Abstract
Background
Human polyomaviruses (HPyVs), JC polyomavirus (JCPyV) and BK polyomavirus (BKPyV), have been found in upper tract urothelial carcinoma UTUC; however, the association of the viral oncogenic factors and clinical characteristics of UTUC remains unclear. This study aimed to investigate the prevalence of JCPyV and BKPyV in UTUC and their correlation with cancer progression among the southwest Taiwanese population from 2020 to 2022.
Methods
A total of 72 paraffin-embedded UTUC tissue samples and 41 adjacent tissue samples were collected from 72 patients. Nested polymerase chain reaction and DNA sequencing were used to detect viral DNA and genotypes. Immunohistochemistry was performed using anti- large T (LT) and anti-p53 monoclonal antibodies to detect the expression of viral early LT protein and cellular p53 protein, respectively.
Results
The overall prevalence of JCPyV and BKPyV were higher in UTUC than in adjacent tissue samples (65.3% [47/72] vs. 17.1% [7/41]). JCPyV and BKPyV were detected in 95.7% (45/47) and 4.3% (2/47) of the HPyVs-positive UTUC samples, respectively. JCPyV-TW-3 was the predominant strain of JCPyV infection. In UTUC samples, the LT protein of JCPyV and BKPyV positivity rate was 65.3%, while that of mutant p53 protein was 52.7%. JCPyV infection and LT protein expression increased the odds ratio (OR) of UTUC by 9.13-fold. The OR of UTUC was higher by 10.34-fold in patients with mutant p53 and by 10.37-fold in those with simultaneous LT and mutant p53 expression. The presence of LT protein in UTUC patients may increase the OR of mutant p53 protein expression by 2.93-fold compared to its absence. Women had a 5.19-fold higher superiority of JCPyV infection and LT expression than men. Patients with chronic kidney disease (CKD) had a 3.15-fold higher OR for mutant p53 protein expression than those without it. In the UTUC advanced stages, the OR of virus and LT expression was 3.18-fold higher compared to those who do not require chemotherapy.
Conclusions
JCPyV infection is highly prevalent in UTUC, and the presence of CKD concurrent with high expressions of LT and mutant p53 proteins in patients may be a useful indicator for chemotherapy and poor prognosis.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12985-025-02643-8.
Keywords: Upper tract urothelial carcinoma, JCPyV infection, LT protein, p53 protein, Chronic kidney disease (CKD)
Background
Urothelial carcinoma, the sixth most prevalent solid tumor, comprises lower urothelial carcinomas (bladder and urethra) and upper tract urothelial carcinomas (UTUCs, including renal calyces, renal pelvis, and ureter). The occurrence of UTUC, similar to that of bladder cancer, is influenced by various factors [1], including arsenic exposure, smoking, analgesic abuse, occupational carcinogens, hypertension, chronic urinary tract obstruction, infections, and kidney disease [2–4]. About two-thirds of patients diagnosed with UTUC have invasive disease. The most prevalent symptom is hematuria (70–80%), with flank pain also common due to clot or tumor obstruction. Systemic symptoms such as anorexia, weight loss, malaise, fatigue, fever, night sweats, and cough suggest a worse prognosis [1]. Treatment options include nephroureterectomy, endoscopic management, and systemic therapies, such as chemotherapy and immunotherapy [1]. According to the tumor- node-metastasis (TNM) staging system [5], chemotherapy is generally recommended for advanced stages, specifically from stage T2 beyond (invasion into the muscle layer), including stages N (lymph node involvement) and M (distant metastasis) [1]. The grade of the tumor, which indicates how much the tumor cells differ from normal cells, is crucial for prognosis. High-grade tumors tend to be more aggressive and exhibit a poorer prognosis [1]. Tissue-tropic microorganisms that remain latent or proliferate in the urinary or reproductive tract for an extended period can cause chronic infection, thereby increasing the risk of related cancers in the urinary or reproductive tract [6, 7]. Pathogens, such as the Epstein–Barr virus (EBV), cytomegalovirus (CMV), human papilloma virus (HPV), and JC polyomavirus (JCPyV), not only infect cells but also disrupt multiple cellular processes by producing oncoproteins that interact with intracellular p53. This disruption leads to unrestricted cell proliferation and tumor formation [6, 7].
At least 14 species of human polyomaviruses (HPyVs) have been identified, with the BK polyomavirus (BKPyV) and JCPyV being the most common. Both share a high degree of similarity, with nearly 72% homology in their entire nucleotide sequences [8]. Their genomes are circular and consist of three major regions, namely early, late and non-coding control regions (NCCR). The early region encodes alternatively spliced transforming proteins, including the large T (LT) and small t (st) antigens. The late region encodes the capsid structural proteins, VP1, VP2, and VP3, along with a small regulatory protein, termed agnoprotein. Meanwhile, the NCCR contains the origin of replication, promoter, and enhancer, driving the bidirectional replication of early and late genes. Most strains show a high degree of sequence conservation in the protein-coding regions of the genome, whereas the NCCR displays significant variation among different HPyV isolates. This variation is attributed to deletions, duplications, and rearrangements of a core set of sequence blocks [9–11]. Primary infections of JCPyV and BKPyV typically occur during childhood, often go unnoticed, and present asymptomatically. Following the initial infection, polyomaviruses establish latency within tissue cells and may later reactivate under immunocompromised conditions, leading to tissue damage [12]. JCPyV is associated with progressive multifocal leukoencephalopathy (PML) [8, 13], while BKPyV is linked to polyoma-associated nephropathy, including hemorrhagic cystitis, interstitial nephritis, and ureteral stenosis [8]. HPyVs have been implicated in many tumors; particularly, JCPyV is linked to brain, colon, esophageal, gastric, bladder, and prostate cancers, while BKPyV is associated with insulinomas, Kaposi’s sarcomas, bladder, and bone tumors [8]. Transformed cells express polyomavirus genomic DNA and early LT antigen, which interact with the tumor suppressor proteins, p53 and pRb, leading to uncontrolled cell proliferation [14–18].
The p53 protein is a crucial tumor suppressor. Viral infections can lead to p53 gene mutations and altered expression, often compromising its tumor suppressor function; the loss of p53 function is represents a crucial step in carcinogenesis [19, 20]. p53 is the most commonly mutated gene in human cancers, with mutations occurring in over 50% of all cases across various cancer types [19]. For instance, HPV, particularly HPV-16 and HPV-18, interact with p53 and retinoblastoma (pRb) through the HPV E6 and E7 protein, which is associated with cervical cancer [21]. Hepatitis B Virus (HBV) and Hepatitis C Virus (HCV)-related hepatocellular carcinoma (HCC) demonstrate that the viruses can integrate into the host genome and induce p53 gene mutations [22]. In nasopharyngeal carcinoma and Burkitt’s lymphoma caused by EBV, EBV is present in 30–50% of cases with varying p53 expression [23, 24]. JCPyV and BKPyV have been found in many human cancers [8]. Over 80% of colorectal cancers harbor JCPyV DNA [7], with some cases showing p53 protein activity, where the virus inactivates p53 through its LT protein binding [15, 25]. However, some tumors demonstrate mutant p53 protein expression regardless of viral infection, such as non-small cell lung cancers harboring p53 mutations in 50–80% [26–28], breast cancer in 20–35% [29, 30], and high-grade serous ovarian carcinomas in over 96% of cases [31]. Therefore, the mutation of p53 protein and their correlation with viral infections vary by cancer type and viral etiology.
