Abstract
To realize the full potential of targeted protein kinase inhibitors for the treatment of cancer, it is important to address the emergence of drug resistance in treated patients. Mutant forms of BCR-ABL, KIT, and the EGF receptor (EGFR) have been found that confer resistance to the drugs imatinib, gefitinib, and erlotinib. The mutations weaken or prevent drug binding, and interestingly, one of the most common sites of mutation in all three kinases is a highly conserved “gatekeeper” threonine residue near the kinase active site. We have identified existing clinical compounds that bind and inhibit drug-resistant mutant variants of ABL, KIT, and EGFR. We found that the Aurora kinase inhibitor VX-680 and the p38 inhibitor BIRB-796 inhibit the imatinib- and BMS-354825-resistant ABL(T315I) kinase. The KIT/FLT3 inhibitor SU-11248 potently inhibits the imatinib-resistant KIT(V559D/T670I) kinase, consistent with the clinical efficacy of SU-11248 against imatinib-resistant gastrointestinal tumors, and the EGFR inhibitors EKB-569 and CI-1033, but not GW-572016 and ZD-6474, potently inhibit the gefitinib- and erlotinib-resistant EGFR(L858R/T790M) kinase. EKB-569 and CI-1033 are already in clinical trials, and our results suggest that they should be considered for testing in the treatment of gefitinib/erlotinib-resistant non-small cell lung cancer. The results highlight the strategy of screening existing clinical compounds against newly identified drug-resistant mutant variants to find compounds that may serve as starting points for the development of next-generation drugs, or that could be used directly to treat patients that have acquired resistance to first-generation targeted therapy.
Keywords: drug resistance, gatekeeper mutation, kinase inhibitor
Targeted protein-tyrosine kinase inhibitors represent a major advance in cancer treatment (1, 2). Although these drugs have been extremely effective in specific patient populations with tumors containing mutated, oncogenic forms of tyrosine kinases, the accumulating clinical experience suggests that most patients will develop resistance (3). Resistance can be caused by amplification of the oncogenic protein kinase gene (4) or other mechanisms, but in a significant fraction of cases, resistance can be traced to the selection of cancer cells with secondary mutations in the targeted kinase. The resistance mutations often appear in the kinase catalytic domain and directly prevent or weaken the interaction with the inhibitor. Resistance mutations have been observed in the kinase domain of BCR-ABL, KIT, and the platelet-derived growth factor receptor in patients treated with imatinib (5-9), and in the EGF receptor (EGFR) in patients treated with gefitinib or erlotinib (10, 11). Consequently, it is important to develop efficient strategies to identify and rapidly develop alternative compounds that will be effective against mutated targets resistant to first-line inhibitors (12, 13). Such compounds also provide the opportunity for developing multidrug therapies to delay or prevent the appearance of resistant kinase variants, analogous to the successful use of drug cocktails for treatment of HIV infections.
One strategy to identify inhibitors that could be brought to the clinic quickly is to screen compounds already approved by the Food and Drug Administration or in clinical development against kinases with newly identified mutations that confer drug resistance. Most protein kinase inhibitors inhibit other “off-target” kinases in addition to their intended target, but other targets are generally not predictable and must be identified experimentally (14). Although off-target activities may lead to side effects, they can also lead to the expansion of a drug's clinical utility. A good example of this idea is the rapid development of imatinib as a treatment for gastrointestinal stromal tumors (GIST) after the discovery that the drug is an effective inhibitor of KIT as well as ABL (15-17). Screening clinical compounds against drug-resistant, mutated kinases takes advantage of the propensity of protein kinase inhibitors to hit multiple targets, and provides potentially new uses for drugs and new treatment options for patients.
Here we apply this strategy to the therapeutically important tyrosine kinases BCR-ABL, KIT, and EGFR. Interestingly, some of the common drug-resistant mutations in these kinases are structurally related: BCR-ABL(T315I), KIT(T670I), and EGFR(T790M) contain homologous mutations of the conserved “gatekeeper” threonine residue (7, 10, 11). Mutations of this residue can have profound effects on small molecule binding in the context of different kinases (13, 18), yet the mutations do not inactivate kinase function. Based on the behavior of imatinib, BMS-354825 (also known as dasatinib) (19) and other ABL inhibitors (20-22), all of which share a significant loss of affinity for ABL(T315I) relative to other ABL variants, one might conclude that it is particularly difficult to inhibit ABL(T315I) with an ATP-competitive compound. We have shown previously that the p38 inhibitor BIRB-796 (23) binds tightly (Kd = 40 nM) to ABL(T315I) (14), and here identify VX-680, an Aurora kinase inhibitor chemically unrelated to BIRB-796 (24), as a second high-affinity binder to ABL(T315I) (Kd = 5 nM). We show further that the KIT/FLT3 inhibitor SU-11248 (25), which is in late stage clinical trials for imatinib-resistant GIST, is a potent inhibitor of imatinib-resistant KIT(V559D/T670I). This result is consistent with the clinical efficacy of the compound (Pharmaprojects database, www.pjbpubs.com/pharmaprojects/index.htm), but the direct interaction has not been demonstrated to our knowledge. Finally, we find that EKB-569 and CI-1033, EGFR inhibitors that have completed phase I clinical trials (26, 27) (Pharmaprojects database), potently inhibit the erlotinib- and gefitinib-resistant EGFR(L858R/T790M) variant.
Based on our results, VX-680 and BIRB-796 should be further explored as the basis for a possible treatment of imatinib-resistant chronic myeloid leukemia, and EKB-569 and CI-1033 should be tested in patients with EGFR(T790M)-mediated resistance to gefitinib or erlotinib.
Methods
Compounds. Imatinib, PD-180970, VX-680, BIRB-796, SU-11248, MLN-518, gefitinib, erlotinib, GW-572016, EKB-569, CI-1033, ZD-6474, PKI-166, and SU-11464 were custom synthesized. BMS-354825 was provided by Bristol-Myers Squibb. CL-387785 was purchased from Calbiochem.
