Abstract
There are 16 approved human immunodeficiency virus type 1 (HIV-1) drugs belonging to three mechanistic classes: protease inhibitors, nucleoside and nucleotide reverse transcriptase (RT) inhibitors, and nonnucleoside RT inhibitors. HIV-1 resistance to these drugs is caused by mutations in the protease and RT enzymes, the molecular targets of these drugs. Drug resistance mutations arise most often in treated individuals, resulting from selective drug pressure in the presence of incompletely suppressed virus replication. HIV-1 isolates with drug resistance mutations, however, may also be transmitted to newly infected individuals. Three expert panels have recommended that HIV-1 protease and RT susceptibility testing should be used to help select HIV drug therapy. Although genotypic testing is more complex than typical antimicrobial susceptibility tests, there is a rich literature supporting the prognostic value of HIV-1 protease and RT mutations. This review describes the genetic mechanisms of HIV-1 drug resistance and summarizes published data linking individual RT and protease mutations to in vitro and in vivo resistance to the currently available HIV drugs.
INTRODUCTION
Sixteen antiretroviral drugs have been approved for the treatment of human immunodeficiency virus type 1 (HIV-1) infection: seven nucleoside/nucleotide reverse transcription (RT) inhibitors (NRTI), six protease inhibitors (PIs), and three nonnucleoside RT inhibitors (NNRTI). In previously untreated individuals with drug-susceptible HIV-1 strains, combinations of three or more drugs from two drug classes can lead to prolonged virus suppression and immunologic reconstitution. However, the margin of success for achieving and maintaining virus suppression is narrow. Extraordinary patient effort is required to adhere to drug regimens that are expensive, inconvenient, and often associated with dose-limiting side effects. In addition to these hurdles, the development of drug resistance looms as both a cause and consequence of incomplete virus suppression that threatens the success of future treatment regimens.
RATIONALE FOR HIV-1 DRUG RESISTANCE TESTING
An increasing number of studies are showing that the presence of drug resistance before starting a new drug regimen is an independent predictor of virologic response to that regimen (reviewed in references 72, 75, 128, and 138). In addition, several prospective controlled studies have shown that patients whose physicians have access to drug resistance data, particularly genotypic resistance data, respond better to therapy than control patients whose physicians do not have access to these assays (19, 47a, 50a, 92, 139, 380a; Melnick, D., J. Rosenthal, M. Cameron, M. Snyder, S. Griffith-Howard, K. Hertogs, W. Verbiest, N. Graham, and S. Pham, Abstract 786, 7th Conference on Retroviruses and Opportunistic Infections, San Francisco, Calif., 2000; Meynard, J. L., M. Vray, L. Monard-Joubert, S. Matheron, G. Peytavin, F. Clavel, F. Brun-Vezinet, and P. M. Girard, 40th Interscience Conference on Antimicrobial Agents and Chemotherapy, Toronto, Canada, abstract 698, p. 294, 2000). The accumulation of such retrospective and prospective data has led three expert panels to recommend the use of resistance testing in the treatment of HIV-infected patients (101, 150; U.S. Department of Health and Human Services Panel on Clinical Practices for Treatment of HIV Infection, Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents, 28 January 2000, http://www.hivatis.org/trtgdlns.html) (Tables 1 and 2).
TABLE 1.
Expert-panel recommendations on HIV drug resistance testing
| Case type | Recommendationsa
|
||
|---|---|---|---|
| IAS-USAb | DHHS | EuroGuidelinesc | |
| Primary HIV-1 infection | Consider testing: detect transmission of drug-resistant virus; modify therapy to optimize response; treatment should not be delayed pending the genotype results. | Consider testing | Strongly consider testing particularly if transmission rate is high or if transmission from a treated individual is suspected; treatment should not be delayed pending genotype results. |
| Established HIV-1 infection in untreated individuals | Consider testing: detect prior transmission of drug-resistant HIV, although this may not always be possible with current tests. | Testing not generally recommended: Uncertain prevalence of resistant virus; current assays may not detect minor drug-resistant species. | Consider testing (the reliability of negative findings may be a function of time since infection). |
| First regimen failure | Recommend testing: document drug(s) to which there is resistance. | Recommend testing | Recommend testing |
| Multiple regimen failures | Recommend testing to optimize the number of active drugs in the next regimen; exclude drugs to which response is unlikely | Recommend testing | Recommend testing |
| Suboptimal viral suppression after initiation of HAART | Not addressed | Recommend testing | Not addressed |
| Pregnancy | Recommend testing to optimize maternal treatment and prophylaxis for neonate. | Not addressed | Recommend testing if the mother has a detectable virus load; recommend testing HIV-1-infected children born to infected mothers while on treatment. |
| Postexposure prophylaxis | Not addressed | Not addressed | Recommend testing but do not delay treatment waiting for test result; but if a sample from index case is available, test and modify treatment of recipient accordingly. |
IAS-USA, International AIDS Society-USA; DHHS, Department of Health and Human Services web site (http://www.hivatis.org/trtgdlns.html).
See reference 150.
See reference 101.
TABLE 2.
Prospective intervention studies comparing HIV resistance testing to physician-guided therapy
| Studya (reference) | Previous treatment | No. of patients | Study duration (wk) | RNA change (log10 copies/ml)
|
Comment | ||
|---|---|---|---|---|---|---|---|
| PGTb | Genotypic | Phenotypic | |||||
| GART (19) | ≥16 wk of 2 NRTIs and 1 PI | 153 | 12 | −0.61 | 1.19 | ND | |
| Havana (380a) | ≥24wk of heavy treatment | 326 | 24 | −0.63 | −0.84 | ND | Expert advice added benefit to genotypic testing |
| −0.99 | −1.45 | ND | |||||
| VIRA 3001 (50a) | ≥2 NRTIs and 1 PI | 272 | 16 | −0.87 | ND | −1.23 | |
| VIRADAPT (92) | ≥24wk of NRTI and 12wk of PIs | 108 | 24 | −0.67 | −1.15 | ND | Benefit maintained for 9-12 months |
| Kaiser | >12wk of heavy treatment, NNRTI naïve | 115 | 16 | NSD | NSD | ||
| NARVAL | Heavy treatment (median, 7 drugs) | 541 | 12 | NSD | NSD | NSD | Genotypic testing had significant benefit at 24 wk |
| ARGENTAc (47a) | Variable treatment (41% received NRTIs, PIs, and NNRTIs) | 174 | 24 | −0.39 | −0.57 | ND | Genotypic testing more effective in patients with plasma HIV-1 RNA <10,000 copies/ml |
| CCTG 575 (139) | ≥24 wk of treatment, 1-2 prior PIs, 76% NNRTI naive | 238 | 24 | −0.69 | ND | −0.71 | Phenotypic testing was associated with better outcome in a subgroup with >5 yr of treatment |
GART, Genotypic Antiretroviral Resistance Testing; ARGENTA, Antiretroviral Genotypic Resistance and Patient Reported Adherence Study. Kaiser study, Melnick et al., 7th Conf. on Retroviruses, abstr. 786, 2000; NARVAL, Meynard et al., 40th ICAAC, abstr. 698, 2000. The Virco Antivirogram was used in the VIRA 3001 and Kaiser studies. A different recombinant virus assay was used in NARVAL.
PGT, physician-guided therapy. NSD, no significant difference. ND, not determined.
At week 12, genotypic more effective than PGT (27 versus 12%, P = 0.02), but at week 24, no significant difference.
Genotypic testing is the most commonly used method of detecting resistant HIV-1 isolates and is one of the earliest applications of gene sequencing for clinical purposes. Although genotypic tests are more complex than typical antimicrobial susceptibility tests, their ability to detect mutations present as mixtures, even if the mutation is present at a level too low to affect drug susceptibility in a phenotypic assay, provides insight into the potential for resistance to emerge. They are also advantageous because they can detect transitional mutations that do not cause drug resistance by themselves but indicate the presence of selective drug pressure.
Genotypic testing has been shown to be clinically useful in four of five prospective randomized studies (19, 47a, 92, 380a; Meynard, J. L., M. Vray, L. Monard-Joubert, S. Matheron, G. Peytavin, F. Clavel, F. Brun-Vezinet, and P. M. Girard, 40th Interscience Conference on Antimicrobial Agents and Chemotherapy, Toronto, Canada, abstract 698, p. 294, 2000); in contrast, phenotypic testing has been shown to be clinically useful in just one of four prospective randomized studies (50a, 139, 256; Melnick, D., J. Rosenthal, M. Cameron, M. Snyder, S. Griffith-Howard, K. Hertogs, W. Verbiest, N. Graham, and S. Pham, Abstract 786, 7th Conference on Retroviruses and Opportunistic Infections, San Francisco, Calif., 2000).
Several reviews on the genetic basis of HIV-1 drug resistance have recently been published (69, 127, 128, 235, 260, 337). This review will focus on how knowledge of the genetic basis of HIV-1 drug resistance can be exploited to test for HIV drug resistance in clinical settings. HIV drug resistance is an interdisciplinary field; important data have been derived from structural biology, biochemistry, virology, and clinical studies. This review will integrate data from these different disciplines that are relevant to the development of new HIV drugs and to the optimal use of those HIV drugs that are already available.
EVOLUTION OF HIV-1 DRUG RESISTANCE
The evolution of HIV-1 drug resistance within an individual depends on the generation of genetic variation in the virus and on the selection of drug-resistant variants during therapy. HIV-1 genetic variability is a result of the inability of HIV-1 RT to proofread nucleotide sequences during replication (242). It is exacerbated by the high rate of HIV-1 replication in vivo, the accumulation of proviral variants during the course of HIV-1 infection, and genetic recombination when viruses with different sequences infect the same cell. As a result, innumerable genetically distinct variants (quasispecies) evolve in individuals in the months following primary infection (50).
The HIV-1 quasispecies within an individual undergo continuous genetic variation, competition, and selection. Development of drug resistance depends on the size and heterogeneity of the HIV-1 population within an individual, the extent to which virus replication continues during drug therapy, the ease of acquisition of a particular mutation (or set of mutations), and the effect of drug resistance mutations on drug susceptibility and virus fitness. Some mutations selected during drug therapy confer measurable phenotypic resistance by themselves, whereas other mutations increase resistance only when present with other mutations or compensate for the diminished replicative activity that can be associated with drug resistance.
It has been estimated that every possible single point mutation occurs between 104 and 105 times per day in an untreated HIV-1-infected individual and that double mutants also occur commonly (50). It is not known, however, whether multidrug-resistant viruses already exist at low frequencies in untreated persons or if they are generated by residual viral replication during therapy (304). Answers to this question depend on the effective population number of HIV-1 in vivo. Some authors have argued in favor of a high effective population number and a deterministic model of HIV-1 evolution in which chance effects play a small role (313); others have argued in favor of a lower effective population number and a stochastic model of HIV-1 evolution (31, 34, 107).
Resistant virus strains can also be transmitted between individuals. In the United States and Europe, about 10% of new infections are with HIV-1 strains harboring resistance to at least one of three classes of anti-HIV drugs (16, 23, 30, 93, 144, 233, 234, 317, 357, 372, 384, 416; Grant, R. M., F. Hecht, C. Petropoulos, N. Hellmann, M. Warmerdam, N. I. Bandrapalli, T. Gittens, M. Chesney, and J. Kahn, abstract 142, Antivir. Ther. 4[Suppl. 1]:98-99, 1999; Harzic, M., C. Deveau, I. Pellegrin, b. Dubeaux, P. Sageat, N. Ngo, H. Fleury, B. Hoen, D. Sereni, and J. F. Delfraissy, abstract 91, Antivir. Ther. 4[Suppl. 1]:61-62, 1999) (Table 3).
TABLE 3.
Antiretroviral drug resistance in individuals with primary HIV-1 infection or seroconversion within the preceding 12 monthsa
| Study | Years | Country (cities) | No. of patients | Resistance test | % Resistance
|
|||
|---|---|---|---|---|---|---|---|---|
| NRTI | NNRTI | PI | MDR | |||||
| Balotta (16) | 1994-1997 | Italy (Milan) | 38 | Genotype | 21 | 3 | 0 | 0 |
| Boden (23) | 1995-1999 | USA (Los Angeles, New York) | 80 | Genotype with confirmatory phenotype on selected isolates | 13 | 8 | 3 | 4 |
| Grantb | 1996-1999 | USA (San Francisco) | 118 | Genotype with confirmatory phenotype on selected isolates | 11 | 4 | 4 | NA |
| Harzicc | 1996-1998 | France | 158 | Genotype | 6 | 1 | 2 | NA |
| Yerly (416) | 1996-1998 | Switzerland | 82 | Genotype with confirmatory phenotype on selected isolates | 10 | 2 | 4 | 4 |
| Little (234) | 1989-1998 | USA (Boston, Dallas, Denver, Los Angeles, San Diego) | 141 | Phenotype with confirmatory genotype on selected isolates | ≥3 | ≥1 | ≥1 | ≥1 |
| Salomon (317) | 1997-1999 | Canada (Montreal) | 81 | Genotype with confirmatory phenotype on selected isolates | 6 | 4 | 4 | 5 |
| Tamalet (372) | 1995-1998 | France (Marseilles, Toulouse) | 48 | Genotype | 17 | 0 | 2 | 2 |
| Duwe (93) | 1996-1999 | Germany | 64 | Genotype with confirmatory phenotype on selected isolates | 14 | 0 | 3 | 0 |
| Briones (30) | 1997-1999 | Spain | 30 | Genotype with confirmatory phenotype on selected isolates | 23 | 3 | 7 | 7 |
| Pillay (384) | 2000 | United Kingdom | 26 | Genotype | 19 | 11 | 4 | 7 |
| Simon (357) | 1999-2000 | New York | 61 | Genotype with confirmatory phenotype | 26 | 5 | 7 | 5 |
Abbreviations: MDR, multidrug resistance, resistance within more than one class of drugs; NA, not available. In these studies, the following mutations were detected and considered genotypic evidence of resistance: for the NRTIs, M41L, D67N, T69D, K70R, L74V, M184V, L210W, and T215Y/F/S/D/C; for the NNRTIs, L100I, K101E, K103N, Y181C, and G190A; for the PIs, D30N, M46I/L/V, G48V, I54V, V82A, I84V, and L90M. In the study by Little et al., not all isolates had genotypic testing done, and the percent resistance represents a lower limit to the prevalence of genotypic resistance in that study.
Grant et al., Antivir. Ther. 4(Suppl. 1):98-99, abstr. 142, 1999.
Harzic et al., Antivir. Ther. 4(Suppl. 1):68-69, abstr. 91, 1999.
IDENTIFYING AND CHARACTERIZING DRUG RESISTANCE MUTATIONS
HIV drug resistance is mediated by mutations in the molecular targets of drug therapy. Drug-resistant viruses are usually first identified by in vitro passage experiments in which viral isolates are cultured in the presence of increasing concentrations of an antiviral compound. Isolates identified in this manner are further characterized by sequencing to identify genetic changes arising during selective drug pressure and by in vitro susceptibility testing. In some cases, HIV-1 constructs containing specific mutations have been created using site-directed mutagenesis to directly assess the effect of specific mutations on drug susceptibility.
Drug susceptibility testing involves culturing a fixed inoculum of HIV-1 in the presence of serial dilutions of an inhibitory drug. The concentration of drug required to inhibit virus replication by 50% (IC50) or 90% (IC90) is the most commonly used measure of drug susceptibility. Drug susceptibility assays are not designed to determine the amount of drug required to inhibit virus replication in vivo, but rather to compare the drug concentration required to inhibit a fixed inoculum of the isolated virus with the concentration required to inhibit the same inoculum of wild-type virus.
Drug susceptibility results depend on multiple unstandardized factors including the inoculum of virus tested, the cells used for virus replication, and the means of assessing virus replication. Susceptibility testing of NRTIs is further complicated by the fact that NRTIs are triphosphorylated to their active form at different rates in different cell lines. The dynamic susceptibility range between wild-type and the most drug-resistant isolates depends on the drug tested and the susceptibility assay used. It is as low as 10-fold for some drugs and as high as 1,000-fold for others. The dynamic susceptibility range does not necessarily correlate with the potency of a drug; rather it provides a useful context for interpreting an individual susceptibility result. For example, a 10-fold reduction in susceptibility to a drug would be considered high-level resistance if the dynamic susceptibility range for that drug is 10-fold but not if it is 1,000-fold.
The process of identifying drug resistance mutations using virus passage studies and characterizing their impact by testing the susceptibility of site-directed mutants containing the same amino acid changes is highly rigorous but has several limitations. First, the spectrum of mutations developing during in vitro passage experiments is narrower than in isolates from treated patients. This is particularly true for patients receiving combinations of drugs targeting the same enzyme. Second, site-directed mutagenesis studies cannot capture the complicated patterns of mutations observed in clinical isolates and cannot account for the impact of background polymorphisms that may influence the viability and extent of resistance in isolates containing known drug resistance mutations. Finally, clinical data often provide additional insight into which mutations are the most significant in vivo.
To characterize the mutations responsible for drug resistance, it is therefore necessary to also study HIV-1 isolates from patients receiving treatment. Specifically, three additional types of data must be collected: correlations between mutations and drug susceptibility in clinical HIV isolates, correlations between mutations and the drug treatment histories of persons from whom the sequenced isolates have been obtained, and correlations between mutations and the virologic response to a new HIV drug regimen. HIV-1 isolates from persons failing drug therapy are crucial observations of HIV-1 evolution that show which mutations the virus uses to escape from drug suppression in vivo. Such data are particularly important for elucidating the genetic mechanisms of resistance to drugs that are difficult to test in vitro susceptibility tests.
Correlations between genotype and virologic response to a new regimen are essential for demonstrating the clinical significance of drug resistance mutations. Because drug resistance mutations arise in the enzymes targeted by therapy, many of these mutations compromise enzymatic function. Although, the fitness of these variants can be tested in vitro, such fitness tests are not standardized and are unable to detect subtle changes in replication or the likelihood that certain defects in fitness may be readily compensated for by other genetic changes in the virus while other defects may be more crippling. How a mutant virus responds to a new drug regimen in vivo, therefore provides the most meaningful test of virus fitness.
Because insertions and deletions are uncommon in HIV-1 RT and protease, researchers have been able to develop a standardized numbering system for HIV-1 drug resistance mutations. The most commonly used wild type reference sequence is the subtype B consensus sequence. This sequence was originally derived from alignments in the HIV Sequence Database at Los Alamos (205) and can now be found on the HIV RT and Protease Sequence Database (339). The standardized numbering system and reference sequence have led to the development of a shorthand for mutations in which a letter indicating the consensus B wild-type amino acid is followed by the amino acid residue number, followed by a letter indicating the mutation (e.g., T215Y).
So many mutations in both the protease and RT have been associated with drug resistance that it has become customary to label some mutations either primary (or less commonly major) and other mutations secondary (or minor). The term primary is used to indicate mutations that reduce drug susceptibility by themselves whereas the term secondary is used to indicate mutations that reduce drug susceptibility or improve the replicative fitness of isolates with a primary mutation. However, the labels primary and secondary are not strictly defined. For example, some mutations might be considered to be primary for one drug but secondary for another drug. Moreover, secondary mutations commonly arise before primary mutations.
PI RESISTANCE
HIV-1 Protease
The HIV-1 protease enzyme is responsible for the posttranslational processing of the viral gag- and gag-pol-encoded polyproteins to yield the structural proteins and enzymes of the virus. The enzyme is an aspartic protease composed of two noncovalently associated, structurally identical monomers 99 amino acids in length (Fig. 1). Its active site resembles that of other aspartic proteases and contains the conserved triad, Asp-Thr-Gly, at positions 25 to 27. The hydrophobic substrate cleft recognizes and cleaves 9 different peptide sequences to produce the matrix, capsid, nucleocapsid, and p6 proteins from the gag polyprotein and the protease, RT, and integrase proteins from the gag-pol polyprotein. The enzyme contains a flexible flap region that closes down on the active site upon substrate binding.
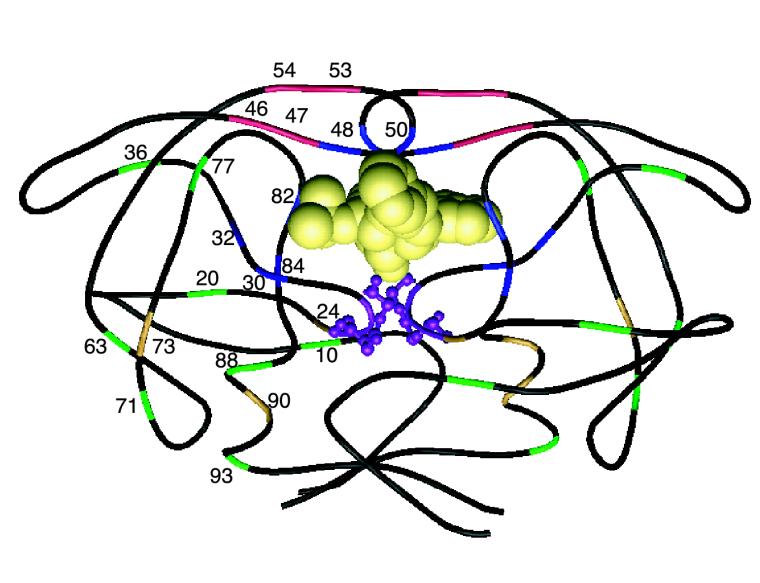
FIG. 1.
Structural model of HIV-1 protease homodimer labeled with protease inhibitor resistance mutations. The polypeptide backbone of both protease subunits (positions 1 to 99) is shown. The active site, made up of positions 25 to 27 from both subunits, is displayed in ball and stick mode. The protease inhibitor resistance mutations are shown for the subunit on the left but not for the subunit on the right. The protease was cocrystallized with indinavir, which is displayed in space-fill mode. This drawing is based on a structure published by Chen et al. (46).
Resistance is mediated by at least two different types of mechanisms. Mutations in the substrate cleft cause resistance by reducing the binding affinity between the inhibitor and the mutant protease enzyme. Other mutations either compensate for the decreased kinetics of enzymes with active site mutations or cause resistance by altering enzyme catalysis, dimer stability, inhibitor binding kinetics, or active site reshaping through long-range structural perturbations (97). The three-dimensional structures of wild-type HIV-1 protease and several drug-resistant mutant forms bound to various inhibitors have been determined by crystallography (1, 2, 15, 47, 238, 239).
PI resistance usually develops gradually from the accumulation of multiple primary and secondary mutations. Most primary mutations, by themselves, cause a two- to fivefold reduction in susceptibility to one or more PIs. However, this level of resistance is often insufficient to interfere with the antiviral activity of these drugs. Higher levels of resistance are resulting from the accumulation of additional primary and secondary mutations are often required for clinically significant reductions in drug susceptibility. This requirement for multiple mutations to overcome the activity of PI inhibitors has been referred to as a “genetic barrier” to drug resistance (54, 188, 265).
Sequence analyses of drug resistant isolates has shown that mutations at several of the protease cleavage sites are also selected during treatment with protease inhibitors (60, 87, 180, 187, 237, 240, 420). Growth kinetic studies have shown that cleavage site mutations in some circumstances improve the kinetics of protease enzymes containing drug resistance mutations and that the cleavage site mutations are compensatory rather than primary. Moreover, there have been no reports that changes at cleavage sites alone can cause PI resistance.
PIs
There are six FDA-approved PIs: amprenavir, indinavir, lopinavir (manufactured in combination with ritonavir), nelfinavir, ritonavir, and saquinavir. The spectrum of mutations developing during therapy with indinavir, nelfinavir, saquinavir, and ritonavir have been well characterized (9, 24, 54, 61, 265, 281, 320, 334), but fewer data are available for amprenavir (237a) and lopinavir (35). The dynamic susceptibility range for indinavir, ritonavir, saquinavir, nelfinavir, and lopinavir is about 100-fold in most drug susceptibility assays (148, 149, 290, 393; Brun, S., D. Kempf, J. Isaacson, A. Molla, H. Mo, C. Benson, and E. Sun, abstract 452, 8th Conference on Retroviruses and Opportunistic Infections, Chicago, Ill., 2001). The dynamic susceptibility range for amprenavir is about 10- to 20-fold. In patients receiving PI combinations or in patients maintaining high PI levels, virologic rebound requires multiple mutations and high-levels of phenotypic resistance.
Pharmacologic factors influence the clinical efficacy of PIs more than that of the other two classes of HIV drugs. Virologic response is highly correlated with the ratio of the trough drug concentration divided by the inhibitory concentration of the drug (e.g., the IC50 in a standardized assay), a ratio that is commonly referred to as the inhibitory quotient (IQ) (153). Drug levels achieved during PI monotherapy can vary greatly among individuals, often resulting in low IQs. This has led to the practice of administering subtherapeutic doses of ritonavir (a P450 enzyme inhibitor) in combination with other PIs to increase their drug levels—a practice known as PI boosting. Lopinavir is formulated in a fixed combination with ritonavir; and saquinavir, indinavir, and amprenavir are also increasingly likely to be administered with low-dose ritonavir (162). Boosted PIs require higher levels of resistance than PIs given as monotherapy before significant loss of antiviral activity and virologic rebound occur.
Protease Substrate Cleft Mutations
V82A/T/F/S mutations occur predominantly in HIV-1 isolates from patients receiving treatment with indinavir and ritonavir (54, 265). V82A also occurs in isolates from patients receiving prolonged therapy with saquinavir following the development of the mutation G48V (330, 409). By themselves, mutations at codon 82 confer decreased in vitro susceptibility to indinavir, ritonavir, and lopinavir (54, 188, 265, 345) but not to nelfinavir, saquinavir, or amprenavir. However, when present with other PI mutations, V82A/T/F/S contributes phenotypic and clinical resistance to each of the PIs (102, 188, 343, 345, 409; Kempf, D., S. Brun, R. Rode, J. Isaacson, M. King, Y. Xu, K. Real, A. Hsu, R. Granneman, Y. Lie, N. Hellmann, B. Bernstein, and E. Sun, abstract 89, Antivir. Ther. 5[Suppl. 3]:70-71, 2000). V82I occurs in about 1% of untreated individuals with subtype B HIV-1 and in 5 to 10% of untreated individuals with non-B isolates (114). Preliminary data suggest that V82I confers minimal or no resistance to the available PIs (79, 193; Brown, A. J., H. M. Precious, J. Whitcomb, V. Simon, E. S. Daar, R. D'Aquila, P. Keiser, E. Connick, N. Hellmann, C. Petropoulos, M. Markowitz, D. Richman, and S. J. Little, abstract 424, 8th Conference on Retroviruses and Opportunistic Infections, Chicago, Ill., 2001).
I84V has been reported in patients receiving indinavir, ritonavir, saquinavir, and amprenavir 54, 61, 148, 237a, 265, 330) and causes phenotypic 42, 54, 188, 277, 280, 282, 378 387) and clinical (278, 423; Kempf, D., S. Brun, R. Rode, J. Isaacson, M. King, Y. Xu, K. Real, A. Hsu, R. Granneman, Y. Lie, N. Hellmann, B. Bernstein, and E. Sun, abstract 89, Antivir. Ther. 5[Suppl. 3]:70-71, 2000) resistance to each PI. I84V tends to develop in isolates that already have the mutation L90M and is rarely the first major mutation to develop in patients receiving a PI (178).
G48V occurs primarily in patients receiving saquinavir and rarely in patients receiving indinavir. This mutation causes 10-fold resistance to saquinavir and about 3-fold resistance to indinavir, ritonavir, and nelfinavir (149, 172, 282, 409). Isolates with a combination of mutations at codons 48, 54, and 82 have been tested against each of the PIs except lopinavir and found to have high-level resistance to each (277, 343).
D30N occurs solely in patients receiving nelfinavir and confers no in vitro or clinical cross-resistance to the other PIs (243, 282, 409). Cross-resistance to indinavir, ritonavir, and saquinavir has been observed in isolates that have D30N along with mutations at positions 88 and 90 (279).
I50V has been reported only in patients receiving amprenavir as their first PI (237a). In addition to causing reduced amprenavir susceptibility, it has been shown to increased ki values to ritonavir, indinavir, and nelfinavir in biochemical studies (Xu, R., W. Andrews, A. Spaltenstein, D. Danger, W. Dallas, L. Carter, M. Hanlon, L. Wright, and E. Furfine, abstract 54, Antivir. Ther. 6[Suppl. 1]:43, 2001) and to cause in vitro cross-resistance to ritonavir and lopinavir (279, 280, 378). Possibly because of the rarity of this mutation, there have been few reports of multidrug-resistant isolates containing this mutation.
V32I occurs in patients receiving indinavir, ritonavir, and amprenavir. It usually occurs only in association with other PI resistance mutations in the substrate cleft or flap and by itself appears to cause minimal resistance to any one drug. R8K and R8Q are substrate cleft mutation that cause high-level resistance to one of the precursors of ritonavir (A-77003) (124, 152), but they have not been reported with the current PIs.
Protease Flap Mutations
The protease flap region (positions 45 to 56) extends over the substrate-binding cleft and must be flexible to allow entry and exit of the polypeptide substrates and products (346). In addition to G48V and I50V, which are also in the substrate cleft, mutations at positions 46, 47, 53, and 54 make important contributions to drug resistance. Mutations at position 54 (generally I54V, less commonly I54T/L/M) contribute resistance to each of the six approved PIs and have been commonly reported during therapy with indinavir, ritonavir, amprenavir and saquinavir, and lopinavir (55, 237a, 265, 281, 320). I54L and I54 M are particularly common in persons receiving amprenavir and have been shown to have a greater effect on amprenavir than the mutation I54V (237a).
Mutations at position 46 contribute to resistance to each of the PIs except saquinavir and have been commonly reported during therapy with indinavir, ritonavir, amprenavir, and nelfinavir (55, 237a, 265, 281, 320). Mutations at codon 47 have been reported in patients receiving amprenavir, indinavir, and ritonavir, and often occur in conjunction with the nearby substrate cleft mutation, V32I. F53L has been reported rarely in patients receiving PI monotherapy, but it occurs in more than 10% of patients treated with multiple PIs (178). It has most recently come to attention as one of the mutations associated with phenotypic resistance to lopinavir in multivariate analyses (188).
