Abstract
To clarify the chromatin-based imprinting mechanism of the p57KIP2/LIT1 subdomain at chromosome 11p15.5 and the mouse ortholog at chromosome 7F5, we investigated the histone-modification status at a differentially CpG methylated region of Lit1/LIT1 (DMR-Lit1/LIT1), which is an imprinting control region for the subdomain and is demethylated in half of patients with Beckwith-Wiedemann syndrome (BWS). Chromatin-immunoprecipitation assays revealed that, in both species, DMR-Lit1/LIT1 with the CpG-methylated, maternally derived inactive allele showed histone H3 Lys9 methylation, whereas the CpG-unmethylated, paternally active allele was acetylated on histone H3/H4 and methylated on H3 Lys4. We have also investigated the relationship between CpG methylation and histone H3 Lys9 methylation at DMR-LIT1 in patients with BWS. In a normal individual and in patients with BWS with normal DMR-LIT1 methylation, histone H3 Lys9 methylation was detected on the maternal allele; however, it disappeared completely in the patients with the DMR-LIT1 imprinting defect. These findings suggest that the histone-modification status at DMR-Lit1/LIT1 plays an important role in imprinting control within the subdomain and that loss of histone H3 Lys9 methylation, together with CpG demethylation on the maternal allele, may lead to the BWS phenotype.
Genomic imprinting is a specific example of an epigenetic phenomenon that occurs, prior to fertilization, during gametogenesis. Imprinting can lead to parental-allele–specific expression of genes, which ensures the functional inequality of paternal and maternal genomes in somatic cells (Barlow 1995; Tilghman 1999). It has been shown that imprinted genes play an important role in development because of the highly restricted developmental potential of androgenotes with two paternal genomes and of gynogenotes or parthenogenotes with two maternal genomes (McGrath and Solter 1984; Surani et al. 1984). Aberrant expression of imprinted genes is also associated with several childhood and adult tumors and genetic diseases. The Beckwith-Wiedemann syndrome (BWS [MIM 130650]) region, on human chromosome 11p15.5, is a major example of the role that genomic imprinting plays in the pathogenesis of human disease (Maher and Reik 2000).
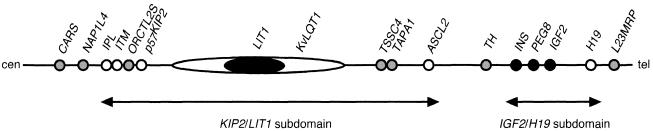
BWS is a congenital syndrome characterized by pre- and postnatal overgrowth, macroglossia, and anterior abdominal-wall defects. Additional but variable complications include organomegaly, hypoglycemia in infancy, hemihypertrophy, genitourinary abnormalities, and, in ∼5% of children, embryonal tumors (most frequently Wilms tumors) (Maher and Reik 2000). It has been known that at least 12 imprinted genes are clustered within the human chromosome 11p15.5 and form an imprinted domain (fig. 1). BWS has a complex genetic etiology and can arise from paternal uniparental disomy (UPD), paternal duplication of 11p15.5, maternally inherited coding mutations in the p57KIP2 (KIP2 [MIM 600856]) gene, or maternal chromosome rearrangements (Reik and Maher 1997; Li et al. 1998). However, the most common mechanism that results in BWS is imprinting defect, with loss of CpG methylation of the maternal DMR-LIT1 (Lee et al. 1999; Mitsuya et al. 1999; Smilinich et al. 1999).
Figure 1.
Imprinted domain at human 11p15.5. The relative positions of the genes are shown. The imprinting status is shown by blackened circles (paternally expressed), unblackened circles (maternally expressed), and gray circles (biallelic expression).
The imprinted domain at 11p15.5 and its mouse orthologous domain, located at chromosome 7F5, can be separated into two subdomains: IGF2/H19 (IGF2 [MIM 147470]; H19 [MIM 103280]) and KIP2/LIT1 (LIT1 [MIM 604115]) (Caspary et al. 1998; Lee et al. 1999; Mitsuya et al. 1999; Smilinich et al. 1999; Paulsen et al. 2000) (fig. 1). In the Igf2/H19 subdomain, the differentially CpG-methylated region (DMR) upstream of H19, which is established in sperm (Davis et al. 2000; Kerjean et al. 2000; Lucifero et al. 2002), controls the imprinting of Ins2, Igf2, and H19 by functioning, in part, as a chromatin insulator that restricts access to shared enhancers (Bell and Felsenfeld 2000; Hark et al. 2000). On the other hand, DMR-Lit1 in the Kip2/Lit1 subdomain is a gametic CpG-methylated region in oocytes but not in sperm (Engemann et al. 2000; Yatsuki et al. 2002). And it was shown to be the imprinting-control region (ICR) (or imprinting center [IC]) of the Kip2/Lit1 subdomain that regulates the neighboring imprinted genes between Ipl and Ascl2 by targeted deletion of DMR-Lit1 in mice (Fitzpatrick et al. 2002). The evidence in mouse suggested the importance of the DMR-LIT1 in human as an imprinting-regulator element for the KIP2/LIT1 subdomain and the pathogenesis of BWS. However, the regulatory mechanism(s) of the subdomain has not been elucidated completely (Kanduri et al. 2002; Mancini-DiNardo et al. 2003; Thakur et al. 2003).
