Abstract
The hereditary spastic paraplegias (HSPs) are genetically heterogeneous disorders characterized by progressive lower-extremity weakness and spasticity. The molecular pathogenesis is poorly understood. We report discovery of a dominant negative mutation in the NIPA1 gene in a kindred with autosomal dominant HSP (ADHSP), linked to chromosome 15q11-q13 (SPG6 locus); and precisely the same mutation in an unrelated kindred with ADHSP that was too small for meaningful linkage analysis. NIPA1 is highly expressed in neuronal tissues and encodes a putative membrane transporter or receptor. Identification of the NIPA1 function and ligand will aid an understanding of axonal neurodegeneration in HSP and may have important therapeutic implications.
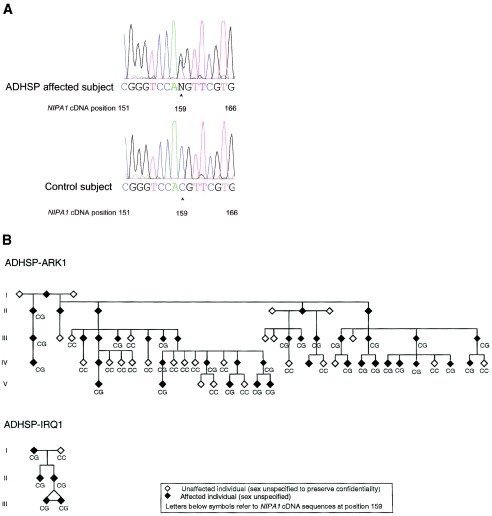
Ten loci for autosomal dominant hereditary spastic paraplegia (ADHSP) have been mapped, and four ADHSP genes have been identified: SPG4/spastin, SPG3A/atlastin, SPG13/chaperonin 60, and SPG10/KIF5A (Hazan et al. 1999; Zhao et al. 2001; Hansen et al. 2002; Reid et al. 2002). (For reviews of hereditary spastic paraplegia [HSP] [“Strümpell-Lorrain syndrome,” MIM 182600], see Fink and Hedera 1999; Fink 2001, 2002; Reid 2003.) Despite these advances, the molecular pathophysiology of the ADHSPs is largely unknown. Elsewhere, we identified a locus for uncomplicated ADHSP in chromosome 15q (SPG6) (Fink et al. 1995a; 1995b). We now report identification of disease-specific mutations in a novel gene, NIPA1 (fig. 1a) in the original SPG6-linked kindred (Fink et al. 1995a) and in an unrelated kindred with ADHSP (fig. 1b).
Figure 1.
a, Representative SPG6 sequence of normal and affected subjects from the ADHSP-ARK and ADHSP-IRQ kindreds. PCR products (from 50 ng of DNA) were purified through Sephadex G-50 columns, were analyzed by agarose gel electrophoresis, and were sequenced either by use of the ABI PRISM dRhodamine Terminator Cycle Sequencing Ready Reaction Kit (PE Applied Biosystems) on an ABI PRISM 3100 Genetic Analyzer, according to the manufacturer’s instructions, or through the University of Pennsylvania DNA sequencing facility. The arrows mark the position of the NIPA1 mutation (cDNA nucleotide 159). b, Pedigrees of ADHSP-ARK1 and ADHSP-IRQ1. Letters refer to sequence at nucleotide 159 of the NIPA1 cDNA sequence. Clinical features (Fink et al. 1995a), muscle biopsy (Hedera et al. 2000 ), and genetic features (Fink et al. 1995b) of ADHSP-ARK1 have been reported elsewhere. The ADHSP-IRQ1 kindred was ascertained through the University of Michigan Neurology Outpatient Clinic.