Competition Binding Assays. To assess small molecule binding to wild-type and mutant kinases, we used ATP site-dependent competition binding assays. Assays were developed, validated, and performed as described (14).
In Vitro Enzyme Activity Assays. Upstate Biotechnology's KinaseProfiler service was used to measure small molecule inhibition of ABL and ABL(T315I) in vitro. For experimental details, see Supporting Text, which is published as supporting information on the PNAS web site.
Cell-Based Assays for BCR-ABL Inhibition. To assess the ability of small molecules to inhibit BCR-ABL in cells, we used Ba/F3 cells that express wild-type or mutated BCR-ABL. Assays were performed as described (28).
Cell-Based Assays for KIT Inhibition. Full-length KIT(V559D) and KIT(V559D/T670I) were expressed in HEK-293 cells. Cells were treated with compound or DMSO vehicle for 2 hours, and levels of total KIT protein and KIT phosphorylated at tyrosine 823 were measured by using an ELISA (Biosource, Camarillo, CA). See Supporting Text for experimental details.
Cell-Based Assays for EGFR Inhibition. To measure cell proliferation, H1975 cells were treated with vehicle or compound for 48 h and viable cells were quantitated. To measure EGFR autophosphorylation, cells were treated with vehicle or compound for 2 h and stimulated with EGF for 5 min, and levels of total EGFR protein and EGFR phosphorylated at tyrosine 1173 were measured by using an ELISA (Biosource). See Supporting Text for experimental details.
Results
Inhibition of Drug-Resistant Forms of ABL and KIT. To test existing inhibitors against drug-resistant mutants of ABL and KIT, we developed competition binding assays for a panel of clinically important mutant isoforms: wild-type and eight imatinib-resistant mutant variants of ABL (E255K, H396P, M351T, Q252H, T315I, Y253F, as described in ref. 14, plus F359V and T315N) (5), two variants of KIT with activating mutations found in GIST (V559D, N822K) (29, 30), as well as one double-mutant variant of KIT with an imatinib-resistant secondary mutation introduced in the context of an activating mutation (V559D/T670I) (7). We then tested seven compounds for binding to this panel of 12 kinase variants (Table 2, which is published as supporting information on the PNAS web site). Imatinib, BMS-354825, and PD-180970 are potent inhibitors of wild-type and various mutant forms of BCR-ABL (19, 21, 31), but not BCR-ABL(T315I). BMS-354825 is in clinical development for imatinib-resistant chronic myeloid leukemia (19, 32). BIRB-796 is a p38 inhibitor that has been in clinical trials for inflammatory disease (23). MLN-518 and SU-11248 are inhibitors of wild-type and activated KIT and FLT3 (33-36), and both have been in clinical trials for treatment of acute myeloid leukemia (25, 37) (Pharmaprojects database). SU-11248 is also in late-stage clinical trials for treatment of imatinib-resistant GIST. The Aurora kinase inhibitor VX-680 is in phase I clinical development for solid tumors (www.clinicaltrials.gov) (Pharmaprojects database), and is also known to inhibit FLT3 (24). VX-680 was included in this study because many FLT3 inhibitors, such as SU-11248 and MLN-518, also inhibit KIT.
The binding affinity of imatinib for imatinib-resistant ABL variants correlates well with results from cell-based inhibition experiments, as described (Table 1) (14). BMS-354825 binds ABL with 4-fold greater affinity than imatinib, consistent with the significantly higher potency of BMS-354825 compared to imatinib in cell-based assays (19). Although BMS-354825, PD-180970, and a number of other compounds have been described as effective inhibitors of multiple imatinib-resistant ABL variants, none of these compounds are effective against ABL(T315I) (13, 20). Indeed, the affinity of BMS-354825 and PD-180970 for ABL(T315I) and ABL(T315N) is down at least 80-fold relative to wild type ABL (Table 1). In contrast, BIRB-796 binds with good affinity to ABL(T315I) (Kd = 40 nM), but has significantly weaker affinity for wild-type and other imatinib-resistant forms of ABL, with Kd values >1 μM (14) (Table 1). Therefore, BIRB-796 has a binding profile opposite, or complementary to, that of imatinib, BMS-354825, and the other known ABL inhibitors. This observation raised the possibility that perhaps ATP-competitive compounds will bind only wild-type or T315I mutant ABL, but not both. However, this does not appear to be the case based on our finding that VX-680 binds tightly, with a Kd of ≈20 nM or lower, to wild-type ABL and most of the ABL variants, including T315I (Kd = 5 nM) (Table 1). The only ABL variant tested with somewhat lower affinity for VX-680 was ABL(T315N), with a binding constant ≈5-fold higher than for wild-type ABL. Our strategy has identified two existing, chemically unrelated, protein kinase inhibitors, VX-680 and BIRB-796, that bind ABL(T315I) with high affinity, and it will be of great interest to define the structural basis for the different binding profiles of imatinib, BMS-354835, BIRB-796, and VX-680.
Table 1. Binding affinity of existing kinase inhibitors for drug-resistant kinase variants.
|
Kd, nanomolar
|
|||||||
|---|---|---|---|---|---|---|---|
| Kinase variant | Imatinib | BMS-354825 | PD-180970 | BIRD-796 | VX-680 | SU-11248 | MLN-518 |
| ABL1 | 2* | 0.5 | 1 | 2,000* | 20 | 1,000* | >10,000* |
| ABL1(Q252H) | 20* | 1 | 2 | 4,000* | 10 | 2,000* | >10,000* |
| ABL1(Y253F) | 40* | 1 | 1 | 2,000* | 20 | 700* | >10,000* |
| ABL1(E255K) | 100* | 2 | 4 | >10,000* | 50 | >10,000* | >10,000* |
| ABL1(M351T) | 10* | 0.7 | 0.7 | 2,000* | 8 | 500* | >10,000* |
| ABL1(F359V) | 20 | 0.3 | 1 | 8,000 | 20 | 1,000 | 7,000 |
| ABL1(H396P) | 60* | 1 | 1 | >10,000* | 7 | 900* | >10,000* |
| ABL1(T3151) | 6,000* | 600 | 600 | 40* | 5 | 200* | >10,000* |
| ABL1(T315N) | >10,000 | 40 | 300 | >10,000 | 100 | 400 | >10,000 |
| KIT(N822K) | 3 | 0.4 | 4 | 200 | 100 | 3 | 5 |
| KIT(V559D) | 20 | 0.7 | 1 | 200 | 300 | 0.4 | 4 |
| KIT(V559D, T6701) | 3,000 | >10,000 | 3,000 | 300 | 600 | 0.3 | 1,000 |
Each binding constant was measured at least in duplicate, and average values are shown.