Protease Mutations at Other Conserved Residues
L90M has been reported in isolates from patients treated with saquinavir, nelfinavir, indinavir, and ritonavir. L90M either contributes to or directly confers in vitro resistance to each of the six approved PIs and plays a role in causing clinical cross-resistance to each of the PIs (88, 102, 148, 188, 227, 278, 423; Kempf, D., S. Brun, R. Rode, J. Isaacson, M. King, Y. Xu, K. Real, A. Hsu, R. Granneman, Y. Lie, N. Hellmann, B. Bernstein, and E. Sun, abstract 89, Antivir. Ther. 5[Suppl. 3]:70-71, 2000). Crystal structures with and without the mutant have shown that the Leu90 side chain lies next to Leu24 and Thr26 on either side of the catalytic Asp25 (238, 239, 274) but the mechanism by which L90M causes PI resistance is not known.
Mutations at codon 73, particularly G73S, have been reported in 10% of patients receiving indinavir and saquinavir monotherapy and occasionally during nelfinavir monotherapy (178, 334). However, this mutation occurs most commonly in patients failing multiple PIs, usually in conjunction with L90M. Mutations at position 88 (N88D and N88S) commonly occur in patients receiving nelfinavir and occasionally in patients receiving indinavir. By itself, a mutation at this position causes low-level nelfinavir resistance. However, a mutation at this position causes high-level nelfinavir resistance in the presence of D30N or M46I (290, 421). N88S (but not N88D) has been shown to hypersensitize isolates to amprenavir (421), but the clinical significance of this finding is not known. L24I has been reported only in HIV-1 isolates from patients receiving indinavir (55) and has not been shown to confer cross-resistance to other PIs, except possibly lopinavir (188).
Polymorphic Sites Contributing to Resistance
Amino acid variants at seven polymorphic positions, including codons 10, 20, 36, 63, 71, 77, and 93, also make frequent contributions to drug resistance. These mutations do not cause drug resistance by themselves. Some contribute to drug resistance when present together with other protease mutations; whereas others compensate for the decrease in catalytic efficiency caused by other mutations (56, 241, 246, 272, 310).
Mutations at codons 10, 20, 36, and 71 occur in up to 5 to 10% of untreated persons infected with subtype B viruses. However, in heavily treated patients harboring isolates with multiple mutations in the substrate cleft, flap, or at codon 90, the prevalence of mutations at these positions increases dramatically. Mutations at codon 10 and 71 increase to 60 to 80%, whereas mutations at codons 20 and 36 increase to 30 to 40% (148, 177). Codon 63 is the most polymorphic protease position. In untreated persons about 45% of isolates have 63L (considered the subtype B consensus), about 45% have 63P, and about 10% have other residues at this position. However, the prevalence of amino acids other than L increases to 90% in heavily treated patients (177, 413). Mutations at codons 77 and 93 double in prevalence from 15 to 20% in untreated persons to 30 to 40% in heavily treated persons (177).
In some HIV-1 subtypes, mutations at codons 10, 20, and 36 occur at higher rates than they do in subtype B isolates (58, 114, 293, 331). It has been hypothesized that individuals harboring isolates containing multiple accessory mutations may be at a greater risk of virologic failure during PI therapy (288, 289). However, most studies to date have not supported this hypothesis (3a, 25, 106, 208, 288, 289, 329).
PI Cross-Resistance Patterns
Most PI resistance mutations confer resistance to multiple PIs and should be considered class-specific rather than drug-specific mutations. In a study of over 6,000 HIV-1 isolates tested for susceptibility to indinavir, nelfinavir, ritonavir, and saquinavir, 59% to 80% of isolates with a 10-fold decrease in susceptibility to one PI also had a 10-fold decrease in susceptibility to at least one other PI (148). In another study of 3000 HIV-1 isolates, susceptibility to indinavir, ritonavir, and lopinavir were highly correlated (279). Isolates that were resistant to these drugs were generally also resistant to nelfinavir; however, isolates resistant to nelfinavir due to D30N were generally not cross-resistant to other drugs. Susceptibilities to saquinavir and amprenavir were less well correlated to one another or to the other PIs (279). Similar cross-resistance patterns among the PIs have been reported in other smaller studies (185, 300, 321, 322).
Patients in whom nelfinavir-resistant isolates arise after nelfinavir treatment often respond to a regimen containing a different PI because D30N and N88D/S confer little cross-resistance to other PIs (185, 423). But because as many as 15% of nelfinavir failures may be associated with mutations at codons 46 and/or 90, virologic failure while receiving nelfinavir does not guarantee susceptibility to other PIs (9, 281). Nelfinavir is usually unsuccessful as salvage therapy because most of the mutations that confer resistance to other PIs confer cross-resistance to nelfinavir (148, 227, 399).
In vitro drug susceptibility studies suggest that patients failing other PIs often have isolates that retain susceptibility to amprenavir and saquinavir (300, 321). But neither drug has demonstrated usefulness when administered as salvage therapy without ritonavir boosting (82, 92a, 102). In a study of ritonavir/saquinavir salvage therapy, the number of mutations at positions 46, 48, 54, 82, 84, and 90 predicted the virologic response at 4, 12, and 24 weeks. Patients with three or more of these mutations had no response to salvage (423). Data on salvage therapy with ritonavir-boosted amprenavir are not yet available.
In a study of salvage therapy with a regimen containing lopinavir and efavirenz, the number of mutations at positions 10, 20, 24, 46, 53, 54, 63, 71, 82, 84, and 90 predicted the level of phenotypic resistance and the virologic response after 24 weeks of therapy (188; Kempf, D., S. Brun, R. Rode, J. Isaacson, M. King, Y. Xu, K. Real, A. Hsu, R. Granneman, Y. Lie, N. Hellmann, B. Bernstein, and E. Sun, abstract 89, Antivir. Ther. 5[Suppl. 3]:70-71, 2000). A decreased response to therapy was observed only in those patients that had ≥6 of the listed mutations. Subsequent analyses have suggested that mutations at positions 10, 20, 46, 54, and 82 may be more predictive than the others listed (40, 264) and that other mutations, including I50V and G73S may contribute to resistance in different patient cohorts (135, 279). Nonetheless, the large number of mutations required to interfere with a clinical response to therapy demonstrates the high genetic barrier to resistance associated with a drug that achieves high levels in vivo.
In summary, clinical studies have shown that most patients developing virologic failure during treatment with one PI have a diminished virologic response to treatment with a second PI (Table 4). Indeed, most of the successful cases of salvage therapy in patients failing a PI regimen have included regimens with dual PIs or a change to a new PI in combination with an NNRTI (20a, 92a, 294, 423; Kempf, D., S. Brun, R. Rode, J. Isaacson, M. King, Y. Xu, K. Real, A. Hsu, R. Granneman, Y. Lie, N. Hellmann, B. Bernstein, and E. Sun, abstract 89, Antivir. Ther. 5[Suppl. 3]:70-71, 2000). There continues to be great interest in discovering ways to use genotypic data to help switch from one PI to another, although the second PI is increasingly being given as part of a boosted regimen.
TABLE 4.
Correlations between HIV-1 PI resistance mutations and response to a new PI-containing regimena
| Study | Previous regimen | Follow-up regimen | Wk | Effect of baseline mutations on response to follow-up ART |
|---|---|---|---|---|
| Harrigan (132) | ≥1 PI | RTV+SQV | 66 | In the papers by Harrigan and Zolopa, the number of mutations at codons 46, 48, 54, 82, 84, and 90 correlated with a worse response to RTV+SQV. In the paper by Zolopa, virologic response occurred in patients with ≤3 of the mutations. The presence of D30N did not affect response to RTV+SQV. In the paper by Deeks, only 4 of 18 patients had a sustained decrease in RNA of 0.5 log10 at week 24. |
| Deeks (70) | IDV or RTV | RTV+SQV | 24 | |
| Tebas (375) | NFV | RTV+SQV | 24 | |
| Zolopa (423) | ≥1 PI | RTV+SQV | 26 | |
| Para (278) | SQV | IDV | 8 | Mutations at codons 10, 20, 48, 82, 84, and 90 predicted a poor response to IDV salvage therapy. |
| Condra (55) | NFV | IDV | 24 | L90M predicted a higher risk of virologic failure than D30N. |
| Lawrence (227) | SQV | NFV | 16 | L90M predicted virologic failure with NFV. |
| Walmsley (399) | ≥1 PI | NFV (± NNRTI) | 63 | 41% and 22% had RNA declines of ≥1 0.5 log at 24 and 48 wk, respectively. The presence of mutations at codons 48, 82, 84, and 90 correlated with a poor virologic response. |
| Kleinb | ≥1 PI | APV | 12 | I84V and L90M predicted virologic failure; D30N did not. |
| Falloon (102) | ≥1 PI | APV | 16 | 9 heavily treated patients harboring PI mutations at codons 82 and 90 together with mutations at codons 46 and/or 54 had no virologic response to salvage therapy with an APV-containing regimen. |
| Descamps (82) | ≥1 PI | APV | 46 | The presence of ≥4 of the following mutations differentiated those with an RNA decrease of ≥1.0 log at week 12: L10I, V32I, M46IL, I47V, I54V, G73S, V82ATFS, I84V, L90M. |
| Cosado (43) | NRTI, IDV ± RTV | NFV + SQV + NVP + d4T | 31 | 35% and 56% of patients had RNA <50 copies/ml after 6 and 12 months, respectively. L90M decreased the rate of response (43% vs. 0%) but not V82A (36% vs. 38%). |
| Kempfc | ≥1 PI | RTV/LPV (+EFV) | 24 | Mutations at 11 positions were associated with drug resistance (codons 10, 20, 24, 46, 53, 54, 63, 71, 82, 84, and 90). Among 122 NNRTI-naive patients, 24 of 25 with 0-5 of the above mutations, 16 of 21 with 6-7 mutations, and 2 of 6 with 8-10 had plasma HIV-1 RNA <400 copies/ml at wk 24. |
Abbreviations: APV, amprenavir; d4T, starudine; EFV, efavirenz, IDV, indinavir, LPV, lopinavir, NFV, nelfinavir, RTV, ritonavir, SQV, saquinavir.
Klein et al., Antivir. Ther. 5(Suppl. 2):4, abstr. 3, 2000.
Kempf et al., Antivir. Ther. 5(Suppl. 3):70-71, abstr. 89, 2000.
Investigational PIs
The most advanced of the new PI are BMS-232,632 and tipranavir. BMS-232,632 is a highly potent inhibitor of HIV-1 protease with a favorable pharmacokinetic profile that allows once daily dosing. In phase I/II studies it has demonstrated anti-HIV activity similar to that of each of the approved PIs (308; Squires, K., J. Gatell, P. Piliero, I. Sanne, R. Wood, and S. M. Schnittman, abstract 15, 8th Conference on Retroviruses and Opportunistic Infections, Chicago, Ill., 2001). During in vitro passage experiments BMS-232,632 resistant isolates develop mutations at positions 32, 50, 84, and/or 88, a pattern of mutations that is different from that developing in patients treated with other PIs (113). But isolates developing resistance during treatment with other PIs and containing mutations at positions 82, 84, or 90, together with mutations in the protease flap (e.g., positions 46 and 54) are usually cross-resistant to BMS-232,632 (277 Colonno, R. J., K. Hertogs, B. Larder, K. Limoli, G. Heilek-Snyder, and N. Parkin, abstract 8, Antivir. Ther. 5[Suppl. 3]:7, 2000). The drug's potency and pharmacokinetic profile make it a promising candidate for approval. But because its resistance profile overlaps with that of the other approved PIs its usefulness as a salvage therapy is uncertain.
Tipranavir is a nonpeptidomimetic PI with greater flexibility in conforming to enzyme variants with PI resistance mutations (219, 260, 383). However, tipranavir is less potent than other PIs both in vitro and in vivo, and has a narrower dynamic susceptibility range compared with other PIs (14, 219, 316). The narrow dynamic susceptibility range makes it difficult to assess the clinical significance of the decreased cross-resistance between tipranavir and the currently approved PIs. Data are pending on its activity when used in combination with ritonavir.
NRTI RESISTANCE
HIV-1 RT
The RT enzyme is responsible for RNA-dependent DNA polymerization and DNA-dependent DNA polymerization. RT is a heterodimer consisting of p66 and p51 subunits. The p51 subunit is composed of the first 440 amino acids of the RT gene. The p66 subunit is composed of all 560 amino acids of the RT gene. Although the p51 and p66 subunits share 440 amino acids, their relative arrangements are significantly different. The p66 subunit contains the DNA-binding groove and the active site; the p51 subunit displays no enzymatic activity and functions as a scaffold for the enzymatically active p66 subunit. The p66 subunit has five subdomains, including the fingers, palm, and thumb subdomains that participate in polymerization, and the connection and RNase H subdomains.
Most RT inhibitor resistance mutations are in the 5′ polymerase coding regions, particularly in the “fingers” and “palm” subdomains (Fig. 2). Structural information for RT is available from X-ray crystallographic studies of RT bound to an NNRTI (198), unliganded RT (309), and RT bound to double-stranded DNA (158, 171). However, only one structure exists that enables visualization of the interaction between the catalytic complex and the incoming deoxynucleoside triphosphate (dNTP) (158). There have been fewer structural determinations of mutant RT enzymes than of mutant protease enzymes (302, 318).
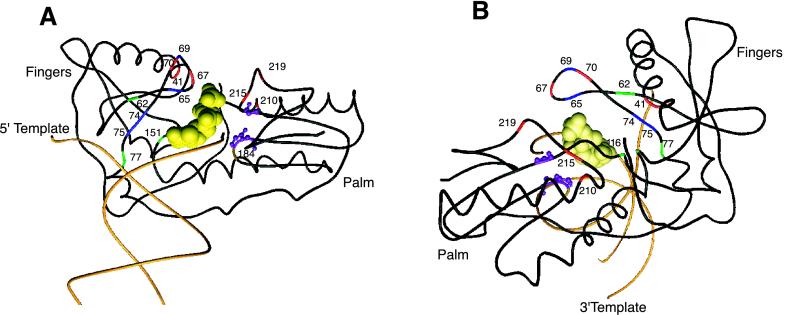
FIG. 2.
Structural model of HIV-1 RT labeled with NRTI resistance mutations. The polypeptide backbone of the fingers and palm domain (positions 1 to 235) and DNA primer and template strands are shown. The active-site positions (110, 185, and 186) are displayed in ball and stick mode. The incoming nucleotide is displayed in space-fill mode. These drawings are based on the structure published by Huang et al. (158) and are shown in front (A) and back (B) views.
NRTIs
Six nucleoside analogs and one nucleotide analog have been approved by the Food and Drug Administration (FDA). The nucleoside analogs include zidovudine, didanosine, zalcitibine, stavudine, lamivudine, and abacavir. Abacavir, which was approved in 1998 is the most recently approved nucleoside. Tenofovir, which was approved in late 2001, is the only FDA-approved nucleotide analog. Both nucleoside and nucleotide analogs are prodrugs that must be phosphorylated by host cellular enzymes. Nucleosides must be tri-phosphorylated; nucleotides, because they already have one phosphate moiety, must be di-phosphorylated. Phosphorylated NRTIs compete with natural dNTPs for incorporation into the newly synthesized DNA chains where they cause chain termination. Because both nucleoside and nucleotide analog RT inhibitors act by a similar mechanism, the abbreviation NRTIs will be used for both classes of compounds.
There are two biochemical mechanisms of NRTI drug resistance. The first mechanism is mediated by mutations that allow the RT enzyme to discriminate against NRTIs during synthesis, thereby preventing their addition to the growing DNA chain (158, 226, 319). The second mechanism is mediated by nucleotide excision mutations (NEMs) that increase the rate of hydrolytic removal of the chain-terminating NRTI and enable continued DNA synthesis (6, 7, 252, 254).
In most drug susceptibility assays, the dynamic susceptibility range is >100-fold for zidovudine and lamivudine and 15- to 20-fold for didanosine, stavudine, zalcitibine, abacavir, and tenofovir (393). Mutant isolates from patients failing therapy with zidovudine, lamivudine, and abacavir usually have measurable phenotypic drug resistance. In contrast, mutant isolates from patients failing therapy with stavudine or didanosine are often found to be drug susceptible in phenotypic assays. Because tenofovir has only recently been approved, there are fewer data on the genotypic correlates of drug resistance and on how well these changes can be detected in phenotypic assays. The difficulty in detecting didanosine resistance is thought to be an artifact of susceptibility testing caused by the inefficient conversion of didanosine to the active compound ddATP when stimulated lymphocytes are used for susceptibility testing (111). The difficulty in detecting stavudine resistance may also be an artifact of the current susceptibility tests that rely on stimulated lymphocytes (230, 253).
NEMs
The most common mutations occurring in clinical HIV-1 samples obtained from patients receiving NRTIs were originally identified for their role in causing zidovudine resistance. Various combinations of these mutations which occur at codons 41, 67, 70, 210, 215, and 219 (133, 156, 182, 222), have been shown to mediate both ATP and pyrophosphate (PP)-dependent hydrolytic removal of zidovudine and stavudine monophosphate from a terminated cDNA chain (6, 252, 254) and cause a compensatory increase in RT processivity (6, 8, 39). ATP-dependent hydrolytic removal of ddNTP, which traps the unblocked ddNTP in an inactive dinucleoside polyphosphate moiety, is more clinically significant than pyrophosphate-dependent hydrolytic removal, which merely regenerates an active ddNTP(262).
In a ddNTP-terminated primer, the presence of the dNTP that would have been incorporated next, had the primer been free for elongation, results in the formation of a stable “dead-end” catalytic complex between RT, primer, template, and dNTP (29, 230, 262, 379). The formation of such a dead-end complex may interfere with the ability of NEMs to facilitate the resumption of virus DNA chain elongation. Biochemical and structural modeling studies have suggested that the bulky azido group of zidovudine may interfere with the formation of a dead-end catalytic complex by sterically preventing the addition of the next dNTP (29, 230). This observation may explain, at least in part, why the NEMs cause the highest levels of phenotypic resistance to zidovudine, despite the fact that biochemical studies have shown that some combinations of NEMs elevate ATP-dependent removal of blocked stavudine-monophosphate (MP) to the same degree as zidovudine-MP (230, 253).
The structural basis underlying the NEMs mechanism of action is not yet understood. Two crystallographic studies have described possibly different roles for the NEMs depending on the particular mutant enzyme studied. One study suggested that positions 215 and 219 give rise to changes that propagate to the active site residues via adjacent residues in the enzyme (302). Whereas the second study suggested that in some mutant structures, T215Y may make direct contact with the dNTP substrate (Stammers, D. K., J. Ren, C. Nichols, P. Chamberlain, L. Douglas, J. Lennerstrand, B. Larder, and D. I. Stuart, abstract 72, Antivir. Ther. 6[Suppl. 1]:54-55, 2001).
During the past few years, several studies have shown that the NEMs are associated with resistance not just to zidovudine, but also to stavudine, abacavir, and to a lesser extent, to didanosine, zalcitibine, and tenofovir (259, 262, 398). The NEMs are selected primarily in patients treated with zidovudine or stavudine alone or in combination with other NRTIs (27, 49, 169, 203, 232, 266, 285, 291, 305, 311, 335, 353). They occur in about 10% of patients treated with didanosine monotherapy (77, 410; Winters, M. A., M. Hughes, S. Lustgarten, and D. A. Katzenstein, abstract 131, Antivir. Ther. 6[Suppl. 1]:96-97, 2001) but do not appear to occur during abacavir monotherapy (261). There are few data on the development of NEMs in patients receiving zalcitibine or tenofovir without other NRTIs.
K70R causes low-level (four- to eightfold) zidovudine resistance and is usually the first drug resistance mutation to develop in patients receiving zidovudine monotherapy (27, 64). T215Y/F results from a two base-pair mutation and causes intermediate (10- to 20-fold) zidovudine resistance. It arises in patients receiving dual NRTI therapy, as well as, in those receiving zidovudine monotherapy (207, 224, 335). T215S/C/D are transitional mutations between wild-type and Y or F that do not cause reduced drug susceptibility but rather indicate the presence of previous selective drug pressure (67, 221, 417). Mutations at positions 70 and 215 are antagonistic in their effect on zidovudine resistance and these two mutations rarely occur together unless additional NEMs are also present (27).
Mutations at positions 41 and 210 usually occur with mutations at position 215 (133, 156, 414). Mutations at positions 67 and 219 may occur with mutations at position 70 or with mutations at position 215. T215Y and K219Q are associated with increased processivity. L210W is strongly associated with M41L and T215F/Y and decreases the susceptibilities of isolates with these mutations by several fold. L210W may stabilize the interaction of 215YF with the dNTP binding pocket (262, 414). In patients failing multiple dual nucleoside therapy it is not unusual for isolates to have four, five, or even all six NEMs.
Clinical studies have shown that the NEMs, particularly mutations at position 215 interfere with the clinical response to zidovudine (203, 303), stavudine (353), abacavir (102, 191, 214), didanosine (155, 173), and most dual NRTI combinations (169, 266; Costagliola, D., D. Descamps, V. Calvez, B. Masquelier, A. Ruffault, F. Telles, J. L. Meynard, and F. Brun-Vizinet, abstract 7, Antivir. Ther. 6:S8, 2001; Mayers, D., T. Merigan, and P. Gilbert, abstract 129, 6th Conference on Retroviruses and Opportunistic Infections, Chicago, Ill., 1999) (Table 5). Complete loss of response to abacavir appears to require the combination of three or more NEMs together with the mutation M184V (214; Costagliola, D., D. Descamps, V. Calvez, B. Masquelier, A. Ruffault, F. Telles, J. L. Meynard, and F. Brun-Vizinet, abstract 7, Antivir. Ther. 6:S8, 2001). The extent to which NEMs interfere with response to tenofovir is not known; however, preliminary data presented to the FDA have shown that tenofovir usually retains antiviral activity even in patients with extensive previous NRTI therapy.
TABLE 5.
Correlations between HIV-1 NRTI resistance mutations and response to a treatment regimena
| Study | Previous regimen | Follow-up regimen | Wk | Effect of baseline mutations on response to follow-up ART |
|---|---|---|---|---|
| Holodniy (155) | AZT | AZT+ddI | 30 | The presence of AZT resistance mutations, particularly T215Y, predicted a poor outcome in patients receiving salvage therapy with AZT+ddI, AZT+ddI+NVP, d4T+ddI, AZT+3TC, and d4T+3TC. It did not appear to limit the effectiveness of AZT+3TC+RTV and AZT+3TC+IDV. |
| Mayersb | AZT | AZT+ddI or AZT+ddI+NVP | 24 | |
| Izopet (169) | AZT+ddC | d4T+ddI | 24 | |
| Japour (173) | AZT | AZT or ddI | 52 | |
| Montaner (266) | AZT | D4T+3TC | 48 | |
| Kuritzkes (208) | AZT | AZT+3TC+RTV | 48 | |
| Gulick (123) | AZT | AZT+3TC+IDV | 156 | |
| Havlir (143) | AZT ± ddI, ddC, d4T | AZT/3TC/IDV followed by AZT/3TC | ≥24 | T215Y did not limit the effectiveness of AZT/3TC/IDV but was strongly associated with virologic failure during AZT/3TC maintenance. |
| Shulman (353) | AZT | d4T | 12 | K70R alone did not prevent a subsequent virologic response to d4T. All other combinations of AZT mutations did interfere with a subsequent response. |
| Rusconi (315) | NRTI (including 3TC), PI ± NNRTI | Change from 3TC to ddI | 8 | In 6 of 8 patients infected with HIV-1 isolates containing M184V + multiple classical AZT resistance mutations, RNA decreased ≥0.5 log. |
| Albrecht (3) | NRTI | NRTI+NFV vs. NRTI+EFV vs. NRTI+NFV+EFV | 195 | Addition of 3TC was associated with an improved virologic response in patients without M184V. |
| Katlama (179) | NRTI, NNRTI, PI | Addition of ABC | 16 | M184V did not preclude an antiviral response; 73% of subjects with M184V had a ≥1.0 log reduction in plasma HIV-1 RNA. |
| Lanierc | NRTI, NNRTI, PI | Addition of ABC | 12-24 | The presence of ≥3 AZT resistance mutations, particularly when present with M184V, was associated with a poor virologic response. The presence of M184V alone was not. |
Abbreviations: ABC, abacavir, AZT, zidovudine, ddC, zalcitibine; ddI, didanosine, d4T, stavudine, IDV, indinavir, NVP, nevirapine, 3TC, lamivudine.
Mayers et al., 6th Conf. on Retroviruses, abstr 129, 1999.
Lanier et al., Antivir. Ther. 4(Suppl. 1):56, abstr. 82, 1999.
The NEMs reduce zidovudine susceptibility more than any other drug. Both K70R and T215Y cause reproducible reductions in drug susceptibility regardless of the susceptibility assay used. Phenotypic resistance to other NRTIs generally requires multiple NEMs. The presence of four or more NEMs will typically cause >100-fold decreased susceptibility to zidovudine, five- to sevenfold decreased susceptibility to abacavir, but usually not more than two- to threefold decreased susceptibility to stavudine, didanosine, zalcitibine, and tenofovir (134, 217, 230, 250, 259, 262, 354, 398).
The NEMs cause minimal lamivudine resistance and do not greatly compromise lamivudine activity (3) except to the extent that they interfere with the synergism between lamivudine and zidovudine and lamivudine and stavudine. One abstract that correlated the presence of NEMs with low-level lamivudine resistance (Skowron, G., J. Whitcomb, M. Wesley, C. Petropoulos, N. Hellmann, M. Holodniy, J. Kolberg, J. Detmer, M. T. Wrin, and K. Frost, abstract 81, Antivir. Ther. 4[Suppl. 1]:55, 1999) relied on a point mutation assay and did not account for other RT mutations which were likely to have explained the results (e.g., codons 44 and 118 [147]).
M184V
M184V causes high-level (>100-fold) lamivudine resistance and emerges rapidly in patients receiving lamivudine monotherapy (26, 326, 377). This mutation is also usually the first to develop in isolates from patients receiving incompletely suppressive lamivudine-containing regimens (81, 142, 236, 266a). M184V is also selected during therapy with abacavir (134, 261, 376) and less commonly during therapy with zalcitibine and didanosine (122, 351, 410). M184V causes about 2-fold resistance to these drugs (122, 263, 290, 376, 410).
M184I results from a G to A mutation (ATG to ATA) and usually develops before M184V in patients receiving lamivudine because HIV-1 RT is more prone to G to A mutations than to A to G mutations (ATG to GTA) (174, 189). Although M184I also causes high-level resistance to lamivudine, the enzymatic efficiency of M184I is less than that of M184V and nearly all patients with mutations at this position eventually also develop M184V (107).
M184V alone renders lamivudine ineffective but may not significantly compromise virologic response to treatment with abacavir (145, 179, 391, 399a). However, M184V in combination with multiple zidovudine resistance or in combination with mutations at positions 65, 74, or 115 leads to both in vitro and in vivo abacavir resistance (134, 179, 277, 343; Lanier, R., J. Scott, H. Steel, B. Hetherington, M. Ait-Khaled, G. Pearce, W. Spreen, and S. Lafon, abstract 82, Antivir. Ther. 4[Suppl. 1]:56, 1999). The effect of M184V on the virologic response to didanosine-containing regimens has been less well studied though in one small observational study showed that in heavily treated patients infected with isolates containing multiple NEMs and M184V, a change from lamivudine to didanosine was usually associated with an RNA decrease of ≥0.5 log10 RNA (315).
Position 184 is in a conserved part of the RT close to the active site. M184V sterically hinders certain NRTIs, particularly lamivudine, while still allowing the enzyme to function (318). The possibility that isolates containing M184V are compromised was suggested by the initial lamivudine monotherapy studies which showed that RNA levels remained about 0.5 log copies below their starting value in patients receiving lamivudine for 6 to 12 months despite the presence of lamivudine-resistant isolates containing M184V (98, 167, 296). Several studies have shown that in vitro RT enzymes with M184V displayed increased fidelity (89, 275, 396) and others decreased processivity (12, 13, 28, 348). The clinical significance of these biochemical studies is not known and the increased fidelity does not appear to limit the ability of HIV to develop new mutations under continued selective drug pressure (176, 190).
M184V reverses T215Y-mediated zidovudine resistance (26, 223, 377); in plaque-forming assays, HIV-1 isolates containing M41L/T215Y displayed 64-fold resistance, while isolates containing M41L/T215Y and M184V were just 4-fold resistant. Resensitization may be due to the ability of M184V to impair the rescue of chain-terminated DNA synthesis (115) and does not appear to apply to zidovudine resistance caused by Q151M (342). This resensitization is probably clinically significant and explains the slow evolution of phenotypic zidovudine resistance in patients receiving zidovudine plus lamivudine (209, 223, 247). Resensitization, however, can be overcome by the presence of four or more zidovudine resistance mutations (343, 377). M184V also appears to reverse the effect of the classical zidovudine mutations on resistance to stavudine and tenofovir, but not abacavir (89, 257, 270, 277).
Mutations at Codons 65, 69, 74, and 75
Positions 64 to 72 form a loop between the β2 and β3 strands in the fingers region of the RT, which makes important contacts with the incoming dNTP during polymerization (158, 319). In addition to the zidovudine-resistance mutations at codons 67 and 70, this region contains several other NRTI-resistance mutations. The most common mutations in this region occur at position 69 and include T69D/N/S/A, as well as single and double amino acid insertions.
T69D was initially identified as causing resistance to zalcitibine (104) but substitutions at this position have since been reported after treatment with each of the available NRTIs. In site-directed mutagenesis studies, other mutations at this position including T69N, T69S, and T69A have been shown to confer resistance to zidovudine, didanosine, zalcitibine, and stavudine (404a). It also appears likely that mutations at position 69 may contribute to resistance to each of the NRTIs when they occur together with NEMs (149, 257, 397, 404a).
