Abstract
Translin is a human octameric protein that specifically binds the single-stranded microsatellite repeats d(GT)n and the corresponding transcripts (GU)n. It also binds, with lesser affinities, other single-stranded G-rich DNA and RNA sequences. TRAX is a human protein that bears a homology to Translin and interacts with it. Translin and TRAX have been proposed to be involved in DNA recombination, chromosomal translocation and mRNA transport and translation. Both proteins are highly conserved in eukaryotes, including the fission yeast Schizosaccharomyces pombe, which is amenable to genetic analysis. Here, we report the first study of the S.pombe Translin and TRAX homologs. We have deleted the genes encoding Translin and TRAX in S.pombe and found that the proliferation of the mutant cells was slightly stimulated, suggesting that these genes are not essential for the fission yeast. We have also shown that the S.pombe Translin and TRAX interact. Biochemical analysis of the S.pombe Translin, which was cloned and expressed in Escherichia coli, revealed that it is octameric and that it selectively binds d(GT)n and d(GTT)n microsatellite repeats. However, unlike the human protein, it has much higher affinities for the homologous RNA sequences (GU)n and (GUU)n. These data suggest that the S.pombe Translin is primarily involved in functions related to RNA metabolism.
INTRODUCTION
The human protein Translin has been identified in cell extracts by virtue of its ability to bind single-stranded oligodeoxynucleotides consisting of consensus sequences adjacent to chromosomal translocations in leukemia and lymphoma cells (1). The Translin gene (designated tsn) has been cloned and found to encode a polypeptide consisting of 228 amino acids. The gene was expressed in Escherichia coli and eight molecules of the recombinant polypeptide were found to assemble into an octamer, which forms a ring structure. It was this structure that was shown to bind single-stranded DNA (2,3).
We have previously discovered in nuclear extracts of human fibroblasts a protein that specifically binds the single-stranded microsatellite repeats, d(GT)n, and with lower affinities, other G-rich DNA sequences (4). This protein was later shown to be identical, or closely related, to Translin (5). Recent studies carried out in our laboratory revealed that recombinant Translin binds single-stranded d(GT)n microsatellites and G-strand telomeric repeats, d(TTAGGG)n, with higher affinities (Kdis ≅ 2 nM and Kdis ≅ 12.5 nM, respectively) than the affinity with which it binds a prototypical sequence flanking translocation sites (Kdis ≅ 23 nM). However, it does not bind the corresponding double-stranded DNAs [(5,6); E. Jacob, N. Baran and H. Manor, unpublished data).
Translin has been highly conserved throughout eukaryotic evolution. Of the 228 amino acids in the mouse Translin (also designated TB-RBP), 225 amino acids are identical to corresponding amino acids in the human protein (7). Translin-like proteins in chicken, Xenopus, Drosophila and the fission yeast Schizosaccharomyces pombe share 86, 81, 53 and 36% identities, respectively, with the human Translin and also have similar lengths (8–10). However, the budding yeast Saccharomyces cerevisiae does not contain a Translin ortholog.
The 3D structures of the mouse and the human Translins have been determined by X-ray crystallography and found to be virtually identical (11,12). The monomer has a highly helical secondary structure with seven α helices, which constitute >70% of the amino acid residues. Both crystal structures reported had tetramers as asymmetric units, which by using crystal symmetry operations show that the actual crystallized complex is that of an octamer. However, the octamers visualized in the crystal structures are neither consistent with the assembly of the subunits into octameric rings as seen in electron microscopic studies (3), nor do they have a pore large enough to encompass single-stranded DNA or RNA.
A yeast two-hybrid screen of a human cDNA library led to the discovery of a 33 kDa protein that forms a specific complex with Translin. This protein, whose sequence has 28% identity with that of Translin, was designated Translin-associated factor X (TRAX) (13). There are homologous TRAX-like proteins in the various species that also contain Translin orthologs. The human and the mouse TRAX proteins, which share 90% amino acids identities, contain identical bipartite nuclear targeting sequence near their N-terminus, and the mouse protein targeting sequence was shown to be functional (14). TRAX does not bind DNA or RNA (15).
The data about the strong affinity of the human Translin to the d(GT)n and d(TTAGGG)n repeats indicated that it might play a role in the metabolism of d(GT)n.d(AC)n microsatellites and telomeres (5). It could also be involved in chromosomal translocations, as implied previously (2). Other studies, which were carried out with the mouse Translin, indicated that the mouse protein binds specific RNA sequences in the 3′-untranslated regions of mRNAs purified from testis and brain (16). Based on these and subsequent studies, it has been suggested that complexes including Translin and TRAX might be involved in the control of mRNA translation and transport (17–21). Thus, it appears likely that Translin and TRAX are multifunctional proteins that could play various roles in both DNA and RNA metabolism. However, a definition of their functions in precise molecular terms is lacking. We reasoned that the use of S.pombe as a model organism would be advantageous for such functional analysis, because S.pombe can be genetically manipulated by techniques that cannot be easily adopted in research performed on higher eukaryotic systems.