In all eukaryotes, the covalent modification of histone N-terminal tails plays an important role in the regulation of transcription, mitosis, and heterochromatin formation. The “histone code” hypothesis (Strahl and Allis 2000; Jenuwein and Allis 2001) predicts that different modifications of specific amino acids in histones or their combinations are translated into functionally distinct effects on the nuclear process. It has been reported that histone H3 Lys9 (H3K9) methylation was associated with the formation of stably silenced chromatin regions in mammals (Lachner et al. 2001), that histone acetylation presumably changed chromatin structure to increase accessibility to transcriptional factors (Strahl and Allis 2000), and that histone H3 Lys4 (H3K4) methylation was correlated with transcriptional activity (Strahl et al. 1999; Santos-Rosa et al. 2002). To date, histone modifications at several imprinted loci in human and mouse—such as an ICR (IC) for the Prader-Willi syndrome region (PWS [MIM 176270]), DMR-H19, Igf2r, and U2af1-rs1—have been examined (Saitoh and Wada 2000; Xin et al. 2001; Yoshioka et al. 2001; Fournier et al. 2002). However, DMR-Lit1/LIT1, an ICR (IC) for the KIP2/LIT1 subdomain in mouse and human, has not yet been examined for histone modifications, although acetylation of histone H4 was analyzed only in mouse hybrid cells containing human chromosome (Yoshioka et al. 2001).
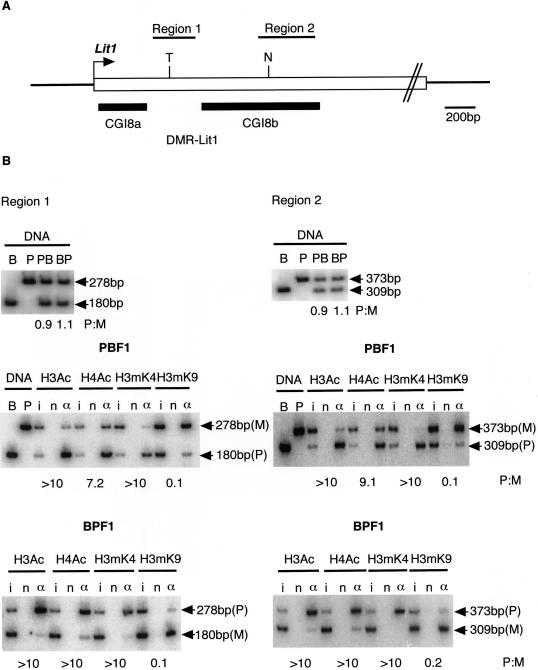
To determine the allele-specific histone-modification status of mouse DMR-Lit1, we performed chromatin immunoprecipitation (ChIP) assay on primary fibroblast cells using antiacetylated histone H3 (H3Ac), antiacetylated histone H4 (H4Ac), anti-Lys4 methylated histone H3 (H3mK4), and our originally developed monoclonal anti-Lys9 methylated histone H3 (H3mK9) antibodies (T. Nakagawachi and H. Soejima, unpublished data). The primary fibroblast cell lines were established from newborn F1 mice, (PWK×C57BL/6)F1 (PBF1) and (C57BL/6×PWK)F1 (BPF1). Normal imprinting of the Kip2/Lit1 subdomain was confirmed, using polymorphisms, by the allelic expression of Lit1 and Kip2 and CpG-methylation status of DMR-Lit1 and Kip2 promoter (data not shown). The regions analyzed by ChIP were region 1, between CpG island 8a (CGI8a) and CpG island 8b (CGI8b), and region 2, within CGI8b (fig. 2A). Chromatin was prepared from 2.0×106 cells and was sonicated to an average size of ∼0.5 kb; it then was immunoprecipitated with each antibody. DNAs recovered from immunoprecipitated complex were subjected to PCR by use of the oligonucleotide primers shown in table 1. Alleles between C57BL/6 and PWK were distinguished by Tsp509I and NciI polymorphisms. We determined the ratio of the paternal to maternal (P:M) band intensities. At both regions 1 and 2, histone H3K9 was clearly hypermethylated on the maternal allele, indicating condensed and inactive chromatin. Histone H3 and H4 were hyperacetylated, and H3K4 was hypermethylated at both regions on the paternal allele, indicating open chromatin and active transcription. (fig. 2B). The result showed that histone modifications at DMR-Lit1 were associated with differential CpG methylation. The allelic ratio for each antibody was corrected by the P:M ratios in input, because the maternal and paternal alleles were not equally represented in input. Since both the parental alleles were amplified equally using genomic DNAs from PBF1 and BPF1 (fig 2B), at the DMR-Lit1 region, the paternal allele was possibly more sensitive to sonication than the maternal allele during ChIP preparation because of relaxed chromatin, shown as hyperacetylation of H3 and H4 and hypermethylation of H3K4.