Insertions at position 69 occur in about 2% of heavily treated HIV-1-infected patients (390). By themselves, these insertions cause low-level resistance to each of the NRTIs, but isolates containing insertions together with T215Y/F and other zidovudine-resistance mutations have high-level resistance to each of the NRTIs (63, 218, 248, 373, 406). Insertions at this position are associated with about 20-fold resistance to tenofovir, which is the highest reported level of resistance to this drug (258). The precise mechanism by which this mutation causes resistance is not known with certainty but one paper suggests that higher levels of resistance occur in the presence of ATP suggesting that this mutation may act in a manner similar to the NEMs by causing ATP-mediated primer unblocking (230). Single amino acid deletions between codons 67 to 70 occur in <1% of heavily treated patients (164 to 166, 405). These deletions contributes to resistance to each of the NRTIs in patients with viruses containing multiple NRTI mutations.
L74V occurs commonly during didanosine and abacavir monotherapy (202, 261, 338, 410) and confers two- to fivefold resistance to didanosine and zalcitibine (368, 410) and two- to threefold resistance to abacavir (376). L74V is sufficient to cause virologic failure in patients receiving didanosine monotherapy (202) but additional mutations may be required to cause virologic failure to abacavir monotherapy. L74V causes hypersensitivity to zidovudine and possibly also to stavudine (368) and is consequently rarely observed in patients receiving dual nucleoside therapy with didanosine/zidovudine or didanosine/stavudine (49, 200, 285, 335, 338). L74V has also been shown to be cause decreased RT processivity in enzymatic studies and decreased replication in cell culture (347, 348).
K65R confers intermediate levels of resistance to didanosine, abacavir, zalcitibine, lamivudine, and tenofovir (120, 121, 259, 261, 290, 359, 367, 376, 397, 419). This mutation has been shown to increase the replication fidelity of HIV-1 RT in vitro and to cause increased enzymatic processivity mediated by a decrease in the rate of template-primer dissociation (5, 344). K65R occurs rarely in vivo (404, 413), and the biological and clinical significance of these biochemical findings are not known.
V75T develops in isolates cultured in the presence of increasing concentrations of stavudine and causes about fivefold resistance to stavudine, didanosine, and zalcitibine (212). Biochemical data and modeling data suggest that mutations at this position cause drug resistance through nucleotide discrimination and possibly also through a non-ATP-mediated mechanism of primer unblocking (230, 327). V75T occurs rarely even in patients receiving stavudine. V75I generally occurs in isolates that also have the multinucleoside resistance mutation, Q151M. The phenotypic effects of other mutations at this position including V75 M/A have not been well-characterized.
Multinucleoside Resistance Due to Q151M
Q151M is a 2-bp change in a conserved RT region that is close to the first nucleotide of the single-stranded nucleotide template (158, 350). This mutation develops in up to 5% of patients who receive dual NRTI therapy with didanosine in combination with zidovudine or stavudine (49, 181, 285, 323, 335, 338, 390). Q151M alone causes intermediate levels of resistance to zidovudine, didanosine, zalcitibine, stavudine, and abacavir (168, 342, 350, 389). Q151M is generally followed by mutations at positions 62, 75, 77, and 116. Isolates with V75I, F77L, F116Y, and Q151M have high-level resistance to each of these NRTIs, low-level resistance to lamivudine and tenofovir (259, 277). HIV-1 isolates with Q151M usually contain few, if any, NEMs.
Other NRTI Resistance Mutations
E44DA and V118I each occur in about 1% of untreated individuals (177). The prevalence of these two mutations is much higher in isolates obtained from patients receiving dual NRTI combinations, particularly in isolates containing multiple zidovudine resistance mutations (74, 177). When present in combination, E44D and V118I cause intermediate lamivudine resistance (147). However, the frequent occurrence of these mutations even in patients who have not received lamivudine suggests a much broader role.
G333E is a polymorphism that has been reported in 4 of 70 (6%) untreated persons and 26 of 212 (12%) of persons receiving NRTIs (109). G333E has been reported to facilitate zidovudine resistance in isolates from patients receiving zidovudine and lamivudine who also have multiple NEMs (184). However, dual resistance to these drugs usually emerges without this change (247, 343). There are no data suggesting that this mutation by itself reduces zidovudine susceptibility. Two abstracts have suggested that in some isolates the common polymorphisms R211K and L214F also facilitate dual zidovudine and lamivudine resistance in the presence of mutations at positions 41, 184, and 215 (262, 380). P157A/S is a rare mutation associated with lamivudine resistance. This mutation was first identified in a feline immunodeficiency virus isolate cultured in the presence of lamivudine and has subsequently shown to be associated with high-level lamivudine resistance even in isolates lacking M184V (291, 362, 363).
NRTI Cross-Resistance Patterns
The NEMs confer some degree of clinically significant resistance to all NRTIs except lamivudine. The lamivudine resistance mutation, M184V, confers some degree of cross-resistance to all NRTIs except zidovudine, stavudine, and tenofovir. Indeed, M184V and several other NRTI-resistance mutations including L74V and possibly K65R (226) interfere with the effect of the NEMs. The mutational antagonism between the NEMs and several of the mutations that act by allowing RT to discriminate against NRTIs probably explains the clinical synergism observed with certain dual NRTI combinations such as zidovudine/lamivudine, stavudine/lamivudine, zidovudine/didanosine, and stavudine/didanosine.
High-level resistance to both drugs in a dual NRTI combination usually requires multiple NRTI resistance mutations. Two genetic mechanisms of multidrug resistance have received much attention: (i) Q151M usually together with V75I, F77L, and F116Y; and (ii) a double amino acid insertion at position 69 in combination with T215Y/F and other NEMs. These two mutational patterns, however, are responsible for only a minority of multidrug resistant isolates. Multidrug resistance more commonly results from a combination of ≥4 NEMs, M184V, and 1 to 2 mutations in the β2-β3 loop, particularly at position 69.
The extent of cross-resistance between one dual NRTI combination and a second dual NRTI combination is currently being evaluated in clinical trials (360). Preliminary data suggest that patients switching from one dual NRTI combination to a second dual NRTI combination will generally have some response as long as high-level resistance to the first combination has not yet emerged. Because of the high-level of cross-resistance between zidovudine and stavudine, it is unlikely that substituting one drug for the other is likely to be highly effective. There appears to be less clinical cross-resistance between lamivudine and didanosine and a salvage regimen that substitutes one of these drugs for the other is likely to have some activity.
The optimal uses of abacavir and tenofovir, the two most recently approved NRTIs have not yet been defined. In previously untreated patients, abacavir is highly potent, reducing plasma HIV-1 RNA levels by ≥1.5 log10 copies/ml. Its activity in treated patients, however, is compromised by the fact that a combination of M184V together with ≥3 NEMs appear to prevent a clinical response to the addition of this drug (214). This would suggest that its main role should be as part of an initial treatment regimen and not for salvage therapy. Preliminary data presented to the FDA from phase III trials in which tenofovir was added to a failing treatment regimen suggest that this drug may be uniquely effective (reducing plasma HIV-1 RNA levels by ≥0.7 log10 copies/ml) in heavily treated patients harboring viruses resistant to most other NRTIs. The usefulness of tenofovir in salvage therapy should not necessarily preclude a possible role in initial therapy.
NNRTI RESISTANCE MUTATIONS
The NNRTIs bind to a hydrophobic pocket in the RT enzyme close to, but not contiguous with, the active site. These compounds inhibit HIV-1 replication allosterically by displacing the catalytic aspartate residues relative to the polymerase binding site (100, 198, 365). The mutations responsible for NNRTI resistance are in the hydrophobic pocket which bind the inhibitors (Fig. 3). A single mutation in this pocket may result in high-level resistance to one or more NNRTIs. Resistance usually emerges rapidly when NNRTIs are administered as monotherapy or in the presence of incomplete virus suppression, suggesting that resistance may be caused by the selection of a pre-existing population of mutant viruses within an individual (57, 141, 170, 400). Like many of the PI and NRTI resistance mutations, some of the NNRTI resistance mutations may also compromise virus replication. Two mechanisms of impaired replication have been proposed: changes in the conformation of the dNTP binding pocket (194, 389) and changes in RNase H activity (4, 112).
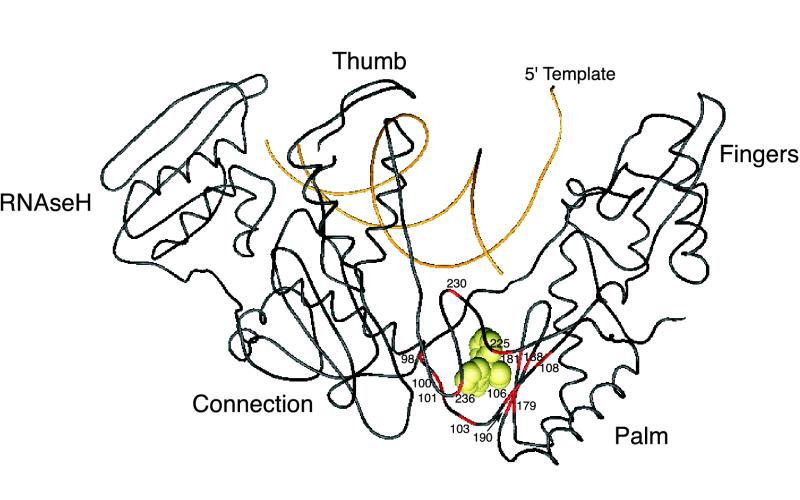
FIG. 3.
Structural model of HIV-1 RT labeled with NNRTI resistance mutations. The polypeptide backbone of the complete p66 subunit (positions 1 to 560) and DNA primer and template strands are shown. This drawing is based on the structure published by Kohlstaedt et al. (199) in which the RT is cocrystallized with nevirapine, which is displayed in space-fill mode. The positions associated with NNRTI resistance are shown surrounding the hydrophobic pocket to which nevirapine and other NNRTIs bind.
NNRTIs
There are three FDA-approved NNRTIs: nevirapine, delavirdine, and efavirenz. The hydrophobic binding pocket to which the NNRTIs bind is less well conserved than the dNTP binding site. Indeed, HIV-1 group O and HIV-2 (80, 151, 349, 415) are intrinsically resistant to most NNRTIs. The FDA-approved NNRTIs are highly active against group M HIV-1 isolates and the dynamic susceptibility range for each of the NNRTIs is greater than 100-fold. Wild-type HIV-1 group M isolates tend to have greater interisolate variability in their susceptibility to NNRTIs than to NRTIs and PIs (33). However, preliminary data suggest that the moderate (<10-fold) decreases in NNRTI susceptibility that have been reported in the absence of previous NNRTI therapy or known NNRTI resistance mutations do not interfere with the virologic response to an NNRTI-containing highly active antiretroviral therapy (HAART) regimen (Bacheler, L., L. Ploughman, K. Hertogs, and B. Larder, abstract 88, Antivir. Ther. 5[Suppl. 3]:70, 2000; Harrigan, P. R., W. Verbiest, B. Larder, K. Hertogs, J. Tilley, J. Raboud, and J. S. Montaner, abstract 86, Antivir. Ther. 5[Suppl. 3]:68-69, 2000).
NNRTI Mutations between Codons 98 and 108
K103N occurs more commonly than any other mutation in patients receiving NNRTIs (11, 57, 69, 78, 129) and causes 20- to 50-fold resistance to each of the available NNRTIs (11a, 78, 290, 418). Although this degree of resistance is less than the highest levels of resistance observed with these drugs, K103N by itself appears sufficient to cause virologic failure with each of the NNRTIs (44, 78, 175, 355). It has been proposed that K103N may have minimal effects on viral fitness and that this mutation can result in a virus that is both resistant and highly fit (69). Structural studies of HIV-1 RT with K103N in both unliganded and bound to an NNRTI have shown that the structure is only minimally changed in that in the unliganded form it forms a network of hydrogen bonds that are not present in the wild-type enzyme (157). These changes are likely to stabilize the closed pocket form of the enzyme and interfere with the ability of inhibitors to bind to the enzyme. A different mutation at position 103, K103R, occurs in 2 to 3% of patients not receiving NNRTIs and has not been reported to cause NNRTI resistance (177).
V106A causes >30-fold resistance to nevirapine, intermediate resistance to delavirdine, and low-level resistance to efavirenz (18, 38, 95, 108, 220, 284, 290, 418). L100I causes intermediate resistance to efavirenz and delavirdine and low-level resistance to nevirapine (37, 38, 108, 290, 403, 418). L100I usually occurs with K103N in patients receiving efavirenz and significantly increases efavirenz resistance in these isolates (11). A98G, K101E, and V108I each cause low-level resistance to each of the NNRTIs (11a, 37, 290, 418).
NNRTI Mutations between Codons 179 and 190
Y181C/I causes >30-fold resistance to nevirapine and delavirdine and 2 to 3-fold resistance to efavirenz (37, 38, 290, 418). Nonetheless, nevirapine-treated patients with isolates containing Y181C generally have only transient virologic responses to efavirenz-containing salvage regimens (355, 398a). It is not known whether virologic failure in this setting is due to low-level Y181C-mediated efavirenz resistance or to the presence of a subpopulation of viruses containing K103N that predominate upon exposure to efavirenz.
Y188C/L/H causes high-level resistance to nevirapine and efavirenz and intermediate resistance to delavirdine (38, 108, 290, 418). G190A/S causes high-level resistance to nevirapine and efavirenz but do not cause in vitro resistance to delavirdine (11a, 108, 290). There are no clinical data, however, on the usefulness of delavirdine in patients harboring isolates with these mutations. V179D causes low-level (about twofold) resistance to each of the NNRTIs (38, 195, 403, 418).
NNRTI Mutations between Codons 225 and 236
P225H causes low-level resistance to efavirenz and possibly nevirapine. By itself, P225H causes delavirdine hypersusceptibility. However, it usually occurs with K103N in patients receiving efavirenz (11, 11a, 284). M230L is a recently identified uncommon mutation that causes about 20-fold resistance to efavirenz, 40-fold resistance to nevirapine, and 60-fold resistance to delavirdine (Huang, W., N. T. Parkin, Y. S. Lie, T. Wrin, R. Haubrich, S. Deeks, N. Hellmann, C. J. Petropoulos, and J. M. Whitcomb, abstract 30, Antivir. Ther. 5[Suppl. 3]:24-25, 2000). P236L is an uncommon mutation that causes high-level resistance to delavirdine and hypersusceptibility to nevirapine (78, 91, 160). P236L causes slowing of both DNA 3′-end- and RNA 5′-end-directed RNase H cleavage possibly explaining the markedly decreased replication of isolates with this mutation (112). F227L and L234I cause resistance to two experimental NNRTI but their effect on current NNRTIs is not known (18, 108).
Other NNRTI Resistance Mutations
Mutations at codon 138 (e.g., E138K) have been shown to confer resistance to an experimental group of NNRTIs, called the TSAO inhibitors (17), but do not cause resistance to the currently approved NNRTIs (283). This mutation exerts its effect via the p51 subunit of HIV-1 RT, which lies close to the NNRTI binding pocket (17). Mutations at position 135 and 283 have been shown to cause low-level resistance to NNRTIs, particularly when present in combination (33). Y318F is a mutation in the NNRTI-binding pocket which causes high-level resistance (about 40-fold) to delavirdine and low-level resistance (<3-fold) resistance to nevirapine and efavirenz (183). However, this mutation rarely occurs in the absence of other major NNRTI resistance mutations.
NNRTI Mutation Interactions
Mutational interactions within the NNRTI class (e.g., hypersusceptibility caused by P225H and P236L) have not yet been shown to be clinically significant in that there has been no demonstrated benefit of using NNRTIs either in combination or in sequence. Mutational interactions between NNRTI resistance mutations and NRTI-resistance mutations, however, will probably prove to be clinically relevant. It has been known for several years that Y181C and L100I hypersensitize HIV-1 to zidovudine (215, 216), and recently it has been shown that some NRTI-resistance mutations appear to hypersensitize HIV-1 to certain NNRTIs (352; Haubrich, R., J. Whitcomb, P. Keiser, C. Kemper, M. Witt, M. Dube, D. Forthal, M. Leibowitz, J. Hwang, A. Rigby, N. Hellmann, J. A. McCutchan, and D. Richman, abstract 87, Antivir. Ther. 5[Suppl. 3]:69, 2000; Whitcomb, J., S. Deeks, D. Huang, T. Wrin, E. Paxinos, K. Limoli, R. Hoh, N. Hellmann, and C. Petropoulos, abstract 234, 7th Conference on Retroviruses and Opportunistic Infection, San Francisco, Calif., 2000). Although multidrug resistance to both NRTIs and NNRTIs occurs commonly (95, 220, 352), the interactions suggest that the number of ways in which HIV-1 can develop simultaneous high-level resistance to both NRTIs and NNRTIs may be restricted. These interactions may also help explain the success of dual NRTI/NNRTI-containing regimens not only as part of initial therapy but also in certain salvage therapy situations (3, 206; Haubrich, R., J. Whitcomb, P. Keiser, C. Kemper, M. Witt, M. Dube, D. Forthal, M. Leibowitz, J. Hwang, A. Rigby, N. Hellmann, J. A. McCutchan, and D. Richman, abstract 87, Antivir. Ther. 5[Suppl. 3]:69, 2000).
HIV-1 FUSION INHIBITORS
The HIV-1 envelope glycoprotein consists of two noncovalently associated subunits, a surface glycoprotein (gp120) and a transmembrane glycoprotein (gp41). Portions of gp120 bind to both the CD4 receptor and to one of the chemokine receptors on target cells. After gp120-CD4-coreceptor binding, the gp41 subunit undergoes a conformational change that promotes fusion of viral and cellular membranes, resulting in entry of the viral core into the cell. This conformational change results in a transient species, termed the prehairpin intermediate in which gp41 exists simultaneously as a membrane protein in both the viral and cellular membranes (94).
Recent crystallographic studies of gp41 fragments show that two heptad repeat domains form a helical bundle containing trimers of each domain (45). The first successful inhibitors of viral entry were synthetic peptides corresponding to predicted alpha helical regions of the HIV-1 gp41 sequences. One of these peptides, T-20 (pentafuside; Trimeris, Durham, N.C.), corresponds to residues 127 to 162 of the outer layer of gp41. When this peptide was administered intravenously over a two-week period the median plasma HIV-1 RNA levels of subjects receiving the higher dose levels (100 mg twice daily) declined 100-fold (192).
The extraviral portion of gp41 is the most conserved region in the HIV-1 envelope glycoprotein, which otherwise displays considerable genetic diversity. Yet HIV-1 isolates resistant to T-20 have been derived by culturing HIV-1 in the presence of increasing concentrations of the peptide. Sequence analysis of the resistant isolates demonstrated that a contiguous 3-amino-acid sequence (codons 36 to 38) within the amino-terminal heptad repeat motif of gp41 is associated with resistance (306). T-1249 is another injectable fusion inhibitor developed by Trimeris that has a longer half-life and retains activity against T-20-resistant isolates. Additional fusion inhibitors in development include other peptides, antibodies, and small molecules that bind to either gp41 or the chemokine receptors CCR5 and CXCR4.
TECHNICAL ASPECTS OF HIV-1 GENOTYPIC TESTING IN CLINICAL SETTINGS
The following subsections review the source of virus used for sequencing in clinical settings, the method of preparing nucleic acid material for sequencing, methods of sequencing, methods of sequence quality control, and the approach to analyzing HIV-1 isolates belonging to non-B subtypes. More detailed reviews of some of these issues have been covered in other recent reviews on the technical aspects of HIV-1 genotypic testing (75, 128, 341, 392).
Source of Virus and Initial Sample Processing
Plasma is the main source of virus used for testing HIV-1 drug resistance in clinical settings. Because the half-life in of HIV-1 in plasma is approximately 6 h, only actively replicating virus can be isolated from this source; thus the sequence of plasma virus represents the quasispecies most recently selected for by antiretroviral drug therapy (287). Plasma is easier to process and store than peripheral blood mononuclear cells and the evolution of HIV-1 drug resistance in peripheral blood mononuclear cell virus lags behind that in plasma (197, 203, 356, 361, 400). Because HIV-1 genotypic testing requires the extraction, reverse transcription, and PCR amplification of a larger segment of the HIV-1 genome (>1 kb) than used for assays designed quantitative assays (about 100 bp) the sensitivity of most genotypic assays is generally reduced compared with quantitative assays with a lower limit of detection of between 100 and 1,000 RNA copies/ml, depending on the assay.
Population-Based versus Clonal Sequencing
Clonal sequencing is performed in research settings to answer questions about the evolution of HIV-1 drug resistance. Direct PCR or population-based sequencing is done in clinical settings because it is quicker and more affordable than sequencing multiple clones. For both population-based and clonal sequencing, the ability to detect minor variants is related to the proportion of the minor variants within the whole virus population. In direct PCR sequencing, a nucleotide mixture can be detected when the least common nucleotide is present in at least 20% of the total virus population. (62, 125, 225, 325, 340).
Dideoxynucleotide Sequencing
Dideoxynucleotide sequencing is the most commonly used method for HIV-1 sequencing. One commercial HIV-1 RT and protease genotyping kit has been approved by the FDA for use in clinical settings (47a, 380a); a second kit is under consideration for FDA approval (61a, 267a). These kits have stronger quality control and validation profiles than home brew methods which will make them preferable in clinical laboratories. However, these kits are more expensive and may not provide the versatility of current home brew methods. The assays in these kits differ technically but are similar in overall complexity. In one recent comparison they had similar performance (96).
Several studies indicate that dideoxynucleotide sequencing is highly reproducible in experienced laboratories. In one study, 13 research laboratories were shipped cell pellets from cultured HIV-1 isolates (76). The sequence concordance among laboratories was 99.7% at all nucleotide positions and 97% at positions associated with zidovudine resistance. Sequencing cultured cell pellets is simpler than sequencing plasma because RNA extraction and reverse transcription are not necessary and because cultured virus is more homogeneous than uncultured virus (73, 210). Nonetheless, the high interlaboratory concordance in this study attests to the intrinsic reliability of the dideoxy method for HIV-1 analysis.
Two large multicenter comparisons of sequence results obtained from samples containing mixtures of plasmid clones (ENVA-1) and spiked plasma samples (ENVA-2) have also been performed (325, 325a). These studies found that the ability of the participating laboratories to detect mutations was directly proportional to the percent of mutant plasmid clones within each mixture. Only a minority of laboratories detected mutations in mixtures in which the mutant clones made up less than 25% of the total.
Two clinical laboratories also assessed the reproducibility of HIV-1 RT and protease sequencing using plasma aliquots obtained from 46 heavily treated HIV-1 infected individuals (333). Although both laboratories used sequencing reagents from Applied Biosystems (Foster City, Calif.), each used a different in-house protocol for plasma HIV-1 RNA extraction, reverse transcription, PCR, and sequencing. Overall sequence concordance between the two laboratories was 99.0%. Approximately 90% of the discordances were partial, defined as one laboratory detecting a mixture while the second laboratory detected only one of the mixture's components. Discordance was significantly more likely to occur in plasma samples with lower plasma HIV-1 RNA levels. Nucleotide mixtures were detected at approximately 1% of the nucleotide positions, and, in every case in which one laboratory detected a mixture, the second laboratory detected either the same mixture or one of the mixture's components. The high concordance in detecting mixtures and the fact that most discordance between the two laboratories was partial suggest that most discordances were due to variation in sampling the HIV-1 quasispecies rather than to technical errors.
Hybridization Methods
Sequencing by hybridization can determine the complete sequence of an unknown DNA molecule or detect specific mutations. The Affymetrix GeneChip is designed to determine the complete sequence of HIV-1 protease and the first 1,200 nucleotides of HIV-1 RT. The INNO-LiPA HIV-1 line probe assays (Innogenetics, Ghent, Belgium) are point mutation assays designed to detect specific HIV-1 protease and RT mutations.
The GeneChip is divided into several thousand segments each containing millions of similar probes designed to interrogate every nucleotide position in a test DNA or RNA molecule. Every nucleotide in the test molecule requires at least four sets of oligonucleotide probes to determine whether that nucleotide is an A, C, G, or U. It is essential that the probe hybridizes perfectly to the nucleotides on either side of the position being interrogated. The design or tiling of Affymetrix gene chips therefore requires prior knowledge of the most commonly expected polymorphisms in a gene. Because of this requirement, this method of sequencing is also referred to as resequencing.
Because of its genetic variability, sequencing HIV-1 by hybridization is challenging. Several studies have compared the performance of the GeneChip to dideoxynucleotide cycle sequencing and most have found that dideoxynucleotide sequencing is more reliable at detecting HIV-1 RT and protease mutations (125, 130, 386, 402). DNA chips are also not capable of detecting insertions or deletions in viral sequences and are unreliable at sequencing viral subtypes other than subtype B—the subtype on which the chip tiling has been based. In addition, genomic regions containing clusters of adjacent mutations can interfere with probe hybridization and result in frank errors (130, 386). Improved microarrays for sequencing isolates belonging to subtypes A-F and for detecting insertions are under development (Myers, T., D. Birch, V. Bodepudi, D. Fong, D. Gelfand, K. L. A., R. Nersesian, R. Shahinian, C. Sigua, N. Schonbrunner, R. Resnick, K. Wu, and T. Ryder, abstract 49, Antivir. Ther. 5[Suppl. 3]:172, 2000).
Point mutation assays are inexpensive and have the potential to be highly sensitive for mutations present in only a small proportion of circulating viruses (328, 388). Because they require only simple laboratory equipment, they may be useful in areas that do not have ready access to sophisticated sequencers. The INNO-LiPA assays have probes for wild-type and mutant alleles of each codon attached to a nitrocellulose strip (48, 371). Biotin-labeled RT-PCR product from the patient sample is hybridized to the strip. An avidin-enzyme complex and the enzyme substrate produce a color change on the paper strip where the PCR product has hybridized with a probe. This assay is limited because it can only detect a subset of drug resistance mutations and has a 10% rate of uninterpretable results due to poor hybridization, which is particularly likely to occur when uncommon mutations are present at key codons (298, 328).
Sequence Quality Control
Sequence quality control should aim at avoiding PCR contamination and sample mix-ups, obtaining high quantities of specific template DNA, and detecting as many mixtures as possible. Laboratories should use standard physical precautions to prevent sample contamination with DNA from other sources (211) and negative controls should be run with each PCR step. Alternate primers for reverse transcription and/or PCR sequencing should be used on samples that cannot be amplified despite plasma HIV-1 RNA levels >1,000 copies/ml. Heat-stable RNase H− RT enzymes can be used to increase the yield of the reverse transcription step. A uracil N-glycosylase (UNG) system can be used to minimize contamination of PCRs with products generated in previous amplifications.
Sequence analyses can detect the possibility of contamination with other samples studied during the same time period (228). These analyses should compare each new sequence to other recently generated sequences to look for unexpectedly high levels of similarity. Phylogenetic trees can also be constructed to visually detect unexpectedly similar isolates. The HIV Sequence Database at Los Alamos National Laboratories has a tutorial to assist with sequence analysis for quality control purposes (204).
Global HIV-1 Isolates
During its spread among humans, group M HIV-1 has evolved into multiple subtypes that differ from one another by 10 to 30% along their genomes (201, 307). In North America and Europe, most HIV-1 isolates belong to subtype B and the available anti-HIV drugs have been developed by drug screening and susceptibility testing using subtype B isolates. However, subtype B accounts for only a small proportion of HIV-1 isolates worldwide and non-B isolates are being identified with increasing frequency, particularly in Europe.
A few studies have tested the in vitro susceptibility of non-subtype B HIV-1 isolates to antiretroviral drugs. Although group O isolates often demonstrate intrinsic resistance to the NNRTIs (80, 299), most studies have shown that non-B group M isolates are as susceptible as subtype B isolates to each of the three anti-HIV drug classes (276, 331, 332, 374). There is no evidence for novel drug resistance mutations in non-B HIV-1 isolates and most available data suggest that drug resistance mutations described in the context of subtype B isolates will exert the same phenotypic effects in all HIV-1 subtypes.
Intersubtype genetic variability may complicate HIV-1 genotyping because primers used for reverse transcription, PCR, and sequencing may have a lower rate of annealing to non-B compared with subtype B templates. But the extent to which this occurs has not been studied. Both the Applied Biosystems ViroSeq HIV-1 Genotyping System have been used for the analysis of non-B isolates, but the primers used in both commercial systems are proprietary (98a, 267a) and the Visible Genetics TRUGENE HIV-1 Genotyping System (213).
GENOTYPIC INTERPRETATION
General Principles
HIV-1 drug resistance is rarely an all-or-none phenomenon. Clinicians treating infected patients usually need the answers to the following two questions: (i) Does the genotype suggest that the patient will respond to a drug in a manner comparable to a patient with a wild-type isolate? (ii) Does the genotype suggest that the patient will obtain any antiviral benefit from the drug? The second question distinguishes antiviral susceptibility testing from anti-bacterial susceptibility testing. In the case of bacteria, it is usually possible to avoid using any drug with reduced susceptibility against a pathogen. This is usually not possible in the case of HIV, however, because of the extent of cross-resistance within each class of HIV drugs. To answer both these questions it is necessary to grade the extent of inferred resistance relative to the wild type and to the most resistant isolates (e.g., low-level, intermediate, and high-level).
Genotypic results bear little resemblance to those of a typical antimicrobial susceptibility assay. Rather than receiving a result such as susceptible or resistant for each of the available HIV drugs, the ordering clinician receives a list of mutations present in the virus isolate. The difficulty in understanding the results of these genotypic assays and the fact that genotypic interpretation is independent of the process of genotyping makes it an ideal application for a computerized expert system. Laboratories doing HIV-1 genotyping can provide physicians with the option of receiving a file with the sequence data (string of nucleotides or list of amino acid differences from consensus). Such data can then be analyzed by interpretation systems other than those used by the sequencing laboratory.
An expert system performs reasoning over representations of human knowledge. It consists of a computerized knowledge base and an inference engine. A computerized knowledge base has benefits for patients, clinicians, and researchers because it can identify gaps in what is known about drugs and drug resistance mutations and homogeneous data, such as genotype-phenotype correlations, are amenable to machine learning algorithms. In contrast, diverse forms of data such as phenotypic and clinical data, are amenable to rules-based algorithms. Table 6 describes the requirements of an expert system for HIV-1 genotypic interpretation: data input, knowledge base, inference engine, and data output.
TABLE 6.