The SPG6 locus extends 6.1 cM between D15S128 and the centromere (Rainier et al. 2000) (fig. 2a), an interval often involved in deletions that result in Prader-Willi syndrome (PWS) or Angelman syndrome (AS). PWS and AS are characterized by genetic imprinting, a phenomenon in which gene expression and the phenotype depend on the sex of the transmitting parent (Nicholls and Knepper 2001). We analyzed a large kindred—ADHSP-ARK1 (fig. 1b), in which ADHSP was linked to the SPG6 locus—and found no evidence of genetic imprinting (Fink et al. 1995a). Therefore, we analyzed as SPG6 candidates the four unique, nonimprinted, and highly evolutionarily conserved genes mapped proximal to the imprinted domain and within the pericentromeric region of chromosome 15q (Chai et al. 2003 [in this issue]). These candidate genes included nonimprinted in Prader-Willi/Angelman loci 1 (NIPA1) (NCBI accession number BK001020) and 2 (NIPA2) (NCBI accession number BK001120) (Chai et al. 2003 [in this issue]), GCP5 (NCBI accession number AF272884) (Murphy et al. 2001), and CYFIP1 (NCBI accession number NM_014608) (Koybayashi et al. 1998; Schenk et al. 2001).
Figure 2.
a, SPG6 occurs in regions deleted in PWS and AS. SPG6 is mapped to chromosome 15q11, proximal to the imprinted PWS and AS domains, within a region containing four nonimprinted genes. These genes, including NIPA1, map within class I deletions in patients with PWS and AS but are intact in class II deletions (Chai et al. 2003 [in this issue]). BP = breakpoint hotspot; cen = centromere; tel = telomere. b, NIPA1 secondary structure analysis. Nine predicted transmembrane (TM) domains (Chai et al. 2003 [in this issue]) are shown. The NIPA1 T45R mutation occurs at the interface of TM1 and the first putative outside loop. c, Expression of NIPA1 by northern blot analysis. NIPA1 transcripts of 2.2 kb and 7.5 kb are found in all tissues but are greatly enriched in neuronal tissues. Probes were prepared from TA-cloned RT-PCR products of human NIPA1 (167 bp) and exons 2–3 of the control SNURF-SNRPN gene, were labeled with α-32P-dCTP, and were hybridized in ExpressHyb (Clontech) at 65°C to northern blots (Clontech), according to the manufacturer’s protocol.
We identified a nucleotide substitution at position 159 of the NIPA1 cDNA (159C→G; fig. 1a), which resulted in an amino acid substitution at position 45 of the NIPA1 protein (T45R) in each affected subject (n=28) in ADHSP-ARK1 (fig. 1b). In contrast, each unaffected subject (n=17) had only C at this position (fig. 1b), which agrees with the known human genomic sequence (NCBI accession number NT_024668). We examined 105 control subjects (ascertained through the Elderly Subjects Program of the University of Michigan Institute of Gerontology). Each control subject had only C at position 159 of the NIPA1 cDNA (data not shown). We analyzed the coding sequence of the other three nonimprinted genes (GCP5, CYFIP1, and NIPA2) in two affected members of ADHSP-ARK1 and identified no disease-specific mutations (data not shown). We then analyzed the NIPA1 coding sequence in affected probands from 62 kindreds with ADHSP, 6 kindreds with probable autosomal recessive HSP, and 13 subjects with all the signs and symptoms of HSP but no family history (“apparently sporadic” spastic paraplegia). Affected subjects in one unrelated kindred (ADHSP-IRQ1) (fig. 1b) had precisely the same NIPA1 mutation (159C→G) (fig. 1b) as affected subjects in the ADHSP-ARK1 kindred. Unaffected subjects from ADHSP-IRQ1 kindred showed only the normal nucleotide (159C). Whereas the ADHSP-ARK1 kindred was linked to the SPG6 locus (Fink et al. 1995b), the ADHSP-IRQ1 kindred was too small for meaningful linkage analysis. Clinical features of the ADHSP-ARK1 affected individuals are typical of uncomplicated HSP of late-teenage to early-adult symptom onset (Fink et al. 1995a). Clinical features of ADHSP-IRQ1 were similar: onset of insidiously progressive spastic weakness in both legs that began in late teen-age years and was associated with urinary urgency and mild vibratory sensation impairment in the toes.
Kindreds ADHSP-ARK1 and ADHSP-IRQ1 are of Irish and Iraqi ancestry, respectively. Analysis of haplotypes for polymorphic markers linked to this locus (D15S541, D15S542, D15S646, D15S817, and D15S1021) showed no evidence of haplotype sharing between ADHSP-ARK1 and ADHSP-IRQ1 kindreds (data not shown). This indicates that these two kindreds with ADHSP are not closely related and suggests that the same NIPA1 mutation arose independently in each of them.