Figure 2.
Histone-modification status at DMR-Lit1 in mouse. A, Schematic structure of DMR-Lit1 containing CpG island 8a and 8b (CGI8a and CGI8b, respectively). The positions of CGI8a and CGI8b are shown as thick horizontal lines. Regions 1 and 2, analyzed by ChIP, are indicated in the upper horizontal bars. An arrow indicates the transcription start site of Lit1. T = polymorphic Tsp509I site in C57BL/6; N = polymorphic NciI site in C57BL/6. B, The status of histone modification at DMR-Lit1. The chromatin of primary fibroblasts derived from (PWK×C57BL/6)F1 (PBF1) and (C57BL/6×PWK)F1 (BPF1) were analyzed by ChIP with the antibodies against H3Ac, H4Ac, H3mK4 (Upstate Biotechnology), and originally developed H3mK9. To generate monoclonal H3mK9 antibody, synthesized peptides for histone H3 N-tail containing dimethylated K9 was used to immunize mice. Splenic lymphocytes were fused to the myeloma cell line, and then monoclonal lines, which produced the antibody, were cloned. Three independent experiments confirmed the specificity of the antibody, which reacted with the dimethylated H3K9 peptides but not with dimethylated H3K4, H3K27, and H4K20 at all (T. Nakagawachi and H. Soejima, unpublished data). ChIP DNAs extracted from input (i), no antibody (n), and antibody (α) fractions were subjected to amplification with the Hot-stop PCR method to eliminate the problem of heteroduplex formation (Uejima et al. 2000; Higashimoto et al. 2002). After performing a number of PCR cycles sufficient to detect a product, a primer labeled by (γ-32P)ATP was added to the mixture, and then the PCR step was performed only once. The PCR product was digested with the appropriate restriction endonuclease and was electrophoresed. The intensity of the PCR products was measured with a BAS 2000 bioimaging analyzer (Fujifilm). The results from C57BL/6 (B), PWK (P), PBF1 (PB), and BPF1 (BP) genomic DNAs were shown as controls for complete digestion. PB and BP genomic DNA showed the same intensity of each parental allele. Ratios of paternal intensity to maternal intensity (P:M), corrected by P:M in input chromatin, are indicated below each lane. ChIP was performed independently twice. All primer pairs used in this study are shown in table 1.
Table 1.
Primers and PCR Conditions Used in This Study[Note]
| Procedures and Primersa | Primer Sequence (5′→3′) | Annealing Temperature [PCR Cycle No.](°C) |
| ChIP assay: | ||
| m Lit1-1F | CGATCCTCCTCAGTGTTTGT | |
| m Lit1-1R | GCTGAACAGAAAAGCTCTCC | 57 [34] |
| m Lit1-2F | GCCGAGTCAGAACGCACTGG | |
| m Lit1-2R | TTCCCAATCCCCCACACCTG | 64 [33] |
| m G6pd-F | ATTTTCAAGGCACCGCATC | |
| m G6pd-R | CTAGTTTGGCTTCGGAGCTG | 62 [33] |
| m D13Mit55-F | TCAATATTAACTGCTAGCATGGTT | |
| m D13Mit55-R | GCTTTTCCTCCCCAAACATT | 55 [34] |
| h LIT1-1F | CCGGGGCTCCTCAGCACGAT | |
| h LIT1-1R | GGAGAACCGCGCCGAAAAGC | 68 [33] |
| h LIT1-2F | CAGGGTCGAGGTCCGAGTTC | |
| h LIT1-2R | CCCAATTCGGGCTTTGACTC | 66 [33] |
| h 16CEN-F | GTCTCTTTCTTGTTTTTAAGCTGGG | |
| h 16CEN-R | TGAGCTCATTGAGACATTTGG | 65 [33] |
| h GAPDH-F | GCATCACCCGGAGGAGAAATCGG | |
| h GAPDH-R | GTCACGTGTCGCAGAGGAGC | 65 [32] |
| RT-PCR: | ||
| h LIT1 RT-F | AAGAAAGTGTTGAGTGGTAA | |
| h LIT1 RT-R | GATGATCTGAAAATGGAAAAA | 54 [40] |
| h H19 RT-F | GCGGGTCTGTTTCTTTACTT | |
| h H19 RT-R | CGATGGTGTCTTTGATGTTG | 60 [40] |
| m β-actin RT-F | CCTAAGGCCAACCGTGAAAAG | |
| m β-actin RT-R | TCTTCATGGTGCTAGGAGCCA | 60 [20] |
| COBRA: | ||
| h LIT1 BS-F | GTGTTAIGGIGGTGGAGATTTTGT | |
| h LIT1 BS-R | AACCIAAAACACIAACCAATTCTCTA | 57 [50] |
Note.— All PCR reactions were performed with an initial denaturation at 95°C for 5 min, followed by appropriate amplification cycles of 95°C for 20 s, appropriate annealing temperature for 20 s, 72°C for 30 s, and a final extension at 72°C for 5 min.