Expert-system features for HIV-1 genotypic interpretation
| Parameter | Options | Comments |
|---|---|---|
| Data input | Nucleic acid sequence | Raw nucleotide sequence data are not necessary for interpretation but are necessary for quality control purposes to exclude PCR contamination and sample mix-up. |
| List of amino acid differences from a consensus reference sequence | A list of mutations is sufficient for sequence interpretation because silent mutations have not been shown to affect drug susceptibility. An amino acid sequence alone may be inadequate because it is not possible to represent mixtures in a “one-dimensional” string of amino acids. | |
| Knowledge base | Correlations between genotype and phenotype (laboratory and clinical isolates) | Machine learning algorithms are possible because the data are homogeneous and quantitative. Such algorithms, however, cannot consider other forms of data, such as associations between mutations and treatment history or clinical outcome. |
| Correlations between genotype and clinical data (treatment history and clinical outcome) | There is generally insufficient clinical outcome data for most mutations. An approach that uses both phenotypic and clinical data is the most complete, but machine learning algorithms have not yet been developed for such heterogeneous data. | |
| Algorithm | Drug-based rules: resistance to drug X if the following mutations are present or if the following combinations of mutations are present | One advantage of this type of rule is that mutation interactions can be taken into account. However, many rules based on different mutation patterns must be encoded to represent multiple patterns of mutations and multiple levels of drug resistance. |
| Mutation-based rules: each mutation contributes some degree of resistance to one or more drugs. Drug resistance interpretations are derived by combining contributions of individual mutations | This approach is easy to encode and to update, but interactions between specific mutations require the addition of drug-based rules. | |
| Machine learning: pattern matching, neural network, decision trees, etc. | These can be implemented if the knowledge base consists of homogeneous data. | |
| Data output | Number of levels of drug resistance and their explanation | At least four levels of drug resistance are probably necessary: susceptible, low-level resistance, intermediate resistance, high-level resistance |
| Comment | Comments could report the presence of atypical findings, mutation interactions, and the degree of confidence associated with each drug interpretation. |
Table 7 describes several of the most commonly used systems for HIV-1 genotypic interpretation. During the next one to two years, these algorithms will evolve and most likely converge through an ongoing process of interalgorithm comparison and validation using clinical data sets. This is because there is probably more concordance among clinical virologists than is currently reflected in published algorithms. It is unlikely that algorithms will remain proprietary because there is no precedent for basing important medical decisions on proprietary unpublished data. The following two sections explore two algorithms in detail: VirtualPhenotype and the HIVDB algorithm.
TABLE 7.
Algorithms for interpreting HIV-1 protease and RT sequences
| Algorithm | Availability | Description |
|---|---|---|
| Resistance Collaborative Group (72) | Public | Table of rules developed for a standardized reanalysis of eight published studies linking drug resistance mutations and clinical outcome. This algorithm is primarily of historical interest because it is no longer being updated. |
| HIV RT and Protease Sequence Database (336) | Public | Mutations are assigned drug penalties. Drug penalties are added and drugs are assigned an inferred level of resistance. Drug penalties are hyperlinked to primary data linking mutation and drug. Program can be found at http://hivdb.stanford.edu. |
| French National Agency for AIDS Research (312) | Public | Table of rules listing mutations conferring genotypic resistance or possible genotypic resistance to anti-HIV drugs. |
| Retrogram (Virology Networks) | Proprietary | Comprehensive set of drug-based rules. Updated regularly by an expert panel. This interpretation system was used in the Havana clinical trial. |
| GuideLines (Visible Genetics) | Public | Drug-based rules. Updated regularly by an expert panel. |
| VirtualPhenotype (Virco; Mechelin, Belgium)a | Proprietary | Pattern matching algorithm that uses a large genotype-phenotype correlative database to infer phenotypic properties based on sequence data. |
Verbiest et al., Antivir. Ther. 5(Suppl. 3):62, abstr. 81, 2000.
VirtualPhenotype
The VirtualPhenotype (Virco, Cambridge, United Kingdom, and Mechelin, Belgium) is a pattern-matching algorithm that uses a large genotypic-phenotypic correlative database to infer phenotypic properties based on sequence data (Verbiest, W., M. Peeters, K. Hertogs, P. Schel, S. Bloor, A. Rinehart, N. Graham, C. Cohen, and B. A. Larder, abstract 81, Antivir. Ther. 5[Suppl. 3]:62, 2000). The analysis includes a tabulation of the number of matches in the database for each drug, and the distribution of phenotypes (fold increase in IC50) for the matching samples. The mean IC50 of the matching samples is interpreted using a biologically defined, drug-specific cutoff value, providing a quantitative prediction of drug resistance. Although the VirtualPhenotype has been described in several abstracts, there is no publication that describes the workings of this approach in its entirety. Specifically, it is not known which mutations are used to match a new sequence to those sequences that are already in the database.
The VirtualPhenotype has been shown to have a high correlation with results from Virco's recombinant phenotypic assay (Graham, N., M. Peeters, W. Verbeist, R. Harrigan, and B. Larder, abstract 524, 8th Conference on Retroviruses and Opportunistic Infections, Chicago, Ill., 2001). It has also been compared to rule-based algorithms using the data set from a completed clinical trial (126). Further studies examining the predictive value of the VirtualPhenotype using data from other clinical trials is planned.
HIV RT and Protease Sequence Database
The HIV RT and Protease Sequence Database at Stanford University is an online database (http://hivdb.stanford.edu) that links sequence data to the HIV drug treatments of the patients from whom the sequenced isolates were obtained and to drug susceptibility results. The database also contains two sequence analysis programs. HIV-SEQ accepts user-submitted RT and protease sequences, compares them to a reference sequence, and uses the differences as query parameters for interrogating the sequence database (336). This allows users to detect unusual sequence results immediately so that the person doing the sequencing can check the primary sequence output while it is still on the desktop. In addition, unexpected associations between sequences or isolates can be discovered by immediately retrieving data on isolates sharing one or more mutations with the sequence.
The second program, Drug Resistance Interpretation, is an expert system that accepts user-submitted protease and RT sequences and returns inferred levels of resistance to the 16 FDA-approved anti-HIV drugs. Each drug resistance mutation is assigned a drug penalty score; the total score for a drug is derived by adding the scores of each mutation associated with resistance to that drug. Using the total drug score, the program reports one of the following levels of inferred drug resistance: susceptible, potential low-level resistance, low-level resistance, intermediate resistance, and high-level resistance. Genotypic interpretations do not necessarily correlate with the inferred level of phenotypic resistance because the genotypic interpretation also uses correlations between genotype and clinical outcome in deciding how a drug's susceptibility should be graded. A listing of all mutation/drug score pairs can be found with the program's release notes (http://hiv-4.stanford.edu/cgi-test/hivtest-web.pl).
LIMITATIONS OF HIV-1 DRUG RESISTANCE TESTING
Several factors limit the utility of both genotypic and phenotypic testing. (i) There is a complex relationship between drug resistance and clinical failure, often making it difficult to discern the contribution of drug resistance to virologic failure. (ii) The HIV-1 population within an individual consists of innumerable variants and minor variants often go undetected. (iii) Because of extensive cross-resistance within each drug class, the results of resistance testing often leave clinicians with few options for treatment.
Complex Relationship between Drug Resistance and Disease Progression
Drug resistance is not the only cause of treatment failure. Nonadherence, the use of insufficiently potent treatment regimens, and pharmacokinetic factors that decrease the levels of one or more drugs in a treatment regimen also contribute to treatment failure. In addition, the natural history of HIV-1 is highly variable and dependent on a complex set of host-virus interactions (273). In the absence of therapy some patients progress to advanced immunodeficiency within 3 years following infection, whereas other patients remain healthy for more than 15 years. It is likely that the same host-virus interactions that so greatly influence disease progression in the absence of drug therapy also influence the risk of virologic failure in patients receiving anti-HIV therapy.
Two recent observations underscore the complexity of the relationship between drug resistance and disease progression. The first is that patients developing virologic failure on their first treatment regimen are usually found to have HIV-1 isolates with resistance to only one of the drugs in the regimen (81, 110, 142, 236, 267, 268). The drugs to which resistance most commonly develops in this situation are lamivudine and the NNRTIs; resistance to PIs and NRTIs is less common in patients with initial virologic failure. The fact that virus becomes detectable and replication ensues despite the fact that the replicating virus remains sensitive to at least two drugs suggests that factors in addition to drug resistance are contributing to virologic failure. Possibly the remaining drugs in the regimen are not potent enough to fully suppress virus even though they remain active. Alternatively, one of the presumably “effective” drugs in the regimen may be present at insufficient levels because of nonadherence or pharmacokinetic factors.
The second observation is that virologic failure in patients receiving HAART is not always followed by immunologic and clinical deterioration (20, 71, 229, 295). This may be because the immunologic benefits of virus suppression persist beyond the period of virus suppression or because multidrug-resistant viruses may be less virulent, particularly when they first emerge and are associated with fewer compensatory mutations (83, 292).
Several lines of evidence suggest that drug-resistant viruses are less fit than drug-susceptible viruses. First, in vitro experiments have consistently shown that isolates containing protease and/or RT drug resistance mutations replicate less well in cell culture and that purified enzymes with these mutations usually have less activity than wild-type enzymes (reviewed in reference 271). There are conflicting data, however, on whether multidrug-resistant variants are less cytopathic in specific types of cells (e.g., thymus) (231, 286, 370). Second, drug resistance mutations are often replaced in vivo by wild-type variants within weeks to months after removal of selective drug pressure (83, 84, 116, 117, 395). The rate at which this occurs depends on the extent to which archived wild type viruses exist within an individual patient. If there are no archived wild-type viruses, a significant interlocking of primary and compensatory mutations may limit reversion to wild type (21).
Finally, one clinical trial in patients with detectable viremia and mulitdrug-resistant virus showed that in those patients randomized to discontinuing HIV therapy, RNA decreased by 0.84 logs and CD4 cell counts decreased by 128 cells/μl. This study suggests that the decreases in the fitness of drug-resistant viruses seen in vitro are clinically significant and that continuing drug therapy in the face of resistance may have utility in patients with few other therapeutic options. However, the possibility that many of the isolates in this study retained some degree of susceptibility to one or more drugs in treatment regimens that were used, cannot be excluded.
Quasispecies Nature of HIV-1
The inability to reliably detect minor drug-resistant HIV-1 variants is a recognized limitation of HIV-1 drug susceptibility testing using either genotypic or phenotypic methods. This is particularly troublesome in patients with complicated treatment histories or in patients who have discontinued one or more antiretroviral drugs (84, 395). To maximize the likelihood that a sequence will identify mutations present within the virus population of a patient, it is important to obtain plasma samples for resistance testing before stopping or changing antiretroviral drugs and to consider a patient's treatment history when interpreting the results of resistance testing.
In some patients, the treatment history can be used to infer the presence of archived drug resistance mutations. For example, if a patient previously received lamivudine as part of an incompletely suppressive treatment regimen, it is likely that M184V exists within the virus population of that patient even if it is not detected at the time of genotyping. The same principle would apply to patients who received NNRTIs as part of an incompletely suppressive treatment regimen; however, in this situation, it would not be possible to know specifically which NNRTI mutations are likely to be archived. In contrast, patients receiving lamivudine and NNRTIs as part of completely suppressive treatment regimens are not expected to harbor variants resistant to these drugs.
If a patient once harbored drug-resistant variants, these variants may persist at low levels in latently infected cells even if a subsequent treatment regimen brings about complete virus suppression (103, 146, 163, 245, 411). In patients in whom previous resistance tests have documented extensive drug resistance, the clinical usefulness of repeated resistance testing is likely to be minimal, because many resistant variants selected by previous treatment regimens will go undetected in future tests, yet are likely to emerge during attempts at salvage therapy.
Cross-Resistance
Most mutations arising during drug therapy contribute resistance to multiple drugs within the same drug class. This is particularly problematic considering that there are just three drug classes and that combinations of drugs from at least two classes are usually required to achieve durable HIV-1 suppression. Genotypic assays frequently do not identify enough fully active non-cross-resistant drugs to completely block HIV-1 replication and many patients changing regimens because of virologic failure will have to use a regimen containing drugs that are partially compromised at the start of therapy.
Although cross-resistance is not a direct limitation of genotypic or phenotypic testing, it limits the utility of resistance testing particularly in heavily treated patients. Nonetheless, resistance assays have a role even in heavily treated patients because they provide prognostic data and help avoid unnecessary drugs. Rather than including fully active drugs, salvage therapy in heavily treated patients will have to include drug combinations that exploit antagonistic mutational interactions or generate high in vivo drug levels.
Acknowledgments
I am supported by NIH grants AI-27666-12 and AI46148-01 and received unrestricted educational grants from various pharmaceutical and diagnostic companies for maintenance of the HIV RT and Protease Sequence Database. A list of the companies who have supported the database can be found at http://hivdb.stanford.edu/hiv/acknowledgments.html.
REFERENCES
- 1.Ala, P. J., E. E. Huston, R. M. Klabe, P. K. Jadhav, P. Y. Lam, and C. H. Chang. 1998. Counteracting HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with XV638 and SD146, cyclic urea amides with broad specificities. Biochemistry 37:15042-15049. [DOI] [PubMed] [Google Scholar]
- 2.Ala, P. J., E. E. Huston, R. M. Klabe, D. D. McCabe, J. L. Duke, C. J. Rizzo, B. D. Korant, R. J. DeLoskey, P. Y. Lam, C. N. Hodge, and C. H. Chang. 1997. Molecular basis of HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with cyclic urea inhibitors. Biochemistry 36:1573-1580. [DOI] [PubMed] [Google Scholar]
- 3.Albrecht, M. A., R. J. Bosch, S. M. Hammer, S. H. Liou, H. Kessler, M. F. Para, J. Eron, H. Valdez, M. Dehlinger, and D. A. Katzenstein. 2001. Nelfinavir, efavirenz, or both after the failure of nucleoside treatment of HIV infection. N. Engl. J. Med. 345:398-407. [DOI] [PubMed] [Google Scholar]
- 3a.Alexander, C. S., W. Dong, K. Chan, N. Jahnke, M. V. O’Shaughnessy, T. Mo, M. A. Piaseczny, J. S. Montaner, and P. R. Harrigan. 2001. HIV protease and reverse transcriptase variation and therapy outcome in antiretroviral-naive individuals from a large North American cohort. AIDS 15:601-617. [DOI] [PubMed] [Google Scholar]
- 4.Archer, R. H., C. Dykes, P. Gerondelis, A. Lloyd, P. Fay, R. C. Reichman, R. A. Bambara, and L. M. Demeter. 2000. Mutants of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase resistant to nonnucleoside reverse transcriptase inhibitors demonstrate altered rates of RNase H cleavage that correlate with HIV-1 replication fitness in cell culture. J. Virol. 74:8390-8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arion, D., G. Borkow, Z. Gu, M. A. Wainberg, and M. A. Parniak. 1996. The K65R mutation confers increased DNA polymerase processivity to HIV-1 reverse transcriptase. J. Biol. Chem. 271:19860-19864. [DOI] [PubMed] [Google Scholar]
- 6.Arion, D., N. Kaushik, S. McCormick, G. Borkow, and M. A. Parniak. 1998. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry 37:15908-15917. [DOI] [PubMed] [Google Scholar]
- 7.Arion, D., N. Sluis-Cremer, and M. A. Parniak. 2000. Mechanism by which phosphonoformic acid resistance mutations restore 3′-azido-3′-deoxythymidine (AZT) sensitivity to AZT-resistant HIV-1 reverse transcriptase. J. Biol. Chem. 275:9251-9255. [DOI] [PubMed] [Google Scholar]
- 8.Arts, E. J., M. E. Quinones-Mateu, J. L. Albright, J. P. Marois, C. Hough, Z. Gu, and M. A. Wainberg. 1998. 3′-Azido-3′-deoxythymidine (AZT) mediates cross-resistance to nucleoside analogs in the case of AZT-resistant human immunodeficiency virus type 1 variants. J. Virol. 72:4858-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atkinson, B., J. Isaacson, M. Knowles, E. Mazabel, and A. K. Patick. 2000. Correlation between human immunodeficiency virus genotypic resistance and virologic response in patients receiving nelfinavir monotherapy or nelfinavir with lamivudine and zidovudine. J. Infect. Dis. 182:420-427. [DOI] [PubMed] [Google Scholar]
- 10.Reference deleted.
- 11.Bacheler, L. T., E. D. Anton, P. Kudish, D. Baker, J. Bunville, K. Krakowski, L. Bolling, M. Aujay, X. V. Wang, D. Ellis, M. F. Becker, A. L. Lasut, H. J. George, D. R. Spalding, G. Hollis, and K. Abremski. 2000. Human immunodeficiency virus type 1 mutations selected in patients failing efavirenz combination therapy. Antimicrob. Agents Chemother. 44:2475-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Bacheler, L., S. Jeffrey, G. Hanna, R. D’Aquila, L. Wallace, K. Logue, B. Cordova, K. Hertogs, B. Larder, R. Buckery, D. Baker, K. Gallagher, H. Scarnati, R. Tritch, and C. Rizzo. 2001. Genotypic correlates of phenotypic resistance to efavirenz in virus isolates from patients failing nonnucleoside reverse transcriptase inhibitor therapy. J. Virol. 75:4999-5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Back, N. K., and B. Berkhout. 1997. Limiting deoxynucleoside triphosphate concentrations emphasize the processivity defect of lamivudine-resistant variants of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 41:2484-2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Back, N. K., M. Nijhuis, W. Keulen, C. A. Boucher, B. O. Oude Essink, A. B. van, Kuilenburg, A. H. van Gennip, and B. Berkhout. 1996. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 15:4040-4049. [PMC free article] [PubMed] [Google Scholar]
- 14.Back, N. K., A. van Wijk, D. Remmerswaal, M. van Monfort, M. Nijhuis, R. Schuurman, and C. A. Boucher. 2000. In-vitro tipranavir susceptibility of HIV-1 isolates with reduced susceptibility to other protease inhibitors. AIDS 14:101-102. [DOI] [PubMed] [Google Scholar]
- 15.Baldwin, E. T., T. N. Bhat, S. Gulnik, B. Liu, I. A. Topol, Y. Kiso, T. Mimoto, H. Mitsuya, and J. W. Erickson. 1995. Structure of HIV-1 protease with KNI-272, a tight-binding transition-state analog containing allophenylnorstatine. Structure 3:581-590. [DOI] [PubMed] [Google Scholar]
- 16.Balotta, C., A. Berlusconi, A. Pan, M. Violin, C. Riva, M. C. Colombo, A. Gori, L. Papagno, S. Corvasce, R. Mazzucchelli, G. Facchi, R. Velleca, G. Saporetti, M. Galli, S. Rusconi, and M. Moroni. 2000. Prevalence of transmitted nucleoside analogue-resistant HIV-1 strains and pre-existing mutations in pol reverse transcriptase and protease region: outcome after treatment in recently infected individuals. Antivir. Ther. 5:7-14. [PubMed] [Google Scholar]
- 17.Balzarini, J., A. Karlsson, V. V. Sardana, E. A. Emini, M. J. Camarasa, and E. De Clercq. 1994. Human immunodeficiency virus 1 (HIV-1)-specific reverse transcriptase (RT) inhibitors may suppress the replication of specific drug-resistant (E138K)RT HIV-1 mutants or select for highly resistant (Y181C→C181I)RT HIV-1 mutants. Proc. Natl. Acad. Sci. USA 91:6599-6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balzarini, J., H. Pelemans, R. Esnouf, and E. De Clercq. 1998. A novel mutation (F227L) arises in the reverse transcriptase of human immunodeficiency virus type 1 on dose-escalating treatment of HIV type 1-infected cell cultures with the nonnucleoside reverse transcriptase inhibitor thiocarboxanilide UC-781. AIDS Res. Hum. Retroviruses 14:255-260. [DOI] [PubMed] [Google Scholar]
- 19.Baxter, J. D., D. L. Mayers, D. N. Wentworth, J. D. Neaton, M. L. Hoover, M. A. Winters, S. B. Mannheimer, M. A. Thompson, D. I. Abrams, B. J. Brizz, J. P. Ioannidis, and T. C. Merigan. 2000. A randomized study of antiretroviral management based on plasma genotypic antiretroviral resistance testing in patients failing therapy. CPCRA 046 Study Team for the Terry Beirn Community Programs for Clinical Research on AIDS. AIDS 14:F83-F93. [DOI] [PubMed] [Google Scholar]
- 20.Belec, L., C. Piketty, A. Si-Mohamed, C. Goujon, M. C. Hallouin, S. Cotigny, L. Weiss, and M. D. Kazatchkine. 2000. High levels of drug-resistant human immunodeficiency virus variants in patients exhibiting increasing CD4+ T cell counts despite virologic failure of protease inhibitor-containing antiretroviral combination therapy. J. Infect. Dis. 181:1808-1812. [DOI] [PubMed] [Google Scholar]
- 20a.Benson, C. A., S. G. Deeks, S. C. Brun, R. M. Gulick, J. J. Eron, H. A. Kessler, R. L. Murphy, C. Hicks, M. King, D. Wheeler, J. Feinberg, R. Stryker, P. E. Sax, S. Riddler, M. Thompson, K. Real, A. Hsu, D. Kempf, A. J. Japour, and E. Sun. 2002. Safety and antiviral activity at 48 weeks of lopinavir/ritonavir plus nevirapine and 2 nucleoside reverse-transcriptase inhibitors in human immunodeficiency virus type 1-infected protease inhibitor-experienced patients. J. Infect. Dis. 185:599-607. [DOI] [PubMed] [Google Scholar]
- 21.Bleiber, G., M. Munoz, A. Ciuffi, P. Meylan, and A. Telenti. 2001. Individual contributions of mutant protease and reverse transcriptase to viral infectivity, replication, and protein maturation of antiretroviral drug-resistant human immunodeficiency virus type 1. J. Virol. 75:3291-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reference deleted.
- 23.Boden, D., A. Hurley, L. Zhang, Y. Cao, Y. Guo, E. Jones, J. Tsay, J. Ip, C. Farthing, K. Limoli, N. Parkin, and M. Markowitz. 1999. HIV-1 drug resistance in newly infected individuals. JAMA 282:1135-1141. [DOI] [PubMed] [Google Scholar]
- 24.Boden, D., and M. Markowitz. 1998. Resistance to human immunodeficiency virus type 1 protease inhibitors. Antimicrob. Agents Chemother. 42:2775-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bossi, P., M. Mouroux, A. Yvon, F. Bricaire, H. Agut, J. M. Huraux, C. Katlama, and V. Calvez. 1999. Polymorphism of the human immunodeficiency virus type 1 (HIV-1) protease gene and response of HIV-1-infected patients to a protease inhibitor. J. Clin. Microbiol. 37:2910-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boucher, C. A., N. Cammack, P. Schipper, R. Schuurman, P. Rouse, M. A. Wainberg, and J. M. Cameron. 1993. High-level resistance to (-) enantiomeric 2′-deoxy-3′-thiacytidine in vitro is due to one amino acid substitution in the catalytic site of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 37:2231-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boucher, C. A., E. O'Sullivan, J. W. Mulder, C. Ramautarsing, P. Kellam, G. Darby, J. M. Lange, J. Goudsmit, and B. A. Larder. 1992. Ordered appearance of zidovudine resistance mutations during treatment of 18 human immunodeficiency virus-positive subjects. J. Infect. Dis. 165:105-110. [DOI] [PubMed] [Google Scholar]
- 28.Boyer, P. L., and S. H. Hughes. 1995. Analysis of mutations at position 184 in reverse transcriptase of human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 39:1624-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boyer, P. L., S. G. Sarafianos, E. Arnold, and S. H. Hughes. 2001. Selective excision of aztmp by drug-resistant human immunodeficiency virus reverse transcriptase. J. Virol. 75:4832-4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Briones, C., M. Perez-Olmeda, C. Rodriguez, J. del Romero, K. Hertogs, and V. Soriano. 2001. Primary genotypic and phenotypic HIV-1 drug resistance in recent seroconverters in Madrid. J. Acquir. Immune Defic. Syndr. 26:145-150. [DOI] [PubMed] [Google Scholar]
- 31.Brown, A. J. 1997. Analysis of HIV-1 env gene sequences reveals evidence for a low effective number in the viral population. Proc. Natl. Acad. Sci. USA 94:1862-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reference deleted.
- 33.Brown, A. J., H. M. Precious, J. M. Whitcomb, J. K. Wong, M. Quigg, W. Huang, E. S. Daar, R. T. D'Aquila, P. H. Keiser, E. Connick, N. S. Hellmann, C. J. Petropoulos, D. D. Richman, and S. J. Little. 2000. Reduced susceptibility of human immunodeficiency virus type 1 (HIV-1) from patients with primary HIV infection to nonnucleoside reverse transcriptase inhibitors is associated with variation at novel amino acid sites. J. Virol. 74:10269-10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown, A. J., and D. D. Richman. 1997. HIV-1: gambling on the evolution of drug resistance? Nat. Med. 3:268-271. [DOI] [PubMed] [Google Scholar]
- 35.Reference deleted.
- 36.Reference deleted.
- 37.Byrnes, V. W., E. A. Emini, W. A. Schleif, J. H. Condra, C. L. Schneider, W. J. Long, J. A. Wolfgang, D. J. Graham, L. Gotlib, and A. J. Schlabach. 1994. Susceptibilities of human immunodeficiency virus type 1 enzyme and viral variants expressing multiple resistance-engendering amino acid substitutions to reserve transcriptase inhibitors. Antimicrob. Agents Chemother. 38:1404-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Byrnes, V. W., V. V. Sardana, W. A. Schleif, J. H. Condra, J. A. Waterbury, J. A. Wolfgang, W. J. Long, C. L. Schneider, A. J. Schlabach, and B. S. Wolanski. 1993. Comprehensive mutant enzyme and viral variant assessment of human immunodeficiency virus type 1 reverse transcriptase resistance to nonnucleoside inhibitors. Antimicrob. Agents Chemother. 37:1576-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caliendo, A. M., A. Savara, D. An, K. DeVore, J. C. Kaplan, and R. T. D'Aquila. 1996. Effects of zidovudine-selected human immunodeficiency virus type 1 reverse transcriptase amino acid substitutions on processive DNA synthesis and viral replication. J. Virol. 70:2146-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Calvez, V., I. Cohen-Codar, A. G. Marcelin, D. Descamps, C. Tamalet, J. Ritter, M. Segondy, H. Peigue-Lafeuille, F. Brun-Vezinet, E. Guillevic, J. Isaacson, R. Rode, B. Bernstein, E. Sun, D. Kempf, and J. P. Chauvin. 2001. Identification of individual mutations in HIV protease associated with virological response to lopinavir/ritonavir therapy. Antivir. Ther. 6:64. [Google Scholar]
- 41.Reference deleted.
- 42.Carrillo, A., K. D. Stewart, H. L. Sham, D. W. Norbeck, W. E. Kohlbrenner, J. M. Leonard, D. J. Kempf, and A. Molla. 1998. In vitro selection and characterization of human immunodeficiency virus type 1 variants with increased resistance to ABT-378, a novel protease inhibitor. J. Virol. 72:7532-7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Casado, J. L., F. Dronda, K. Hertogs, R. Sabido, A. Antela, P. Marti-Belda, P. Dehertogh, and S. Moreno. 2001. Efficacy, tolerance, and pharmacokinetics of the combination of stavudine, nevirapine, nelfinavir, and saquinavir as salvage regimen after ritonavir or indinavir failure. AIDS Res. Hum. Retrovirus. 17:93-98. [DOI] [PubMed] [Google Scholar]
- 44.Casado, J. L., K. Hertogs, L. Ruiz, F. Dronda, A. Van Cauwenberge, A. Arno, I. Garcia-Arata, S. Bloor, A. Bonjoch, J. Blazquez, B. Clotet, and B. Larder. 2000. Non-nucleoside reverse transcriptase inhibitor resistance among patients failing a nevirapine plus protease inhibitor-containing regimen. AIDS 14:F1-7. [DOI] [PubMed] [Google Scholar]
- 45.Chan, D. C., C. T. Chutkowski, and P. S. Kim. 1998. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proc. Natl. Acad. Sci. USA 95:15613-15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen, Z., Y. Li, E. Chen, D. L. Hall, P. L. Darke, C. Culberson, J. A. Shafer, and L. C. Kuo. 1994. Crystal structure at 1.9-A resolution of human immunodeficiency virus (HIV) II protease complexed with L-735,524, an orally bioavailable inhibitor of the HIV proteases. J. Biol. Chem. 269:26344-26348. [PubMed] [Google Scholar]
- 47.Chen, Z., Y. Li, H. B. Schock, D. Hall, E. Chen, and L. C. Kuo. 1995. Three-dimensional structure of a mutant HIV-1 protease displaying cross-resistance to all protease inhibitors in clinical trials. J. Biol. Chem. 270:21433-21436. [DOI] [PubMed] [Google Scholar]
- 47a.Cingolani, A., A. Antinori, M. G. Rizzo, R. Murri, A. Ammassari, F. Baldini, S. Di Giambenedetto, R. Cauda, and A. De Luca. 2002. Usefulness of monitoring HIV drug resistance and adherence in individuals failing highly active antiretroviral therapy: a randomized study (ARGENTA). AIDS 16:369-379. [DOI] [PubMed] [Google Scholar]
- 48.Clarke, J. R., S. Kaye, A. G. Babiker, M. H. Hooker, R. Tedder, and J. N. Weber. 2000. Comparison of a point mutation assay with a line probe assay for the detection of the major mutations in the HIV-1 reverse transcriptase gene associated with reduced susceptibility to nucleoside analogues. J. Virol. Methods 88:117-124. [DOI] [PubMed] [Google Scholar]
- 49.Coakley, E. P., J. M. Gillis, and S. M. Hammer. 2000. Phenotypic and genotypic resistance patterns of HIV-1 isolates derived from individuals treated with didanosine and stavudine. AIDS 14:F9-F15. [DOI] [PubMed] [Google Scholar]
- 50.Coffin, J. M. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267:483-489. [DOI] [PubMed] [Google Scholar]
- 50a.Cohen, C J., S. Hunt, M. Sension, C. Farthing, M. Conant, S. Jacobson, J. Nadler, W. Verbiest, K. Hertogs, M. Ames, A. R. Rinehart, and N. M. Graham. 2002. A randomized trial assessing the impact of phenotypic resistance testing on antiretroviral therapy. AIDS 16:579-588. [DOI] [PubMed] [Google Scholar]
- 51.Reference deleted.