In the current epidemiological investigation of UTUC in Taiwan, an unusual prevalence of UTUC [32, 33] was found to be associated with Blackfoot disease in southern Taiwan, as well as arsenic-contaminated water consumption [34, 35], sex, tumor staging, and chronic kidney disease (CKD) [32, 36]. Although the association of UTUC and viruses remains poorly studied, HPyVs have been detected in urothelial carcinoma [37–41]. However, no study has explored the mutation of p53 protein and its association with human polyomavirus infection in UTUC. Moreover, the relationship between the expression of the viral LT antigen and mutant p53 protein following viral infection and the clinicopathological characteristics and progression of UTUC remains unclear.
Thus, this study aimed to investigate the prevalence of JCPyV and BKPyV infections in the southwestern Taiwanese population from 2020 to 2022 and their association with UTUC. Moreover, we evaluated the association between the presence of JCPyV/BKPyV and the clinical characteristics in patients with UTUC.
Methods
Determining JCPyV and BKPyV DNA positivity rates in UTUC and adjacent tissues
Clinical specimens
To examine the polyomavirus genome and viral protein expression, 113 formalin-fixed, paraffin-embedded (FFPE) specimens (72 and 41 specimens from cancer and adjacent tissues, respectively) were obtained from 72 patients diagnosed with UTUC at the Urology Department of Chiayi Christian Hospital, Taiwan between 2020 and 2022 during nephroureterectomy. These specimens were cleaned by phosphate buffered saline, processed independently, and preserved in the pathology department until analysis. All patients included in the study were diagnosed with UTUC after presenting with hematuria. The clinicopathological stage of the cancers ranged from Tx to T4, as defined by the American Joint Committee on Cancer TNM staging system for UTUC [5]. Exclusion criteria comprised: (1) other tumors, (2) with immune system diseases or ongoing immune therapy, and (3) lack of independent consciousness precluding the patient from making decisions based on their own thoughts and senses. All patients provided written informed consent, and their confidentiality was strictly protected. Specimens were collected following thorough review, with the study receiving approval from the institutional review board of Chiayi Christian Hospital.
DNA extraction
Tissue samples were processed according to the methodology described by Tseng et al. [42–44]. Briefly, DNA was extracted from paraffin-embedded tissues following the Gene JET FFPE DNA purification reagent (Thermo Fisher, Vilnius, Lithuania) guidelines. The tissues were deparaffinized using xylene, washed 2–3 times with anhydrous alcohol, rinsed with deionized water to eliminate residual alcohol, and air dried. Subsequently, the samples were treated with 50 µg/mL proteinase K at 50 °C for 16–18 h, heat inactivated by boiling in water for 10 min, and centrifuged at 10,000 rpm for 3 min to collect the 100-microliter supernatant for DNA purification. We used 200 ng of DNA for viral DNA detection through nested polymerase chain reaction (PCR). The amplified DNA was preserved at − 20 °C until further use.
Nested PCR and DNA sequencing
An FFPE cell line carrying JCPyV (JCI) was used as a positive control for both PCR and immunohistochemical staining [43]. Nested PCR was used to confirm the presence of viral DNA in UTUC specimens. To identify changes in polyomavirus infection, the viral genome may undergo rearrangement in its regulatory region, leading to the emergence of new genetic variants [11]. We employed two primer pairs specifically designed to amplify the conserved regulatory regions of both JCPyV and BKPyV [42, 43]. The initial PCR utilized the primer pair JBR1 and JBR2 (nucleotides 5067–5091 of JCPyV TW-3 strain, 5ʹ-CCTCCACGCCCTTACTACTTCTGAG-3ʹ and 279 − 255, 5ʹ-GTGACAGCTGGCGAAGAACCATGGC-3ʹ, respectively) for regulatory region amplification. Subsequently, 5 µL of the initial PCR product served as the template, and the primer pair JBRNS and JBRNAS (nucleotides 5,100-5 of JCPyV TW-3 strain, 5ʹ-GAGGCGGCCTCGGCCTC-3ʹ and 227 − 212, 5ʹ-GGCTCGCAAAACATGT-3ʹ, respectively) was employed for the second PCR, amplified DNA fragments of 243 bp for JCPyV or 289 bp for BKPyV. To confirm the presence of tissue DNA and minimize the risk of false-negative results, concurrent PCR amplification of the human beta-actin (β-actin) 5ʹ UTR region was conducted.
All specimens were analyzed in triplicate, and the resulting secondary PCR products were subjected to 2.5% agarose gel electrophoresis. The positive controls for the JCPyV CY genotype (GenBank accession No. AB038249.1) and BKPyV UT (GenBank accession No. M 34049.1) were incorporated, resulting in bands of approximately 243 bp and 289 bp in size, respectively. To further characterize the genetic variations, the purified PCR DNA fragments were ligated into the sequencing plasmid T-vector (Promega, Madison, WI, USA). Subsequent sequencing of the inserts within the T vector was performed using M13 primers (Mingxin Biotechnology, Taipei City, Taiwan). All the obtained sequences were compared with the JCPyV-CY archetype (GenBank accession No. AB038249.1) NCCR sequence, which contains the complete sequence without mutations from − 6 to 237 nucleotides, and the BKPyV-UT prototype (GenBank accession No. M34049.1), with its unmutated NCCR sequence spanning 1–289 nucleotides. This comparison was conducted to confirm whether the regulatory regions underwent rearrangement and to determine the variants.
Detection of viral LT protein and cellular p53 protein expressions in UTUC tissues by immunohistochemistry staining
To test the potential contribution of JCPyV and BKPyV to UTUC tumorigenesis, the expressions of early viral protein and cellular p53 proteins were assessed using immunohistochemistry (IHC) [45–47]. IHC staining was performed using the avidin–biotin–peroxidase complex system (Vectastain ABC peroxidase kit, Vector Laboratories, Burlingame, CA, USA), as previously described [42] with minor modifications. Briefly, a 3 μm FFPE tissue sections were deparaffinized in xylene, rehydrated through a gradient of 70–100% ethanol, and subjected to antigen retrieval. Anti-SV40 LT (MA1-90661; Thermo Fisher, Vilnius, Lithuania) and anti-p53 (ab131442; Abcam, Cambridge, USA) primary monoclonal antibodies were used to detect the expression of early viral protein and cellular p53 proteins. Tissue sections were incubated overnight in a humid chamber at 37 °C, followed by sequential addition of biotinylated secondary antibodies and avidin-biotin complexes for 1 h each. After color development with diaminobenzidine substrate (Sigma-Aldrich, St. Louis, Missouri, USA), sections were counterstained with hematoxylin. The slides were mounted and observed under a microscope. Scores for each strain of LT protein are defined as: negative (-), for < 10% positive cells and positive results, including (+) for 10–30%, (++) for 31–60%, and (+++) for > 60% positive cells, respectively [42]. In p53 IHC staining, we distinguish between wild-type (WT) and mutant (MT) p53 primarily based on the intensity, distribution, and localization of the staining. The proportion of wild-type positive cells is typically 10–30%, with moderate to weak nuclear staining intensity. The staining is localized, with expression observed in only some cells and not exhibiting uniformly strong intensity throughout the tumor. Mutant staining patterns can be categorized into overexpression, cytoplasmic, and null patterns. The overexpression pattern is characterized by intense nuclear staining, with staining intensity significantly higher than that of normal tissue controls. The proportion of positive cells often exceeds 70%, with strong and evenly distributed nuclear staining. This demonstrates consistent and robust nuclear staining, originating in the basal layer of the urothelium and extending into suprabasal layers. The cytoplasmic pattern involves abnormal cytoplasmic staining of the p53 protein, sometimes accompanied by nuclear staining. The staining intensity is typically strong, and the proportion of positive cells in tumors is generally > 30%. The null pattern is defined by the complete absence of p53 expression in tumor cells, presenting with no or extremely weak staining. In this pattern, the proportion of positive cells is typically < 10% [45–47].