Previously published binding constants (14), shown here for comparison.
To determine whether binding of VX-680 and BIRB-796 to ABL(T315I) leads to inhibition of the kinase, we tested the compounds in in vitro enzyme activity assays. In the enzyme activity assays, VX-680 potently inhibited wild-type ABL with an IC50 value of 10 nM and ABL(T315I) with an IC50 value of 30 nM. These results confirm that VX-680 can potently inhibit the enzymatic activity of ABL(T315I). BIRB-796 inhibited ABL(T315I) in vitro, but the IC50 value (4 μM) was higher than the affinity measured in our binding assays (inhibition of the enzymatic activity of wild-type ABL was not tested with BIRB-796). By comparison, the IC50 for inhibition by imatinib was 0.4 μM for wild-type ABL, and no significant inhibition was observed at 10 μM for ABL(T315I). For the binding assays, the ABL protein is produced at low concentration in bacteria (14), and the enzyme activity assays use purified protein expressed in insect cells (see Supporting Text). The weaker activity of BIRB-796 and imatinib in the in vitro enzymatic assay may therefore be due to different activation states of the kinase in the binding and activity assays and the propensity of both compounds to bind more tightly to the unactivated kinase conformation (23, 38).
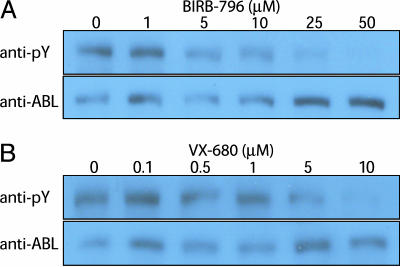
To further explore the relationship between binding and inhibition, we determined the effects of BIRB-796 and VX-680 in Ba/F3 cells expressing wild-type or mutant BCR-ABL. BIRB-796 inhibited proliferation of cells expressing BCR-ABL(T315I) (IC50 ≈2-3 μM) more potently than cells expressing BCR-ABL with no resistance mutation (IC50 > 10 μM) (Fig. 5, which is published as supporting information on the PNAS web site). VX-680 is a known inhibitor of Aurora kinases, which are required for cellular proliferation (24), and indeed, VX-680 inhibited proliferation of cells expressing either form of BCR-ABL as well as the parental Ba/F3 cell line (IC50 = 100-200 nM) (data not shown). To more directly assess inhibition of BCR-ABL enzymatic activity in cells, we measured BCR-ABL autophosphorylation. BIRB-796 inhibited BCR-ABL(T315I) autophosphorylation in Ba/F3 cells with an IC50 value of 1-2 μM (Fig. 1A), consistent with the results of the cell proliferation assay and the in vitro activity assay (see above), and confirming that this compound is an inhibitor of ABL(T315I). The IC50 for inhibition of BCR-ABL(T315I) autophosphorylation in Ba/F3 cells by VX-680 was ≈5 μM (Fig. 1B), significantly higher than the binding constant (Table 1) and the IC50 for inhibition of ABL enzymatic activity measured in vitro. By comparison, there is no significant inhibition of BCR-ABL(T315I) by imatinib in this assay, even at 10 μM concentration (28). We do not yet fully understand the reason for the quantitative discrepancy between cell-based results in the Ba/F3 system and in vitro results for VX-680, and additional studies will be necessary to resolve this question. Possible explanations include unusual kinase conformations and/or nonnative phosphorylation patterns induced by overexpression of the protein in Ba/F3 cells.
Fig. 1.
BIRB-796 and VX-680 inhibit imatinib-resistant BCR-ABL(T315I). Ba/F3 cells expressing BCR-ABL(T315I) were treated for 2 hours with compound, whole-cell lysates prepared, and total protein analyzed by Western blot using anti-ABL and anti-phosphotyrosine antibodies as described (28). (A) Inhibition of BCR-ABL(T315I) by BIRB-796. (B) Inhibition of BCR-ABL(T315I) by VX-680.
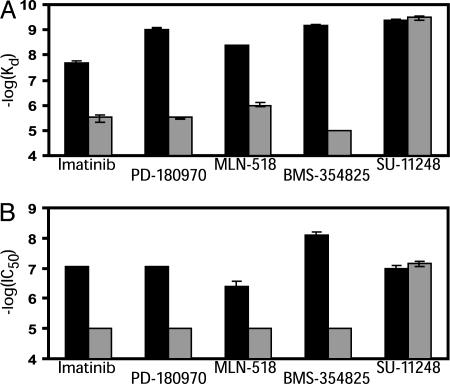
Based on our findings with ABL, we decided to explore other kinases that are targeted by existing drugs and in which drug-resistant “gatekeeper” mutations have been found. The five known KIT inhibitors, imatinib, BMS-354825, PD-180970, MLN-518, and SU-11248, bind both KIT variants with activating mutations (V559D and N822K) with high affinity (Kd ≤ 20 nM) (Table 1) (Fig. 2A). The affinity for imatinib was decreased >100-fold by the T670I mutation, and a similar pattern was observed for BMS-354825 (decreased >10,000-fold), MLN-518 (decreased >100-fold), and PD-180970 (decreased >1000-fold) (Table 1) (Fig. 2A). The effect of the T670I mutation in KIT is therefore similar to that of the T315I mutation in ABL. However, binding of SU-11248 to KIT was not significantly affected by the T670I mutation (Table 1) (Fig. 2A), again illustrating that it is possible to identify compounds that can bind both wild-type and “gatekeeper” mutant variants of the same kinase. VX-680 and BIRB-796 did not bind with high affinity to any of the KIT variants tested.