- 52.Reference deleted.
- 53.Reference deleted.
- 54.Condra, J. H., D. J. Holder, W. A. Schleif, O. M. Blahy, R. M. Danovich, L. J. Gabryelski, D. J. Graham, D. Laird, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, T. Yang, J. A. Chodakewitz, P. J. Deutsch, R. Y. Leavitt, F. E. Massari, J. W. Mellors, K. E. Squires, R. T. Steigbigel, H. Teppler, and E. A. Emini. 1996. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J. Virol. 70:8270-8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Condra, J. H., C. J. Petropoulos, R. Ziermann, W. A. Schleif, M. Shivaprakash, and E. A. Emini. 2000. Drug resistance and predicted virologic responses to human immunodeficiency virus type 1 protease inhibitor therapy. J. Infect. Dis. 182:758-765. [DOI] [PubMed] [Google Scholar]
- 56.Condra, J. H., W. A. Schleif, O. M. Blahy, L. J. Gabryelski, D. J. Graham, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, and M. Shivaprakash. 1995. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature 374:569-571. [DOI] [PubMed] [Google Scholar]
- 57.Conway, B., M. A. Wainberg, D. Hall, M. Harris, P. Reiss, D. Cooper, S. Vella, R. Curry, P. Robinson, J. M. Lange, and J. S. Montaner. 2001. Development of drug resistance in patients receiving combinations of zidovudine, didanosine and nevirapine. AIDS 15:1269-1274. [DOI] [PubMed] [Google Scholar]
- 58.Cornelissen, M., d. B. van, F. Zorgdrager, V. Lukashov, and J. Goudsmit. 1997. pol gene diversity of five human immunodeficiency virus type 1 subtypes: evidence for naturally occurring mutations that contribute to drug resistance, limited recombination patterns, and common ancestry for subtypes B and D. J. Virol. 71:6348-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reference deleted.
- 60.Cote, H. C., Z. L. Brumme, and P. R. Harrigan. 2001. Human immunodeficiency virus type 1 protease cleavage site mutations associated with protease inhibitor cross-resistance selected by indinavir, ritonavir, and/or saquinavir. J. Virol. 75:589-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Craig, C., E. Race, J. Sheldon, L. Whittaker, S. Gilbert, A. Moffatt, J. Rose, S. Dissanayeke, G. W. Chirn, I. B. Duncan, and N. Cammack. 1998. HIV protease genotype and viral sensitivity to HIV protease inhibitors following saquinavir therapy. AIDS 12:1611-1618. [DOI] [PubMed] [Google Scholar]
- 61a.Cunningham, S., B. Ank, D. Lewis, W. Lu, M. Wantman, J. A. Dileanis, J. B. Jackson, P. Palumbo, P. Krogstad, and S. H. Eshleman. 2001. Performance of the applied biosystems ViroSeq human immunodeficiency virus type 1 (HIV-1) genotyping system for sequence-based analysis of HIV-1 in pediatric plasma samples. J. Clin. Microbial. 39:1254-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.D'Aquila, R. T. 2000. Limits of resistance testing. Antivir. Ther. 5:71-76. [PubMed] [Google Scholar]
- 63.de Jong, J. J., J. Goudsmit, V. V. Lukashov, M. E. Hillebrand, E. Baan, R. Huismans, S. A. Danner, J. H. ten Veen, F. de Wolf, and S. Jurriaans. 1999. Insertion of two amino acids combined with changes in reverse transcriptase containing tyrosine-215 of HIV-1 resistant to multiple nucleoside analogs. AIDS 13:75-80. [DOI] [PubMed] [Google Scholar]
- 64.de Jong, M. D., J. Veenstra, N. I. Stilianakis, R. Schuurman, J. M. Lange, R. J. de Boer, and C. A. Boucher. 1996. Host-parasite dynamics and outgrowth of virus containing a single K70R amino acid change in reverse transcriptase are responsible for the loss of human immunodeficiency virus type 1 RNA load suppression by zidovudine. Proc. Natl. Acad. Sci. USA 93:5501-5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reference deleted.
- 66.Reference deleted.
- 67.de Ronde, A., M. van Dooren, L. van Der Hoek, D. Bouwhuis, E. de Rooij, B. van Gemen, R. de Boer, and J. Goudsmit. 2001. Establishment of new transmissible and drug-sensitive human immunodeficiency virus type 1 wild types due to transmission of nucleoside analogue-resistant virus. J. Virol. 75:595-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reference deleted.
- 69.Deeks, S. G. 2001. Nonnucleoside reverse transcriptase inhibitor resistance. J. Acquir. Immune Defic. Syndr. 26[Suppl. 1]:S25-33. [DOI] [PubMed] [Google Scholar]
- 70.Deeks, S. G., R. M. Grant, G. W. Beatty, C. Horton, J. Detmer, and S. Eastman. 1998. Activity of a ritonavir plus saquinavir-containing regimen in patients with virologic evidence of indinavir or ritonavir failure. AIDS 12:F97-F102. [DOI] [PubMed] [Google Scholar]
- 71.Deeks, S. G., T. Wrin, T. Liegler, R. Hoh, M. Hayden, J. D. Barbour, N. S. Hellmann, C. J. Petropoulos, J. M. McCune, M. K. Hellerstein, and R. M. Grant. 2001. Virologic and immunologic consequences of discontinuing combination antiretroviral-drug therapy in HIV-infected patients with detectable viremia. N. Engl. J. Med. 344:472-480. [DOI] [PubMed] [Google Scholar]
- 72.DeGruttola, V., L. Dix, R. D'Aquila, D. Holder, A. Phillips, M. Ait-Khaled, J. Baxter, P. Clevenbergh, S. Hammer, R. Harrigan, D. Katzenstein, R. Lanier, M. Miller, M. Para, S. Yerly, A. Zolopa, J. Murray, A. Patick, V. Miller, S. Castillo, L. Pedneault, and J. Mellors. 2000. The relation between baseline HIV drug resistance and response to antiretroviral therapy: reanalysis of retrospective and prospective studies using a standardized data analysis plan. Antivir. Ther. 5:41-48. [DOI] [PubMed] [Google Scholar]
- 73.Delassus, S., R. Cheynier, and S. Wain-Hobson. 1991. Evolution of human immunodeficiency virus type 1 nef and long terminal repeat sequences over 4 years in vivo and in vitro. J. Virol. 65:225-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Delaugerre, C., M. Mouroux, A. Yvon-Groussin, A. Simon, F. Angleraud, J. M. Huraux, H. Agut, C. Katlama, and V. Calvez. 2001. Prevalence and conditions of selection of E44D/A and V118I human immunodeficiency virus type 1 reverse transcriptase mutations in clinical practice. Antimicrob. Agents Chemother. 45:946-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Demeter, L., and R. Haubrich. 2001. Phenotypic and genotypic resistance assays: methodology, reliability, and interpretations. J. Acquir. Immune Defic. Syndr. 26[Suppl. 1]:S3-9. [DOI] [PubMed] [Google Scholar]
- 76.Demeter, L. M., R. D'Aquila, O. Weislow, E. Lorenzo, A. Erice, J. Fitzgibbon, R. Shafer, D. Richman, T. M. Howard, Y. Zhao, E. Fisher, D. Huang, D. Mayers, S. Sylvester, M. Arens, K. Sannerud, S. Rasheed, V. Johnson, D. Kuritzkes, P. Reichelderfer, and A. Japour. 1998. Interlaboratory concordance of DNA sequence analysis to detect reverse transcriptase mutations in HIV-1 proviral DNA. ACTG Sequencing Working group. AIDS Clinical Trials group. J. Virol. Med. 75:93-104. [DOI] [PubMed] [Google Scholar]
- 77.Demeter, L. M., T. Nawaz, G. Morse, R. Dolin, A. Dexter, P. Gerondelis, and R. C. Reichman. 1995. Development of zidovudine resistance mutations in patients receiving prolonged didanosine monotherapy. J. Infect. Dis. 172:1480-1485. [DOI] [PubMed] [Google Scholar]
- 78.Demeter, L. M., R. W. Shafer, P. M. Meehan, J. Holden-Wiltse, M. A. Fischl, W. W. Freimuth, M. F. Para, and R. C. Reichman. 2000. Delavirdine susceptibilities and associated reverse transcriptase mutations in human immunodeficiency virus type 1 isolates from patients in a phase I/II trial of delavirdine monotherapy (ACTG 260). Antimicrob. Agents Chemother. 44:794-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Descamps, D., C. Apetrei, G. Collin, F. Damond, F. Simon, and F. Brun-Vezinet. 1998. Naturally occurring decreased susceptibility of HIV-1 subtype G to protease inhibitors. AIDS 12:1109-1111. [PubMed] [Google Scholar]
- 80.Descamps, D., G. Collin, F. Letourneur, C. Apetrei, F. Damond, I. Loussert-Ajaka, F. Simon, S. Saragosti, and F. Brun-Vezinet. 1997. Susceptibility of human immunodeficiency virus type 1 group O isolates to antiretroviral agents: in vitro phenotypic and genotypic analyses. J. Virol. 71:8893-8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Descamps, D., P. Flandre, V. Calvez, G. Peytavin, V. Meiffredy, G. Collin, C. Delaugerre, S. Robert-Delmas, B. Bazin, J. P. Aboulker, G. Pialoux, F. Raffi, and F. Brun-Vezinet. 2000. Mechanisms of virologic failure in previously untreated HIV-infected patients from a trial of induction-maintenance therapy. Trilege (Agence Nationale de Recherches sur le SIDA 072) Study Team. JAMA 283:205-211. [DOI] [PubMed] [Google Scholar]
- 82.Descamps, D., B. Masquelier, J. P. Mamet, C. Calvez, A. Ruffault, F. Telles, A. Goetschel, P. M. Girard, F. Brun-Vezinet, and D. Costagliola. 2001. A genotypic sensitivity score for amprenavir based genotype at baseline and virological response. Antivir. Ther. 6:103. [Google Scholar]
- 83.Devereux, H. L., V. C. Emery, M. A. Johnson, and C. Loveday. 2001. Replicative fitness in vivo of HIV-1 variants with multiple drug resistance-associated mutations. J. Med. Virol. 65:218-224. [DOI] [PubMed] [Google Scholar]
- 84.Devereux, H. L., M. Youle, M. A. Johnson, and C. Loveday. 1999. Rapid decline in detectability of HIV-1 drug resistance mutations after stopping therapy. AIDS 13:F123-F127. [PubMed] [Google Scholar]
- 85.Reference deleted.
- 86.Reference deleted.
- 87.Doyon, L., G. Croteau, D. Thibeault, F. Poulin, L. Pilote, and D. Lamarre. 1996. Second locus involved in human immunodeficiency virus type 1 resistance to protease inhibitors. J. Virol. 70:3763-3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dronda, F., J. L. Casado, S. Moreno, K. Hertogs, I. Garcia-Arata, A. Antela, M. J. Perez-Elias, L. Ruiz, and B. Larder. 2001. Phenotypic cross-resistance to nelfinavir: the role of prior antiretroviral therapy and the number of mutations in the protease gene. AIDS Res. Hum. Retrovirus. 17:211-215. [DOI] [PubMed] [Google Scholar]
- 89.Drosopoulos, W. C., and V. R. Prasad. 1998. Increased misincorporation fidelity observed for nucleoside analog resistance mutations M184V and E89G in human immunodeficiency virus type 1 reverse transcriptase does not correlate with the overall error rate measured in vitro. J. Virol. 72:4224-4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89a.Duan, C., V. Poticha, T. C. Stoeckli, C. J. Petropoulos, J. m. Whitcomb, C. S. McHenry, and N. R. Kuritzkes. 2001. Inhibition of purified recombinant reverse transcriptase from wild-type and zidovudine-resistant clinical isolates of human immunodeficiency virus type 1 by zidovudine, stavudine, and lamivudine triphosphates. J. Infect. Dis. 184:1336-1340. [DOI] [PubMed] [Google Scholar]
- 90.Reference deleted.
- 91.Dueweke, T. J., T. Pushkarskaya, S. M. Poppe, S. M. Swaney, J. Q. Zhao, I. S. Chen, M. Stevenson, and W. G. Tarpley. 1993. A mutation in reverse transcriptase of bis(heteroaryl)piperazine-resistant human immunodeficiency virus type 1 that confers increased sensitivity to other nonnucleoside inhibitors. Proc. Natl. Acad. Sci. USA 90:4713-4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Durant, J., P. Clevenbergh, P. Halfon, P. Delgiudice, S. Porsin, P. Simonet, N. Montagne, C. A. Boucher, J. M. Schapiro, and P. Dellamonica. 1999. Drug resistance genotyping in HIV-1 therapy: the VIRADAPT randomised controlled trial. Lancet 353:2195-2199. [DOI] [PubMed] [Google Scholar]
- 92a.Duval, X., C. Lamotte, E. Race, D. Descamps, F. Damond, F. Clavel, C. Leport, G. Peytavin, and J. L. Vilde. 2002. Amprenavir inhibitory quotient and virological response in human immunodeficiency virus-infected patients on an amprenavir-containing salvage regimen without or with ritonavir. Antimicrob. Agents. Chemother. 46:570-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Duwe, S., M. Brunn, D. Altmann, O. Hamouda, B. Schmidt, H. Walter, G. Pauli, and C. Kucherer. 2001. Frequency of genotypic and phenotypic drug-resistant HIV-1 among therapy-naive patients of the german seroconverter study. J. Acquir. Immune Defic. Syndr. 26:266-273. [DOI] [PubMed] [Google Scholar]
- 94.Eckert, D. M., V. N. Malashkevich, L. H. Hong, P. A. Carr, and P. S. Kim. 1999. Inhibiting HIV-1 entry: discovery of D-peptide inhibitors that target the gp41 coiled-coil pocket. Cell 99:103-115. [DOI] [PubMed] [Google Scholar]
- 95.Emini, E. A., D. J. Graham, L. Gotlib, J. H. Condra, V. W. Byrnes, and W. A. Schleif. 1993. HIV and multidrug resistance. Nature 364:679.. [DOI] [PubMed] [Google Scholar]
- 96.Erali, M., S. Page, L. G. Reimer, and D. R. Hillyard. 2001. Human immunodeficiency virus type 1 drug resistance testing: a comparison of three sequence-based methods. J. Clin. Microbiol. 39:2157-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Erickson, J. W., S. V. Gulnik, and M. Markowitz. 1999. Protease inhibitors: resistance, cross-resistance, fitness and the choice of initial and salvage therapies. AIDS 13[Suppl. A]:S189-S204. [PubMed] [Google Scholar]
- 98.Eron, J. J., S. L. Benoit, J. Jemsek, R. D. MacArthur, J. Santana, J. B. Quinn, D. R. Kuritzkes, M. A. Fallon, and M. Rubin. 1995. Treatment with lamivudine, zidovudine, or both in HIV-positive patients with 200 to 500 CD4+ cells per cubic millimeter. North American HIV Working Party. N. Engl. J. Med. 333:1662-1669. [DOI] [PubMed] [Google Scholar]
- 98a.Eshleman, S. H., M. Mracna, L. A. Guay, M. Deseyve, S. Cunningham, M. Mirochnick, P. Musoke, T. Fleming, M. Glenn Fowler, L. M. Mofenson, F. Mmiro, and J. B. Jackson. 2001. Selection and fading of resistance mutations in women and infants receiving nevirapine to prevent HIV-1 vertical transmission (HIVNET 012). AIDS 15:1951-1957. [DOI] [PubMed] [Google Scholar]
- 99.Reference deleted.
- 100.Esnouf, R., J. Ren, C. Ross, Y. Jones, D. Stammers, and D. Stuart. 1995. Mechanism of inhibition of HIV-1 reverse transcriptase by non-nucleoside inhibitors. Nat. Struct. Biol. 2:303-308. [DOI] [PubMed] [Google Scholar]
- 101.EuroGuidelines group for HIV Resistance. 2001. Clinical and laboratory guidelines for the use of HIV-1 drug resistance testing as part of treatment management: recommendations for the European setting. AIDS 15:309-320. [DOI] [PubMed] [Google Scholar]
- 102.Falloon, J., S. Piscitelli, S. Vogel, B. Sadler, H. Mitsuya, M. F. Kavlick, K. Yoshimura, M. Rogers, S. LaFon, D. J. Manion, H. C. Lane, and H. Masur. 2000. Combination therapy with amprenavir, abacavir, and efavirenz in human immunodeficiency virus (HIV)-infected patients failing a protease-inhibitor regimen: pharmacokinetic drug interactions and antiviral activity. Clin. Infect. Dis. 30:313-318. [DOI] [PubMed] [Google Scholar]
- 103.Finzi, D., J. Blankson, J. D. Siliciano, J. B. Margolick, K. Chadwick, T. Pierson, K. Smith, J. Lisziewicz, F. Lori, C. Flexner, T. C. Quinn, R. E. Chaisson, E. Rosenberg, B. Walker, S. Gange, J. Gallant, and R. F. Siliciano. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5:512-517. [DOI] [PubMed] [Google Scholar]
- 104.Fitzgibbon, J. E., R. M. Howell, C. A. Haberzettl, S. J. Sperber, D. J. Gocke, and D. T. Dubin. 1992. Human immunodeficiency virus type 1 pol gene mutations which cause decreased susceptibility to 2′,3′-dideoxycytidine. Antimicrob. Agents Chemother. 36:153-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Reference deleted.
- 106.Frater, A. J., A. Beardall, K. Ariyoshi, D. Churchill, S. Galpin, J. R. Clarke, J. N. Weber, and M. O. McClure. 2001. Impact of baseline polymorphisms in RT and protease on outcome of highly active antiretroviral therapy in HIV-1-infected African patients. AIDS 15:1493-1502. [DOI] [PubMed] [Google Scholar]
- 107.Frost, S. D., M. Nijhuis, R. Schuurman, C. A. Boucher, and A. J. Brown. 2000. Evolution of lamivudine resistance in human immunodeficiency virus type 1-infected individuals: the relative roles of drift and selection. J. Virol. 74:6262-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fujiwara, T., A. Sato, M. el-Farrash, S. Miki, K. Abe, Y. Isaka, M. Kodama, Y. Wu, L. B. Chen, H. Harada, H. Sugimoto, M. Hatanaka, and Y. Hinuma. 1998. S-1153 inhibits replication of known drug-resistant strains of human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 42:1340-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gallego, O., A. Corral, and V. Soriano. 2001. Prevalence of the codon 333 mutation in pre-treated patients experiencing virologic failure. Antivir. Ther. 6[Suppl. 1]:S17-S18. [Google Scholar]
- 110.Gallego, O., C. de Mendoza, M. J. Perez-Elias, J. M. Guardiola, J. Pedreira, D. Dalmau, J. Gonzalez, A. Moreno, J. R. Arribas, A. Rubio, I. Garcia-Arata, M. Leal, P. Domingo, and V. Soriano. 2001. Drug resistance in patients experiencing early virological failure under a triple combination including indinavir. AIDS 15:1701-1706. [DOI] [PubMed] [Google Scholar]
- 111.Gao, W. Y., T. Shirasaka, D. G. Johns, S. Broder, and H. Mitsuya. 1993. Differential phosphorylation of azidothymidine, dideoxycytidine, and dideoxyinosine in resting and activated peripheral blood mononuclear cells. J. Clin. Investig. 91:2326-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gerondelis, P., R. H. Archer, C. Palaniappan, R. C. Reichman, P. J. Fay, R. A. Bambara, and L. M. Demeter. 1999. The P236L delavirdine-resistant human immunodeficiency virus type 1 mutant is replication defective and demonstrates alterations in both RNA 5′-end- and DNA 3′-end-directed RNase H activities. J. Virol. 73:5803-5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gong, Y. F., B. S. Robinson, R. E. Rose, C. Deminie, T. P. Spicer, D. Stock, R. J. Colonno, and P. F. Lin. 2000. In vitro resistance profile of the human immunodeficiency virus type 1 protease inhibitor BMS-232632. Antimicrob. Agents Chemother. 44:2319-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gonzales, M. J., R. N. Machekano, and R. W. Shafer. 2001. Human immunodeficiency virus type 1 reverse-transcriptase and protease subtypes: classification, amino acid mutation patterns, and prevalence in a Northern California clinic-based population. J. Infect. Dis. 184:998-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gotte, M., D. Arion, M. A. Parniak, and M. A. Wainberg. 2000. The M184V mutation in the reverse transcriptase of human immunodeficiency virus type 1 impairs rescue of chain-terminated DNA synthesis. J. Virol. 74:3579-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Goudsmit, J., R. A. de Ronde, E. de Rooij, and R. de Boer. 1997. Broad spectrum of in vivo fitness of human immunodeficiency virus type 1 subpopulations differing at reverse transcriptase codons 41 and 215. J. Virol. 71:4479-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Goudsmit, J., R. A. de Ronde, D. D. Ho, and A. S. Perelson. 1996. Human immunodeficiency virus fitness in vivo: calculations based on a single zidovudine resistance mutation at codon 215 of reverse transcriptase. J. Virol. 70:5662-5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Reference deleted.
- 119.Reference deleted.
- 120.Gu, Z., E. J. Arts, M. A. Parniak, and M. A. Wainberg. 1995. Mutated K65R recombinant reverse transcriptase of human immunodeficiency virus type 1 shows diminished chain termination in the presence of 2′,3′-dideoxycytidine 5′-triphosphate and other drugs. Proc. Natl. Acad. Sci. USA 92:2760-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gu, Z., Q. Gao, H. Fang, H. Salomon, M. A. Parniak, E. Goldberg, J. Cameron, and M. A. Wainberg. 1994. Identification of a mutation at codon 65 in the IKKK motif of reverse transcriptase that encodes human immunodeficiency virus resistance to 2′,3′-dideoxycytidine and 2′,3′-dideoxy-3′-thiacytidine. Antimicrob. Agents Chemother. 38:275-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gu, Z., Q. Gao, X. Li, M. A. Parniak, and M. A. Wainberg. 1992. Novel mutation in the human immunodeficiency virus type 1 reverse transcriptase gene that encodes cross-resistance to 2′,3′-dideoxyinosine and 2′,3′-dideoxycytidine. J. Virol. 66:7128-7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gulick, R. M., J. W. Mellors, D. Havlir, J. J. Eron, A. Meibohm, J. H. Condra, F. T. Valentine, D. McMahon, C. Gonzalez, L. Jonas, E. A. Emini, J. A. Chodakewitz, R. Isaacs, and D. D. Richman. 2000. 3-year suppression of HIV viremia with indinavir, zidovudine, and lamivudine. Ann. Intern. Med. 133:35-39. [DOI] [PubMed] [Google Scholar]
- 124.Gulnik, S. V., L. I. Suvorov, B. Liu, B. Yu, B. Anderson, H. Mitsuya, and J. W. Erickson. 1995. Kinetic characterization and cross-resistance patterns of HIV-1 protease mutants selected under drug pressure. Biochemistry 34:9282-9287. [DOI] [PubMed] [Google Scholar]
- 125.Gunthard, H. F., J. K. Wong, C. C. Ignacio, D. V. Havlir, and D. D. Richman. 1998. Comparative performance of high-density oligonucleotide sequencing and dideoxynucleotide sequencing of HIV type 1 pol from clinical samples. AIDS Res. Hum. Retroviruses 14:869-876. [DOI] [PubMed] [Google Scholar]
- 126.Hammer, S., M. Peeters, R. Harrigan, and B. Larder. 2001. Virtual phenotype is predictive of treatment failure in treatment-experienced patients. Antivir. Ther. 6:107. [Google Scholar]
- 127.Hammond, J., C. Calef, B. Larder, R. F. Schinazi, and J. Mellors. 1999. Mutations in retroviral genes associated with drug resistance. In C. L. Kuiken, B. Foley, B. H. Hahn, P. Marx, F. E. McCutchan, J. Mellors, J. I. Mullins, S. Wolanski, and B. Korber (ed.), Human retroviruses and AIDS: a compilation and analysis of nucleic and amino acid sequences. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 128.Hanna, G. J., and R. T. D'Aquila. 2001. Clinical use of genotypic and phenotypic drug resistance testing to monitor antiretroviral chemotherapy. Clin. Infect. Dis. 32:774-782. [DOI] [PubMed] [Google Scholar]
- 129.Hanna, G. J., V. A. Johnson, D. R. Kuritzkes, D. D. Richman, A. J. Brown, A. V. Savara, J. D. Hazelwood, and R. T. D'Aquila. 2000. Patterns of resistance mutations selected by treatment of human immunodeficiency virus type 1 infection with zidovudine, didanosine, and nevirapine. J. Infect. Dis. 181:904-911. [DOI] [PubMed] [Google Scholar]
- 130.Hanna, G. J., V. A. Johnson, D. R. Kuritzkes, D. D. Richman, J. Martinez-Picado, L. Sutton, J. D. Hazelwood, and R. T. D'Aquila. 2000. Comparison of sequencing by hybridization and cycle sequencing for genotyping of human immunodeficiency virus type 1 reverse transcriptase. J. Clin. Microbiol. 38:2715-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Reference deleted.
- 132.Harrigan, P. R., K. Hertogs, W. Verbiest, R. Pauwels, B. Larder, S. Kemp, S. Bloor, B. Yip, R. Hogg, C. Alexander, and J. S. Montaner. 1999. Baseline HIV drug resistance profile predicts response to ritonavir-saquinavir protease inhibitor therapy in a community setting. AIDS 13:1863-1871. [DOI] [PubMed] [Google Scholar]
- 133.Harrigan, P. R., I. Kinghorn, S. Bloor, S. D. Kemp, I. Najera, A. Kohli, and B. A. Larder. 1996. Significance of amino acid variation at human immunodeficiency virus type 1 reverse transcriptase residue 210 for zidovudine susceptibility. J. Virol. 70:5930-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Harrigan, P. R., C. Stone, P. Griffin, I. Najera, S. Bloor, S. Kemp, M. Tisdale, and B. Larder. 2000. Resistance profile of the human immunodeficiency virus type 1 reverse transcriptase inhibitor abacavir (1592U89) after monotherapy and combination therapy. CNA2001 Investigative group. J. Infect. Dis. 181:912-920. [DOI] [PubMed] [Google Scholar]
- 135.Harrigan, P. R., C. Van Den Eynde, and B. A. Larder. 2001. Quantitation of lopinavir resistance and cross-resistance and phenotypic contribution of mutations shared with other protease inhibitors. Antivir. Ther. 6:40. [Google Scholar]
- 136.Reference deleted.
- 137.Reference deleted.
- 138.Haubrich, R., and L. Demeter. 2001. Clinical utility of resistance testing: retrospective and prospective data supporting use and current recommendations. J. Acquir. Immune Defic. Syndr. 26[Suppl. 1]:S51-59. [DOI] [PubMed] [Google Scholar]
- 139.Haubrich, R., P. Keiser, C. Kemper, M. Witt, J. Leedom, D. Forthal, M. Leibowitz, J. Hwang, E. Seefried, J. A. McCutchan, N. Hellemann, and D. Richman. 2001. CCTG 575: a randomized, prospective study of phenotype testing versus standard of care for patients failing antiretroviral therapy. Antivir. Ther. 6:63.11417763 [Google Scholar]
- 140.Reference deleted.
- 141.Havlir, D. V., S. Eastman, A. Gamst, and D. D. Richman. 1996. Nevirapine-resistant human immunodeficiency virus: kinetics of replication and estimated prevalence in untreated patients. J. Virol. 70:7894-7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Havlir, D. V., N. S. Hellmann, C. J. Petropoulos, J. M. Whitcomb, A. C. Collier, M. S. Hirsch, P. Tebas, J. P. Sommadossi, and D. D. Richman. 2000. Drug susceptibility in HIV infection after viral rebound in patients receiving indinavir-containing regimens. JAMA 283:229-234. [DOI] [PubMed] [Google Scholar]
- 143.Havlir, D. V., I. C. Marschner, M. S. Hirsch, A. C. Collier, P. Tebas, R. L. Bassett, J. P. Ioannidis, M. K. Holohan, R. Leavitt, G. Boone, and D. D. Richman. 1998. Maintenance antiretroviral therapies in HIV infected patients with undetectable plasma HIV RNA after triple-drug therapy. AIDS Clinical Trials group Study 343 Team. N. Engl. J. Med. 339:1261-1268. [DOI] [PubMed] [Google Scholar]
- 144.Hecht, F. M., R. M. Grant, C. J. Petropoulos, B. Dillon, M. A. Chesney, H. Tian, N. S. Hellmann, N. I. Bandrapalli, L. Digilio, B. Branson, and J. O. Kahn. 1998. Sexual transmission of an HIV-1 variant resistant to multiple reverse-transcriptase and protease inhibitors. N. Engl. J. Med. 339:307-311. [DOI] [PubMed] [Google Scholar]
- 145.Henry, K., R. J. Wallace, P. C. Bellman, D. Norris, R. L. Fisher, L. L. Ross, Q. Liao, and M. S. Shaefer. 2001. Twice-daily triple nucleoside intensification treatment with lamivudine-zidovudine plus abacavir sustains suppression of human immunodeficiency virus type 1: results of the TARGET Study. J. Infect. Dis. 183:571-578. [DOI] [PubMed] [Google Scholar]
- 146.Hermankova, M., S. C. Ray, C. Ruff, M. Powell-Davis, R. Ingersoll, R. T. D'Aquila, T. C. Quinn, J. D. Siliciano, R. F. Siliciano, and D. Persaud. 2001. HIV-1 drug resistance profiles in children and adults with viral load of <50 copies/ml receiving combination therapy. JAMA 286:196-207. [DOI] [PubMed] [Google Scholar]
- 147.Hertogs, K., S. Bloor, V. De Vroey, C. van Den Eynde, P. Dehertogh, A. van Cauwenberge, M. Sturmer, T. Alcorn, S. Wegner, M. van Houtte, V. Miller, and B. A. Larder. 2000. A novel human immunodeficiency virus type 1 reverse transcriptase mutational pattern confers phenotypic lamivudine resistance in the absence of mutation 184V. Antimicrob. Agents Chemother. 44:568-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hertogs, K., S. Bloor, S. D. Kemp, C. Van den Eynde, T. M. Alcorn, R. Pauwels, M. Van Houtte, S. Staszewski, V. Miller, and B. A. Larder. 2000. Phenotypic and genotypic analysis of clinical HIV-1 isolates reveals extensive protease inhibitor cross-resistance: a survey of over 6000 samples. AIDS 14:1203-1210. [DOI] [PubMed] [Google Scholar]
- 149.Hertogs, K., M. P. de Bethune, V. Miller, T. Ivens, P. Schel, A. Van Cauwenberge, C. Van Den Eynde, V. Van Gerwen, H. Azijn, M. Van Houtte, F. Peeters, S. Staszewski, M. Conant, S. Bloor, S. Kemp, B. Larder, and R. Pauwels. 1998. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob. Agents Chemother. 42:269-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hirsch, M. S., F. Brun-Vezinet, R. T. D'Aquila, S. M. Hammer, V. A. Johnson, D. R. Kuritzkes, C. Loveday, J. W. Mellors, B. Clotet, B. Conway, L. M. Demeter, S. Vella, D. M. Jacobsen, and D. D. Richman. 2000. Antiretroviral drug resistance testing in adult HIV-1 infection: recommendations of an International AIDS Society-USA Panel. JAMA 283:2417-2426. [DOI] [PubMed] [Google Scholar]
- 151.Hizi, A., R. Tal, M. Shaharabany, M. J. Currens, M. R. Boyd, S. H. Hughes, and J. B. McMahon. 1993. Specific inhibition of the reverse transcriptase of human immunodeficiency virus type 1 and the chimeric enzymes of human immunodeficiency virus type 1 and type 2 by nonnucleoside inhibitors. Antimicrob. Agents Chemother. 37:1037-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ho, D. D., T. Toyoshima, H. Mo, D. J. Kempf, D. Norbeck, C. M. Chen, N. E. Wideburg, S. K. Burt, J. W. Erickson, and M. K. Singh. 1994. Characterization of human immunodeficiency virus type 1 variants with increased resistance to a C2-symmetric protease inhibitor. J. Virol. 68:2016-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hoetelmans, R. M. 2001. Pharamacological exposure and the development of drug resistance in HIV. Antivir. Ther. 6[Suppl. 2]:37-47. [PubMed] [Google Scholar]
- 154.Reference deleted.
