 analysis, yet it is instructive that the single strongest finding from the updated stage 1 analysis of 110 multiplex families appears to have been a false-positive signal. Although a number of putative disease loci detected in the early stages of analysis were supported in the follow-up analysis, it is interesting that evidence for linkage did not obviously increase with the increase in sample size for any single locus. Because further incremental increases in sample size will challenge the capabilities of any single research group, the need for large collaborative studies to pool families and perform dense genotyping becomes clear. However, we must also recognize the possibility that even dramatic increases in sample size may fail to detect linkage, or association, if the diagnosis of autism or ASD does not significantly increase the likelihood of carrying any single genetic variant (Weiss and Terwilliger 2000). Thus, it will be important to identify and characterize autism-related quantitative traits and/or endophenotypes that are more directly correlated with underlying genetic variation.
Acknowledgments
We gratefully acknowledge the AGRE families, whose participation made this study possible; the Cure Autism Now Foundation, which founded and continues to support the AGRE program; and the scientists who provided oversight to the AGRE consortium (listed in the paragraph below). We thank Iresha Abeyhayake and Miguel Brito for technical support in determining genotypes. Special thanks to Ms. Nancy Hart for her help with the AGRE database. We especially thank Amdec Core Facility for use of their powerful computational technologies, as well as Hans-Erik G. Aronson and Jason M. Yonan for their technical help, which enabled us to complete the simulations. This work was supported by National Institutes of Health grants MH63749 (to J.D.T.) and MH64547 (to D.H.G. and T.C.G.) and by a generous donation from Dr. Judith P. Sulzberger.
Members of AGRE Consortium are Daniel H. Geschwind, University of California at Los Angeles, Los Angeles; Maja Bucan, University of Pennsylvania, Philadelphia; W. Ted Brown, New York State Institute for Basic Research in Developmental Disabilities, Staten Island; Joseph D. Buxbaum, Mt. Sinai School of Medicine, New York; T. Conrad Gilliam, Columbia Genome Center, New York; David A. Greenberg, Mt. Sinai Medical Center, New York; David H. Ledbetter, University of Chicago, Chicago; Bruce L. Miller, University of California at San Francisco, San Francisco; Stanley F. Nelson, University of California at Los Angeles School of Medicine, Los Angeles; Jonathan Pevsner, Kennedy Krieger Institute, Baltimore; Gerard D. Schellenberg, University of Washington and Veterans Affairs Medical Center, Seattle; Carole Samango-Sprouse, Children’s National Medical Center, Baltimore; Rudolph E. Tanzi, Massachusetts General Hospital, Boston; and Kirk C. Wilhelmsen, University of California at San Francisco, San Francisco.
Electronic-Database Information
URLs for data presented herein are as follows:
- Autism Genetic Resource Exchange, http://www.agre.org/ (for full diagnostic protocol for AGRE families)
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/
- Cure Autism Now Foundation, http://www.canfoundation.org/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
- UCSC Genome Bioinformatics, http://genome.cse.ucsc.edu/
References
- Adams P (1994) LABMAN and LINKMAN: a data management system specifically designed for genome searches of complex diseases. Genet Epidemiol 11:87–98 [DOI] [PubMed] [Google Scholar]
- Aita VM, Liu J, Knowles JA, Terwilliger JD, Baltazar R, Grunn A, Loth JE, Kanyas K, Lerer B, Endicott J, Wang Z, Penchaszadeh G, Gilliam TC, Baron M (1999) A comprehensive linkage analysis of chromosome 21q22 supports prior evidence for a putative bipolar affective disorder locus. Am J Hum Genet 64:210–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcón M, Cantor RM, Liu J, Gilliam TC, Geschwind DH, Autism Genetic Research Exchange Consortium (2002) Evidence for a language quantitative trait locus on chromosome 7q in multiplex autism families. Am J Hum Genet 70:60–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association (2000) Diagnostic and statistical manual of mental disorders, 4th ed, text revision. American Psychiatric Association, Washington, DC [Google Scholar]
- Auranen M, Vanhala R, Varilo T, Ayers K, Kempas E, Ylisaukko-Oja T, Sinsheimer JS, Peltonen L, Jarvela I (2002) A genomewide screen for autism-spectrum disorders: evidence for a major susceptibility locus on chromosome 3q25-27. Am J Hum Genet 71:777–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badner JA, Gershon ES (2002) Regional meta-analysis of published data supports linkage of autism with markers on chromosome 7. Mol Psychiatry 7:56–66 [DOI] [PubMed] [Google Scholar]
- Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M (1995) Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med 25:63–77 [DOI] [PubMed] [Google Scholar]
- Barrett S, Beck JC, Bernier R, Bisson E, Braun TA, Casavant TL, Childress D, et al (1999) An autosomal genomic screen for autism: collaborative linkage study of autism. Am J Med Genet 88:609–615 [DOI] [PubMed] [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL (1998) Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet 63:861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Weber JL (1998) Estimation of pairwise relationships in the presence of genotyping errors. Am J Hum Genet 63:1563–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxbaum JD, Silverman JM, Smith CJ, Kilifarski M, Reichert J, Hollander E, Lawlor BA, Fitzgerald M, Greenberg DA, Davis KL (2001) Evidence for a susceptibility gene for autism on chromosome 2 and for genetic heterogeneity. Am J Hum Genet 68:1514–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S, Fombonne E (2001) Pervasive developmental disorders in preschool children. JAMA 285:3093–3099 [DOI] [PubMed] [Google Scholar]
- Charman T (2002) The prevalence of autism spectrum disorders: recent evidence and future challenges. Eur Child Adolesc Psychiatry 11:249–256 [DOI] [PubMed] [Google Scholar]
- Cook EH, Leventhal BL (1996) The serotonin system in autism. Curr Opin Pediatr 8:348–354 [DOI] [PubMed] [Google Scholar]
- Cook EH Jr, Courchesne R, Lord C, Cox NJ, Yan S, Lincoln A, Haas R, Courchesne E, Leventhal BL (1997) Evidence of linkage between the serotonin transporter and autistic disorder. Mol Psychiatry 2:247–250 [DOI] [PubMed] [Google Scholar]
- Cordell HJ, Kawaguchi Y, Todd JA, Farrall M (1995) An extension of the maximum LOD score method to X-linked loci. Ann Hum Genet 59:435–449 [DOI] [PubMed] [Google Scholar]
- Cottingham RW Jr, Idury RM, Schaffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed] [Google Scholar]
- Faraway JJ (1993) Improved sib-pair linkage test for disease susceptibility loci. Genet Epidemiol 10:225–233 [DOI] [PubMed] [Google Scholar]
- Folstein SE, Rosen-Sheidley B (2001) Genetics of autism: complex aetiology for a heterogeneous disorder. Nat Rev Genet 2:943–955 [DOI] [PubMed] [Google Scholar]
- Folstein S, Rutter M (1977) Infantile autism: a genetic study of 21 twin pairs. J Child Psychol Psychiatry 18:297–321 [DOI] [PubMed] [Google Scholar]
- Fombonne E (2002) Epidemiological trends in rates of autism. Mol Psychiatry Suppl 7:S4–S6 [DOI] [PubMed] [Google Scholar]
- Geschwind DH, Sowinski J, Lord C, Iversen P, Shestack J, Jones P, Ducat L, Spence SJ, the AGRE Steering Committee (2001) The autism genetic resource exchange: a resource for the study of autism and related neuropsychiatric conditions. Am J Hum Genet 69:463–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillberg C, Wing L (1999) Autism: not an extremely rare disorder. Acta Psychiatr Scand 99:399–406 [DOI] [PubMed] [Google Scholar]
- Göring HHH, Terwilliger JD (2000a) Linkage analysis in the presence of errors. III. Marker loci and their map as nuisance parameters. Am J Hum Genet 66:1298–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (2000b) Linkage analysis in the presence of errors. IV. Joint pseudomarker analysis of linkage and/or linkage disequilibrium on a mixture of pedigrees and singletons when the mode of inheritance cannot be accurately specified. Am J Hum Genet 66:1310–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariri AR, Mattay VS, Tessitore A, Kolachana B, Fera F, Goldman D, Egan MF, Weinberger DR (2002) Serotonin transporter genetic variation and the response of the human amygdala. Science 297:400–433 [DOI] [PubMed] [Google Scholar]
- Holmans P (1993) Asymptotic properties of affected–sib-pair linkage analysis. Am J Hum Genet 52:362–374 [PMC free article] [PubMed] [Google Scholar]
- International Molecular Genetic Study of Autism Consortium (IMGSAC) (1998) A full genome screen for autism with evidence for linkage to a region on chromosome 7q. Hum Mol Genet 7:571–578 [DOI] [PubMed] [Google Scholar]
- ——— (2001a) Further characterization of the autism susceptibility locus AUTS1 on chromosome 7q. Hum Mol Genet 10:973–982 [DOI] [PubMed] [Google Scholar]
- ——— (2001b) A genomewide screen for autism: strong evidence for linkage to chromosomes 2q, 7q, and 16p. Am J Hum Genet 69:570–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Cox N, Courchesne R, Lord C, Corsello C, Akshoomoff N, Guter S, Leventhal BL, Courchesne E, Cook EH Jr (2002) Transmission disequilibrium mapping at the serotonin transporter gene (SLC6A4) region in autistic disorder. Mol Psychiatry 7:278–288 [DOI] [PubMed] [Google Scholar]