Hence, we have undertaken a study of the S.pombe Translin and TRAX homologs and present here the first report on their properties. We have deleted the genes encoding Translin and TRAX in S.pombe and found that the proliferation of the mutant cells was slightly stimulated, suggesting that these genes are not essential for the fission yeast. We have also shown that the S.pombe Translin and TRAX interact. Biochemical analysis of the S.pombe Translin, which was cloned and expressed in E.coli, revealed that it is octameric and that it selectively binds single-stranded DNA microsatellite repeats and the corresponding RNA sequences, but that it does not bind double-stranded DNA. However, it differs from the human Translin in having a much higher affinity for the RNA repeats. These data suggest that the S.pombe Translin is primarily involved in functions related to RNA metabolism.
MATERIALS AND METHODS
Materials
Oligodeoxynucleotides, oligoribonucleotides and [γ-32P]ATP were purchased from Sigma, Dharmacon and New England Nuclear, respectively. An S.pombe cDNA library was obtained from Dr A. Cohen of the Department of Molecular Biology, Hebrew University-Hadassah Medical School, Jerusalem, Israel. The vector expressing human Translin was obtained from Dr M. Kasai of the Department of Immunology, National institute of Infectious Diseases, Tokyo, Japan.
S.pombe culture procedures and construction of new strains
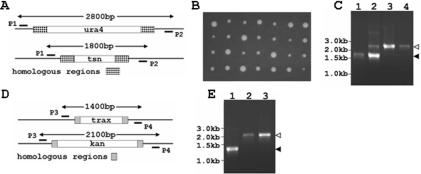
The S.pombe culture procedures described by Moreno et al. (22) were used. YE medium (0.5% yeast extract and 3% glucose) was used for vegetative growth. The strains constructed in this study are listed in Table 1 along with strains obtained from other laboratories. The strain SP1124f, a ura4− derivate of SP1124, was selected by growth of the latter strain on YES plates (plates of YE solid medium) containing 5-Fluoroorotic acid (5FOA). The strains AP137 and SP1124f were crossed on ME plates (3% malt extract) and diploid cells were selected on EMM plates (0.3% potassium hydrogen phthalate, 0.2% Na2HPO4, 0.5% NH4Cl and 2% glucose) lacking adenine. Diploid cells were transformed with a linear DNA fragment containing the ura4 open reading frame (ORF) flanked by sequences that also flank the tsn gene. Transformed cells were selected on EMM plates lacking adenine and uracil. Translin heterozygote strains were screened by PCR and the diploid ura4+ strain Spmd49 was found to harbor the correct gene replacement, as shown in Figure 4. Spmd49 was then induced to sporulate on ME plates. Asci were dissected and four viable spores were obtained in all tetrads. The strain Spm14d was selected from the sporulation offspring colonies on EMM plates lacking uracil. The correct gene replacement was confirmed by PCR analysis. The Δtrax strain was obtained from Dr A. Decottignies (23). Spm14d and Δtrax were allowed to mate and sporulate on ME plates. Asci were dissected and four viable spores were obtained in all tetrads. The strain Spm28a was selected from the sporulation offspring colonies by growth on EMM plates lacking uracil and on YES plates containing 100 mg/l of the antibiotic G418.
Table 1.
S.pombe strains used in the present study
| Strain | Genotype | Source |
|---|---|---|
| FY370 | h+, ade6-DN/N, ura4-D18 | (44) |
| AP137 | mat1-PΔ17::leu2, leu1-32, ura4-D18, ade6-210, his2 | (45) |
| SP1124 | mat1-Msmt0, L(XbaI) ::ura4+, ade-216, ura4-D18 | (45) |
| SP1124fa | mat1-Msmt0, L(XbaI) ::ura4−, ade-216, ura4-D18 | This study |
| ΔTRAX | h−, Δtraxb::kanR, leu1-32, ura4-D18, ade6-216 | (23) |
| Spmd49c | mat1-PΔ17::leu2, mat1-Msmt0, leu1-32, ura4-D18, L(XbaI)::ura4−, Δtsn::ura4+, ade6-210, ade-216, his2 | This study |
| Spm14d | mat1-PΔ17::leu2, Δtsn::ura4+, leu1-32, ura4-D18, ade6-210 | This study |
| Spm28a | h−, Δtsn::ura4, Δtrax::kanR, leu1-32, ura4-D18, ade6-210 | This study |
aSelected from SP1124 by growth in medium containing 5FOA.
btrax designates the gene encoding the TRAX protein. In (23) the TRAX open reading frame is designated spcc736.09c.
cTranslin heterozygous diploid.