- 155.Holodniy, M., D. Katzenstein, L. Mole, M. Winters, and T. Merigan. 1996. Human immunodeficiency virus reverse transcriptase codon 215 mutations diminish virologic response to didanosine-zidovudine therapy in subjects with non-syncytium-inducing phenotype. J. Infect. Dis. 174:854-857. [DOI] [PubMed] [Google Scholar]
- 156.Hooker, D. J., G. Tachedjian, A. E. Solomon, A. D. Gurusinghe, S. Land, C. Birch, J. L. Anderson, B. M. Roy, E. Arnold, and N. J. Deacon. 1996. An in vivo mutation from leucine to tryptophan at position 210 in human immunodeficiency virus type 1 reverse transcriptase contributes to high-level resistance to 3′-azido-3′-deoxythymidine. J. Virol. 70:8010-8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hsiou, Y., J. Ding, K. Das, A. D. Clark, Jr., P. L. Boyer, P. Lewi, P. A. Janssen, J. P. Kleim, M. Rosner, S. H. Hughes, and E. Arnold. 2001. The Lys103Asn mutation of HIV-1 RT: a novel mechanism of drug resistance. J. Mol. Biol. 309:437-445. [DOI] [PubMed] [Google Scholar]
- 158.Huang, H., R. Chopra, G. L. Verdine, and S. C. Harrison. 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282:1669-1675. [DOI] [PubMed] [Google Scholar]
- 159.Reference deleted.
- 160.Reference deleted.
- 161.Reference deleted.
- 162.Hurst, M., and D. Faulds. 2000. Lopinavir. Drugs 60:1371-1381. [DOI] [PubMed] [Google Scholar]
- 163.Imamichi, H., K. A. Crandall, V. Natarajan, M. K. Jiang, R. L. Dewar, S. Berg, A. Gaddam, M. Bosche, J. A. Metcalf, R. T. Davey Jr, and H. C. Lane. 2001. Human immunodeficiency virus type 1 quasi species that rebound after discontinuation of highly active antiretroviral therapy are similar to the viral quasi species present before initiation of therapy. J. Infect. Dis. 183:36-50. [DOI] [PubMed] [Google Scholar]
- 164.Imamichi, T., S. C. Berg, H. Imamichi, J. C. Lopez, J. A. Metcalf, J. Falloon, and H. C. Lane. 2000. Relative replication fitness of a high-level 3′-azido-3′-deoxythymidine-resistant variant of human immunodeficiency virus type 1 possessing an amino acid deletion at codon 67 and a novel substitution (Thr→Gly) at codon 69. J. Virol. 74:10958-10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Imamichi, T., M. A. Murphy, H. Imamichi, and H. C. Lane. 2001. Amino acid deletion at codon 67 and Thr-to-Gly change at codon 69 of human immunodeficiency virus type 1 reverse transcriptase confer novel drug resistance profiles. J. Virol. 75:3988-3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Imamichi, T., T. Sinha, H. Imamichi, Y. M. Zhang, J. A. Metcalf, J. Falloon, and H. C. Lane. 2000. High-level resistance to 3′-azido-3′-deoxythimidine due to a deletion in the reverse transcriptase gene of human immunodeficiency virus type 1. J. Virol. 74:1023-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ingrand, D., J. Weber, C. A. Boucher, C. Loveday, C. Robert, A. Hill, and N. Cammack. 1995. Phase I/II study of 3TC (lamivudine) in HIV-positive, asymptomatic or mild AIDS-related complex patients: sustained reduction in viral markers. The Lamivudine European HIV Working group. AIDS 9:1323-1329. [DOI] [PubMed] [Google Scholar]
- 168.Iversen, A. K., R. W. Shafer, K. Wehrly, M. A. Winters, J. I. Mullins, B. Chesebro, and T. C. Merigan. 1996. Multidrug-resistant human immunodeficiency virus type 1 strains resulting from combination antiretroviral therapy. J. Virol. 70:1086-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Izopet, J., A. Bicart-See, C. Pasquier, K. Sandres, E. Bonnet, B. Marchou, J. Puel, and P. Massip. 1999. Mutations conferring resistance to zidovudine diminish the antiviral effect of stavudine plus didanosine. J. Med. Virol. 59:507-511. [PubMed] [Google Scholar]
- 170.Jackson, J. B., G. Becker-Pergola, L. A. Guay, P. Musoke, M. Mracna, M. G. Fowler, L. M. Mofenson, M. Mirochnick, F. Mmiro, and S. H. Eshleman. 2000. Identification of the K103N resistance mutation in Ugandan women receiving nevirapine to prevent HIV-1 vertical transmission. AIDS 14:F111-F115. [DOI] [PubMed] [Google Scholar]
- 171.Jacobo-Molina, A., J. Ding, R. G. Nanni, A. D. Clark, Jr., X. Lu, C. Tantillo, R. L. Williams, G. Kamer, A. L. Ferris, P. Clark, A. Hizi, S. H. Hughes, and E. Arnold. 1993. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc. Natl. Acad. Sci. USA 90:6320-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Jacobsen, H., K. Yasargil, D. L. Winslow, J. C. Craig, A. Krohn, I. B. Duncan, and J. Mous. 1995. Characterization of human immunodeficiency virus type 1 mutants with decreased sensitivity to proteinase inhibitor Ro 31-8959. Virology 206:527-534. [DOI] [PubMed] [Google Scholar]
- 173.Japour, A. J., S. Welles, R. T. D'Aquila, V. A. Johnson, D. D. Richman, R. W. Coombs, P. S. Reichelderfer, J. O. Kahn, C. S. Crumpacker, and D. R. Kuritzkes. 1995. Prevalence and clinical significance of zidovudine resistance mutations in human immunodeficiency virus isolated from patients after long-term zidovudine treatment. AIDS Clinical Trials group 116B/117 Study Team and the Virology Committee Resistance Working group. J. Infect. Dis. 171:1172-1179. [DOI] [PubMed] [Google Scholar]
- 174.Ji, J., and L. A. Loeb. 1994. Fidelity of HIV-1 reverse transcriptase copying a hypervariable region of the HIV-1 env gene. Virology 199:323-330. [DOI] [PubMed] [Google Scholar]
- 175.Joly, V., M. Moroni, E. Concia, A. Lazzarin, B. Hirschel, J. Jost, F. Chiodo, Z. Bentwich, W. C. Love, D. A. Hawkins, E. G. Wilkins, A. J. Gatell, N. Vetter, C. Greenwald, W. W. Freimuth, and W. de Cian. 2000. Delavirdine in combination with zidovudine in treatment of human immunodeficiency virus type 1-infected patients: evaluation of efficacy and emergence of viral resistance in a randomized, comparative phase III trial. Antimicrob. Agents Chemother. 44:3155-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Jonckheere, H., M. Witvrouw, E. De Clercq, and J. Anne. 1998. Lamivudine resistance of HIV type 1 does not delay development of resistance to nonnucleoside HIV type 1-specific reverse transcriptase inhibitors as compared with wild-type HIV type 1. AIDS Res. Hum. Retroviruses. 14:249-253. [DOI] [PubMed] [Google Scholar]
- 176a.Kantor, R., W. J. Fessel, A. R. Zolopa, D. Israelski, N. Shulman, J. Montoya, M. Harbour, J. Schapiro, and R. W. Shafer. 2002. Evolution of primary protease inhibitor resistance mutations during protease inhibitor therapy. Antimicrob. Agents Chemother., in press. [DOI] [PMC free article] [PubMed]
- 177.Kantor, R., R. Machekano, M. J. Gonzales, B. S. Dupnik, J. M. Schapiro, and R. W. Shafer. 2001. Human immunodeficiency virus reverse transcriptase and protease sequence database: An expanded model integrating natural language text and sequence analysis. Nucleic Acids Res. 29:296-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kantor, R., A. R. Zolopa, D. Israelski, N. Shulman, J. Montoya, M. Harbour, J. Fessel, and R. W. Shafer. Evolution of IAS-USA primary protease resistance mutations during protease inhibitor salvage therapy. Antimicrob. Agents Chemother., in press. [DOI] [PMC free article] [PubMed]
- 179.Katlama, C., B. Clotet, A. Plettenberg, J. Jost, K. Arasteh, E. Bernasconi, V. Jeantils, A. Cutrell, C. Stone, M. Ait-Khaled, and S. Purdon. 2000. The role of abacavir (ABC, 1592) in antiretroviral therapy-experienced patients: results from a randomized, double-blind, trial. CNA3002 European Study Team. AIDS 14:781-789. [DOI] [PubMed] [Google Scholar]
- 180.Kaufmann, G. R., K. Suzuki, P. Cunningham, M. Mukaide, M. Kondo, M. Imai, J. Zaunders, and D. A. Cooper. 2001. Impact of HIV type 1 protease, reverse transcriptase, cleavage site, and p6 mutations on the virological response to quadruple therapy with saquinavir, ritonavir, and two nucleoside analogs. AIDS Res. Hum. Retrovirus. 17:487-497. [DOI] [PubMed] [Google Scholar]
- 181.Kavlick, M. F., K. Wyvill, R. Yarchoan, and H. Mitsuya. 1998. Emergence of multi-dideoxynucleoside-resistant human immunodeficiency virus type 1 variants, viral sequence variation, and disease progression in patients receiving antiretroviral chemotherapy. J. Infect. Dis. 177:1506-1513. [DOI] [PubMed] [Google Scholar]
- 182.Kellam, P., C. A. Boucher, and B. A. Larder. 1992. Fifth mutation in human immunodeficiency virus type 1 reverse transcriptase contributes to the development of high-level resistance to zidovudine. Proc. Natl. Acad. Sci. USA 89:1934-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kemp, S. D., M. Salin, D. Stammers, B. Wynhoven, B. A. Larder, and P. R. Harrigan. 2001. A mutation in HIV-1 reverse transcriptase at codon 318 (Y-F) confers high-level non-nucleoside reverse transcriptase inhibitor resistance in clinical samples. Antivir. Ther. 6:36. [Google Scholar]
- 184.Kemp, S. D., C. Shi, S. Bloor, P. R. Harrigan, J. W. Mellors, and B. A. Larder. 1998. A novel polymorphism at codon 333 of human immunodeficiency virus type 1 reverse transcriptase can facilitate dual resistance to zidovudine and L-2′,3′-dideoxy-3′-thiacytidine. J. Virol. 72:5093-5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kemper, C. A., M. D. Witt, P. H. Keiser, M. P. Dube, D. N. Forthal, M. Leibowitz, D. S. Smith, A. Rigby, N. S. Hellmann, Y. S. Lie, J. Leedom, D. Richman, J. A. McCutchan, and R. Haubrich. 2001. Sequencing of protease inhibitor therapy: insights from an analysis of HIV phenotypic resistance in patients failing protease inhibitors. AIDS 15:609-615. [DOI] [PubMed] [Google Scholar]
- 186.Reference deleted.
- 187.Kempf, D., M. King, S. Brun, J. Sylte, T. Marsh, K. Stewart, and E. Sun. 2001. Association of the nucleocapsid-p1 (NC-p1) Gag cleavage site alanine to valine mutation with mutational patterns in HIV protease producing protease inhibitor resistance. Antivir. Ther. 6:46. [Google Scholar]
- 188.Kempf, D. J., J. D. Isaacson, M. S. King, S. C. Brun, Y. Xu, K. Real, B. M. Bernstein, A. J. Japour, E. Sun, and R. A. Rode. 2001. Identification of genotypic changes in human immunodeficiency virus protease that correlate with reduced susceptibility to the protease inhibitor lopinavir among viral isolates from protease inhibitor-experienced patients. J. Virol. 75:7462-7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Keulen, W., N. K. Back, W. A. van, C. A. Boucher, and B. Berkhout. 1997. Initial appearance of the 184Ile variant in lamivudine-treated patients is caused by the mutational bias of human immunodeficiency virus type 1 reverse transcriptase. J. Virol. 71:3346-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Keulen, W., A. van Wijk, R. Schuurman, B. Berkhout, and C. A. Boucher. 1999. Increased polymerase fidelity of lamivudine-resistant HIV-1 variants does not limit their evolutionary potential. AIDS 13:1343-1349. [DOI] [PubMed] [Google Scholar]
- 191.Khanna, N., T. Klimkait, V. Schiffer, J. Irigoyen, A. Telenti, B. Hirschel, and M. Battegay. 2000. Salvage therapy with abacavir plus a non-nucleoside reverse transcriptase inhibitor and a protease inhibitor in heavily pre-treated HIV-1 infected patients. Swiss HIV Cohort Study. AIDS 14:791-799. [DOI] [PubMed] [Google Scholar]
- 192.Kilby, J. M., S. Hopkins, T. M. Venetta, B. DiMassimo, G. A. Cloud, J. Y. Lee, L. Alldredge, E. Hunter, D. Lambert, D. Bolognesi, T. Matthews, M. R. Johnson, M. A. Nowak, G. M. Shaw, and M. S. Saag. 1998. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 4:1302-1307. [DOI] [PubMed] [Google Scholar]
- 193.King, R. W., D. L. Winslow, S. Garber, H. T. Scarnati, L. Bachelor, S. Stack, and M. J. Otto. 1995. Identification of a clinical isolate of HIV-1 with an isoleucine at position 82 of the protease which retains susceptibility to protease inhibitors. Antivir. Res. 28:13-24. [DOI] [PubMed] [Google Scholar]
- 194.Kleim, J. P., R. Bender, R. Kirsch, C. Meichsner, A. Paessens, and G. Riess. 1994. Mutational analysis of residue 190 of human immunodeficiency virus type 1 reverse transcriptase. Virology 200:696-701. [DOI] [PubMed] [Google Scholar]
- 195.Kleim, J. P., M. Rosner, I. Winkler, A. Paessens, R. Kirsch, Y. Hsiou, E. Arnold, and G. Riess. 1996. Selective pressure of a quinoxaline nonnucleoside inhibitor of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) on HIV-1 replication results in the emergence of nucleoside RT-inhibitor-specific (RT Leu-74→Val or Ile and Val-75→Leu or Ile) HIV-1 mutants. Proc. Natl. Acad. Sci. USA 93:34-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Reference deleted.
- 197.Koch, N., N. Yahi, F. Ariasi, J. Fantini, and C. Tamalet. 1999. Comparison of human immunodeficiency virus type 1 (HIV-1) protease mutations in HIV-1 genomes detected in plasma and in peripheral blood mononuclear cells from patients receiving combination drug therapy. J. Clin. Microbiol. 37:1595-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kohlstaedt, L. A., and T. A. Steitz. 1992. Reverse transcriptase of human immunodeficiency virus can use either human tRNA(3Lys) or Escherichia coli tRNA(2Gln) as a primer in an in vitro primer-utilization assay. Proc. Natl. Acad. Sci. USA 89:9652-9656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Kohlstaedt, L. A., J. Wang, J. M. Friedman, P. A. Rice, and T. A. Steitz. 1992. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 256:1783-1790. [DOI] [PubMed] [Google Scholar]
- 200.Kojima, E., T. Shirasaka, B. D. Anderson, S. Chokekijchai, S. M. Steinberg, S. Broder, R. Yarchoan, and H. Mitsuya. 1995. Human immunodeficiency virus type 1 (HIV-1) viremia changes and development of drug-related mutations in patients with symptomatic HIV-1 infection receiving alternating or simultaneous zidovudine and didanosine therapy. J. Infect. Dis. 171:1152-1158. [DOI] [PubMed] [Google Scholar]
- 201.Korber, B., M. Muldoon, J. Theiler, F. Gao, R. Gupta, A. Lapedes, B. H. Hahn, S. Wolinsky, and T. Bhattacharya. 2000. Timing the ancestor of the HIV-1 pandemic strains. Science 288:1789-1796. [DOI] [PubMed] [Google Scholar]
- 202.Kozal, M. J., K. Kroodsma, M. A. Winters, R. W. Shafer, B. Efron, D. A. Katzenstein, and T. C. Merigan. 1994. Didanosine resistance in HIV-infected patients switched from zidovudine to didanosine monotherapy. Ann. Intern. Med. 121:263-268. [DOI] [PubMed] [Google Scholar]
- 203.Kozal, M. J., R. W. Shafer, M. A. Winters, D. A. Katzenstein, and T. C. Merigan. 1993. A mutation in human immunodeficiency virus reverse transcriptase and decline in CD4 lymphocyte numbers in long-term zidovudine recipients. J. Infect. Dis. 167:526-532. [DOI] [PubMed] [Google Scholar]
- 204.Kuiken, C. L., B. Foley, B. Hahn, B. Korber, F. McCutchan, P. A. Marx, J. W. Mellors, J. I. Mullins, J. Sodroski, and S. Wolinksy. 1999. Human retroviruses and AIDS 1999: a compilation and analysis of nucleic and amino acid sequences. Theoretical Biology and Biophysics group, Los Alamos National Laboratory, Los Alamos, N. Mex.
- 205.Reference deleted.
- 206.Kuritzkes, D. R., R. L. Bassett, V. A. Johnson, I. C. Marschner, J. J. Eron, J. P. Sommadossi, E. P. Acosta, R. L. Murphy, K. Fife, K. Wood, D. Bell, A. Martinez, and C. B. Pettinelli. 2000. Continued lamivudine versus delavirdine in combination with indinavir and zidovudine or stavudine in lamivudine-experienced patients: results of Adult AIDS Clinical Trials group protocol 370. AIDS 14:1553-1561. [DOI] [PubMed] [Google Scholar]
- 207.Kuritzkes, D. R., J. B. Quinn, S. L. Benoit, D. L. Shugarts, A. Griffin, M. Bakhtiari, D. Poticha, J. J. Eron, M. A. Fallon, and M. Rubin. 1996. Drug resistance and virologic response in NUCA 3001, a randomized trial of lamivudine (3TC) versus zidovudine (ZDV) versus ZDV plus 3TC in previously untreated patients. AIDS 10:975-981. [DOI] [PubMed] [Google Scholar]
- 208.Kuritzkes, D. R., A. Sevin, B. Young, M. Bakhtiari, H. Wu, M. St Clair, E. Connick, A. Landay, J. Spritzler, H. Kessler, and M. M. Lederman. 2000. Effect of zidovudine resistance mutations on virologic response to treatment with zidovudine-lamivudine-ritonavir: genotypic analysis of human immunodeficiency virus type 1 isolates from AIDS clinical trials group protocol 315.ACTG Protocol 315 Team. J. Infect. Dis. 181:491-497. [DOI] [PubMed] [Google Scholar]
- 209.Kuritzkes, D. R., D. Shugarts, M. Bakhtiari, D. Poticha, J. Johnson, M. Rubin, T. R. Gingeras, M. Kennedy, and J. J. Eron. 2000. Emergence of dual resistance to zidovudine and lamivudine in HIV-1-infected patients treated with zidovudine plus lamivudine as initial therapy. J. Acquir. Immune Defic. Syndr. 23:26-34. [DOI] [PubMed] [Google Scholar]
- 210.Kusumi, K., B. Conway, S. Cunningham, A. Berson, C. Evans, A. K. Iversen, D. Colvin, M. V. Gallo, S. Coutre, E. G. Shpaer, and et al. 1992. Human immunodeficiency virus type 1 envelope gene structure and diversity in vivo and after cocultivation in vitro. J. Virol. 66:875-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Kwok, S., and R. Higuchi. 1989. Avoiding false positives with PCR. Nature 339:237-238. [DOI] [PubMed] [Google Scholar]
- 212.Lacey, S. F., and B. A. Larder. 1994. Novel mutation (V75T) in human immunodeficiency virus type 1 reverse transcriptase confers resistance to 2′,3′-didehydro-2′,3′-dideoxythymidine in cell culture. Antimicrob. Agents Chemother. 38:1428-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Reference deleted.
- 214.Reference deleted.
- 215.Larder, B. A. 1992. 3′-Azido-3′-deoxythymidine resistance suppressed by a mutation conferring human immunodeficiency virus type 1 resistance to nonnucleoside reverse transcriptase inhibitors. Antimicrob. Agents Chemother. 36:2664-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Larder, B. A. 1994. Interactions between drug resistance mutations in human immunodeficiency virus type 1 reverse transcriptase. J. Gen. Virol. 75:951-957. [DOI] [PubMed] [Google Scholar]
- 217.Larder, B. A., and S. Bloor. 2001. Analysis of clinical isolates and site-directed mutants reveals the genetic determinants of didanosine resistance. Antivir. Ther. 6:38. [Google Scholar]
- 218.Larder, B. A., S. Bloor, S. D. Kemp, K. Hertogs, R. L. Desmet, V. Miller, M. Sturmer, S. Staszewski, J. Ren, D. K. Stammers, D. I. Stuart, and R. Pauwels. 1999. A family of insertion mutations between codons 67 and 70 of human immunodeficiency virus type 1 reverse transcriptase confer multinucleoside analog resistance. Antimicrob. Agents Chemother. 43: 1961-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Larder, B. A., K. Hertogs, S. Bloor, C. H. van den Eynde, W. DeCian, Y. Wang, W. W. Freimuth, and G. Tarpley. 2000. Tipranavir inhibits broadly protease inhibitor-resistant HIV-1 clinical samples. AIDS 14:1943-1948. [DOI] [PubMed] [Google Scholar]
- 220.Larder, B. A., P. Kellam, and S. D. Kemp. 1993. Convergent combination therapy can select viable multidrug-resistant HIV-1 in vitro. Nature 365:451-453. [DOI] [PubMed] [Google Scholar]
- 221.Larder, B. A., P. Kellam, and S. D. Kemp. 1991. Zidovudine resistance predicted by direct detection of mutations in DNA from HIV-infected lymphocytes. AIDS 5:137-144. [DOI] [PubMed] [Google Scholar]
- 222.Larder, B. A., and S. D. Kemp. 1989. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT). Science 246:1155-1158. [DOI] [PubMed] [Google Scholar]
- 223.Larder, B. A., S. D. Kemp, and P. R. Harrigan. 1995. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science 269:696-699. [DOI] [PubMed] [Google Scholar]
- 224.Larder, B. A., A. Kohli, S. Bloor, S. D. Kemp, P. R. Harrigan, R. T. Schooley, J. M. Lange, K. N. Pennington, and St. 1996. Human immunodeficiency virus type 1 drug susceptibility during zidovudine (AZT) monotherapy compared with AZT plus 2′,3′-dideoxyinosine or AZT plus 2′,3′-dideoxycytidine combination therapy. The Protocol 34,225-02 Collaborative group. J. Virol. 70:5922-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Larder, B. A., A. Kohli, P. Kellam, S. D. Kemp, M. Kronick, and R. D. Henfrey. 1993. Quantitative detection of HIV-1 drug resistance mutations by automated DNA sequencing. Nature 365:671-673. [DOI] [PubMed] [Google Scholar]
- 226.Larder, B. A., and D. K. Stammers. 1999. Closing in on HIV drug resistance. Nat. Struct. Biol. 6:103-106. [DOI] [PubMed] [Google Scholar]
- 227.Lawrence, J., J. Schapiro, M. Winters, J. Montoya, A. Zolopa, R. Pesano, B. Efron, D. Winslow, and T. C. Merigan. 1999. Clinical resistance patterns and responses to two sequential protease inhibitor regimens in saquinavir and reverse transcriptase inhibitor-experienced persons. J. Infect. Dis. 179:1356-1364. [DOI] [PubMed] [Google Scholar]
- 228.Learn, G. H., Jr, B. T. Korber, B. Foley, B. H. Hahn, S. M. Wolinsky, and J. I. Mullins. 1996. Maintaining the integrity of human immunodeficiency virus sequence databases. J. Virol. 70:5720-5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Lecossier, D., F. Bouchonnet, P. Schneider, F. Clavel, and A. J. Hance. 2001. Discordant increases in CD4+ t cells in human immunodeficiency virus-infected patients experiencing virologic treatment failure: role of changes in thymic output and t cell death. J. Infect. Dis. 183:1009-1016. [DOI] [PubMed] [Google Scholar]
- 230.Lennerstrand, J., D. K. Stammers, and B. A. Larder. 2001. Biochemical mechanism of human immunodeficiency virus type 1 reverse transcriptase resistance to stavudine. Antimicrob. Agents Chemother. 45:2144-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Liegler, T. J., M. S. Hayden, K. H. Lee, R. Hoh, S. G. Deeks, and R. M. Granta. 2001. Protease inhibitor-resistant HIV-1 from patients with preserved CD4 cell counts is cytopathic in activated CD4 T lymphocytes. AIDS 15:179-184. [DOI] [PubMed] [Google Scholar]
- 232.Lin, P. F., H. Samanta, R. E. Rose, A. K. Patick, J. Trimble, C. M. Bechtold, D. R. Revie, N. C. Khan, M. E. Federici, and H. Li. 1994. Genotypic and phenotypic analysis of human immunodeficiency virus type 1 isolates from patients on prolonged stavudine therapy. J. Infect. Dis. 170:1157-1164. [DOI] [PubMed] [Google Scholar]
- 233.Little, S. J. 2000. Transmission and prevalence of HIV resistance among treatment-naive subjects. Antivir. Ther. 5:33-40. [DOI] [PubMed] [Google Scholar]
- 234.Little, S. J., E. S. Daar, R. T. D'Aquila, P. H. Keiser, E. Connick, J. M. Whitcomb, N. S. Hellmann, C. J. Petropoulos, L. Sutton, J. A. Pitt, E. S. Rosenberg, R. A. Koup, B. D. Walker, and D. D. Richman. 1999. Reduced antiretroviral drug susceptibility among patients with primary HIV infection. JAMA 282:1142-1149. [DOI] [PubMed] [Google Scholar]
- 235.Loveday, C. 2001. International perspectives on antiretroviral resistance. Nucleoside reverse transcriptase inhibitor resistance. J. Acquir. Immune Defic. Syndr. 26(Suppl. 1):S10-24. [DOI] [PubMed] [Google Scholar]
- 236.Maguire, M., M. Gartland, S. Moore, A. Hill, M. Tisdale, R. Harrigan, and J. P. Kleim. 2000. Absence of zidovudine resistance in antiretroviral-naive patients following zidovudine/lamivudine/protease inhibitor combination therapy: virological evaluation of the AVANTI 2 and AVANTI 3 studies. AIDS 14:1195-1201. [DOI] [PubMed] [Google Scholar]
- 237.Maguire, M., S. MacManus, P. Griffin, C. Guinea, W. Harris, N. Richard, J. Wolfram, M. Tisdale, W. Snowden, and J.-P. Klein. 2001. Interaction of HIV-1 protease and gag gene mutations in response to amprenavir-selective pressure exerted in amprenavir-treated subjects - contribution of gag p6 changes L449F and P453L. Antivir. Ther. 6:48. [Google Scholar]
- 238.Mahalingam, B., J. M. Louis, J. Hung, R. W. Harrison, and I. T. Weber. 2001. Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes. Proteins 43:455-464. [DOI] [PubMed] [Google Scholar]
- 239.Mahalingam, B., J. M. Louis, C. C. Reed, J. M. Adomat, J. Krouse, Y. F. Wang, R. W. Harrison, and I. T. Weber. 1999. Structural and kinetic analysis of drug resistant mutants of HIV-1 protease. Eur. J. Biochem. 263:238-245. [DOI] [PubMed] [Google Scholar]
- 240.Mammano, F., C. Petit, and F. Clavel. 1998. Resistance-associated loss of viral fitness in human immunodeficiency virus type 1: phenotypic analysis of protease and gag coevolution in protease inhibitor-treated patients. J. Virol. 72:7632-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Mammano, F., V. Trouplin, V. Zennou, and F. Clavel. 2000. Retracing the evolutionary pathways of human immunodeficiency virus type 1 resistance to protease inhibitors: virus fitness in the absence and in the presence of drug. J. Virol. 74:8524-8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Mansky, L. M. 1998. Retrovirus mutation rates and their role in genetic variation. J. Gen. Virol. 79:1337-1345. [DOI] [PubMed] [Google Scholar]
- 243.Markowitz, M., M. Conant, A. Hurley, R. Schluger, M. Duran, J. Peterkin, S. Chapman, A. Patick, A. Hendricks, G. J. Yuen, W. Hoskins, N. Clendeninn, and D. D. Ho. 1998. A preliminary evaluation of nelfinavir mesylate, an inhibitor of human immunodeficiency virus (HIV)-1 protease, to treat HIV infection. J. Infect. Dis. 177:1533-1540. [DOI] [PubMed] [Google Scholar]
- 244.Reference deleted.