- Klauck SM, Poustka F, Benner A, Lesch KP, Poustka A (1997) Serotonin transporter (5-HTT) gene variants associated with autism? Hum Mol Genet 6:2233–2238 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Lander ES (1995) Complete multipoint sib-pair analysis of qualitative and quantitative traits. Am J Hum Genet 57:439–454 [PMC free article] [PubMed] [Google Scholar]
- Lamb JA, Moore J, Bailey A, Monaco AP (2000) Autism: recent molecular genetic advances. Hum Mol Genet 9:861–868 [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, Ott J (1984) Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci USA 81:3443–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauritsen M, Ewald H (2001) The genetics of autism. Acta Psychiatr Scand 103:411–427 [DOI] [PubMed] [Google Scholar]
- Liu J, Juo SH, Holopainen P, Terwilliger J, Tong X, Grunn A, Brito M, Green P, Mustalahti K, Maki M, Gilliam TC, Partanen J (2002) Genomewide linkage analysis of celiac disease in Finnish families. Am J Hum Genet 70:51–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Nyholt DR, Magnussen P, Parano E, Pavone P, Geschwind D, Lord C, Iversen P, Hoh J, Ott J, Gilliam TC, the Autism Genetic Resource Exchange Consortium (2001) A genomewide screen for autism susceptibility loci. Am J Hum Genet 69:327–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C, Leventhal BL, Cook EH Jr (2001) Quantifying the phenotype in autism spectrum disorders. Am J Med Genet 105:36–38 [PubMed] [Google Scholar]
- Lord C, Rutter M, Le Couteur A (1994) Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord 24:659–685 [DOI] [PubMed] [Google Scholar]
- Medrano JF, Aasen E, Sharrow L (1990) DNA extraction from nucleated red blood cells. Biotechniques 8:43 [PubMed] [Google Scholar]
- Nyholt DR (2000) All LODs are not created equal. Am J Hum Genet 67:282–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe A, Martinez M, Guilloud-Bataille M, Gillberg C, Rastam M, Sponheim E, Coleman M, Zappella M, Aschauer H, Van Maldergem L, Penet C, Feingold J, Brice A, Leboyer M, van Malldergerme L (1999) Genome-wide scan for autism susceptibility genes: Paris Autism Research International Sibpair Study. Hum Mol Genet 8:805–812 [DOI] [PubMed] [Google Scholar]
- Risch N (1990a) Linkage strategies for genetically complex traits. II. The power of affected relative pairs. Am J Hum Genet 46:229–241 [PMC free article] [PubMed] [Google Scholar]
- ——— (1990b) Linkage strategies for genetically complex traits. III. The effect of marker polymorphism on analysis of affected relative pairs. Am J Hum Genet 46:242–253 [PMC free article] [PubMed] [Google Scholar]
- Risch N, Spiker D, Lotspeich L, Nouri N, Hinds D, Hallmayer J, Kalaydjieva L, et al. (1999) A genomic screen of autism: evidence for a multilocus etiology. Am J Hum Genet 65:493–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Wolpert CM, Raiford KL, Menold MM, Donnelly SL, Ravan SA, Bass MP, McClain C, von Wendt L, Vance JM, Abramson RH, Wright HH, Ashley-Koch A, Gilbert JR, DeLong RG, Cuccaro ML, Pericak-Vance MA (2002) Genomic screen and follow-up analysis for autistic disorder. Am J Med Genet 114:99–105 [DOI] [PubMed] [Google Scholar]
- Smalley SL (1997) Genetic influences in childhood-onset psychiatric disorders: autism and attention-deficit/hyperactivity disorder. Am J Hum Genet 60:1276–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley SL, Asarnow RF, Spence MA (1988) Autism and genetics: a decade of research. Arch Gen Psychiatry 45:953–961 [DOI] [PubMed] [Google Scholar]
- Steffenburg S, Gillberg C, Hellgren L, Andersson L, Gillberg IC, Jakobsson G, Bohman M (1989) A twin study of autism in Denmark, Finland, Iceland, Norway and Sweden. J Child Psychol Psychiatry 30:405–416 [DOI] [PubMed] [Google Scholar]
- Szatmari P, Jones MB, Zwaigenbaum L, MacLean JE (1998) Genetics of autism: overview and new directions. J Autism Dev Disord 28:351–368 [DOI] [PubMed] [Google Scholar]
- Terwillger JD, Ott J (1994) Handbook of human genetic linkage. Johns Hopkins University Press, Baltimore, pp 245–250 [Google Scholar]
- Weiss KM, Terwilliger JD (2000) How many diseases does it take to map a gene with SNPs? Nat Genet 26:151–157 [DOI] [PubMed] [Google Scholar]
- World Health Organization (1992) International statistical classification of diseases and related health problems, 10th revision. World Health Organization, Geneva [Google Scholar]
- Yeargin-Allsopp M, Rice C, Karapurkar T, Doernberg N, Boyle C, Murphy C (2003) Prevalence of autism in a US metropolitan area. JAMA 289:49–55 [DOI] [PubMed] [Google Scholar]
- Yirmiya N, Pilowsky T, Nemanov L, Arbelle S, Feinsilver T, Fried I, Ebstein RP (2001) Evidence for an association with the serotonin transporter promoter region polymorphism and autism. Am J Med Genet 105:381–386 [DOI] [PubMed] [Google Scholar]