m = mouse primers; h = human primers.
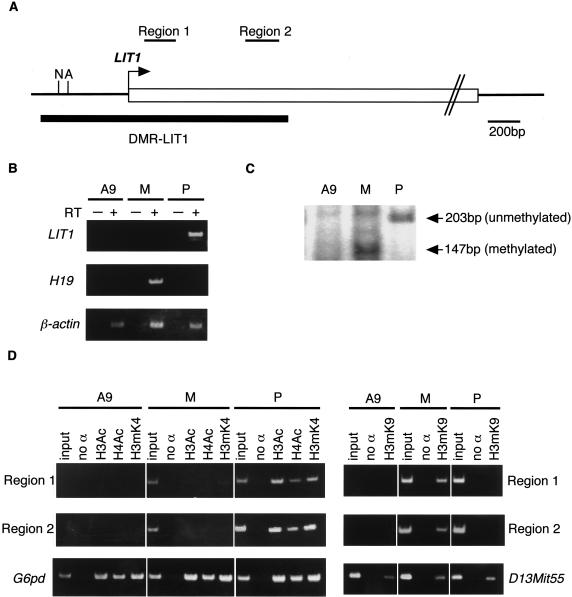
Next, we tried to elucidate the histone-modification status at human DMR-LIT1. Since we could not find any informative normal human cell lines for polymorphisms in DMR-LIT1, mouse A9 hybrid cells containing a paternal or a maternal human chromosome 11 were used as the alternative (Mitsuya et al. 1999). To confirm the normal imprinting status at 11p15.5 in A9 hybrids, the allelic expression of LIT1 and H19 was examined by RT-PCR. Figure 3B shows that LIT1 and H19 were expressed only from the paternal and the maternal chromosome, respectively. Furthermore, the CpG methylation status of DMR-LIT1 was examined by combined bisulfite-restriction analysis (COBRA) (Xiong and Laird 1997). The PCR product amplified from bisulfite-modified genomic DNA was digested with AccII (fig. 3A). The result showed that the PCR products from the A9 hybrid, each containing the maternal and paternal human chromosome 11, were digested and undigested, respectively, and indicated maternal allele-specific CpG methylation at DMR-LIT1 (fig. 3C). Thus, paternal expression and maternal CpG methylation of LIT1 were accurately maintained in the A9 hybrids. We analyzed histone modifications at two regions, region 1 and region 2, in DMR-LIT1 by ChIP (fig. 3A). In A9 hybrids containing a maternal copy, histone H3K9 methylation was clearly detected; both histone H3/H4 acetylation and H3K4 methylation were mostly undetectable. On the other hand, in A9 hybrids containing a paternal copy, histone H3K9 methylation was not detected, and histone H3/H4 acetylation and H3K4 methylation were clearly detected (fig. 3D). These data indicated that the histone-modification status at DMR-Lit1 (LIT1) was well conserved between mouse and human.
Figure 3.