Disease-specific NIPA1 mutations in ADHSP-ARK1 and ADHSP-IRQ1 occur in NIPA1 exon 1 and are predicted to change threonine to arginine at amino acid position 45 (T45R) (fig. 2b). This amino acid is conserved in mouse, chicken, and fish (zebrafish and Fugu) (Chai et al. 2003 [in this issue]; R.D.N., unpublished data) and is predicted to occur at the end of the first of nine transmembrane domains in the NIPA1 polypeptide (fig. 2b). NIPA1 does not contain an AAA domain (like that present in spastin [Hazan et al. 1999]) or a GTPase domain (like that present in atlastin [Zhao et al. 2001]) and does not bear other homology to genes that cause other forms of HSP. Although the NIPA1 function is not known, the clear prediction based on hydrophobicity analysis of an integral membrane-associated protein (Chai et al. 2003 [in this issue]) suggests that NIPA1 functions as a receptor or transporter. Many individuals with PWS or AS have chromosome 15q class I deletions that include NIPA1 (fig. 2a) (Chai et al. 2003 [in this issue]). The fact that such individuals do not exhibit progressive spastic paraplegia indicates that NIPA1 haploinsufficiency does not cause progressive spastic paraplegia. Therefore, we suggest that the NIPA1 T45R missense mutation identified in these kindreds with ADHSP is pathogenic through a dominant negative, gain-of-function mechanism.
NIPA1 mRNA is expressed constitutively at low levels with 2.2-kb and 7.5-kb transcripts in all human tissues but shows significant enrichment in the brain (fig. 2c). The latter expression pattern is found throughout the CNS, whereas spinal cord shows equal expression of the two NIPA1 mRNAs (fig. 2c). The alternative mRNA isoforms arise from alternative polyadenylation within NIPA1 exon 5, and equivalent expression patterns are found for mouse (Chai et al. 2003 [in this issue]).
Our observations of the same NIPA1 gene mutation (159 C→G; T45R) in two unrelated kindreds with ADHSP that disrupts an interspecies conserved amino acid and was absent in control subjects (n=105) strongly support the pathogenic significance of this mutation. Discovery of NIPA1 mutations as the cause of SPG6-linked HSP improves the ability to diagnose HSP and to provide genetic counseling. The presence of a dominant negative NIPA1 mutation in a putative membrane receptor or transporter suggests that SPG6 arises from either altered signal transduction or small-molecule transport through a membrane. We suggest that this property makes NIPA1 and its unknown ligand an attractive target for therapeutic intervention in SPG6 and, perhaps, in other spastic paraplegias. Identification of the NIPA1 cellular and subcellular localization, function, and ligand will therefore aid an understanding of axonal neurodegeneration in HSP and may have important therapeutic implications.
Acknowledgments
This research is supported by grants from the Veterans Affairs Merit Review (to J.K.F.), the Muscular Dystrophy Association (to R.D.N.), and the National Institutes of Health (NINDS R01NS33645, R01NS36177, and R01NS38713 to J.K.F. and HD31491 and ES10631 to R.D.N.). We gratefully acknowledge Dr. Gregory B. Sharp and Dr. Bernadette M. Lange (University of Arkansas) for ascertaining and evaluating ADHSP-ARK; the technical assistance of Colin Delaney and Donald Thomas; the expert secretarial assistance of Lynette Girbach; and the participation of subjects with HSP and their families, without whom our investigations of HSP would not be possible.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- National Center for Biotechnology Information (NCBI), http://www.ncbi.nlm.nih.gov/entrez/query.fcgi
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
References