Statistical analysis
Differences in viral DNA and LT antigen and p53 protein expression between the UTUC and normal tissues were examined using the Fisher’s exact test or the Mann–Whitney U test, where appropriate. The association between clinical patient characteristics and JCPyV/BKPyV expression in UTUC cells was assessed using the χ² test or Fisher’s exact test. Odds ratio (OR) were used to describe the risk of UTUC according to presence of HPyV infection. All statistical analyses were performed using the Statistical Package for Social Sciences version 20.0 software (SPSS for Windows Inc., Chicago, IL, USA), and a P-value < 0.05 indicated statistical significance.
Results
Clinical characteristics of patients with UTUC
Table 1 outlines the patient characteristics. We included 28 males (38.9%) and 44 females (61.1%) for this study; 32 (44.4%) and 40 (55.6%) patients were aged < 65 years and ≥ 65 years, respectively. We categorized the patients based on the presence of CKD: non-CKD (41.7%) and CKD (58.3%). CKD is defined as persistent abnormalities in kidney structure or function, a glomerular filtration rate [GFR] < 60 mL/min/1.73 m², or albuminuria ≥ 30 mg/24 hours, for more than 3 months [48]. Regarding cancer stage according to the need for chemotherapy, 39 (54.2%) patients had Ta-T2 disease and did not require chemotherapy, while 33 (45.8%) had T3-T4 disease and required chemotherapy. For tumor grading, five (6.9%) and 67 (93.1%) patients had low and high grading, respectively.
Table 1.
Patient characteristics
| Variable | UTUC |
|---|---|
| N = 72 (100%) | |
| Sex | |
| Male | 28 (38.9%) |
| Female | 44 (61.1%) |
| Age (years) | |
| < 65 | 32 (44.4%) |
| ≥ 65 | 40 (55.6%) |
| Disease | |
| Non-CKD | 30 (41.7%) |
| CKD | 42 (58.3%) |
| TNM staging | |
| Ta-T2 | 39 (54.2%) |
| T3-T4 | 33 (45.8%) |
| Tumor grading | |
| Low | 5 (6.9%) |
| High | 67 (93.1%) |
CKD, chronic kidney disease; UTUC, upper tract urothelial carcinoma, TNM, tumor- node-metastasis
Viral DNA in UTUC and normal tissue samples
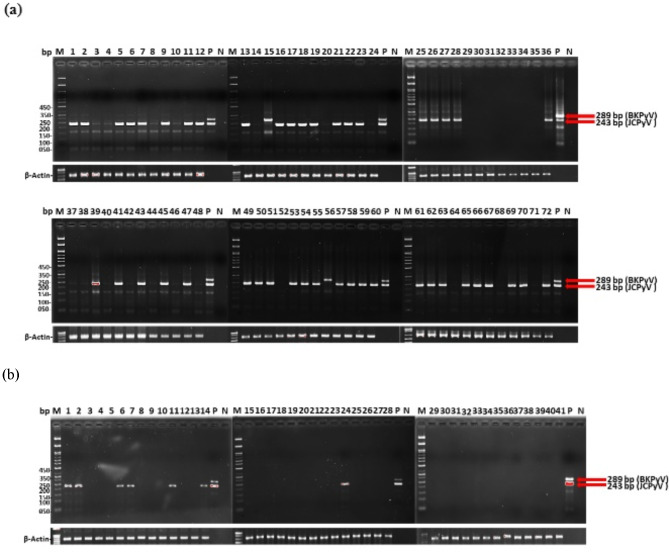
The results of the nested PCR targeting the regulatory regions of JCPyV and BKPyV are shown in Fig. 1; Table 2. Among the 72 UTUC specimens, 47 exhibited amplifications of a 243-bp DNA fragment, consistent with the size of JCPyV (Fig. 1a). In two specimens, a 289-bp DNA fragment was amplified, indicating the presence of BKPyV. PCR analysis revealed that only seven of the 41 adjacent tissue specimens tested positive for infection (Fig. 1b). Subsequent validation through DNA sequencing confirmed that 45 of the 47 UTUC-positive specimens were positive for JCPyV, whereas two were positive for BKPyV, yielding a positivity rate of 65.3% (47/72) for viral DNA in UTUC tissues. All seven adjacent tissue samples that tested positive contained JCPyV, yielding a positivity rate of 17.1% (7/41). The prevalence of JCPyV and BKPyV DNA was higher in UTUC tissues than in adjacent tissues (Table 2).
Fig. 1.
Gel electrophoresis analysis of PCR-amplified products from UTUC and adjacent tissue samples. A set of conserved primers targeting the regulatory regions of JCPyV and BKPyV is used for amplification, followed by gel analysis on a 2.5% agarose gel. Lane M contains DNA markers with molecular weights of 50, 100, 150, 200,250, 350, and 450 bp. (a) Lane numbers correspond to tissue sample numbers No. 1–72 from UTUC samples. (b) Lane numbers represent tissue sample numbers No. 1–41 from adjacent tissue samples. A positive control (P) is included using JCPyV CY and BKPyV UT genomic DNA templates, resulting in bands of approximately 243 bp and 289 bp in size, respectively. Negative control (N) indicates that PCR is performed without the DNA template. Additionally, β-actin is included as an internal control for PCR analysis. BKPyV, BK polyomavirus; JCPyV, JC polyomavirus; PCR, polymerase chain reaction; UTUC, upper tract urothelial carcinoma
Table 2.