Histone-modification status at DMR-LIT1 in human, with A9 hybrid cells containing only one parental human chromosome 11. A, Schematic structure of DMR-LIT1. The position of DMR-LIT1 is shown as a thick horizontal line. Regions 1 and 2, analyzed by ChIP, are indicated as upper horizontal bars. An arrow indicates a putative transcription start site of LIT1 (M. Oshimura, unpublished data). A = AccII; N = NotI. B, Allele-specific expression of LIT1 and H19. A plus sign (+) = reverse transcriptase added; a minus sign (−) = no reverse transcriptase. cDNAs from mouse A9 (A9), A9 hybrids containing a paternal (P) or maternal (M) chromosome 11 were amplified. C, Maternal CpG methylation of DMR-LIT1. After bisulfite-treated DNAs were amplified by PCR, the products were digested with AccII. The PCR product generated from methylated DNA must be digested by AccII. D, Histone-modification status at DMR-LIT1. no α = no antibody; H3Ac = histone H3 acetylation; H4Ac = H4 acetylation; H3mK4 = H3 Lys4 methylation; H3mK9 = H3 Lys9 methylation. Mouse G6pd sequence was amplified as a positive control for H3Ac, H4Ac, and H3mK4. D13Mit55 was a positive control for H3mK9. ChIP was performed independently twice. All primer pairs used in this study are shown in table 1.
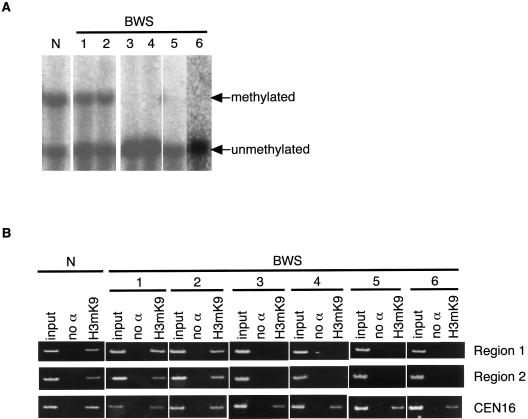
Since approximately one-half of patients with BWS show loss of maternal CpG methylation at DMR-LIT1 (Lee et al. 1999; Mitsuya et al. 1999; Smilinich et al. 1999), we investigated whether methylation of H3K9 alters along with the CpG methylation status in patients with BWS. A total of seven lymphoblastoid cell lines were investigated from six patients with BWS and one normal individual. The CpG methylation status at DMR-LIT1 was examined by methylation-sensitive Southern blotting (fig. 4A), as described elsewhere (Mitsuya et al. 1999). Two BWS cell lines and a normal cell line showed differential CpG methylation, indicating normal DMR-LIT1 methylation, whereas the other four BWS lines showed complete demethylation at DMR-LIT1, indicating the DMR-LIT1 imprinting defect. We confirmed that these four BWS lines were not mosaics for paternal UPD and that they did have two equal parental alleles (data not shown). Histone H3K9 methylation was detected in the normal individual and in patients with BWS with normal DMR-LIT1 methylation, whereas it was not detected in any patients with the DMR-LIT1 imprinting defect (fig. 4B). Thus, loss of histone H3K9 methylation was completely associated with loss of CpG methylation at DMR-LIT1 in BWS.
Figure 4.
The correlation between loss of histone H3K9 methylation and loss of CpG methylation at DMR-LIT1 in BWS. A, CpG methylation at DMR-LIT1 was analyzed by methylation-sensitive Southern blotting in BWS. The DNAs extracted from lymphoblastoid cells derived from normal individual (N) and six patients with BWS were double digested with BamHI plus NotI. Two patients with BWS (patients 1 and 2) showed normal DMR-LIT1 methylation, whereas four patients with BWS (patients 3, 4, 5, and 6) showed DMR-LIT1 imprinting defect. B, Histone H3K9 methylation in BWS. The same cells shown above were analyzed by ChIP with the antibody against methylated histone H3K9. The human chromosome 16 centromere region was amplified as a positive control. Regions 1 and 2 were the same as shown in figure 3. ChIP was performed independently twice. All primer pairs used in this study are shown in table 1.
Histone-modification status at several ICRs (ICs) and imprinting regulatory elements in human and mouse were reported recently (Saitoh and Wada 2000; Xin et al. 2001; Yoshioka et al. 2001; Fournier et al. 2002). These regulatory regions on the maternal allele with differential CpG hypermethylation were marked by hypermethylation of H3K9. The regions on the opposite paternal allele without CpG methylation were acetylated on H3/H4 and methylated on H3K4. Our results were consistent with these previous observations and suggested the existence of a common regulation mechanism between mouse and human—that maternally CpG-methylated imprinting regulatory regions were associated with H3K9 hypermethylation and that paternally CpG-unmethylated region were associated with hyperacetylation of H3/H4 and hypermethylation of H3K4. It was also suggested that histone methylation and acetylation were involved in at least the somatic maintenance of imprinting and that they might be related to the establishment of imprinting.