Fig. 2.
SU-11248 inhibits imatinib-resistant KIT. (A) Binding affinities of known inhibitors of KIT with the activating V559D mutation compared to the imatinib-resistant V559D/T670I double mutant variant. Quantitative binding constants are shown in Table 1. (B) Cell-based assays confirm the results from in vitro binding experiments. KIT variants were expressed in HEK-293 cells and KIT autophosphorylation levels determined after treating for 2 h with each compound. Results for KIT(V559D) are shown in black, and for KIT(V559D/T670I) in gray. Binding and inhibition constants are plotted as -log(Kd) or -log(IC50), such that higher bars indicate higher affinity binding or more potent inhibition. Kd and IC50 values were measured at least twice and average values are plotted. Error bars represent the range of values obtained in independent replicate measurements.
To confirm that binding interactions observed in vitro are predictive of kinase inhibition in cells, we expressed KIT(V559D) and KIT(V559D/T670I) in HEK-293 cells and measured KIT autophosphorylation (Fig. 2B). Consistent with results from the binding assays, imatinib, BMS-354825, PD-180970, MLN-518, and SU-11248 effectively inhibited KIT(V559D), but only SU-11248 was able to inhibit KIT(V559D/T670I) (Fig. 2B) (see Table 3, which is published as supporting information on the PNAS web site, for IC50 values). Interestingly, although BMS-354825, SU-11248, and PD-180970 bind KIT(V559D) with similar affinity (Table 1), the cellular activity of BMS-354825 was >10-fold greater than that of SU-11248 or PD-180970 (Table 3), implying that additional pharmaceutical properties may also contribute to the greater cellular potency of BMS-354825.
Inhibition of Drug-Resistant EGFR. Mutations in the kinase domain of EGFR have been found in a significant fraction of patients who have responded to gefitinib and erlotinib. The L858R mutation is one of the most common mutations associated with a therapeutic response to the EGFR inhibitors gefitinib and erlotinib (>40% of EGFR mutations in non-small cell lung cancer), whereas the T790M mutation results in resistance to both drugs. The combination of the L858R and T790M mutations has been found in at least one patient who initially responded to gefitinib but relapsed after 9 months with a gefitinib-resistant tumor (11). Small deletions near the EGFR ATP-binding site are also found in patients that respond to gefitinib or erlotinib (39-41), and the T790M mutation also confers resistance in these patients (10, 11).
To identify inhibitors of drug-resistant EGFR(L858R/T790M), we tested 47 known kinase inhibitors for the ability to inhibit proliferation of the non-small cell lung cancer (NSCLC) cell line H1975, which contains L858R and T790M mutations in EGFR (11) (see Table 4, which is published as supporting information on the PNAS web site). We found three compounds that showed a significant effect on proliferation (≥85% inhibition at 2 μM), CL-387785, EKB-569, and CI-1033 (Fig. 6, which is published as supporting information on the PNAS web site). CL-387785 is a commercially available research compound, and is the only previously known inhibitor of both wild-type EGFR and EGFR with the T790M mutation (10). EKB-569 and CI-1033 are EGFR inhibitors that have completed phase I clinical trials for NSCLC (Pharmaprojects database).
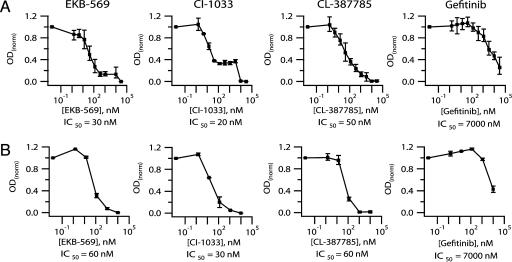
To confirm and extend the results, we measured IC50 values for inhibition of H1975 cell proliferation (Fig. 3A and Table 5, which is published as supporting information on the PNAS web site). CL-387785, EKB-569, and CI-1033 were >100-fold more effective at inhibiting H1975 growth than gefitinib (Fig. 3A) or erlotinib (Table 5), with IC50 values of 20-50 nM. None of the other clinical EGFR inhibitors tested, including GW-572016 (lapatinib) and ZD-6474 (Table 2), inhibited H1975 proliferation with IC50 values <1 μM (Table 5). The growth inhibition curve for CI-1033 shows a significant second transition at higher concentration that was not observed, or was at least much less pronounced, with EKB-569 or CL-387785. To measure a biochemical correlate of EGFR tyrosine kinase activity, we determined EGFR autophosphorylation levels in H1975 cells (Fig. 3B). The IC50 values for inhibition of autophosphorylation were consistent with the IC50 values for growth inhibition, confirming that the three compounds are potent inhibitors of the activity of EGFR(L858R/T790M). No significant second transition was observed for inhibition of EGFR autophosphorylation by CI-1033, and we do not yet know what causes the difference in the shape of the curves for inhibition of growth and autophosphorylation for this compound.
Fig. 3.
EKB-569 and CI-1033 inhibit gefitinib-resistant EGFR. (A) Cellular proliferation assay. For each concentration point, viable cells were quantitated after 48 h of exposure to compound. (B) Autophosphorylation assay. EGFR autophosphorylation levels in H1975 cells were determined by ELISA after 2 h of exposure to compound followed by EGF stimulation. Results for both the proliferation assay and the autophosphorylation assay were normalized relative to vehicle control. IC50 values were measured at least in duplicate, and average values are shown. The graphs show the average of replicate measurements for each concentration point. Error bars represent the range of values obtained.