Positivity rate of human polyomavirus infection in UTUC and adjacent tissue samples
| UTUC [N = 72 (100%)] | Adjacent [N = 41(100%)] | |
|---|---|---|
| Variables | Positivity rate (%) | Positivity rate (%) |
| Viral DNA | 47/72 (65.3%) | 7/41 (17.07%) |
| LT protein | 47/72 (65.3%) | 7/41 (17.07%) |
| Mt p53 protein | 38/72 (52.78%) | 4/41 (9.75%) |
| LT + Mt p53 | 25/72 (34.72%) | 2/41 (4.88%) |
LT, large T; UTUC, upper tract urothelial carcinoma, Mt p53, mutant p53
Viral DNA genotype in UTUC tissue samples
Both archetypal-like and rearranged-like genotypes of JCPyV and BKPyV were identified in the tissues (Supplementary Fig. 1). Most of the samples exhibit deletions in the pentanucleotide (GGGAA) at nucleotides 195–199 and pentanucleotide (AAAGC) at nucleotides 224–228 compared to the JCPyV-CY archetype. This suggests similarity to the JCPyV TW-3-like strain (GenBank accession No. U61771). Among the UTUC specimens, 37 were JCPyV TW3-like strains, three resembled the JCPyV CY-like strain, five were JCPyV SK3-like strains (GenBank accession No. AB118652.1), and two exhibited BKPyV UT-like strains. All seven adjacent specimens that tested positive resembled the JCPyV TW3-like strains. The genotyping results indicated that JCPyV was the predominant viral strain in the UTUC-positive samples, accounting for 95.7% (45/47). Among these, 6.7% were CY-like (3/45), 82.2% were TW3-like (37/45), and 11.1% were SK3-like (5/45). The BKPyV-containing genotypes were UT-like, accounting for 4.3% (2/47). In the adjacent tissue samples, all seven JCPyV-positive samples exhibited TW3-like genotypes (Table 3). These rearranged variants showed distinct nucleotide mutations from the original sequences (Supplementary Fig. 1).
Table 3.
Genotype of JCPyV/BKPyV in UTUC and adjacent tissue samples
| Genotypes | UTUC sample (n = 47) |
Adjacent sample (n = 7) |
|---|---|---|
| JCPyV | 45 (100%) | 7 (100%) |
| TW3-like | 37 (82.2%) | 7 (100%) |
| CY-like | 3 (6.7%) | 0 (0.0%) |
| SK3-like | 5 (11.1%) | 0 (0.0%) |
| BKPyV | 2 (100%) | 0 (0.0%) |
| UT- like | 2 (100%) | 0 (0.0%) |
BKPyV, BK polyomavirus; JCPyV, JC polyomavirus; UTUC, upper tract urothelial carcinoma
CY, JCPyV-CY strain (GenBank accession No. AB038249); TW3, JCPyV-TW3 strain (GenBank accession No. U61771); SK3, JCPyV-SK3 strain (GenBank accession No. AB118652.1)
UT, BKPyV-UT strain (GenBank accession No. M34049.1)
Viral LT and cellular p53 protein in the UTUC and adjacent tissue samples
Both LT and p53 proteins were detected in the UTUC tissues on IHC. The results of IHC staining of LT protein is presented in Fig. 2, and the statistical positivity rates are shown in Table 2, as well as Supplementary Materials Tables 1 and Table 2. Among the 72 UTUC specimens examined, 47 demonstrated positive LT expression, yielding a positivity rate of 65.3% (Table 2). Meanwhile, only seven specimens with LT protein expression were identified in adjacent tissue samples (Supplementary Table 2). The results of IHC staining of p53 protein is presented in Fig. 3. In 72 UTUC tissue sections, p53 staining revealed eight cases with a WT pattern, 23 cases with a mutant-overexpression pattern, 15 cases with a mutant-null pattern, and no cases with a mutant-cytoplasmic pattern. The remaining sections did not exhibit p53 expression, as confirmed by HE staining (Table 4, Supplementary Table 1). In the adjacent tissue sections, four cases exhibited a mutant-overexpression p53 pattern, three cases exhibited a mutant-null pattern, and the rest were normal tissues without p53 expression, also confirmed by HE staining (Table 4, Supplementary Table 2).
Fig. 2.
Immunohistochemical staining detect the LT proteins in UTUC and adjacent tissues. Sections of UTUC tissues were immunostained using anti-LT monoclonal antibodies (a–f). Positive and negative control staining was performed using human prostate adenocarcinoma tissue sections. Positive control staining is shown in panel (a), while negative control staining is shown in panel (b). In UTUC tissue Sect. 33, positive staining for LT protein is observed (c), whereas in tissue Sect. 54, negative staining is evident (d). Similarly, in adjacent tissue Sect. 14, positive staining for LT protein is observed (e), while negative staining is observed in tissue Sect. 31 (f). Magnification, × 200. LT, large T; UTUC, upper tract urothelial carcinoma
Fig. 3.
Immunohistochemical staining detect the p53 proteins in UTUC tissues. Sections of UTUC tissues were immunostained using anti-p53 monoclonal antibodies (a, a', b, b', c, c'). In UTUC tissue Sect. 18, a scattered pattern of sporadic, scattered p53 nuclear staining within urothelial cells is observed, suggesting a normal/wild-type pattern (panel a, magnification × 40, a'× 200). Two abnormal (mutant) patterns are observed; in tissue Sect. 48, the staining shows diffuse overexpression, with strong nuclear staining extending from the basal layer into the suprabasal layers of urothelial cells (panel b, magnification × 40, panel b'× 200), tissue Sect. 67, null pattern (mutant) is present, with a complete absence of p53 staining in the tumor, while positivity is observed in the background inflammatory cells (panel c, magnification × 40, panel c'× 200)
Table 4.
IHC staining for p53 protein pattern of UTUC samples
| Pattern | UTUC sample (n = 72) |
Adjacent sample (n = 41) |
|---|---|---|
| Wild type | 8 (11.11%) | 0 (0%) |
| mutant | 38 (52.78%) | 4 (9.75%) |
| overexpression | 23 (31.95%) | 1(2.44%) |
| null | 15 (20.83%) | 3 (7.31) |
| cytoplasmic | 0(0.0%) | 0(0.0%) |
| Not-detected | 26 (36.11%) | 0 (0.0%) |
IHC, immunohistochemistry, UTUC, upper tract urothelial carcinoma
Association between viral factors and UTUC
Table 5 presents the ORs of UTUC associated with JCPyV/BKPyV infections. The results suggest that the presence of JCPyV/BKPyV infection may be associated with a 9.13-fold increased likelihood of developing UTUC compared to the group without viral infection (95% confidence interval [CI]: 3.54–23.55, P < 0.001). Additionally, the data reveal that the OR of UTUC may increase by 9.13-fold when the virus expresses the LT protein (95% CI: 3.54–23.55, P < 0.001). Cancer is potentially 10.34-fold more likely to develop when p53 expression is mutant (95% CI: 3.54–23.55, P < 0.001). The combined presence of JCPyV/BKPyV infection and simultaneous expression of LT protein and/or cellular mutant p53 protein may be associated with a 10.37-fold increased likelihood of UTUC (95% CI: 1.90–38.72, P = 0.001). Additionally, we analyze the association between p53 protein and LT protein in patients with UTUC. The results suggest that the presence of LT protein in patients with UTUC may increase the OR of MT p53 protein expression by 2.93-fold (95% CI: 1.33–6.48, P = 0.007), compared to cases without LT protein expression (Table 6). These findings represent preliminary associations and do not establish causation.