Figure 4.
Construction of tsn− and tsn−trax− mutants. S.pombe tsn− strains were generated by first deleting one of the two Translin alleles in a diploid. This was accomplished by replacement of one tsn allele with a ura4 gene, through homologous recombination with a linear DNA fragment containing the ura4 open reading frame flanked on both sides by sequences that also flank the tsn gene. Next, meiosis was induced and a tetrad analysis was performed. Haploid tsn− colonies were found in the tetrads. We obtained a trax− S.pombe strain, which was generated by Decottignies et al. (23), and crossed this strain with a tsn− haploid. Meiosis was induced again and strains, in which both Translin and TRAX have been deleted, were found among the haploid descendents of the diploids generated by the cross. (A) Scheme of the heterozygous diploid, tsn+/tsn−. Primers P1 and P2 were used for PCR analysis to confirm the indicated gene replacement. The expected lengths of the PCR products before and after the gene replacement are indicated. (B) Tetrad analysis of the heterozygous diploid strain tsn+/tsn−. Four viable spores were obtained in all tetrads. The larger colonies were shown to be ura4+,tsn−. (C) PCR analysis of a tsn+haploid (lane 1), a heterozygous diploid tsn+/tsn− (lane 2), a haploid tsn−::ura4 (lane 3) and a haploid tsn−::ura4, trax−::kanR (lane 4). The expected length for a tsn+ product (1.8 kb) and a tsn− product (2.8 kb) are shown by a closed and an open arrowhead, respectively. (D) Scheme for trax gene replacement. The TRAX open reading frame has been replaced in S.pombe by the kanamycin-resistant gene (kanR) (23). We used the primer P3 and P4 for PCR analysis designed to confirm the gene replacement. The expected lengths of the PCR product before and after the gene replacement are indicated. (E) PCR analysis of a haploid strain trax+ (lane 1), a haploid strain trax−::kanR (lane 2) and a haploid strain tsn−::ura4, trax−::kanR (lane 3). The expected length for a trax+ product (1.4 kb) and a trax− product (2.1 kb) are indicated by closed and open arrowheads, respectively.
DNA transfection into S.pombe cells
DNA was transfected into S.pombe cells by electroporation, as described previously (24). Then, the cells were plated on EMM medium supplemented with 225 mg/l Leucine and 225 mg/l adenine. The plates were incubated at 30°C for 3–4 days.
PCR amplification of chromosomal DNA fragments
Chromosomal DNA was extracted from yeast cells, as described previously (25). DNA fragments from such chromosomal DNA preparations were amplified by PCR, using the BioTools DNA polymerase, with the primer pairs: P1—TTATGACAAAGTCATGGTC and P2—AAAAAAACATGCACGCC for analysis of the Translin ORF region in the S.pombe genome, or P3—CACAGCATAAATTCTTGC and P4—GTCCAGCATACAGAATCTC for analysis of the TRAX ORF region in the S.pombe genome.
Cloning and expression of the S.pombe Translin and TRAX cDNAs, and purification of the proteins
The predicted ORFs of the S.pombe tsn and trax genes have been identified by the S.pombe genome sequencing project (10). These ORFs were amplified by PCR from an S.pombe cDNA library and cloned in-frame into a bacterial expression of vector (pQE31; QIAGEN) encoding an N-terminal hexahistidine tag. These expression constructs were used for the expression the proteins in E.coli (26). The recombinant proteins were purified from the transformed bacteria, as described previously (5) (see Supplementary Material).
Gel mobility shift assays
Gel mobility shift assays were performed to assess the binding of Translin with oligodeoxynucleotides and oligoribonucleotides that were labeled with 32P at their 5′-termini. The labeled oligonucleotides were mixed with Translin at 4°C in 20 μl of binding buffer [10 mM Tris–HCl, pH 8.0, 0.1 mM EDTA, 1 mM DTT and 10% glycerol] containing 20 mM NaCl (S.pombe Translin) or 75 mM NaCl (human Translin). In the RNA-binding assays, RNAsine (Promega) at a concentration of 0.2 U/μl was added. The mixtures were incubated for 15 min at 30°C. Other details of the procedure used for these assays have been described previously (5).