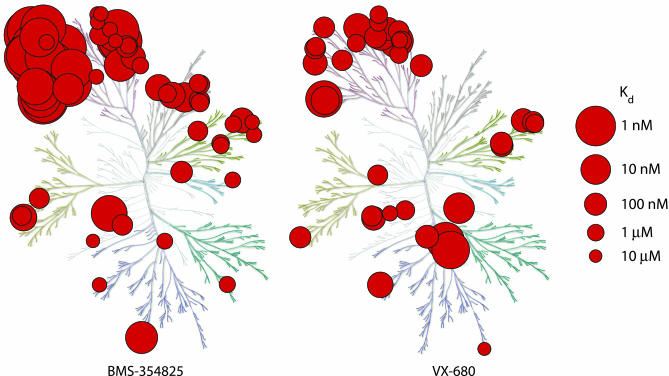
Kinase Interaction Maps for BMS-354825 and VX-680. BMS-354825 was originally evaluated for inhibition of imatinib-resistant BCR-ABL because it had been shown to have broad antiproliferative effects and was known to inhibit multiple kinases, including SRC. The apparent promiscuity of BMS-354825 suggested that the compound might be able to tolerate small changes in a kinase and inhibit mutant forms of ABL (19). Imatinib, by contrast, is a more specific compound that binds a particular conformation of ABL (14, 38). To more fully characterize the overall kinase specificity of BMS-354825, and to compare the specificity to that of VX-680, we constructed interaction maps for both compounds against a large panel of human kinases (Fig. 4) (14). A primary screen at 10 μM identified kinases that bind to the compounds, and a quantitative Kd was measured for each kinase hit in the primary screen (see Table 6, which is published as supporting information on the PNAS web site, for complete quantitative results). We found that BMS-354825 binds 76 of 148 kinases screened at 10 μM, 47 of them with Kd ≤ 200 nM. BMS-354825 binds to a large number of tyrosine kinases with very high affinity (Table 6), consistent with its reported broad antiproliferative activity (31). The interaction map for BMS-354825 is in some ways complementary to that of staurosporine, a highly promiscuous compound that tends to bind with higher affinity to serine-threonine, rather than tyrosine, kinases (14). It is interesting to note that, despite its promiscuity, BMS-354825 seems to be well tolerated in patients (N.P.S. and C.L.S., unpublished observations).
Fig. 4.
Kinase interaction maps for BMS-354825 and VX-680. Each red circle indicates a kinase that binds the compound, and larger circles indicate higher affinity binding (see scale at right). Complete quantitative results are shown in Table 6. With sponsorship from Cell Signaling Technology and Sugen, the dendogram was originally presented as a poster in Science to accompany the first analysis of the human kinkome (52). [Figure adapted with permission from ref. 52 (Copyright 2002, Cell Signaling Technology).]
VX-680 was found to bind tightly to Aurora kinases as well as FLT3 (Fig. 4 and Table 6), consistent with published results (24). However, although VX-680 has been described as a very specific inhibitor, we discovered that the compound binds to 37 of 119 kinases tested at 10 μM, including 19 kinases with Kd values <200 nM. BMS-354825 and VX-680 both bind numerous kinases; however, promiscuity alone does not explain why VX-680 binds ABL(T315I) with much higher affinity than BMS-354825, because BMS-354825 is the more promiscuous compound.
Discussion
The T670I mutation in KIT and the T790M mutation in EGFR are homologous to the T315I mutation in BCR-ABL, and all three mutations confer resistance to clinical-stage ATP-competitive kinase inhibitors. This threonine residue, also called the gatekeeper, is known to be an important determinant of inhibitor binding in the context of multiple kinases (13, 18, 42, 43). Imatinib-resistant mutations at other positions in the ABL kinase tend to weaken binding only moderately, albeit enough to confer clinical resistance. The T315I mutation in BCR-ABL confers resistance not only to imatinib, but to all other second-generation, ATP-competitive BCR-ABL inhibitors described so far, including BMS-354825 and AMN-107 (19, 20). The clinical importance of this mutation may grow considerably, as to date it appears to represent the primary mechanism of resistance to BMS-354825 in patients (N.P.S. and C.L.S., unpublished observations). It has been shown that the Hsp90 inhibitors geldanamycin and 17-allylaminogeldanamycin can induce degradation of BCR-ABL(T315I) (44), but the only direct inhibitor of BCR-ABL(T315I) described so far is a non-ATP-competitive preclinical compound (45). There are currently no effective kinase-targeted treatments for patients with T315I mutations. The IC50 values we have measured for inhibition of BCR-ABL(T315I) in cells by BIRB-796 and VX-680, although relatively high (1-5 μM), are within ≈10-fold of the IC50 value for inhibition of wild-type BCR-ABL by imatinib (≈0.3 μM) (28, 32). The mean trough plasma concentration at steady state in humans for imatinib has been measured as 1.5 μM (400-mg dose) (46), only a fewfold above the IC50 for cellular inhibition of BCR-ABL. Therefore, it may be possible to achieve plasma concentrations of BIRB-796 or VX-680 that are sufficiently high to at least partially inhibit BCR-ABL(T315I). The identification of compounds already in clinical trials that inhibit BCR-ABL(T315I) may offer a new path for the development of a treatment for this imatinib-resistant patient population. At a minimum, our results demonstrate that it is possible to inhibit this kinase with ATP-competitive compounds, and provide a good starting point for medicinal chemistry efforts. The differences we observe between Kd values measured in binding assays, IC50 values measured in in vitro activity assays, and IC50 values in cell-based assays highlight the point that binding or in vitro inhibition of a particular form of an enzyme are generally necessary, but not always sufficient, for cell-based potency. These assays may query different forms of a kinase under different conditions, and therefore provide complementary information, but are not expected to yield identical results in all cases.