Table 5.
Association of HPyVs infection with risk of UTUC
| Cell type | N | Variable | OR (95% CI) | P-value | |
|---|---|---|---|---|---|
| Viral DNA(+) | Viral DNA(-) | ||||
| UTUC | 72 | 47 | 25 | 9.13(3.54–23.55) | < 0.001 |
| Adjacent | 41 | 7 | 34 | 1.00 | |
| Viral LT(+) | Viral LT(-) | ||||
| UTUC | 72 | 47 | 25 | 9.13(3.54–23.55) | < 0.001 |
| Adjacent | 41 | 7 | 34 | 1.00 | |
| Mt-p53 | Others | ||||
| UTUC | 72 | 38 | 34 | 10.34(3.34–32.02) | < 0.001 |
| Adjacent | 41 | 4 | 37 | 1.00 | |
|
LT + Mt p53 |
Others | ||||
| UTUC | 72 | 25 | 47 | 10.37(2.31–46.56) | < 0.001 |
| Adjacent | 41 | 2 | 39 | 1.00 |
P-values are calculated using the χ2 test or Fisher’s exact test, as appropriate
CI, confidence interval; LT, large T; OR, Odds ratio; UTUC, upper tract urothelial carcinoma
Table 6.
ORs of the association between MT p53 protein and viral LT protein in UTUC patients
| N | Variable | OR (95%CI) | P value | ||
|---|---|---|---|---|---|
| Mt p53 | Others | ||||
| Viral LT (+) | 54 | 27 | 27 | 2.93(1.33–6.48) | 0.007 |
| Viral LT (-) | 59 | 15 | 44 | 1.000 | |
P value by χ2 test or Fisher’s exact test when appropriate. CI, confidence interval; OR, odds ratio
Association between JCPyV/BKPyV virus infection and clinical characteristics of UTUC
We analyzed the potential associations between the clinical characteristics of patients with UTUC, including sex, age, CKD, tumor stage, and tumor grade, with JCPyV/BKPyV infection (Table 7). These results showed that, among patients with UTUC, females had a higher OR of JCPyV/BKPyV infection than males did (OR = 5.19, 95% CI: 0.07–0.55, P = 0.001). Similarly, the OR for LT protein expression associated with viral infection was higher in females (OR = 5.19, 95% CI: 1.18–2.92, P = 0.001). The OR for mutant p53 protein expression was significantly higher in patients with UTUC and CKD than in those without CKD (OR = 3.15, 95% CI: 1.17–8.46, P = 0.021). When analyzing UTUC stage based on the need for chemotherapy (Ta-T2 vs. T3-T4), viral infection-associated LT protein expression was linked to a higher OR in the T3-T4 group compared to the Ta-T2 group (OR = 3.18, 95% CI: 1.12–9.06, P = 0.027). No correlation was observed between age and tumor grading.”
Table 7.
Correlations of human polyomavirus infections with UTUC characteristics
| Viral DNA | Viral LT | p53 | LT + p53 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variable | (+) | (-) | OR (95%CI) | (+) | (-) | OR (95%CI) | MT | Others | OR (95%CI) | Both (+) | Others | OR (95%CI) |
| Sex (n = 72) | ||||||||||||
| Male (28) | 12 | 16 | 1.00 | 12 | 16 | 1.00 | 15 | 13 | 1.00 | 6 | 22 | 1.00 |
| Female (44) | 35 | 9 | 5.19 (1.81–14.78) | 35 | 9 | 5.19 (1.81–14.78) | 23 | 21 | 0.95 (0.37–2.45) | 19 | 25 | 2.79 (0.94–8.22) |
| P value | 0.001 | 0.001 | 0.914 | 0.059 | ||||||||
| Age, years (n = 72) | ||||||||||||
| < 65 (32) | 19 | 13 | 1.00 | 19 | 13 | 1.00 | 17 | 15 | 1.00 | 12 | 20 | 1.00 |
| ≥ 65 (40) | 28 | 12 | 1.60 (0.60–4.24) | 28 | 12 | 1.60 (0.60–4.24) | 21 | 19 | 0.98 (0.38–2.48) | 13 | 27 | 0.80 (0.30–2.13) |
| P value | 0.347 | 0.347 | 0.958 | 0.658 | ||||||||
| Disease (n = 72) | ||||||||||||
| Non-CKD (30) | 20 | 8 | 1.00 | 20 | 8 | 1.00 | 10 | 18 | 1.00 | 7 | 21 | 1.00 |
| CKD (42) | 27 | 17 | 0.64 | 27 | 17 | 0.64 | 28 | 16 | 3.15 | 18 | 26 | 2.08 |
| (0.23–1.76) | (0.23–1.76) | (1.17–8.46) | (0.73–5.90) | |||||||||
| P value | 0.382 | 0.382 | 0.021 | 0.167 | ||||||||
| TNM staging (n = 72) | ||||||||||||
| Ta-T2 (39) | 21 | 18 | 1.00 | 21 | 18 | 1.00 | 21 | 18 | 1.00 | 12 | 27 | 1.00 |
| T3-T4 (33) | 26 | 7 | 3.18 (1.12–9.06) | 26 | 7 | 3.18 (1.12–9.06) | 17 | 16 | 0.911(0.36–2.31) | 13 | 20 | 1.46 (0.522–3.88) |
| P value | 0.027 | 0.027 | 0.844 | 0.444 | ||||||||
| Tumor grading (n = 72) | ||||||||||||
| Low (5) | 3 | 2 | 1.00 | 3 | 2 | 1.00 | 1 | 4 | 1.00 | 1 | 4 | 1.00 |
| High (67) | 44 | 23 | 1.28 (0.12–8.18) | 44 | 23 | 1.28 (0.12–8.18) | 37 | 30 | 4.93(0.52–46.51) | 24 | 43 | 2.23(0.24–21.13) |
| P value | 1.000 | 1.000 | 0.182 | 0.652 | ||||||||
P values are calculated using the χ2 test or Fisher’s exact test, as appropriate. CI, confidence interval; CKD, chronic kidney disease; LT, large T; MT, mutant; OR, odds ratio; TNM, tumor- node-metastasis; UTUC, upper tract urothelial carcinoma
Discussion
Although HPyVs have been detected in urothelial carcinoma, research on their significance in UTUC remained limited. In this study, HPyVs were found in 65.3% of the analyzed tissue specimens, with JCPyV infection being predominantly detected. JCPyV-TW-3-like was the predominant strain causing JCPyV infection, followed by JCPyV-SK3-like (11.1%). The JCPyV Taiwan-3 [49, 50], a virus isolated from the urine of an immunosuppressed patient with rheumatoid arthritis, has an enhancer-promoter region of the viral genome that lack a copy of pentanucleotide (GGGAA) at nucleotides 195–199 and pentanucleotide (AAAGC) at nucleotides 224–228 compared to the CY archetype, while JCPyV SK-3 variants (GenBank accession No. AB118652.1) [51] mainly deletes the pentanucleotide (AAAGC) at nucleotides 224–228 in the NCCR region compared to the CY archetype. These results are similar to our previous findings in prostate cancer [42]. We found a high prevalence of JCPyV infection in patients with UTUC in Taiwan between 2020 and 2022. Further, JCPyV infection was observed to be associated with a higher risk of UTUC. In patients with UTUC, the expression of viral LT protein may increase the OR of mutant p53 protein expression by 2.93-fold, compared to cases without LT protein expression. Collectively, these findings suggest that JCPyV/BKPyV infection, LT expression, and MT p53 expression may play a role in the development of UTUC. Additional studies are necessary to validate these associations and explore their biological and clinical relevance.