- 245.Martinez-Picado, J., M. P. DePasquale, N. Kartsonis, G. J. Hanna, J. Wong, D. Finzi, E. Rosenberg, H. F. Gunthard, L. Sutton, A. Savara, C. J. Petropoulos, N. Hellmann, B. D. Walker, D. D. Richman, R. Siliciano, and R. T. D'Aquila. 2000. Antiretroviral resistance during successful therapy of HIV type 1 infection. Proc. Natl. Acad. Sci. USA 97:10948-10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Martinez-Picado, J., A. V. Savara, L. Sutton, and R. T. D'Aquila. 1999. Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J. Virol. 73:3744-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Masquelier, B., D. Descamps, I. Carriere, F. Ferchal, G. Collin, M. Denayrolles, A. Ruffault, B. Chanzy, J. Izopet, C. Buffet-Janvresse, M. P. Schmitt, E. Race, H. J. Fleury, J. P. Aboulker, P. Yeni, and F. Brun-Vezinet. 1999. Zidovudine resensitization and dual HIV-1 resistance to zidovudine and lamivudine in the delta lamivudine roll-over study. Antivir. Ther. 4:69-77. [PubMed] [Google Scholar]
- 248.Masquelier, B., E. Race, C. Tamalet, D. Descamps, J. Izopet, C. Buffet-Janvresse, A. Ruffault, A. S. Mohammed, J. Cottalorda, A. Schmuck, V. Calvez, E. Dam, H. Fleury, and F. Brun-Vezinet. 2001. Genotypic and phenotypic resistance patterns of human immunodeficiency virus type 1 variants with insertions or deletions in the reverse transcriptase (RT): multicenter study of patients treated with RT inhibitors. Antimicrob. Agents Chemother. 45:1836-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Reference deleted.
- 250.Mayers, D. L., A. J. Japour, J. M. Arduino, S. M. Hammer, R. Reichman, K. F. Wagner, R. Chung, J. Lane, C. S. Crumpacker, G. X. McLeod, et al. 1994. Dideoxynucleoside resistance emerges with prolonged zidovudine monotherapy. The RV43 Study group. Antimicrob. Agents Chemother. 38:307-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Reference deleted.
- 252.Meyer, P. R., S. E. Matsuura, A. M. Mian, A. G. So, and W. A. Scott. 1999. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol. Cell 4:35-43. [DOI] [PubMed] [Google Scholar]
- 253.Meyer, P. R., S. E. Matsuura, R. F. Schinazi, A. G. So, and W. A. Scott. 2000. Differential removal of thymidine nucleotide analogues from blocked DNA chains by human immunodeficiency virus reverse transcriptase in the presence of physiological concentrations of 2′-deoxynucleoside triphosphates. Antimicrob. Agents Chemother. 44:3465-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Meyer, P. R., S. E. Matsuura, A. G. So, and W. A. Scott. 1998. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc. Natl. Acad. Sci. USA 95: 13471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Reference deleted.
- 256.Reference deleted.
- 257.Miller, M. D., K. E. Anton, A. S. Mulato, P. D. Lamy, and J. M. Cherrington. 1999. Human immunodeficiency virus type 1 expressing the lamivudine-associated M184V mutation in reverse transcriptase shows increased susceptibility to adefovir and decreased replication capability in vitro. J. Infect. Dis. 179:92-100. [DOI] [PubMed] [Google Scholar]
- 258.Reference deleted.
- 259.Miller, M. D., N. A. Margot, K. Hertogs, B. Larder, and V. Miller. 2001. Antiviral activity of tenofovir (PMPA) against nucleoside-resistant clinical HIV samples. Nucleosides Nucleotides Nucleic Acids 20:1025-1028. [DOI] [PubMed] [Google Scholar]
- 260.Miller, V. 2001. Resistance to protease inhibitors. J. Acquir. Immune Defic. Syndr. 26[Suppl. 1]:S34-50. [DOI] [PubMed] [Google Scholar]
- 261.Miller, V., M. Ait-Khaled, C. Stone, P. Griffin, D. Mesogiti, A. Cutrell, R. Harrigan, S. Staszewski, C. Katlama, G. Pearce, and M. Tisdale. 2000. HIV-1 reverse transcriptase (RT) genotype and susceptibility to RT inhibitors during abacavir monotherapy and combination therapy. AIDS 14:163-171. [DOI] [PubMed] [Google Scholar]
- 262.Miller, V., and B. A. Larder. 2001. Mutational patterns in the HIV genome and cross-resistance following nucleoside and nucleotide analogue drug exposure. Antivir. Ther. 6[Suppl. 3]:25-44. [PubMed] [Google Scholar]
- 263.Miller, V., M. Sturmer, S. Staszewski, B. Groschel, K. Hertogs, B. M. de, R. Pauwels, P. R. Harrigan, S. Bloor, S. D. Kemp, and B. A. Larder. 1998. The M184V mutation in HIV-1 reverse transcriptase (RT) conferring lamivudine resistance does not result in broad cross-resistance to nucleoside analogue RT inhibitors. AIDS 12:705-712. [DOI] [PubMed] [Google Scholar]
- 264.Molla, A., S. Brun, K. Garren, H. Mo, B. Richards, T. Marsh, J. Sylte, M. King, L. Han, E. Sun, and D. Kempf. 2001. Patterns of resistance to lopinavir in protease inhibitor-experienced patients following viral rebound during lopinavir/ritonavir therapy. Antivir. Ther. 6:49. [Google Scholar]
- 265.Molla, A., M. Korneyeva, Q. Gao, S. Vasavanonda, P. J. Schipper, H. M. Mo, M. Markowitz, T. Chernyavskiy, P. Niu, N. Lyons, A. Hsu, G. R. Granneman, D. D. Ho, C. A. Boucher, J. M. Leonard, D. W. Norbeck, and D. J. Kempf. 1996. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat. Med. 2:760-766. [DOI] [PubMed] [Google Scholar]
- 266.Montaner, J. S., T. Mo, J. M. Raboud, S. Rae, C. S. Alexander, C. Zala, D. Rouleau, and P. R. Harrigan. 2000. Human immunodeficiency virus-infected persons with mutations conferring resistance to zidovudine show reduced virologic responses to hydroxyurea and stavudine-lamivudine. J. Infect. Dis. 181:729-732. [DOI] [PubMed] [Google Scholar]
- 266a.Mouroux, M., D. Descamps, J. Izopet, A. Yvon, C. Delaugerre, S. Matheron, A. Coutellier, M. A. Valantin, M. Bonmarchand, H. Agut, P. Massip, D. Costagliola, C. Katlama, F. Brun-Vezinet, and V. Calvez. 2001. Low-rate emergence of thymidine analogue mutations and multi-drug resistance mutations in the HIV-1 reverse transcriptase gene in therapy-naive patients receiving stavudine plus lamivudine combination therapy. Antivir. Ther. 6:179-183. [PubMed] [Google Scholar]
- 267.Mouroux, M., A. Yvon-Groussin, G. Peytavin, C. Delaugerre, M. Legrand, P. Bossi, B. Do, A. Trylesinski, B. Diquet, E. Dohin, J. F. Delfraissy, C. Katlama, and V. Calvez. 2000. Early virological failure in naive human immunodeficiency virus patients receiving saquinavir (soft gel capsule)-stavudine-zalcitabine (MIKADO trial) is not associated with mutations conferring viral resistance. J. Clin. Microbiol. 38:2726-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267a.Mracna, M., G. Becker-Pergola, J. Dileanis, L. A. Guay, S. Cunningham, J. B. Jackson, and S. H. Eshleman. 2001. Performance of Applied Biosystems ViroSeq HIV-1 Genotyping System for sequence-based analysis of non-subtype B human immunodeficiency virus type 1 from Uganda. J. Clin. Microbiol. 39:4323-4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Murphy, R. L., R. M. Gulick, V. DeGruttola, R. T. D'Aquila, J. J. Eron, J. P. Sommadossi, J. S. Currier, L. Smeaton, I. Frank, A. M. Caliendo, J. G. Gerber, R. Tung, and D. R. Kuritzkes. 1999. Treatment with amprenavir alone or amprenavir with zidovudine and lamivudine in adults with human immunodeficiency virus infection. AIDS Clinical Trials group 347 Study Team. J. Infect. Dis. 179:808-816. [DOI] [PubMed] [Google Scholar]
- 269.Reference deleted.
- 270.Naeger, L. K., N. A. Margot, and M. D. Miller. 2001. Increased drug susceptibility of HIV-1 reverse transcriptase mutants containing M184V and zidovudine-associated mutations: analysis of enzyme processivity, chain-terminator removal and viral replication. Antivir. Ther. 6:115-126. [PubMed] [Google Scholar]
- 271.Nijhuis, M., S. Deeks, and C. Boucher. 2001. Implications of antiretroviral resistance on viral fitness. Curr. Opin. Infect. Dis. 14:23-28. [DOI] [PubMed] [Google Scholar]
- 272.Nijhuis, M., R. Schuurman, D. de Jong, J. Erickson, E. Gustchina, J. Albert, P. Schipper, S. Gulnik, and C. A. Boucher. 1999. Increased fitness of drug resistant HIV-1 protease as a result of acquisition of compensatory mutations during suboptimal therapy. AIDS 13:2349-2359. [DOI] [PubMed] [Google Scholar]
- 273.O'Brien, S. J., G. W. Nelson, C. A. Winkler, and M. W. Smith. 2000. Polygenic and multifactorial disease gene association in man: Lessons from AIDS. Annu. Rev. Genet. 34:563-591. [DOI] [PubMed] [Google Scholar]
- 274.Olsen, D. B., M. W. Stahlhut, C. A. Rutkowski, H. B. Schock, A. L. vanOlden, and L. C. Kuo. 1999. Non-active site changes elicit broad-based cross-resistance of the HIV-1 protease to inhibitors. J. Biol. Chem. 274:23699-23701. [DOI] [PubMed] [Google Scholar]
- 275.Oude Essink, B. B., N. K. Back, and B. Berkhout. 1997. Increased polymerase fidelity of the 3TC-resistant variants of HIV-1 reverse transcriptase. Nucleic Acids Res. 25:3212-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Palmer, S., A. Alaeus, J. Albert, and S. Cox. 1998. Drug susceptibility of subtypes A,B,C,D, and E human immunodeficiency virus type 1 primary isolates. AIDS Res. Hum. Retroviruses 14:157-162. [DOI] [PubMed] [Google Scholar]
- 277.Palmer, S., R. W. Shafer, and T. C. Merigan. 1999. Highly drug-resistant HIV-1 clinical isolates are cross-resistant to many antiretroviral compounds in current clinical development. AIDS 13:661-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Para, M. F., D. V. Glidden, R. Coombs, A. Collier, J. Condra, C. Craig, R. Bassett, R. Leavitt, S. Snyder, V. J. McAuliffe, and C. Boucher. 2000. Baseline human immunodeficiency virus type I phenotype, genotype, and RNA response after switching from long-term hard-capsule saquinavir to indinavir or soft-gel-capsule saquinavir in AIDS clinical trials group protocol 333. J. Infect. Dis. 182:733-743. [DOI] [PubMed] [Google Scholar]
- 279.Parkin, N. T., C. Chappey, M. Maranta, N. Whitehurst, and C. Petropoulos. 2001. Genotypic and phenotypic analysis of a large database of patient samples reveals distinct patterns of cross-resistance. Antivir. Ther. 6:49. [Google Scholar]
- 280.Partaledis, J. A., K. Yamaguchi, M. Tisdale, E. E. Blair, C. Falcione, B. Maschera, R. E. Myers, S. Pazhanisamy, O. Futer, A. B. Cullinan, and et al. 1995. In vitro selection and characterization of human immunodeficiency virus type 1 (HIV-1) isolates with reduced sensitivity to hydroxyethylamino sulfonamide inhibitors of HIV-1 aspartyl protease. J. Virol. 69:5228-5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Patick, A. K., M. Duran, Y. Cao, D. Shugarts, M. R. Keller, E. Mazabel, M. Knowles, S. Chapman, D. R. Kuritzkes, and M. Markowitz. 1998. Genotypic and phenotypic characterization of human immunodeficiency virus type 1 variants isolated from patients treated with the protease inhibitor nelfinavir. Antimicrob. Agents Chemother. 42:2637-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Patick, A. K., H. Mo, M. Markowitz, K. Appelt, B. Wu, L. Musick, V. Kalish, S. Kaldor, S. Reich, D. Ho, and S. Webber. 1996. Antiviral and resistance studies of AG1343, an orally bioavailable inhibitor of human immunodeficiency virus protease. Antimicrob. Agents Chemother. 40:292-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Pelemans, H., A. Aertsen, K. Van Laethem, A. M. Vandamme, E. De Clercq, M. J. Perez-Perez, A. San-Felix, S. Velazquez, M. J. Camarasa, and J. Balzarini. 2001. Site-directed mutagenesis of human immunodeficiency virus type 1 reverse transcriptase at amino acid position 138. Virology 280:97-106. [DOI] [PubMed] [Google Scholar]
- 284.Pelemans, H., R. M. Esnouf, M. A. Parniak, A. M. Vandamme, E. De Clercq, and J. Balzarini. 1998. A proline-to-histidine substitution at position 225 of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) sensitizes HIV-1 RT to BHAP U-90152. J. Gen. Virol. 79:1347-1352. [DOI] [PubMed] [Google Scholar]
- 285.Pellegrin, I., J. Izopet, J. Reynes, M. Denayrolles, B. Montes, J. L. Pellegrin, P. Massip, J. Puel, H. Fleury, and M. Segondy. 1999. Emergence of zidovudine and multidrug resistance mutations in the HIV-1 reverse transcriptase gene in therapy-naive patients receiving stavudine plus didanosine combination therapy. STADI group. AIDS 13:1705-1709. [DOI] [PubMed] [Google Scholar]
- 286.Penn, M. L., M. Myers, D. A. Eckstein, T. J. Liegler, M. Hayden, F. Mammano, F. Clavel, S. G. Deeks, R. M. Grant, and M. A. Goldsmith. 2001. Primary and recombinant HIV type 1 strains resistant to protease inhibitors are pathogenic in mature human lymphoid tissues. AIDS Res. Hum. Retrovirus. 17:517-523. [DOI] [PubMed] [Google Scholar]
- 287.Perelson, A. S., A. U. Neumann, M. Markowitz, J. M. Leonard, and D. D. Ho. 1996. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271:1582-1586. [DOI] [PubMed] [Google Scholar]
- 288.Perez, E. E., S. L. Rose, B. Peyser, S. L. Lamers, B. Burkhardt, B. M. Dunn, A. D. Hutson, J. W. Sleasman, and M. M. Goodenow. 2001. Human immunodeficiency virus type 1 protease genotype predicts immune and viral responses to combination therapy with protease inhibitors (PIs) in PI-naive patients. J. Infect. Dis. 183:579-588. [DOI] [PubMed] [Google Scholar]
- 289.Perno, C. F., A. Cozzi-Lepri, C. Balotta, F. Forbici, M. Violin, A. Bertoli, G. Facchi, P. Pezzotti, G. Cadeo, G. Tositti, S. Pasquinucci, S. Pauluzzi, A. Scalzini, B. Salassa, A. Vincenti, A. N. Phillips, F. Dianzani, A. Appice, G. Angarano, L. Monno, G. Ippolito, M. Moroni, and A. Monforte. 2001. Secondary mutations in the protease region of human immunodeficiency virus and virologic failure in drug-naive patients treated with protease inhibitor-based therapy. J. Infect. Dis. 184:983-991. [DOI] [PubMed] [Google Scholar]
- 290.Petropoulos, C. J., N. T. Parkin, K. L. Limoli, Y. S. Lie, T. Wrin, W. Huang, H. Tian, D. Smith, G. A. Winslow, D. J. Capon, and J. M. Whitcomb. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44:920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Picard, V., E. Angelini, A. Maillard, E. Race, F. Clavel, G. Chene, F. Ferchal, and J. M. Molina. 2001. Comparison of genotypic and phenotypic resistance patterns of human immunodeficiency virus type 1 isolates from patients treated with stavudine and didanosine or zidovudine and lamivudine. J. Infect. Dis. 184:781-784. [DOI] [PubMed] [Google Scholar]
- 292.Picchio, G. R., H. Valdez, R. Sabbe, A. L. Landay, D. R. Kuritzkes, M. M. Lederman, and D. E. Mosier. 2000. Altered viral fitness of HIV-1 following failure of protease inhibitor-based therapy. J. Acquir. Immune Defic. Syndr. 25:289-295. [DOI] [PubMed] [Google Scholar]
- 293.Pieniazek, D., M. Rayfield, D. J. Hu, J. Nkengasong, S. Z. Wiktor, R. Downing, B. Biryahwaho, T. Mastro, A. Tanuri, V. Soriano, R. Lal, and T. Dondero. 2000. Protease sequences from HIV-1 group M subtypes A-H reveal distinct amino acid mutation patterns associated with protease resistance in protease inhibitor-naive individuals worldwide. HIV Variant Working group. AIDS 14:1489-1495. [DOI] [PubMed] [Google Scholar]
- 294.Piketty, C., E. Race, P. Castiel, L. Belec, G. Peytavin, A. Si-Mohamed, G. Gonzalez-Canali, L. Weiss, F. Clavel, and M. D. Kazatchkine. 1999. Efficacy of a five-drug combination including ritonavir, saquinavir and efavirenz in patients who failed on a conventional triple-drug regimen: phenotypic resistance to protease inhibitors predicts outcome of therapy. AIDS 13:F71-77. [DOI] [PubMed] [Google Scholar]
- 295.Piketty, C., L. Weiss, F. Thomas, A. S. Mohamed, L. Belec, and M. D. Kazatchkine. 2001. Long-term clinical outcome of human immunodeficiency virus-infected patients with discordant immunologic and virologic responses to a protease inhibitor-containing regimen. J. Infect. Dis. 183:1328-1335. [DOI] [PubMed] [Google Scholar]
- 296.Pluda, J. M., T. P. Cooley, J. S. Montaner, L. E. Shay, N. E. Reinhalter, S. N. Warthan, J. Ruedy, H. M. Hirst, C. A. Vicary, J. B. Quinn, and et al. 1995. A phase I/II study of 2′-deoxy-3′-thiacytidine (lamivudine) in patients with advanced human immunodeficiency virus infection. J. Infect. Dis. 171:1438-1447. [DOI] [PubMed] [Google Scholar]
- 297.Reference deleted.
- 298.Puchhammer-Stockl, E., B. Schmied, C. W. Mandl, N. Vetter, and F. X. Heinz. 1999. Comparison of line probe assay (LIPA) and sequence analysis for detection of HIV-1 drug resistance. J. Med. Virol. 57:283-289. [DOI] [PubMed] [Google Scholar]
- 299.Quinones-Mateu, M. E., V. Soriano, E. Domingo, and L. Menendez-Arias. 1997. Characterization of the reverse transcriptase of a human immunodeficiency virus type 1 group O isolate. Virology 236:364-373. [DOI] [PubMed] [Google Scholar]
- 300.Race, E., E. Dam, V. Obry, S. Paulous, and F. Clavel. 1999. Analysis of HIV cross-resistance to protease inhibitors using a rapid single-cycle recombinant virus assay for patients failing on combination therapies. AIDS 13:2061-2068. [DOI] [PubMed] [Google Scholar]
- 301.Reference deleted.
- 302.Ren, J., R. M. Esnouf, A. L. Hopkins, E. Y. Jones, I. Kirby, J. Keeling, C. K. Ross, B. A. Larder, D. I. Stuart, and D. K. Stammers. 1998. 3′-Azido-3′-deoxythymidine drug resistance mutations in HIV-1 reverse transcriptase can induce long range conformational changes. Proc. Natl. Acad. Sci. USA 95:9518-9523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Rey, D., M. Hughes, J. T. Pi, M. Winters, T. C. Merigan, and D. A. Katzenstein. 1998. HIV-1 reverse transcriptase codon 215 mutation in plasma RNA: immunologic and virologic responses to zidovudine. The AIDS Clinical Trials group Study 175 Virology Team. J. Acquir. Immune Defic. Syndr. 17:203-208. [DOI] [PubMed] [Google Scholar]
- 304.Ribeiro, R. M., and S. Bonhoeffer. 2000. Production of resistant HIV mutants during antiretroviral therapy. Proc. Natl. Acad. Sci. USA 97:7681-7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Richman, D. D., J. C. Guatelli, J. Grimes, A. Tsiatis, and T. Gingeras. 1991. Detection of mutations associated with zidovudine resistance in human immunodeficiency virus by use of the polymerase chain reaction. J. Infect. Dis. 164:1075-1081. [DOI] [PubMed] [Google Scholar]
- 306.Rimsky, L. T., D. C. Shugars, and T. J. Matthews. 1998. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J. Virol. 72:986-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Robertson, D. L., J. L. Anderson, J. A. Bradac, J. K. Carr, B. Foley, R. K. Funkhouser, F. Gao, B. H. Hahn, M. L. Kalish, C. L. Kuiken, G. H. Learn, Jr, T. Leitner, F. E. McCutchan, S. Osmanov, M. Peeters, D. Pieniazek, M. O. Salminen, P. M. Sharp, S. Wolinsky, and B. Korber. 2000. HIV-1 nomenclature proposal: A reference guide to HIV-1 classification, p. 492-505. In C. L. Kuiken, B. Foley, B. H. Hahn, P. Marx, F. E. McCutchan, J. Mellors, J. I. Mullins, S. Wolanski, and B. Korber (ed.), Human retroviruses and AIDS: a compilation and analysis of nucleic and amino acid sequences. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 308.Robinson, B. S., K. A. Riccardi, Y. F. Gong, Q. Guo, D. A. Stock, W. S. Blair, B. J. Terry, C. A. Deminie, F. Djang, R. J. Colonno, and P. F. Lin. 2000. BMS-232632, a highly potent human immunodeficiency virus protease inhibitor that can be used in combination with other available antiretroviral agents. Antimicrob. Agents Chemother. 44:2093-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Rodgers, D. W., S. J. Gamblin, B. A. Harris, S. Ray, J. S. Culp, B. Hellmig, D. J. Woolf, C. Debouck, and S. C. Harrison. 1995. The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 92:1222-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Rose, R. E., Y. F. Gong, J. A. Greytok, C. M. Bechtold, B. J. Terry, B. S. Robinson, M. Alam, R. J. Colonno, and P. F. Lin. 1996. Human immunodeficiency virus type 1 viral background plays a major role in development of resistance to protease inhibitors. Proc. Natl. Acad. Sci. USA 93:1648-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Ross, L., A. Scarsella, S. Raffanti, K. Henry, S. Becker, R. Fisher, Q. Liao, A. Hirani, N. Graham, M. St Clair, and J. Hernandez. 2001. Thymidine analog and multinucleoside resistance mutations are associated with decreased phenotypic susceptibility to stavudine in hiv type 1 isolated from zidovudine-naive patients experiencing viremia on stavudine-containing regimens. AIDS Res. Hum. Retrovirus 17:1107-1115. [DOI] [PubMed] [Google Scholar]
- 312.Rousseau, M. N., L. Vergne, B. Montes, M. Peeters, J. Reynes, E. Delaporte, and M. Segondy. 2001. Patterns of resistance mutations to antiretroviral drugs in extensively treated HIV-1-infected patients with failure of highly active antiretroviral therapy. J. Acquir. Immune Defic. Syndr. 26:36-43. [DOI] [PubMed] [Google Scholar]
- 313.Rouzine, I. M., and J. M. Coffin. 1999. Linkage disequilibrium test implies a large effective population number for HIV in vivo. Proc. Natl. Acad. Sci. USA 96:10758-10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Reference deleted.
- 315.Rusconi, S., S. La Seta Catamancio, P. Citterio, E. Bulgheroni, S. Kurtagic, M. Galazzi, F. Croce, M. Moroni, and M. Galli. 2001. Virological response in multidrug-experienced HIV-1-infected subjects failing highly active combination regimens after shifting from lamivudine to didanosine. Antivir. Ther. 6:41-46. [PubMed] [Google Scholar]
- 316.Rusconi, S., S. La Seta Catamancio, P. Citterio, S. Kurtagic, M. Violin, C. Balotta, M. Moroni, M. Galli, and A. d'Arminio-Monforte. 2000. Susceptibility to PNU-140690 (Tipranavir) of human immunodeficiency virus type 1 isolates derived from patients with multidrug resistance to other protease inhibitors. Antimicrob. Agents Chemother. 44:1328-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Salomon, H., M. A. Wainberg, B. Brenner, Y. Quan, D. Rouleau, P. Cote, R. LeBlanc, E. Lefebvre, B. Spira, C. Tsoukas, R. P. Sekaly, B. Conway, D. Mayers, and J. P. Routy. 2000. Prevalence of HIV-1 resistant to antiretroviral drugs in 81 individuals newly infected by sexual contact or injecting drug use. Investigators of the Quebec Primary Infection Study. AIDS 14:F17-23. [DOI] [PubMed] [Google Scholar]
- 318.Sarafianos, S. G., K. Das, A. D. Clark, Jr., J. Ding, P. L. Boyer, S. H. Hughes, and E. Arnold. 1999. Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with beta-branched amino acids. Proc. Natl. Acad. Sci. USA 96:10027-10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Sarafianos, S. G., K. Das, J. Ding, P. L. Boyer, S. H. Hughes, and E. Arnold. 1999. Touching the heart of HIV-1 drug resistance: the fingers close down on the dNTP at the polymerase active site. Chem. Biol. 6:R137-146. [DOI] [PubMed] [Google Scholar]
- 320.Schapiro, J. M., M. A. Winters, F. Stewart, B. Efron, J. Norris, M. J. Kozal, and T. C. Merigan. 1996. The effect of high-dose saquinavir on viral load and CD4+ T-cell counts in HIV-infected patients. Ann. Intern. Med. 124:1039-1050. [DOI] [PubMed] [Google Scholar]
- 321.Schmidt, B., K. Korn, B. Moschik, C. Paatz, K. Uberla, and H. Walter. 2000. Low level of cross-resistance to amprenavir (141W94) in samples from patients pretreated with other protease inhibitors. Antimicrob. Agents Chemother. 44:3213-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Schmidt, B., H. Walter, B. Moschik, C. Paatz, K. van Vaerenbergh, A. M. Vandamme, M. Schmitt, T. Harrer, K. Uberla, and K. Korn. 2000. Simple algorithm derived from a geno-/phenotypic database to predict HIV-1 protease inhibitor resistance. AIDS 14:1731-1738. [DOI] [PubMed] [Google Scholar]
- 323.Schmit, J. C., L. K. Van, L. Ruiz, P. Hermans, S. Sprecher, A. Sonnerborg, M. Leal, T. Harrer, B. Clotet, V. Arendt, E. Lissen, M. Witvrouw, J. Desmyter, C. E. De, and A. M. Vandamme. 1998. Multiple dideoxynucleoside analogue-resistant (MddNR) HIV-1 strains isolated from patients from different European countries. AIDS 12:2007-2015. [DOI] [PubMed] [Google Scholar]
- 324.Reference deleted.