- Chai J-H, Locke DP, Greally JM, Knoll JHM, Ohta T, Dunai J, Yavor A, Eichler EE, Nicholls R D (2003) Identification of four highly conserved genes between breakpoint hotspots BP1 and BP2 of the Prader-Willi/Angelman syndromes deletion region that have undergone evolutionary transposition mediated by flanking duplicons. Am J Hum Genet 73:898–925 (in this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink JK (2001) Hereditary spastic paraplegia. In: Rimoin DPRCJ, Emery KB (eds) Emery & Rimoin's principles and practice of medical genetics. Harcourt Publishers Limited UK, London, pp 3124–3145 [Google Scholar]
- Fink JK (2002) Hereditary spastic paraplegia: the pace quickens. Ann Neurol 51:669–672 [DOI] [PubMed] [Google Scholar]
- Fink JK, Hedera P (1999) Hereditary spastic paraplegia: genetic heterogeneity and genotype-phenotype correlation. Semin Neurol 19:301–310 [DOI] [PubMed] [Google Scholar]
- Fink JK, Sharp G, Lange B, Wu C-TB, Haley T, Otterud B, Peacock M, Leppert M (1995a) Autosomal dominant hereditary spastic paraparesis, type I: clinical and genetic analysis of a large North American family. Neurology 45:325–331 [DOI] [PubMed] [Google Scholar]
- Fink JK, Wu C-TB, Jones SM, Sharp GB, Lange BM, Lesicki A, Reinglass T, Varvil T, Otterud B, Leppert M (1995b) Autosomal dominant familial spastic paraplegia: tight linkage to chromosome 15q. Am J Hum Genet 56:188–192 [PMC free article] [PubMed] [Google Scholar]
- Hansen JJ, Durr A, Cournu-Rebeix I, Georgopoulous C, Ang D, Nielsen MN, Davoine C-S, Brice A, Fontaine B, Gregersen N, Bross P (2002) Hereditary spastic paraplegia SPG13 is associated with a mutation in the gene encoding the mitochondrial chaperonin Hsp60. Am J Hum Genet 70:1328–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazan J, Fonknechten N, Mavel D, Paternotte C, Samson D, Artiguenave F, Davoine C-S, Cruaud C, Durr A, Wincker P, Brottier P, Cattolico L, Barbe V, Burgunder JM, Prudhomme JF, Brice A, Fontaine B, Heilig B, Weissenbach J (1999) Spastin, a new AAA protein, is altered in the most frequent form of autosomal dominant spastic paraplegia. Nat Genet 23:296–303 [DOI] [PubMed] [Google Scholar]
- Hedera P, DiMauro S, Bonilla E, Wald JJ, Fink JK (2000) Mitochondrial analysis in autosomal dominant hereditary spastic paraplegia. Neurology 55:1591–1592 [DOI] [PubMed] [Google Scholar]
- Koybayashi K, Kuroda S, Fukata M (1998) p140Sra-1 (specifically Rac1-associated protein) is a novel specific target for Rac1 small GTPase. J Biol Chem 273:291–295 [DOI] [PubMed] [Google Scholar]
- Murphy SM, Preble AM, Patel UK, O’Connell KL, Dias DP, Moritz M, Agard D, Stults JT, Stearns T (2001) GCP5 and GCP6: two new members of the human gamma-tubulin complex. Mol Biol Cell 12:3340–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls RD, Knepper JL (2001) Genome organization, function, and imprinting in Prader-Willi and Angelman syndromes. Ann Rev Genomics Hum Genet 2:153–175 [DOI] [PubMed] [Google Scholar]
- Rainier S, Bui M, Jones SM, Fink JK (2000) Chromosome 15q linked autosomal dominant hereditary spastic paraplegia: new mapping information and candidate gene analysis. Am J Hum Genet Suppl 67:391 [Google Scholar]
- Reid E (2003) Science in motion: common molecular pathological themes emerge in the hereditary spastic paraplegias. J Med Genet 40:81–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid E, Kloos M, Ashley-Koch A, Hughes L, Bevan S, Svenson IK, Graham FL, Gaskell PC, Dearlove A, Pericak-Vance MA, Rubinsztein DC, Marchuk DA (2002) A kinesin heavy chain (KIF5A) mutation in hereditary spastic paraplegia (SPG10). Am J Hum Genet 71:1189–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk A, Bardoni, B, Moro A (2001) A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P. Proc Natl Acad Sci USA 98:8844–8849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Alvarado D, Rainier S, Lemons R, Hedera P, Weber C, Tukel T, Apak M, Heiman-Patterson T, Ming L, Bui M, Fink JK (2001) Mutations in a novel GTPase cause autosomal dominant hereditary spastic paraplegia. Nat Genet 29:326–331 [DOI] [PubMed] [Google Scholar]