With respect to the relationship between JCPyV infection and the characteristics in patients with UTUC, we found that female patients were 5.19-fold more likely to be infected with JCPyV. Patients with CKD had a 3.15-fold higher likelihood of MT p53 protein expression than non-CKD patients did. Additionally, compared to patients with stage Ta-T2 disease not requiring chemotherapy, those with stage T3-T4 disease who required chemotherapy had a 3.18-fold higher viral LT protein expression. These associations may imply that the high prevalence of JCPyV infection in female patients with CKD and UTUC, who require chemotherapy and express LT and MT p53 proteins, may provide important information about disease progression and poor outcomes.
An epidemiological investigation of UTUC found a higher prevalence among males than among females, with smoking and occupational exposure being the main reasons [52]. However, studies in Taiwan have reported a higher incidence in females than in males [32, 33, 36], and consistent findings were observed in the current study. This discrepancy may be linked to chronic disease factors in the past. In our results, female patients with CKD were more prevalent than male patients. Nonetheless, further research is needed to elucidate the causes of UTUC. Additionally, atypical incidence rates of UTUC have been observed in Taiwan [33], with potential associations with Blackfoot disease in southern Taiwan and consumption of arsenic-contaminated water [34, 35]. Significant correlations with younger age in females, higher T stage, and elevated pretreatment levels of serum lactate dehydrogenase and creatinine have also been found [33, 36].
From the perspective of viral infections, we found that this high prevalence rate might be related to JCPyV infection, with females being more susceptible to JCPyV infection and expressing more LT protein than males. Additionally, MT p53 protein was found in 52.8% of UTUC samples, and LT and p53 were detected at a higher rate in tumors with a higher T stage. This suggests that these virus-associated phenomena may be important indicators of UTUC progression and treatment efficacy. Regarding age and tumor grading, our results did not reveal a direct relation to viral infections.
JCPyV and BKPyV are widespread in humans. Under conditions such as immunosuppression or pregnancy, these viruses can reactivate, resulting in increased urinary viral shedding [8]. A retrospective study analyzing 19 immunosuppressed patients with renal and bladder cancer revealed that 11 cases of post-transplant urothelial carcinoma, primarily following kidney or heart transplantation, exhibited high aggressiveness and expressed significant levels of LT and p53 proteins, suggesting potential viral involvement [53]. Another study reported that 31% of renal transplant patients with high-grade urothelial carcinoma tested positive for polyomavirus LT antigen, particularly among younger individuals [54]. Similarly, long-term follow-up revealed BKV-related hematuria 14 years after kidney transplantation, further associating BKV with urinary tract cancers [55]. However, it remains unclear whether viral reactivation drives carcinogenesis or if cancer itself promotes viral activation. Our study aimed to investigate the association of JCPyV and BKPyV with UTUC, specifically excluding patients with immune-related diseases or those undergoing immunosuppressive therapy. Our results identified JCPyV as the predominant strain in patients with UTUC, with LT protein expression detected in tumors. These findings suggest that JCPyV may contribute to tumor progression in UTUC under normal immune conditions.
Additionally, four patients exhibited MT p53 protein in adjacent UTUC tissues. The results remained consistent on triple staining. Analysis of the clinical features showed that these patients were aged 62–68 years, and two were aged > 65 years. Previous studies of basal cell carcinoma have shown MT p53 in adjacent tissues. p53 protein expression in the adjacent cells of older patients may be associated with early cancer progression [56]. These results align with our observations and provide further evidence of the potential role of p53 expression in early cancer development.
Limitations
This study collected UTUC specimens from 2020 to 2022. Due to the exclusion of patients with other cancers and immune diseases, the number of cases recruited was limited. PCR, immunohistochemistry (IHC), and sequencing were used to analyze the association between viruses and clinical symptoms, which leaned more toward observational research. While these methods provide valuable insights into potential correlations, they cannot confirm direct interactions between JCPyV or BKPyV and tumorigenesis in UTUC. The pathogenic mechanisms of these viruses in UTUC have not yet been conclusively determined. Although the study’s descriptive nature limits definitive conclusions, it highlights the importance of investigating these viral infections further. Future studies should employ advanced molecular techniques, such as RNA in situ hybridization (RNA-ISH) and fluorescence in situ hybridization (FISH), to clarify these interactions.
Conclusions
In conclusion, we analyzed JCPyV and BKPyV infections in 72 cases of UTUC in Taiwan from 2020 to 2022, identifying JCPyV-TW3 as the predominant strain. Patients with advanced-stage UTUC requiring chemotherapy (T3-T4) and CKD exhibited significant correlations with JCPyV infection, positive LT expression, MT p53 protein expression in tissues. These findings suggest that JCPyV infection and the interplay between LT and MT p53 may contribute to UTUC progression and serve as potential markers for chemotherapy decisions and prognosis. Although BKPyV was also analyzed, its role in UTUC remains inconclusive and requires further investigation. This study provides preliminary evidence, laying the foundation for research on the association between viral infections and UTUC. Further investigation is needed to explore the roles of viral infections, p53 expression, and immune regulation in the development of UTUC.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We express our deep appreciation to Dr. Chang of the Institute of Molecular Biology at the National Chung Cheng University, Taiwan.
Abbreviations
- UTUC
Upper tract urothelial carcinoma
- JCPyV
Human polyomavirus John Cunningham virus
- BKPyV
Human polyomavirus BK virus
- TW3
JCPyV-TW3 strain (GenBank accession No. U61771)
- SK3
JCPyV-SK3 strain (GenBank accession No. AB118652.1)
- UT
BKPyV-UT strain (GenBank accession No. M34049.1)
- CI
Confidence interval
- CMV
Cytomegalovirus
- EBV
Epstein-Barr virus
- FFPE
Formalin-fixed, paraffin-embedded
- HPV
Human papilloma virus
- HPyVs
Human polyomaviruses
- IHC
Immunohistochemistry
- LT
Large T antigen
- OR
Odds ratio
- PCR
Polymerase chain reaction
- PML
Progressive multifocal leukoencephalopathy
Author contributions
PLC and CHS designed the experiments and wrote the main manuscript. CNC and CFH conceived, performed the experiments and prepared Figs. 1 and 2. WHL and CLT collected the clinical samples prepared Tables 1, 2, 3 and 4. WYY and KWY conducted the experiments and prepared Table 5, and 6. ML and YYL analyzed the statistical data and prepared supplementary Fig. 1. All the authors have read and approved the final manuscript.