- 325.Schuurman, R., L. Demeter, P. Reichelderfer, J. Tijnagel, T. de Groot, and C. Boucher. 1999. Worldwide evaluation of DNA sequencing approaches for identification of drug resistance mutations in the human immunodeficiency virus type 1 reverse transcriptase. J. Clin. Microbiol. 37:2291-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325a.Schuurman, R., D. Brambilla, T. de Groot, D. Huang, S. Land, J. Bremer, I. Benders, and C. A. Boucher. 2002. Underestimation of HIV type 1 drug resistance mutations: results from the ENVA-2 genotyping proficiency program. AIDS Res. Hum. Retrovir. 18:243-248. [DOI] [PubMed] [Google Scholar]
- 326.Schuurman, R., M. Nijhuis, R. van Leeuwen, P. Schipper, D. de Jong, P. Collis, S. A. Danner, J. Mulder, C. Loveday, and C. Christopherson. 1995. Rapid changes in human immunodeficiency virus type 1 RNA load and appearance of drug-resistant virus populations in persons treated with lamivudine (3TC). J. Infect. Dis. 171:1411-1419. [DOI] [PubMed] [Google Scholar]
- 327.Selmi, B., J. Boretto, J. M. Navarro, J. Sire, S. Longhi, C. Guerreiro, L. Mulard, S. Sarfati, and B. Canard. 2001. The valine-to-threonine 75 substitution in human immunodeficiency virus type 1 reverse transcriptase and its relation with stavudine resistance. J. Biol. Chem. 276:13965-13974. [DOI] [PubMed] [Google Scholar]
- 328.Servais, J., C. Lambert, E. Fontaine, J. M. Plesseria, I. Robert, V. Arendt, T. Staub, F. Schneider, R. Hemmer, G. Burtonboy, and J. C. Schmit. 2001. Comparison of DNA sequencing and a line probe assay for detection of human immunodeficiency virus type 1 drug resistance mutations in patients failing highly active antiretroviral therapy. J. Clin. Microbiol. 39:454-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Servais, J., C. Lambert, E. Fontaine, J. M. Plesseria, I. Robert, V. Arendt, T. Staub, F. Schneider, R. Hemmer, G. Burtonboy, and J. C. Schmit. 2001. Variant human immunodeficiency virus type 1 proteases and response to combination therapy including a protease inhibitor. Antimicrob. Agents Chemother. 45:893-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Sevin, A. D., V. DeGruttola, M. Nijhuis, J. M. Schapiro, A. S. Foulkes, M. F. Para, and C. A. Boucher. 2000. Methods for investigation of the relationship between drug-susceptibility phenotype and human immunodeficiency virus type 1 genotype with applications to AIDS clinical trials group 333. J. Infect. Dis. 182:59-67. [DOI] [PubMed] [Google Scholar]
- 331.Shafer, R. W., T. K. Chuang, P. Hsu, C. B. White, and D. A. Katzenstein. 1999. Sequence and drug susceptibility of subtype C protease from human immunodeficiency virus type 1 seroconverters in Zimbabwe. AIDS Res. Hum. Retrovirus 15:65-69. [DOI] [PubMed] [Google Scholar]
- 332.Shafer, R. W., J. A. Eisen, T. C. Merigan, and D. A. Katzenstein. 1997. Sequence and drug susceptibility of subtype C reverse transcriptase from human immunodeficiency virus type 1 seroconverters in Zimbabwe. J. Virol. 71:5441-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Shafer, R. W., K. Hertogs, A. R. Zolopa, A. Warford, S. Bloor, B. J. Betts, T. C. Merigan, R. Harrigan, and B. A. Larder. 2001. High degree of interlaboratory reproducibility of human immunodeficiency virus type 1 protease and reverse transcriptase sequencing of plasma samples from heavily treated patients. J. Clin. Microbiol. 39:1522-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Shafer, R. W., P. Hsu, A. K. Patick, C. Craig, and V. Brendel. 1999. Identification of biased amino acid substitution patterns in human immunodeficiency virus type 1 isolates from patients treated with protease inhibitors. J. Virol. 73:6197-6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Shafer, R. W., A. K. Iversen, M. A. Winters, E. Aguiniga, D. A. Katzenstein, and T. C. Merigan. 1995. Drug resistance and heterogeneous long-term virologic responses of human immunodeficiency virus type 1-infected subjects to zidovudine and didanosine combination therapy. The AIDS Clinical Trials group 143 Virology Team. J. Infect. Dis. 172:70-78. [DOI] [PubMed] [Google Scholar]
- 336.Shafer, R. W., D. R. Jung, and B. J. Betts. 2000. Human immunodeficiency virus type 1 reverse transcriptase and protease mutation search engine for queries. Nat. Med. 6:1290-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Shafer, R. W., R. Kantor, and M. J. Gonzales. 2000. The genetic basis of HIV-1 resistance to reverse transcriptase and protease inhibitors. AIDS Rev. 2:211-218. [PMC free article] [PubMed] [Google Scholar]
- 338.Shafer, R. W., M. J. Kozal, M. A. Winters, A. K. Iversen, D. A. Katzenstein, M. V. Ragni, W. A. Meyer, P. Gupta, S. Rasheed, R. Coombs, and T. C. Merigan. 1994. Combination therapy with zidovudine and didanosine selects for drug-resistant human immunodeficiency virus type 1 strains with unique patterns of pol gene mutations. J. Infect. Dis. 169:722-729. [DOI] [PubMed] [Google Scholar]
- 339.Shafer, R. W., D. Stevenson, and B. Chan. 1999. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 27:348-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Shafer, R. W., A. Warford, M. A. Winters, and M. J. Gonzales. 2000. Reproducibility of human immunodeficiency virus type 1 (HIV-1) protease and reverse transcriptase sequencing of plasma samples from heavily treated HIV-1-infected individuals. J. Virol. Med. 86:143-153. [DOI] [PubMed] [Google Scholar]
- 341.Shafer, R. W., M. A. Winters, K. Dupnik, and S. Eshleman. 2001. A guide to HIV reverse transcriptase and protease sequencing for drug resistance studies. In C. L. Kuiken, B. Foley, B. H. Hahn, P. Marx, F. E. McCutchan, J. Mellors, J. I. Mullins, S. Wolanski, and B. Korber (ed.), Human retroviruses and AIDS: a compilation and analysis of nucleic and amino acid sequences. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 342.Shafer, R. W., M. A. Winters, A. K. Iversen, and T. C. Merigan. 1996. Genotypic and phenotypic changes during culture of a multinucleoside-resistant human immunodeficiency virus type 1 strain in the presence and absence of additional reverse transcriptase inhibitors. Antimicrob. Agents Chemother. 40:2887-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Shafer, R. W., M. A. Winters, S. Palmer, and T. C. Merigan. 1998. Multiple concurrent reverse transcriptase and protease mutations and multidrug resistance of HIV-1 isolates from heavily treated patients. Ann. Intern. Med. 128:906-911. [DOI] [PubMed] [Google Scholar]
- 344.Shah, F. S., K. A. Curr, M. E. Hamburgh, M. A. Parniak, H. Mitsuya, J. G. Arnez, and V. R. Prasad. 2000. Differential influence of nucleoside analog-resistance mutations K65R and L74V on the overall mutation rate and error specificity of human immunodeficiency virus type 1 reverse transcriptase. J. Biol. Chem. 275:27037-27044. [DOI] [PubMed] [Google Scholar]
- 345.Sham, H. L., D. J. Kempf, A. Molla, K. C. Marsh, G. N. Kumar, C. M. Chen, W. Kati, K. Stewart, R. Lal, A. Hsu, D. Betebenner, M. Korneyeva, S. Vasavanonda, E. McDonald, A. Saldivar, N. Wideburg, X. Chen, P. Niu, C. Park, V. Jayanti, B. Grabowski, G. R. Granneman, E. Sun, A. J. Japour, and J. M. Leonard. 1998. ABT-378, a highly potent inhibitor of the human immunodeficiency virus protease. Antimicrob. Agents Chemother. 42:3218-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Shao, W., L. Everitt, M. Manchester, D. D. Loeb, C. A. Hutchison, and R. Swanstrom. 1997. Sequence requirements of the HIV-1 protease flap region determined by saturation mutagenesis and kinetic analysis of flap mutants. Proc. Natl. Acad. Sci. USA 94:2243-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Sharma, P. L., and C. S. Crumpacker. 1997. Attenuated replication of human immunodeficiency virus type 1 with a didanosine-selected reverse transcriptase mutation. J. Virol. 71:8846-8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Sharma, P. L., and C. S. Crumpacker. 1999. Decreased processivity of human immunodeficiency virus type 1 reverse transcriptase (RT) containing didanosine-selected mutation Leu74Val: a comparative analysis of RT variants Leu74Val and lamivudine-selected Met184Val. J. Virol. 73:8448-8456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Shih, C. K., J. M. Rose, G. L. Hansen, J. C. Wu, A. Bacolla, and J. A. Griffin. 1991. Chimeric human immunodeficiency virus type 1/type 2 reverse transcriptases display reversed sensitivity to nonnucleoside analog inhibitors. Proc. Natl. Acad. Sci. USA 88:9878-9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Shirasaka, T., M. F. Kavlick, T. Ueno, W. Y. Gao, E. Kojima, M. L. Alcaide, S. Chokekijchai, B. M. Roy, E. Arnold, and R. Yarchoan. 1995. Emergence of human immunodeficiency virus type 1 variants with resistance to multiple dideoxynucleosides in patients receiving therapy with dideoxynucleosides. Proc. Natl. Acad. Sci. USA 92:2398-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Shirasaka, T., R. Yarchoan, M. C. O'Brien, R. N. Husson, B. D. Anderson, E. Kojima, T. Shimada, S. Broder, and H. Mitsuya. 1993. Changes in drug sensitivity of human immunodeficiency virus type 1 during therapy with azidothymidine, dideoxycytidine, and dideoxyinosine: an in vitro comparative study. Proc. Natl. Acad. Sci. USA 90:562-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Shulman, N., A. R. Zolopa, D. Passaro, R. W. Shafer, W. Huang, D. Katzenstein, D. M. Israelski, N. Hellmann, C. Petropoulos, and J. Whitcomb. 2001. Phenotypic hypersusceptibility to non-nucleoside reverse transcriptase inhibitors in treatment-experienced HIV-infected patients: impact on virological response to efavirenz-based therapy. AIDS 15:1125-1132. [DOI] [PubMed] [Google Scholar]
- 353.Shulman, N. S., R. A. Machekano, R. W. Shafer, M. A. Winters, A. R. Zolopa, S. H. Liou, M. Hughes, and D. A. Katzenstein. 2001. Genotypic correlates of a virologic response to stavudine after zidovudine monotherapy. J. Acquir. Immune Defic. Syndr. 27:377-380. [DOI] [PubMed] [Google Scholar]
- 354.Shulman, N. S., M. A. Winters, R. W. Shafer, A. R. Zolopa, S. H. Liou, M. Hughes, J. Whitcomb, C. Petropoulos, N. Hellmann, and D. Katzenstein. 2001. Subtle changes in susceptibility to stavudine predict virological response to stavudine monotherapy after zidovudine treatment. Antivir. Ther. 6:91. [Google Scholar]
- 355.Shulman, N. S., A. R. Zolopa, D. J. Passaro, U. Murlidharan, D. M. Israelski, C. L. Brosgart, M. D. Miller, S. Van Doren, R. W. Shafer, and D. A. Katzenstein. 2000. Efavirenz- and adefovir dipivoxil-based salvage therapy in highly treatment-experienced patients: clinical and genotypic predictors of virologic response. J. Acquir. Immune Defic. Syndr. 23:221-226. [DOI] [PubMed] [Google Scholar]
- 356.Simmonds, P., L. Q. Zhang, F. McOmish, P. Balfe, C. A. Ludlam, and A. J. Brown. 1991. Discontinuous sequence change of human immunodeficiency virus (HIV) type 1 env sequences in plasma viral and lymphocyte-associated proviral populations in vivo: implications for models of HIV pathogenesis. J. Virol. 65:6266-6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Simon, V., J. Vanderhoeven, A. Hurley, B. Ramratnam, D. Boden, M. Louie, R. Kost, K. Dawson, N. Parkin, J. P. Routy, R. P. Sekaly, and M. Markowitz. 2001. Prevalence of drug-resistant HIV-1 variants in newly infected individuals during 1999-2000. In 8th Conference on Retroviruses and Opportunistic Infections, Chicago, Ill.
- 358.Reference deleted.
- 359.Sluis-Cremer, N., D. Arion, N. Kaushik, H. Lim, and M. A. Parniak. 2000. Mutational analysis of Lys65 of HIV-1 reverse transcriptase. Biochem. J. 348:77-82. [PMC free article] [PubMed] [Google Scholar]
- 360.Smeaton, L. M., V. DeGruttola, G. K. Robbins, and R. W. Shafer. 2001. Actg (aids clinical trials group) 384. a strategy trial comparing consecutive treatments for hiv-1. Control. Clin. Trials. 22:142-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Smith, M. S., K. L. Koerber, and J. S. Pagano. 1993. Zidovudine-resistant human immunodeficiency virus type 1 genomes detected in plasma distinct from viral genomes in peripheral blood mononuclear cells. J. Infect. Dis. 167:445-448. [DOI] [PubMed] [Google Scholar]
- 362.Smith, R. A., G. J. Klarmann, K. M. Stray, U. K. von Schwedler, R. F. Schinazi, B. D. Preston, and T. W. North. 1999. A new point mutation (P157S) in the reverse transcriptase of human immunodeficiency virus type 1 confers low-level resistance to (-)-beta-2′,3′-dideoxy-3′-thiacytidine. Antimicrob. Agents Chemother. 43:2077-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Smith, R. A., K. M. Remington, B. D. Preston, R. F. Schinazi, and T. W. North. 1998. A novel point mutation at position 156 of reverse transcriptase from feline immunodeficiency virus confers resistance to the combination of (-)-beta-2′,3′-dideoxy-3′-thiacytidine and 3′-azido-3′-deoxythymidine. J. Virol. 72:2335-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Reference deleted.
- 365.Spence, R. A., W. M. Kati, K. S. Anderson, and K. A. Johnson. 1995. Mechanism of inhibition of HIV-1 reverse transcriptase by nonnucleoside inhibitors. Science 267:988-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Reference deleted.
- 367.Srinivas, R. V., and A. Fridland. 1998. Antiviral activities of 9-R-2-phosphonomethoxypropyl adenine (PMPA) and bis(isopropyloxymethylcarbonyl)PMPA against various drug-resistant human immunodeficiency virus strains. Antimicrob. Agents Chemother. 42:1484-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.St. Clair, M., J. L. Martin, G. Tudor-Williams, M. C. Bach, C. L. Vavro, D. M. King, P. Kellam, S. D. Kemp, and B. A. Larder. 1991. Resistance to ddI and sensitivity to AZT induced by a mutation in HIV-1 reverse transcriptase. Science 253:1557-1559. [DOI] [PubMed] [Google Scholar]
- 369.Reference deleted.
- 370.Stoddart, C. A., T. J. Liegler, F. Mammano, V. D. Linquist-Stepps, M. S. Hayden, S. G. Deeks, R. M. Grant, F. Clavel, and J. M. McCune. 2001. Impaired replication of protease inhibitor-resistant HIV-1 in human thymus. Nat. Med. 7:712-718. [DOI] [PubMed] [Google Scholar]
- 371.Stuyver, L., A. Wyseur, A. Rombout, J. Louwagie, T. Scarcez, C. Verhofstede, D. Rimland, R. F. Schinazi, and R. Rossau. 1997. Line probe assay for rapid detection of drug-selected mutations in the human immunodeficiency virus type 1 reverse transcriptase gene. Antimicrob. Agents Chemother. 41:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Tamalet, C., C. Pasquier, N. Yahi, P. Colson, I. Poizot-Martin, G. Lepeu, H. Gallais, P. Massip, J. Puel, and J. Izopet. 2000. Prevalence of drug resistant mutants and virological response to combination therapy in patients with primary HIV-1 infection. J. Med. Virol. 61:181-186. [DOI] [PubMed] [Google Scholar]
- 373.Tamalet, C., N. Yahi, C. Tourres, P. Colson, A. M. Quinson, I. Poizot-Martin, C. Dhiver, and J. Fantini. 2000. Multidrug resistance genotypes (insertions in the beta3-beta4 finger subdomain and MDR mutations) of HIV-1 reverse transcriptase from extensively treated patients: incidence and association with other resistance mutations. Virology 270:310-316. [DOI] [PubMed] [Google Scholar]
- 374.Tanuri, A., A. C. Vicente, K. Otsuki, C. A. Ramos, O. C. Ferreira, Jr., M. Schechter, L. M. Janini, D. Pieniazek, and M. A. Rayfield. 1999. Genetic variation and susceptibilities to protease inhibitors among subtype B and F isolates in Brazil. Antimicrob. Agents Chemother. 43:253-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Tebas, P., A. K. Patick, E. M. Kane, M. K. Klebert, J. H. Simpson, A. Erice, W. G. Powderly, and K. Henry. 1999. Virologic responses to a ritonavir-saquinavir-containing regimen in patients who had previously failed nelfinavir. AIDS 13:F23-F28. [DOI] [PubMed] [Google Scholar]
- 376.Tisdale, M., T. Alnadaf, and D. Cousens. 1997. Combination of mutations in human immunodeficiency virus type 1 reverse transcriptase required for resistance to the carbocyclic nucleoside 1592U89. Antimicrob. Agents Chemother. 41:1094-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Tisdale, M., S. D. Kemp, N. R. Parry, and B. A. Larder. 1993. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3′-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc. Natl. Acad. Sci. USA 90:5653-5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Tisdale, M., R. E. Myers, B. Maschera, N. R. Parry, N. M. Oliver, and E. D. Blair. 1995. Cross-resistance analysis of human immunodeficiency virus type 1 variants individually selected for resistance to five different protease inhibitors. Antimicrob. Agents Chemother. 39:1704-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Tong, W., C. D. Lu, S. K. Sharma, S. Matsuura, A. G. So, and W. A. Scott. 1997. Nucleotide-induced stable complex formation by HIV-1 reverse transcriptase. Biochemistry 36:5749-5757. [DOI] [PubMed] [Google Scholar]
- 380.Torti, C., Y. Gilleece, K. Hertogs, B. G. Gazzard, and A. L. Pozniak. 2001. R211K and L214F do not invariably confer high level phenotypic resistance to thymidine analogs in zidovudine-naive patients with M184V. J. Acquir. Immune Defic. Syndr. 26:514-515. [DOI] [PubMed] [Google Scholar]
- 380a.Tural, C., L. Ruiz, C. Holtzer, J. Schapiro, P. Viciana, J. Gonzalez, P. Domingo, C. Boucher, C. Rey-Joly, and B. Clotet. 2002. Clinical utility of HIV-1 genotyping and expert advice: the Havana trial. AIDS 16:209-218. [DOI] [PubMed] [Google Scholar]
- 381.Reference deleted.
- 382.Reference deleted.
- 383.Turner, S. R., J. W. Strohbach, R. A. Tommasi, P. A. Aristoff, P. D. Johnson, H. I. Skulnick, L. A. Dolak, E. P. Seest, P. K. Tomich, M. J. Bohanon, M. M. Horng, J. C. Lynn, K. T. Chong, R. R. Hinshaw, K. D. Watenpaugh, M. N. Janakiraman, and S. Thaisrivongs. 1998. Tipranavir (PNU-140690): a potent, orally bioavailable nonpeptidic HIV protease inhibitor of the 5,6-dihydro-4-hydroxy-2-pyrone sulfonamide class. J. Med. Chem. 41:3467-3476. [DOI] [PubMed] [Google Scholar]
- 384.United Kingdom Collaborative group on. Monitoring the Transmission of HIV Drug Resistance. 2001. Analysis of prevalence of HIV-1 drug resistance in primary infections in the United Kingdom. Br. Med. J. 322:1087-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Reference deleted.
- 386.Vahey, M., M. E. Nau, S. Barrick, J. D. Cooley, R. Sawyer, A. A. Sleeker, P. Vickerman, S. Bloor, B. Larder, N. L. Michael, and S. A. Wegner. 1999. Performance of the Affymetrix GeneChip HIV PRT 440 platform for antiretroviral drug resistance genotyping of human immunodeficiency virus type 1 clades and viral isolates with length polymorphisms. J. Clin. Microbiol. 37:2533-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Vaillancourt, M., D. Irlbeck, T. Smith, R. W. Coombs, and R. Swanstrom. 1999. The HIV type 1 protease inhibitor saquinavir can select for multiple mutations that confer increasing resistance. AIDS Res. Hum. Retrovirus. 15:355-363. [DOI] [PubMed] [Google Scholar]
- 388.Van Laethem, K., K. Van Vaerenbergh, J. C. Schmit, S. Sprecher, P. Hermans, V. De Vroey, R. Schuurman, T. Harrer, M. Witvrouw, E. Van Wijngaerden, L. Stuyver, M. Van Ranst, J. Desmyter, E. De Clercq, and A. M. Vandamme. 1999. Phenotypic assays and sequencing are less sensitive than point mutation assays for detection of resistance in mixed HIV-1 genotypic populations. J. Acquir. Immune Defic. Syndr. 22:107-118. [DOI] [PubMed] [Google Scholar]
- 389.Van Laethem, K., M. Witvrouw, J. Balzarini, J. C. Schmit, S. Sprecher, P. Hermans, M. Leal, T. Harrer, L. Ruiz, B. Clotet, M. Van Ranst, J. Desmyter, E. De Clercq, and A. M. Vandamme. 2000. Patient HIV-1 strains carrying the multiple nucleoside resistance mutations are cross-resistant to abacavir. AIDS 14:469-471. [DOI] [PubMed] [Google Scholar]
- 390.Van Vaerenbergh, K., K. Van Laethem, J. Albert, C. A. Boucher, B. Clotet, M. Floridia, J. Gerstoft, B. Hejdeman, C. Nielsen, C. Pannecouque, L. Perrin, M. F. Pirillo, L. Ruiz, J. C. Schmit, F. Schneider, A. Schoolmeester, R. Schuurman, H. J. Stellbrink, L. Stuyver, J. Van Lunzen, B. Van Remoortel, E. Van Wijngaerden, S. Vella, M. Witvrouw, S. Yerly, E. De Clercq, J. Destmyer, and A. M. Vandamme. 2000. Prevalence and characteristics of multinucleoside-resistant human immunodeficiency virus type 1 among European patients receiving combinations of nucleoside analogues. Antimicrob. Agents Chemother. 44:2109-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Van Vaerenbergh, K., K. Van Laethem, E. Van Wijngaerden, J. C. Schmit, F. Schneider, L. Ruiz, B. Clotet, C. Verhofstede, F. Van Wanzeele, G. Muyldermans, P. Simons, L. Stuyver, P. Hermans, C. Evans, E. De Clercq, J. Desmyter, and A. M. Vandamme. 2000. Baseline HIV type 1 genotypic resistance to a newly added nucleoside analog is predictive of virologic failure of the new therapy. AIDS Res. Hum. Retrovirus. 16:529-537. [DOI] [PubMed] [Google Scholar]
- 392.Vandamme, A. M., F. Houyez, D. Banhegyi, B. Clotet, G. De Schrijver, K. A. De Smet, W. W. Hall, R. Harrigan, N. Hellmann, K. Hertogs, C. Holtzer, B. Larder, D. Pillay, E. Race, J. C. Schmit, R. Schuurman, E. Schulse, A. Sonnerborg, and V. Miller. 2001. Laboratory guidelines for the practical use of HIV drug resistance tests in patient follow-up. Antivir. Ther. 6:21-39. [PubMed] [Google Scholar]
- 393.Vandamme, A. M., K. Van Laethem, and E. De Clercq. 1999. Managing resistance to anti-HIV drugs: an important consideration for effective disease management. Drugs 57:337-361. [DOI] [PubMed] [Google Scholar]
- 394.Reference deleted.
- 395.Verhofstede, C., F. V. Wanzeele, B. Van Der Gucht, N. De Cabooter, and J. Plum. 1999. Interruption of reverse transcriptase inhibitors or a switch from reverse transcriptase to protease inhibitors resulted in a fast reappearance of virus strains with a reverse transcriptase inhibitor-sensitive genotype. AIDS 13:2541-2546. [DOI] [PubMed] [Google Scholar]
- 396.Wainberg, M. A., W. C. Drosopoulos, H. Salomon, M. Hsu, G. Borkow, M. Parniak, Z. Gu, Q. Song, J. Manne, S. Islam, G. Castriota, and V. R. Prasad. 1996. Enhanced fidelity of 3TC-selected mutant HIV-1 reverse transcriptase. Science 271:1282-1285. [DOI] [PubMed] [Google Scholar]
- 397.Wainberg, M. A., M. D. Miller, Y. Quan, H. Salomon, A. S. Mulato, P. D. Lamy, N. A. Margot, K. E. Anton, and J. M. Cherrington. 1999. In vitro selection and characterization of HIV-1 with reduced susceptibility to PMPA. Antivir. Ther. 4:87-94. [DOI] [PubMed] [Google Scholar]
- 398.Wainberg, M. A., and A. J. White. 2001. Current insights into reverse transcriptase inhibitor-associated resistance. Antivir. Ther. 6(Suppl. 2):11-19. [PubMed] [Google Scholar]
- 398a.Walmsley, S. L., D. V. Kelly, A. L. Tseng, A. Humar, and P. R. Harrigan. 2001. Non-nucleoside reverse transcriptase inhibitor failure impairs HIV-RNA responses to efavirenz-containing salvage antiretroviral therapy. AIDS 15:1581-1584. [DOI] [PubMed] [Google Scholar]
- 399.Walmsley, S. L., M. I. Becker, M. Zhang, A. Humar, and P. R. Harrigan. 2001. Predictors of virological response in HIV-infected patients to salvage antiretroviral therapy that includes nelfinavir. Antivir. Ther. 6:47-54. [PubMed] [Google Scholar]
- 399a.Walter, H., B. Schmidt, M. Werwein, E. Schwingel, and K. Korn. 2002. Prediction of abacavir resistance from genotypic data: impact of zidovudine and lamivudine resistance in vitro and in vivo. Antimicrob. Agents Chemother. 46:89-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Wei, X., S. K. Ghosh, M. E. Taylor, V. A. Johnson, E. A. Emini, P. Deutsch, J. D. Lifson, S. Bonhoeffer, M. A. Nowak, B. H. Hahn, and G. M. Shaw. 1995. Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373:117-122. [DOI] [PubMed] [Google Scholar]
- 401.Reference deleted.
- 402.Wilson, J. W., P. Bean, T. Robins, F. Graziano, and D. H. Persing. 2000. Comparative evaluation of three human immunodeficiency virus genotyping systems: the HIV-GenotypR Method, the HIV PRT GeneChip Assay, and the HIV-1 RT Line Probe Assay. J. Clin. Microbiol. 38:3022-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Winslow, D. L., S. Garber, C. Reid, H. Scarnati, D. Baker, M. M. Rayner, and E. D. Anton. 1996. Selection conditions affect the evolution of specific mutations in the reverse transcriptase gene associated with resistance to DMP 266. AIDS 10:1205-1209. [DOI] [PubMed] [Google Scholar]
- 404.Winters, M. A., J. D. Baxter, D. L. Mayers, D. N. Wentworth, M. L. Hoover, J. D. Neaton, and T. C. Merigan. 2000. Frequency of antiretroviral drug resistance mutations in HIV-1 strains from patients failing triple drug regimens. The Terry Beirn Community Programs for Clinical Research on AIDS. Antivir. Ther. 5:57-63. [PubMed] [Google Scholar]
- 404a.Winters, M. A., and T. C. Merigan. 2001. Variants other than aspartic acid at codon 69 of the human immunodeficiency virus type 1 reverse transcriptase gene affect susceptibility to nucleoside analogs. Antimicrob. Agents Chemother. 45:2276-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Winters, M. A., K. L. Coolley, P. Cheng, Y. A. Girard, H. Hamdan, L. C. Kovari, and T. C. Merigan. 2000. Genotypic, phenotypic, and modeling studies of a deletion in the beta3-beta4 region of the human immunodeficiency virus type 1 reverse transcriptase gene that is associated with resistance to nucleoside reverse transcriptase inhibitors. J. Virol. 74:10707-10713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Winters, M. A., K. L. Coolley, Y. A. Girard, D. J. Levee, H. Hamdan, R. W. Shafer, D. A. Katzenstein, and T. C. Merigan. 1998. A 6-basepair insert in the reverse transcriptase gene of human immunodeficiency virus type 1 confers resistance to multiple nucleoside inhibitors. J. Clin. Investig. 102:1769-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Reference deleted.
- 408.Reference deleted.
- 409.Winters, M. A., J. M. Schapiro, J. Lawrence, and T. C. Merigan. 1998. Human immunodeficiency virus type 1 protease genotypes and in vitro protease inhibitor susceptibilities of isolates from individuals who were switched to other protease inhibitors after long-term saquinavir treatment. J. Virol. 72:5303-5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Winters, M. A., R. W. Shafer, R. A. Jellinger, G. Mamtora, T. Gingeras, and T. C. Merigan. 1997. Human immunodeficiency virus type 1 reverse transcriptase genotype and drug susceptibility changes in infected individuals receiving dideoxyinosine monotherapy for 1 to 2 years. Antimicrob. Agents Chemother. 41:757-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Wong, J. K., M. Hezareh, H. F. Gunthard, D. V. Havlir, C. C. Ignacio, C. A. Spina, and D. D. Richman. 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278:1291-1295. [DOI] [PubMed] [Google Scholar]
- 412.Reference deleted.
- 413.Yahi, N., C. Tamalet, C. Tourres, N. Tivoli, F. Ariasi, F. Volot, J. A. Gastaut, H. Gallais, J. Moreau, and J. Fantini. 1999. Mutation patterns of the reverse transcriptase and protease genes in human immunodeficiency virus type 1-infected patients undergoing combination therapy: survey of 787 sequences. J. Clin. Microbiol. 37:4099-4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Yahi, N., C. Tamalet, C. Tourres, N. Tivoli, and J. Fantini. 2000. Mutation L210W of HIV-1 Reverse Transcriptase in Patients Receiving Combination Therapy. incidence, association with other mutations, and effects on the structure of mutated reverse transcriptase. J. Biomed. Sci. 7:507-513. [DOI] [PubMed] [Google Scholar]
- 415.Yang, G., Q. Song, M. Charles, W. C. Drosopoulos, E. Arnold, and V. R. Prasad. 1996. Use of chimeric human immunodeficiency virus types 1 and 2 reverse transcriptases for structure-function analysis and for mapping susceptibility to nonnucleoside inhibitors. J. Acquir. Immune Defic. Syndr. 11:326-333. [DOI] [PubMed] [Google Scholar]
- 416.Yerly, S., L. Kaiser, E. Race, J. P. Bru, F. Clavel, and L. Perrin. 1999. Transmission of antiretroviral-drug-resistant HIV-1 variants. Lancet 354:729-733. [DOI] [PubMed] [Google Scholar]
- 417.Yerly, S., A. Rakik, S. K. De Loes, B. Hirschel, D. Descamps, F. Brun-Vezinet, and L. Perrin. 1998. Switch to unusual amino acids at codon 215 of the human immunodeficiency virus type 1 reverse transcriptase gene in seroconvertors infected with zidovudine-resistant variants. J. Virol. 72: 3520-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Young, S. D., S. F. Britcher, L. O. Tran, L. S. Payne, W. C. Lumma, T. A. Lyle, J. R. Huff, P. S. Anderson, D. B. Olsen, S. S. Carroll, and et al. 1995. L-743, 726 (DMP-266): a novel, highly potent nonnucleoside inhibitor of the human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 39:2602-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Zhang, D., A. M. Caliendo, J. J. Eron, K. M. DeVore, J. C. Kaplan, M. S. Hirsch, and R. T. D'Aquila. 1994. Resistance to 2′,3′-dideoxycytidine conferred by a mutation in codon 65 of the human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 38:282-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Zhang, Y. M., H. Imamichi, T. Imamichi, H. C. Lane, J. Falloon, M. B. Vasudevachari, and N. P. Salzman. 1997. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J. Virol. 71:6662-6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Ziermann, R., K. Limoli, K. Das, E. Arnold, C. J. Petropoulos, and N. T. Parkin. 2000. A mutation in human immunodeficiency virus type 1 protease, N88S, that causes in vitro hypersensitivity to amprenavir. J. Virol. 74:4414-4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Reference deleted.
- 423.Zolopa, A. R., R. W. Shafer, A. Warford, J. G. Montoya, P. Hsu, D. Katzenstein, T. C. Merigan, and B. Efron. 1999. HIV-1 genotypic resistance patterns predict response to saquinavir-ritonavir therapy in patients in whom previous protease inhibitor therapy had failed. Ann. Intern. Med. 131:813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]