What role does the H3K9 methylation play in primary imprint? Tamaru and Selker (2001) have shown that CpG methylation requires the Dim-5 H3K9 methyltransferase in Neurospora, and Jackson et al. (2002) have demonstrated that CpNpG methylation is dependent on the H3K9 methyltransferase KRYPTONITE in Arabidopsis. However, little has been known about the hierarchy of CpG or H3K9 methylation in mammals. Mouse PWS-IC shows CpG methylation in oocytes; however, human PWS-IC lacks CpG methylation in oocytes (Shemer et al. 1997; El-Maarri et al. 2001). Mouse ES cells lacking H3K9 methyltransferase, G9a, show loss of maternal CpG methylation of PWS-IC and biallelic expression of Snrpn. In contrast, DNA methyltransferase-deficient ES cells (Dnmt1−/−) lack CpG methylation of PWS-IC but show normal levels of H3K9 methylation of PWS-IC and monoallelic expression of Snrpn (Xin et al. 2003). These findings suggested that histone H3K9 methylation, rather than CpG methylation, would be a primary imprint for PWS-IC. As for DMR-Lit1, the mouse region was CpG methylated in oocytes (Engemann et al. 2000; Yatsuki et al. 2002), but there has been no such study in humans. However, DMR-LIT1 is similar to PWS-IC, in that the maternal CpG methylation in somatic cells, the allelic expression of the gene itself, and bidirectional control on the neighboring imprinted genes, although the transcriptional effect on the neighboring genes on the paternal chromosome is different. Thus, it is likely that H3K9 methylation is of prime importance in establishment or maintenance of imprinting on the maternal DMR-LIT1.
Once CpG methylation is acquired in germ cells, the methylation at the gametic imprint site in the embryo is usually resistant to genomewide CpG demethylation during the preimplantation stage (Warnecke et al. 1998). Maternal CpG methylation at DMR-Lit1 is maintained in the zygote (Engemann et al. 2000); however, it dropped significantly at the two-cell stage, and then preferentially maternal CpG methylation gradually reappeared in blastocysts (Yatsuki et al. 2002). H3K9 methylation of the maternal DMR-Lit1 may provide a mechanism for maintaining the maternal mark during the loss and restoration of CpG methylation of DMR-Lit1 between fertilization and the blastocyst stage. In this context, loss of H3K9 methylation between fertilization and the blastocyst stage by an unknown mechanism may result in lack of methylation of both CpG and H3K9 at DMT-LIT1 on the maternal allele throughout development. This may explain why patients with BWS show the DMR-LIT1 imprinting defect that is observed in approximately one-half of the patients. Studies of the histone-modification status at the two-cell stage in mouse are needed, although a technical difficulty may remain for the current ChIP assay because it needs a considerable number of cells. Also, we should find out whether DMR-LIT1 in the human oocyte has CpG methylation or H3K9 methylation. It has been recently reported that MeCP2, a methyl-CpG binding protein, is associated with methyltransferase activity on H3K9 in vivo, although the identity of the H3K9 methyltransferase was still unknown (Fuks et al. 2003b) and that MBD1, another methyl-CpG binding protein, and DNA methyltransferases (Dnmts), respectively, interact with H3K9 methyltransferase Suv39h1 and methyl-lysine–binding protein HP1 (Fujita et al. 2003; Fuks et al. 2003a); however, further investigations on the interactions among CpG methylation, H3K9 methylation, and their modification enzymes, such as Dnmts and histone H3K9 methyltransferases, at DMR-Lit1/LIT1 and other ICRs (ICs) are also required to elucidate the mechanism for establishment and maintenance of the genomic imprinting.
Acknowledgments
We thank all the member of the Division of Molecular Biology & Genetics, Department of Biomolecular Sciences, Saga Medical School, for their helpful advice and assistance. This work was supported, in part, by a grant-in-aid (13206062) for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan, grant 13770069 from the Japan Society for the Promotion of Science, and a grant from the Uehara Memorial Foundation.