Funding
This study was financially supported by the Ditmanson Medical Foundation Chia-Yi Christian Hospital, Taiwan (grant R110-058), and the Central Taiwan University of Science and Technology (grant CTU111-P-003) for clinical data acquisition, statistical analysis fees, and pathological review of patient samples.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
This study was approved by the review board of the Chia-Yi Christian Hospital (IRB number 2021103). This study was conducted in accordance with the guidelines prescribed in Good Clinical Practice Procedures and the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Chun-Nun Chao, Chi-Feng Hung, Pei-Lain Chen and Cheng-Huang Shen contributed equally to this work.
Contributor Information
Pei-Lain Chen, Email: plchen@ctust.edu.tw.
Cheng-Huang Shen, Email: 01712@cych.org.tw.
References
- 1.Rouprêt M, Seisen T, Birtle AJ, Capoun O, Compérat EM, Dominguez-Escrig JL, et al. European Association of Urology Guidelines on Upper urinary tract Urothelial Carcinoma: 2023 update. Eur Urol. 2023;84:49–64. [DOI] [PubMed] [Google Scholar]
- 2.Colin P, Koenig P, Ouzzane A, Berthon N, Villers A, Biserte J, et al. Environmental factors involved in carcinogenesis of urothelial cell carcinomas of the upper urinary tract. BJU Int. 2009;104:1436–40. [DOI] [PubMed] [Google Scholar]
- 3.Shan H, Tian W, Hong Y, Xu B, Wang C, Yu B, et al. Clinicopathologic characteristics and prognosis of upper tract urothelial carcinoma complicated with aristolochic acid nephropathy after radical nephroureterectomy. BMC Complement Med Ther. 2020;20:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen C-H, Chiou H-Y, Tung M-C, Wu C-C, Kao W-T, Wang Y-H, et al. Clinical and demographic characteristics among patients with urothelial carcinomas of the upper urinary tract and bladder in Taiwan. J Chin Med Assoc. 2017;80:563–8. [DOI] [PubMed] [Google Scholar]
- 5.Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK, et al. The Eighth Edition AJCC Cancer staging Manual: continuing to build a bridge from a population-based to a more personalized approach to cancer staging. CA Cancer J Clin. 2017;67:93–9. [DOI] [PubMed] [Google Scholar]
- 6.Markowski MC, Boorjian SA, Burton JP, Hahn NM, Ingersoll MA, Maleki Vareki S, et al. The Microbiome and Genitourinary Cancer: a collaborative review. Eur Urol. 2019;75:637–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marongiu L, Allgayer H. Viruses in colorectal cancer. Mol Oncol. 2022;16:1423–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prado JCM, Monezi TA, Amorim AT, Lino V, Paladino A, Boccardo E. Human polyomaviruses and cancer: an overview. Clin (Sao Paulo). 2018;73:e558s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moens U, Prezioso C, Pietropaolo V. Genetic diversity of the Noncoding Control Region of the Novel Human polyomaviruses. Viruses. 2020;12:1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.L’Honneur A-S, Leh H, Laurent-Tchenio F, Hazan U, Rozenberg F, Bury-Moné S. Exploring the role of NCCR variation on JC Polyomavirus expression from dual reporter minicircles. PLoS ONE. 2018;13:e0199171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ault GS, Stoner GL. Human polyomavirus JC promoter/enhancer rearrangement patterns from progressive multifocal leukoencephalopathy brain are unique derivatives of a single archetypal structure. J Gen Virol. 1993;74(Pt 8):1499–507. [DOI] [PubMed] [Google Scholar]
- 12.Pinto M, Dobson S. BK and JC virus: a review. J Infect. 2014;68(Suppl 1):S2–8. [DOI] [PubMed] [Google Scholar]
- 13.Padgett BL, Rogers CM, Walker DL. JC virus, a human polyomavirus associated with progressive multifocal leukoencephalopathy: additional biological characteristics and antigenic relationships. Infect Immun. 1977;15:656–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng J, DeCaprio JA, Fluck MM, Schaffhausen BS. Cellular transformation by Simian Virus 40 and murine polyoma virus T antigens. Semin Cancer Biol. 2009;19:218–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kellogg C, Kouznetsova VL, Tsigelny IF. Interactions of large T-Antigen (LT) protein of polyomaviruses with p53 unfold their cancerogenic potential. J Biomol Struct Dyn. 2022;40:5243–52. [DOI] [PubMed] [Google Scholar]
- 16.Szymonowicz KA, Chen J. Biological and clinical aspects of HPV-related cancers. Cancer Biol Med. 2020;17:864–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng H-C, Xue H, Zhang C-Y. The oncogenic roles of JC Polyomavirus in cancer. Front Oncol. 2022;12:976577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hermannstädter A, Ziegler C, Kühl M, Deppert W, Tolstonog GV. Wild-type p53 enhances efficiency of simian virus 40 large-T-antigen-induced cellular transformation. J Virol. 2009;83:10106–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duffy MJ, Synnott NC, Crown J. Mutant p53 as a target for cancer treatment. Eur J Cancer. 2017;83:258–65. [DOI] [PubMed] [Google Scholar]
- 20.Kanapathipillai M. Treating p53 mutant aggregation-Associated Cancer. Cancers (Basel). 2018;10:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balasubramaniam SD, Balakrishnan V, Oon CE, Kaur G. Key molecular events in Cervical Cancer Development. Med (Kaunas). 2019;55:384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levrero M, Zucman-Rossi J. Mechanisms of HBV-induced hepatocellular carcinoma. J Hepatol. 2016;64:S84–101. [DOI] [PubMed] [Google Scholar]
- 23.Chatterjee K, Das P, Chattopadhyay NR, Mal S, Choudhuri T. The interplay between Epstein-Bar virus (EBV) with the p53 and its homologs during EBV associated malignancies. Heliyon. 2019;5:e02624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Queiroga EM, Gualco G, Weiss LM, Dittmer DP, Araujo I, Klumb CEN, et al. Burkitt lymphoma in Brazil is characterized by geographically distinct clinicopathologic features. Am J Clin Pathol. 2008;130:946–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimla LJ, Clark TG, Banerjee S, Campino S. JC Polyomavirus T-antigen protein expression and the risk of colorectal cancer: systematic review and meta-analysis of case-control studies. PLoS ONE. 2023;18:e0283642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramelow J, Brooks CD, Gao L, Almiman AA, Williams TM, Villalona-Calero MA, et al. The oncogenic potential of a mutant TP53 gene explored in two spontaneous lung cancer mice models. BMC Cancer. 2020;20:738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang C, Liu J, Xu D, Zhang T, Hu W, Feng Z. Gain-of-function mutant p53 in cancer progression and therapy. J Mol Cell Biol. 2020;12:674–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saller J, Boyle J. Molecular Pathology of Lung Cancer. Cold Spring Harb Perspect Med. 2022;12:a037812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marvalim C, Datta A, Lee SC. Role of p53 in breast cancer progression: an insight into p53 targeted therapy. Theranostics. 