Although it has been observed that compounds chemically related to SU-11248 can inhibit kinases with mutations at the conserved threonine residue (18), our finding that SU-11248 inhibits KIT(V559D/T670I) with no apparent loss in potency has, to our knowledge, not been described previously, and is of direct clinical relevance because SU-11248 is in late stage trials for imatinib-resistant GIST. It has been reported recently that the trials will be halted ahead of schedule because of the apparent efficacy of the compound (Pharmaprojects database). The mutation status of patients has, to our knowledge, not been reported, and our results suggest that patients with gatekeeper mutations in KIT may be among those treated successfully with SU-11248 for imatinib-resistant GIST.
The recent description of T790M as a secondary clinical gefitinib- and erlotinib-resistance mutation in EGFR was anticipated by in vitro studies with gefitinib in the context of wild-type EGFR (18). The finding that EGFR inhibitors already in clinical trials can inhibit EGFR(L858R/T790M) suggests that refractory or relapsed patients with the T790M mutation may benefit from treatment with EKB-569 or CI-1033. Interestingly, the three inhibitors of EGFR(L858R/T790M) identified in our screen, CL-387785, EKB-569, and CI-1033, are the only compounds we tested that are known to inhibit EGFR irreversibly (47-49). Other clinical EGFR inhibitors that do not act irreversibly, such as GW-572016 and ZD-6474, have only weak activity against EGFR(L858R/T790M). A close derivative of EKB-569, HKI-272, has potent activity against both EGFR and ERBB2 and is in clinical development (50). HKI-272 is also an irreversible inhibitor and may therefore inhibit EGFR with the T790M mutation.
We show here for three different drug-resistant kinase targets that protein kinase inhibitors already in clinical development can inhibit mutant kinase variants resistant to first-line targeted therapies. Our results also indicate that drug resistant gatekeeper mutants are not necessarily more difficult to inhibit with an ATP-site directed small molecule, as suggested by the behavior of compounds such as imatinib and BMS-354825. Although the compounds we identified must be tested further in additional cell and animal models, and their utility in the clinic explored, we hope that this approach can help lead quickly to new treatment options for patients with resistance mutations as well as inform and facilitate the development of next-generation compounds.
Note. While this manuscript was under review, EKB-569 and HKI-272 were independently reported as potent inhibitors of EGFR variants with the T790M mutation (51).
Supplementary Material
Acknowledgments
We thank N. Lydon and T. Hunter for a critical reading of the manuscript and helpful discussions, Bristol-Myers Squibb for providing BMS-354825, P. Ciceri for advice on cell-based assays, Dan Lockhart for writing software tools to facilitate data analysis, and AAAS and Cell Signaling Technology for permission to reproduce the kinase dendogram in Fig. 4. This work was supported by a Career Development Award for Fellows (to N.P.S.), grants from the Leukemia and Lymphoma Society (to N.P.S. and C.L.S.), the CHEST Foundation of the American College of Chest Physicians and the LUNG Foundation (W.P.), and an anonymous donor (H.V.). C.L.S. is an investigator of the Howard Hughes Medical Institute and a Doris Duke Distinguished Clinical Scientist.
Author contributions: T.A.C., L.M.W., N.P.S., M.A.F., D.K.T., H.K.P., W.P., C.L.S., H.V., P.P.Z., and D.J.L. designed research; T.A.C., L.M.W., N.P.S., A.M.V., M.A.F., Z.V.M., C.E.A., W.H.B., P.T.E., M.F., J.M.F., R.M.G., D.E.I., S.A.M., and H.K.P. performed research; W.P. and H.V. contributed new reagents/analytic tools; T.A.C., L.M.W., N.P.S., A.M.V., M.A.F., D.K.T., C.E.A., W.H.B., S.H., C.L.S., P.P.Z., and D.J.L. analyzed data; and T.A.C., S.H., W.P., C.L.S., H.V., P.P.Z., and D.J.L. wrote the paper.
Abbreviations: EGFR, EGF receptor; GIST, gastrointestinal stromal tumor.
References
- 1.Sawyers, C. (2004) Nature 432, 294-297. [DOI] [PubMed] [Google Scholar]
- 2.Druker, B. J. (2004) Oncologist 9, 357-360. [DOI] [PubMed] [Google Scholar]
- 3.Wadleigh, M., DeAngelo, D. J., Griffin, J. D. & Stone, R. M. (2005) Blood 105, 22-30. [DOI] [PubMed] [Google Scholar]
- 4.Gorre, M. E., Mohammed, M., Ellwood, K., Hsu, N., Paquette, R., Rao, P. N. & Sawyers, C. L. (2001) Science 293, 876-880. [DOI] [PubMed] [Google Scholar]
- 5.Deininger, M., Buchdunger, E. & Druker, B. J. (2005) Blood 105, 2640-2653. [DOI] [PubMed] [Google Scholar]