Electronic-Database Information
The URL for data presented herein is as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for BWS, PWS, p57KIP2, IGF2, H19, and LIT1)
References
- Barlow DP (1995) Gametic imprinting in mammals. Science 270:1610–1613 [DOI] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G (2000) Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405:482–485 [DOI] [PubMed] [Google Scholar]
- Caspary T, Cleary MA, Baker CC, Guan XJ, Tilghman SM (1998) Multiple mechanisms regulate imprinting of the mouse distal chromosome 7 gene cluster. Mol Cell Biol 18:3466–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TL, Yang GJ, McCarrey JR, Bartolomei MS (2000) The H19 methylation imprint is erased and re-established differentially on the parental alleles during male germ cell development. Hum Mol Genet 9:2885–2894 [DOI] [PubMed] [Google Scholar]
- El-Maarri O, Buiting K, Peery EG, Kroisel PM, Balaban B, Wagner K, Urman B, Heyd J, Lich C, Brannan CI, Walter J, Horsthemke B (2001) Maternal methylation imprints on human chromosome 15 are established during or after fertilization. Nat Genet 27:341–344 [DOI] [PubMed] [Google Scholar]
- Engemann S, Strodicke M, Paulsen M, Franck O, Reinhardt R, Lane N, Reik W, Walter J (2000) Sequence and functional comparison in the Beckwith-Wiedemann region: implications for a novel imprinting centre and extended imprinting. Hum Mol Genet 9:2691–2706 [DOI] [PubMed] [Google Scholar]
- Fitzpatrick GV, Soloway PD, Higgins MJ (2002) Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat Genet 32:426–431 [DOI] [PubMed] [Google Scholar]
- Fournier C, Goto Y, Ballestar E, Delaval K, Hever AM, Esteller M, Feil R (2002) Allele-specific histone lysine methylation marks regulatory regions at imprinted mouse genes. EMBO J 21:6560–6570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Watanabe S, Ichimura T, Tsuruzoe S, Shinkai Y, Tachibana M, Chiba T, Nakao M (2003) Methyl-CpG binding domain 1 (MBD1) interacts with Suv39h1-HP1 heterochromatic complex for DNA methylation-based transcriptional repression. J Biol Chem 278:24132–24138 [DOI] [PubMed] [Google Scholar]
- Fuks F, Hurd PJ, Deplus R, Kouzarides T (2003a) The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res 31:2305–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T (2003b) The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J Biol Chem 278:4035–4040 [DOI] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM (2000) CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405:486–489 [DOI] [PubMed] [Google Scholar]
- Higashimoto K, Soejima H, Yatsuki H, Joh K, Uchiyama M, Obata Y, Ono R, Wang Y, Xin Z, Zhu X, Masuko S, Ishino F, Hatada I, Jinno Y, Iwasaka T, Katsuki T, Mukai T (2002) Characterization and imprinting status of OBPH1/Obph1 gene: implications for an extended imprinting domain in human and mouse. Genomics 80:575–584 [DOI] [PubMed] [Google Scholar]
- Jackson JP, Lindroth AM, Cao X, Jacobsen SE (2002) Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416:556–560 [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD (2001) Translating the histone code. Science 293:1074–1080 [DOI] [PubMed] [Google Scholar]
- Kanduri C, Fitzpatrick G, Mukhopadhyay R, Kanduri M, Lobanenkov V, Higgins M, Ohlsson R (2002) A differentially methylated imprinting control region within the Kcnq1 locus harbors a methylation-sensitive chromatin insulator. J Biol Chem 277:18106–18110 [DOI] [PubMed] [Google Scholar]
- Kerjean A, Dupont JM, Vasseur C, Le Tessier D, Cuisset L, Paldi A, Jouannet P, Jeanpierre M (2000) Establishment of the paternal methylation imprint of the human H19 and MEST/PEG1 genes during spermatogenesis. Hum Mol Genet 9:2183–2187 [DOI] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116–120 [DOI] [PubMed] [Google Scholar]
- Lee MP, DeBaun MR, Mitsuya K, Galonek HL, Brandenburg S, Oshimura M, Feinberg AP (1999) Loss of imprinting of a paternally expressed transcript, with antisense orientation to KVLQT1, occurs frequently in Beckwith-Wiedemann syndrome and is independent of insulin-like growth factor II imprinting. Proc Natl Acad Sci USA 96:5203–5208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Squire JA, Weksberg R (1998) Molecular genetics of Wiedemann-Beckwith syndrome. Am J Med Genet 79:253–259 [PubMed] [Google Scholar]
- Lucifero D, Mertineit C, Clarke HJ, Bestor TH, Trasler JM (2002) Methylation dynamics of imprinted genes in mouse germ cells. Genomics 79:530–538 [DOI] [PubMed] [Google Scholar]