2023;13:1421–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ungerleider NA, Rao SG, Shahbandi A, Yee D, Niu T, Frey WD, et al. Breast cancer survival predicted by TP53 mutation status differs markedly depending on treatment. Breast Cancer Res. 2018;20:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wallis B, Bowman KR, Lu P, Lim CS. The challenges and prospects of p53-Based therapies in Ovarian Cancer. Biomolecules. 2023;13:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang Y, Hsu W, Lee Y, Chiang C, Yang Y, You S, et al. Trends and sex-specific incidence of upper urinary tract cancer in Taiwan: a birth cohort study. Cancer Med. 2023;12:15350–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang M-H, Chen K-K, Yen C-C, Wang W-S, Chang Y-H, Huang WJ-S, et al. Unusually high incidence of upper urinary tract urothelial carcinoma in Taiwan. Urology. 2002;59:681–7. [DOI] [PubMed] [Google Scholar]
- 34.Chiou HY, Chiou ST, Hsu YH, Chou YL, Tseng CH, Wei ML, et al. Incidence of transitional cell carcinoma and arsenic in drinking water: a follow-up study of 8,102 residents in an arseniasis-endemic area in northeastern Taiwan. Am J Epidemiol. 2001;153:411–8. [DOI] [PubMed] [Google Scholar]
- 35.Christoforidou EP, Riza E, Kales SN, Hadjistavrou K, Stoltidi M, Kastania AN, et al. Bladder cancer and arsenic through drinking water: a systematic review of epidemiologic evidence. J Environ Sci Health Tox Hazard Subst Environ Eng. 2013;48:1764–75. [DOI] [PubMed] [Google Scholar]
- 36.Huang C-C, Su Y-L, Luo H-L, Chen Y-T, Sio TT, Hsu H-C, et al. Gender is a significant prognostic factor for Upper Tract Urothelial Carcinoma: a large hospital-based Cancer Registry Study in an endemic area. Front Oncol. 2019;9:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Llewellyn MA, Gordon NS, Abbotts B, James ND, Zeegers MP, Cheng KK, et al. Defining the frequency of human papillomavirus and polyomavirus infection in urothelial bladder tumours. Sci Rep. 2018;8:11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo HL, Chen YT, Huang SC, Chen TC, Chiang PH, Lin SS et al. Human Polyomavirus Is Associated With Earlier Onset of Upper Urinary Tract Urothelial Carcinoma in Patients After Kidney Transplantation. Transplant Proc. 2017;49:1064–7. [DOI] [PubMed]
- 39.Shen C-H, Wu J-D, Hsu C-D, Jou Y-C, Lin C-T, Wang M, et al. The high incidence of JC virus infection in urothelial carcinoma tissue in Taiwan. J Med Virol. 2011;83:2191–9. [DOI] [PubMed] [Google Scholar]
- 40.Taherkhani R, Farshadpour F. BK and JC polyomaviruses and risk of urothelial bladder carcinoma: a preliminary study in the northern shores of Persian Gulf, Iran. Infect Agent Cancer. 2022;17:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu S-W, Liou J-H, Yeh K-T, Hung T-W, Chang H-R. The prevalence and prognostic significance of Polyomavirus infection in patients with Urothelial Carcinoma of the bladder. Urol J. 2016;13:2773–8. [PubMed] [Google Scholar]
- 42.Shen C, Tung C, Chao C, Jou Y, Huang S, Meng M, et al. The differential presence of human polyomaviruses, JCPyV and BKPyV, in prostate cancer and benign prostate hypertrophy tissues. BMC Cancer. 2021;21:1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tseng CE, Yeh CM, Fang CY, Shay J, Chen PL, Lin MC, et al. Detection of human JCPyV and BKPyV in diffuse large B-cell lymphoma of the GI tract. Eur J Clin Microbiol Infect Dis. 2014;33:665–72. [DOI] [PubMed] [Google Scholar]
- 44.Fang C-Y, Chen S-Y, Hsiao B-X, Huang H-Y, Chen Y-J, Tung C-L, et al. Unusually high incidence of polyomavirus JC infection in the higher grade of colorectal cancer tissues in Taiwan. Eur J Med Res. 2022;27:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Köbel M, Ronnett BM, Singh N, Soslow RA, Gilks CB, McCluggage WG. Interpretation of P53 immunohistochemistry in Endometrial carcinomas: toward increased reproducibility. Int J Gynecol Pathol. 2019;38(Suppl 1):S123–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson EF, Chen J, Huvila J, Pors J, Ren H, Ho J, et al. p53 immunohistochemical patterns in HPV-related neoplasms of the female lower genital tract can be mistaken for TP53 null or missense mutational patterns. Mod Pathol. 2020;33:1649–59. [DOI] [PubMed] [Google Scholar]
- 47.Trias I, Saco A, Marimon L, López Del Campo R, Manzotti C, Ordi O, et al. P53 in Penile squamous cell carcinoma: a pattern-based immunohistochemical Framework with molecular correlation. Cancers (Basel). 2023;15:2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen TK, Knicely DH, Grams ME. Chronic kidney disease diagnosis and management: a review. JAMA. 2019;322:1294–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang D, Tsai R-T, Wang M, Ou W-C. Different genotypes of human polyomaviruses found in patients with autoimmune diseases in Taiwan. J Med Virol. 1996;48:204–9. [DOI] [PubMed] [Google Scholar]
- 50.Ou WC, Tsai RT, Wang M, Fung CY, Hseu TH, Chang D. Genomic cloning and sequence analysis of Taiwan-3 human polyomavirus JC virus. J Formos Med Assoc. 1997;96:511–6. [PubMed] [Google Scholar]
- 51.Zheng H-Y, Zhao P, Suganami H, Ohasi Y, Ikegaya H, Kim J-C, et al. Regional distribution of two related northeast Asian genotypes of JC virus, CY-a and -b: implications for the dispersal of Northeast asians. Microbes Infect. 2004;6:596–603. [DOI] [PubMed] [Google Scholar]
- 52.Miyazaki J, Nishiyama H. Epidemiology of Urothelial Carcinoma. Int J Urol. 2017;24:730–4. [DOI] [PubMed] [Google Scholar]
- 53.Sirohi D, Vaske C, Sanborn Z, Smith SC, Don MD, Lindsey KG, et al. Polyoma virus-associated carcinomas of the urologic tract: a clinicopathologic and molecular study. Mod Pathol. 2018;31:1429–41. [DOI] [PubMed] [Google Scholar]
- 54.Yan L, Salama ME, Lanciault C, Matsumura L, Troxell ML. Polyomavirus large T antigen is prevalent in urothelial carcinoma post-kidney transplant. Hum Pathol. 2016;48:122–31. [DOI] [PubMed] [Google Scholar]
- 55.Jones H, Bhakta A, Jia L, Wojciechowski D, Torrealba J. An unusual presentation of metastatic BK Virus-Associated Urothelial Carcinoma arising in the allograft, persisting after transplant nephrectomy. Int J Surg Pathol. 2023;31:1586–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Demirkan NC, Colakoglu N, Düzcan E. Value of p53 protein in biological behavior of basal cell carcinoma and in normal epithelia adjacent to carcinomas. Pathol Oncol Res. 2000;6:272–4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.