- 6.Chen, L. L., Trent, J. C., Wu, E. F., Fuller, G. N., Ramdas, L., Zhang, W., Raymond, A. K., Prieto, V. G., Oyedeji, C. O., Hunt, K. K., et al. (2004) Cancer Res. 64, 5913-5919. [DOI] [PubMed] [Google Scholar]
- 7.Tamborini, E., Bonadiman, L., Greco, A., Albertini, V., Negri, T., Gronchi, A., Bertulli, R., Colecchia, M., Casali, P. G., Pierotti, M. A., et al. (2004) Gastroenterology 127, 294-299. [DOI] [PubMed] [Google Scholar]
- 8.Debiec-Rychter, M., Cools, J., Dumez, H., Sciot, R., Stul, M., Mentens, N., Vranckx, H., Wasag, B., Prenen, H., Roesel, J., et al. (2005) Gastroenterology 128, 270-279. [DOI] [PubMed] [Google Scholar]
- 9.Cools, J., DeAngelo, D. J., Gotlib, J., Stover, E. H., Legare, R. D., Cortes, J., Kutok, J., Clark, J., Galinsky, I., Griffin, J. D., et al. (2003) N. Engl. J. Med. 348, 1201-1214. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi, S., Boggon, T. J., Dayaram, T., Janne, P. A., Kocher, O., Meyerson, M., Johnson, B. E., Eck, M. J., Tenen, D. G. & Halmos, B. (2005) N. Engl. J. Med. 352, 786-792. [DOI] [PubMed] [Google Scholar]
- 11.Pao, W., Miller, V. A., Politi, K. A., Riely, G. J., Somwar, R., Zakowski, M. F., Kris, M. G. & Varmus, H. (2005) PLoS Med. 2, e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deininger, M. W. & Druker, B. J. (2004) Cancer Cell 6, 108-110. [DOI] [PubMed] [Google Scholar]
- 13.Daub, H., Specht, K. & Ullrich, A. (2004) Nat. Rev. Drug Discov. 3, 1001-1010. [DOI] [PubMed] [Google Scholar]
- 14.Fabian, M. A., Biggs, W. H., Treiber, D. K., Atteridge, C. E., Azimioara, M. D., Benedetti, M. G., Carter, T. A., Ciceri, P., Edeen, P. T., Floyd, M., et al. (2005) Nat. Biotechnol. 23, 329-336. [DOI] [PubMed] [Google Scholar]
- 15.Buchdunger, E., Cioffi, C. L., Law, N., Stover, D., Ohno-Jones, S., Druker, B. J. & Lydon, N. B. (2000) J. Pharmacol. Exp. Ther. 295, 139-145. [PubMed] [Google Scholar]
- 16.Heinrich, M. C., Griffith, D. J., Druker, B. J., Wait, C. L., Ott, K. A. & Zigler, A. J. (2000) Blood 96, 925-932. [PubMed] [Google Scholar]
- 17.Joensuu, H., Roberts, P. J., Sarlomo-Rikala, M., Andersson, L. C., Tervahartiala, P., Tuveson, D., Silberman, S., Capdeville, R., Dimitrijevic, S., Druker, B., et al. (2001) N. Engl. J. Med. 344, 1052-1056. [DOI] [PubMed] [Google Scholar]
- 18.Blencke, S., Zech, B., Engkvist, O., Greff, Z., Orfi, L., Horvath, Z., Keri, G., Ullrich, A. & Daub, H. (2004) Chem. Biol. 11, 691-701. [DOI] [PubMed] [Google Scholar]
- 19.Shah, N. P., Tran, C., Lee, F. Y., Chen, P., Norris, D. & Sawyers, C. L. (2004) Science 305, 399-401. [DOI] [PubMed] [Google Scholar]
- 20.Weisberg, E., Manley, P. W., Breitenstein, W., Bruggen, J., Cowan-Jacob, S. W., Ray, A., Huntly, B., Fabbro, D., Fendrich, G., Hall-Meyers, E., et al. (2005) Cancer Cell 7, 129-141. [DOI] [PubMed] [Google Scholar]
- 21.La Rosee, P., Corbin, A. S., Stoffregen, E. P., Deininger, M. W. & Druker, B. J. (2002) Cancer Res. 62, 7149-7153. [PubMed] [Google Scholar]
- 22.O'Hare, T., Pollock, R., Stoffregen, E. P., Keats, J. A., Abdullah, O. M., Moseson, E. M., Rivera, V. M., Tang, H., Metcalf, C. A., III, Bohacek, R. S., et al. (2004) Blood 104, 2532-2539. [DOI] [PubMed] [Google Scholar]
- 23.Pargellis, C., Tong, L., Churchill, L., Cirillo, P. F., Gilmore, T., Graham, A. G., Grob, P. M., Hickey, E. R., Moss, N., Pav, S., et al. (2002) Nat. Struct. Biol. 9, 268-272. [DOI] [PubMed] [Google Scholar]
- 24.Harrington, E. A., Bebbington, D., Moore, J., Rasmussen, R. K., Ajose-Adeogun, A. O., Nakayama, T., Graham, J. A., Demur, C., Hercend, T., Diu-Hercend, A., et al. (2004) Nat. Med. 10, 262-267. [DOI] [PubMed] [Google Scholar]
- 25.Fiedler, W., Serve, H., Dohner, H., Schwittay, M., Ottmann, O. G., O'Farrell, A. M., Bello, C. L., Allred, R., Manning, W. C., Cherrington, J. M., et al. (2005) Blood 105, 986-993. [DOI] [PubMed] [Google Scholar]
- 26.Calvo, E., Tolcher, A. W., Hammond, L. A., Patnaik, A., de Bono, J. S., Eiseman, I. A., Olson, S. C., Lenehan, P. F., McCreery, H., Lorusso, P., et al. (2004) Clin. Cancer Res. 10, 7112-7120. [DOI] [PubMed] [Google Scholar]
- 27.Tsou, H. R., Overbeek-Klumpers, E. G., Hallett, W. A., Reich, M. F., Floyd, M. B., Johnson, B. D., Michalak, R. S., Nilakantan, R., Discafani, C., Golas, J., et al. (2005) J. Med. Chem. 48, 1107-1131. [DOI] [PubMed] [Google Scholar]
- 28.Shah, N. P., Nicoll, J. M., Nagar, B., Gorre, M. E., Paquette, R. L., Kuriyan, J. & Sawyers, C. L. (2002) Cancer Cell 2, 117-125. [DOI] [PubMed] [Google Scholar]
- 29.Hirota, S., Isozaki, K., Moriyama, Y., Hashimoto, K., Nishida, T., Ishiguro, S., Kawano, K., Hanada, M., Kurata, A., Takeda, M., et al. (1998) Science 279, 577-580. [DOI] [PubMed] [Google Scholar]