- Maher ER, Reik W (2000) Beckwith-Wiedemann syndrome: imprinting in clusters revisited. J Clin Invest 105:247–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini-DiNardo D, Steele SJ, Ingram RS, Tilghman SM (2003) A differentially methylated region within the gene Kcnq1 functions as an imprinted promoter and silencer. Hum Mol Genet 12:283–294 [DOI] [PubMed] [Google Scholar]
- McGrath J, Solter D (1984) Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 37:179–183 [DOI] [PubMed] [Google Scholar]
- Mitsuya K, Meguro M, Lee MP, Katoh M, Schulz TC, Kugoh H, Yoshida MA, Niikawa N, Feinberg AP, Oshimura M (1999) LIT1, an imprinted antisense RNA in the human KvLQT1 locus identified by screening for differentially expressed transcripts using monochromosomal hybrids. Hum Mol Genet 8:1209–1217 [DOI] [PubMed] [Google Scholar]
- Paulsen M, El-Maarri O, Engemann S, Strodicke M, Franck O, Davies K, Reinhardt R, Reik W, Walter J (2000) Sequence conservation and variability of imprinting in the Beckwith-Wiedemann syndrome gene cluster in human and mouse. Hum Mol Genet 9:1829–1841 [DOI] [PubMed] [Google Scholar]
- Reik W, Maher ER (1997) Imprinting in clusters: lessons from Beckwith-Wiedemann syndrome. Trends Genet 13:330–334 [DOI] [PubMed] [Google Scholar]
- Saitoh S, Wada T (2000) Parent-of-origin specific histone acetylation and reactivation of a key imprinted gene locus in Prader-Willi syndrome. Am J Hum Genet 66:1958–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T (2002) Active genes are tri-methylated at K4 of histone H3. Nature 419:407–411 [DOI] [PubMed] [Google Scholar]
- Shemer R, Birger Y, Riggs AD, Razin A (1997) Structure of the imprinted mouse Snrpn gene and establishment of its parental-specific methylation pattern. Proc Natl Acad Sci USA 94:10267–10272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smilinich NJ, Day CD, Fitzpatrick GV, Caldwell GM, Lossie AC, Cooper PR, Smallwood AC, Joyce JA, Schofield PN, Reik W, Nicholls RD, Weksberg R, Driscoll DJ, Maher ER, Shows TB, Higgins MJ (1999) A maternally methylated CpG island in KvLQT1 is associated with an antisense paternal transcript and loss of imprinting in Beckwith-Wiedemann syndrome. Proc Natl Acad Sci USA 96:8064–8069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403:41–45 [DOI] [PubMed] [Google Scholar]
- Strahl BD, Ohba R, Cook RG, Allis CD (1999) Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc Natl Acad Sci USA 96:14967–14972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani MA, Barton SC, Norris ML (1984) Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature 308:548–550 [DOI] [PubMed] [Google Scholar]
- Tamaru H, Selker EU (2001) A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414:277–283 [DOI] [PubMed] [Google Scholar]
- Thakur N, Kanduri M, Holmgren C, Mukhopadhyay R, Kanduri C (2003) Bidirectional silencing and DNA methylation-sensitive methylation-spreading properties of the Kcnq1 imprinting control region Map to the same regions. J Biol Chem 278:9514–9519 [DOI] [PubMed] [Google Scholar]
- Tilghman SM (1999) The sins of the fathers and mothers: genomic imprinting in mammalian development. Cell 96:185–193 [DOI] [PubMed] [Google Scholar]
- Uejima H, Lee MP, Cui H, Feinberg AP (2000) Hot-stop PCR: a simple and general assay for linear quantitation of allele ratios. Nat Genet 25:375–376 [DOI] [PubMed] [Google Scholar]
- Warnecke PM, Mann JR, Frommer M, Clark SJ (1998) Bisulfite sequencing in preimplantation embryos: DNA methylation profile of the upstream region of the mouse imprinted H19 gene. Genomics 51:182–190 [DOI] [PubMed] [Google Scholar]
- Xin Z, Allis CD, Wagstaff J (2001) Parent-specific complementary patterns of histone H3 lysine 9 and H3 lysine 4 methylation at the Prader-Willi syndrome imprinting center. Am J Hum Genet 69:1389–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Z, Tachibana M, Guggiari M, Heard E, Shinkai Y, Wagstaff J (2003) Role of histone methyltransferase G9a in CpG methylation of the Prader-Willi syndrome imprinting center. J Biol Chem 278:14996–15000 [DOI] [PubMed] [Google Scholar]
- Xiong Z, Laird PW (1997) COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res 25:2532–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsuki H, Joh K, Higashimoto K, Soejima H, Arai Y, Wang Y, Hatada I, Obata Y, Morisaki H, Zhang Z, Nakagawachi T, Satoh Y, Mukai T (2002) Domain regulation of imprinting cluster in Kip2/Lit1 subdomain on mouse chromosome 7F4/F5: large-scale DNA methylation analysis reveals that DMR-Lit1 is a putative imprinting control region. Genome Res 12:1860–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka H, Shirayoshi Y, Oshimura M. (2001) A novel in vitro system for analyzing parental allele-specific histone acetylation in genomic imprinting. J Hum Genet 46:626–632 [DOI] [PubMed] [Google Scholar]