- 30.Rubin, B. P., Singer, S., Tsao, C., Duensing, A., Lux, M. L., Ruiz, R., Hibbard, M. K., Chen, C. J., Xiao, S., Tuveson, D. A., et al. (2001) Cancer Res. 61, 8118-8121. [PubMed] [Google Scholar]
- 31.Lombardo, L. J., Lee, F. Y., Chen, P., Norris, D., Barrish, J. C., Behnia, K., Castaneda, S., Cornelius, L. A., Das, J., Doweyko, A. M., et al. (2004) J. Med. Chem. 47, 6658-6661. [DOI] [PubMed] [Google Scholar]
- 32.Burgess, M. R., Skaggs, B. J., Shah, N. P., Lee, F. Y. & Sawyers, C. L. (2005) Proc. Natl. Acad. Sci. USA 102, 3395-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corbin, A. S., Griswold, I. J., La Rosee, P., Yee, K. W., Heinrich, M. C., Reimer, C. L., Druker, B. J. & Deininger, M. W. (2004) Blood 104, 3754-3757. [DOI] [PubMed] [Google Scholar]
- 34.O'Farrell, A. M., Abrams, T. J., Yuen, H. A., Ngai, T. J., Louie, S. G., Yee, K. W., Wong, L. M., Hong, W., Lee, L. B., Town, A., et al. (2003) Blood 101, 3597-3605. [DOI] [PubMed] [Google Scholar]
- 35.Abrams, T. J., Lee, L. B., Murray, L. J., Pryer, N. K. & Cherrington, J. M. (2003) Mol. Cancer Ther. 2, 471-478. [PubMed] [Google Scholar]
- 36.Kelly, L. M., Yu, J. C., Boulton, C. L., Apatira, M., Li, J., Sullivan, C. M., Williams, I., Amaral, S. M., Curley, D. P., Duclos, N., et al. (2002) Cancer Cell 1, 421-432. [DOI] [PubMed] [Google Scholar]
- 37.Griswold, I. J., Shen, L. J., La Rosee, P., Demehri, S., Heinrich, M. C., Braziel, R. M., McGreevey, L., Haley, A. D., Giese, N., Druker, B. J., et al. (2004) Blood 104, 2912-2918. [DOI] [PubMed] [Google Scholar]
- 38.Schindler, T., Bornmann, W., Pellicena, P., Miller, W. T., Clarkson, B. & Kuriyan, J. (2000) Science 289, 1938-1942. [DOI] [PubMed] [Google Scholar]
- 39.Lynch, T. J., Bell, D. W., Sordella, R., Gurubhagavatula, S., Okimoto, R. A., Brannigan, B. W., Harris, P. L., Haserlat, S. M., Supko, J. G., Haluska, F. G., et al. (2004) N. Engl. J. Med. 350, 2129-2139. [DOI] [PubMed] [Google Scholar]
- 40.Paez, J. G., Janne, P. A., Lee, J. C., Tracy, S., Greulich, H., Gabriel, S., Herman, P., Kaye, F. J., Lindeman, N., Boggon, T. J., et al. (2004) Science 304, 1497-1500. [DOI] [PubMed] [Google Scholar]
- 41.Pao, W., Miller, V., Zakowski, M., Doherty, J., Politi, K., Sarkaria, I., Singh, B., Heelan, R., Rusch, V., Fulton, L., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 13306-13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eyers, P. A., Craxton, M., Morrice, N., Cohen, P. & Goedert, M. (1998) Chem. Biol. 5, 321-328. [DOI] [PubMed] [Google Scholar]
- 43.Fox, T., Coll, J. T., Xie, X., Ford, P. J., Germann, U. A., Porter, M. D., Pazhanisamy, S., Fleming, M. A., Galullo, V., Su, M. S., et al. (1998) Protein Sci. 7, 2249-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gorre, M. E., Ellwood-Yen, K., Chiosis, G., Rosen, N. & Sawyers, C. L. (2002) Blood 100, 3041-3044. [DOI] [PubMed] [Google Scholar]
- 45.Gumireddy, K., Baker, S. J., Cosenza, S. C., John, P., Kang, A. D., Robell, K. A., Reddy, M. V. & Reddy, E. P. (2005) Proc. Natl. Acad. Sci. USA 102, 1992-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Druker, B. J., Talpaz, M., Resta, D. J., Peng, B., Buchdunger, E., Ford, J. M., Lydon, N. B., Kantarjian, H., Capdeville, R., Ohno-Jones, S., et al. (2001) N. Engl. J. Med. 344, 1031-1037. [DOI] [PubMed] [Google Scholar]
- 47.Smaill, J. B., Rewcastle, G. W., Loo, J. A., Greis, K. D., Chan, O. H., Reyner, E. L., Lipka, E., Showalter, H. D., Vincent, P. W., Elliott, W. L., et al. (2000) J. Med. Chem. 43, 1380-1397. [DOI] [PubMed] [Google Scholar]
- 48.Wissner, A., Overbeek, E., Reich, M. F., Floyd, M. B., Johnson, B. D., Mamuya, N., Rosfjord, E. C., Discafani, C., Davis, R., Shi, X., et al. (2003) J. Med. Chem. 46, 49-63. [DOI] [PubMed] [Google Scholar]
- 49.Discafani, C. M., Carroll, M. L., Floyd, M. B., Jr., Hollander, I. J., Husain, Z., Johnson, B. D., Kitchen, D., May, M. K., Malo, M. S., Minnick, A. A., Jr., et al. (1999) Biochem. Pharmacol. 57, 917-925. [DOI] [PubMed] [Google Scholar]
- 50.Rabindran, S. K., Discafani, C. M., Rosfjord, E. C., Baxter, M., Floyd, M. B., Golas, J., Hallett, W. A., Johnson, B. D., Nilakantan, R., Overbeek, E., et al. (2004) Cancer Res. 64, 3958-3965. [DOI] [PubMed] [Google Scholar]
- 51.Kwak, E. L., Sordella, R., Bell, D. W., Godin-Heymann, N., Okimoto, R. A., Brannigan, B. W., Harris, P. L., Driscoll, D. R., Fidias, P., Lynch, T. J., et al. (2005) Proc. Natl. Acad. Sci. USA 102, 7665-7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manning, G., Whyte, D. B., Martinez, R., Hunter, T. & Sudarsanam, S. (2002) Science 298, 1912-1934. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.