Glycerol gradient centrifugation
Recombinant S.pombe Translin was loaded on a 10 ml linear 20–40% (v/v) glycerol gradient containing 20 mM Tris–HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA and 2 mM DTT. The tubes were centrifuged for 24 h in a Sorvall ultracentrifuge in the TH-641 rotor at 38 000 r.p.m. at 8°C. Fractions of 0.25 ml were collected from the bottom of the gradient. The position attained by the protein was determined by SDS–PAGE of aliquots withdrawn from each fraction, followed by Coomassie blue staining of the gel. The positions of the RNA- and the DNA-binding activities were determined by gel mobility shift assays of other aliquots withdrawn from the same gradient fractions, using 32P-labeled (GU)12 and d(GT)12 as probes and phosphorimaging of the gels. For centrifugation of preformed complexes, recombinant Translin (20 μg) was preincubated with the 32P-labeled (GU)12 probe (2 ng) in 100 μl binding buffer for 15 min at 30°C. Following centrifugation, aliquots from each fraction were used to determine the position attained by the protein, as described above. Other aliquots were electrophoresed in a native 6% polyacrylamide gel and 32P-labeled RNA–protein complexes were detected by the use of the PhosphorImager. Estimations of the sedimentation constant and the molecular weight of Translin were obtained by comparison with sedimentation constants of known protein markers, as described previously (27).
Yeast two-hybrid assays
The cDNAs encoding the complete ORFs of S.pombe Translin and TRAX were subcloned into the BamHI/SalI sites of the pGAD-C and pGBD-C vectors (28). Pairs of binding domain and activation domain constructs were cotransfected into the S.cerevisiae strain Y1183 (mata, ura3-52, his3-200, ade2-101, lys2-801, trp1-901, leu2,3-112, gal4-542, gal80-538, lys2::gal1UAS-gal1TATA-his3, ura3::gal417mers(x3)-cyc1TATA-lacZ, gal2-ade2; Y. Kassir and N. Guttmann, personal communication). Co-transformants were selected on SD medium [0.67% nitrogen base without amino acids (Difco) and 2% glucose] lacking leucine and tryptophan. Protein–protein interactions were detected by growth on SD medium lacking leucine, tryptophan and adenine and by growth on SD medium lacking leucine, tryptophan and histidine supplemented with 15 mM 3-amino-1,2,4-Triazole (Sigma).
Western blotting
Antibodies directed against the purified recombinant S.pombe Translin were raised in rabbits using standard procedures (29). S.pombe strains were grown to mid-log phase (A595 = 0.4–0.6) and washed once with ice-cold STOP buffer (150 mM NaCl, 50 mM NaF, 10 mM EDTA, 1 mM NaN3, pH 8.0). Pellets were resuspended in a buffer containing 20 mM Tris–HCl, pH 7.5, 1 mM EDTA, 1 mM DTT, 0.4% NP-40, 40 mM β-glycerophosphate and 0.1 mM sodium orthovanadate supplemented with protease inhibitor cocktail (Sigma). Cells were disrupted in a French pressure cell press and extracts were cleared by centrifugation in an Eppendorf microfuge (20 min, 10 000 g, 4°C). Protein concentrations were estimated by using the Bradford protein assay (Bio-Rad). An aliquot of 150 μg of total protein from each strain were loaded on a 12% SDS–PAGE gel, electrophoresed and transferred to Imobilon P membrane (Millipore). The membrane was blocked in Tris-buffered saline buffer containing 20% skim milk and 0.1% Tween-20 at room temperature for 1 h and then probed with polyclonal anti-Translin antibodies (1:500) in the same buffer for half an hour. Peroxidase conjugated anti-rabbit IgG (1:5000) (Sigma) and an enhanced chemiluminescence detection kit (Amersham Biosciences) were used to detect Translin. Equal sample loading was confirmed by Coomassie blue staining of the membrane.
RESULTS
Comparison between the sequences and structures of the S.pombe and the human Translin and TRAX
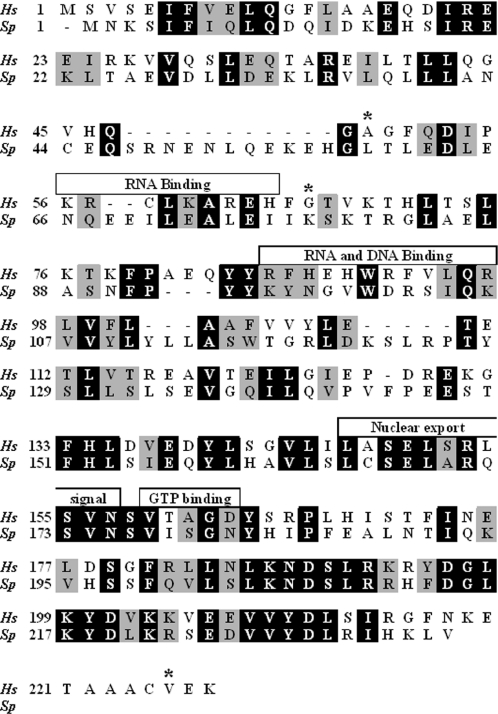
Figure 1 presents a BLAST alignment of the amino acid sequences of the human Translin, which consists of 228 amino acids, and the S.pombe Translin, which consists of 236 amino acids. The mouse Translin, which also consists of 228 amino acids, differs from the human Translin by three amino acids, whose positions are indicated in Figure 1 by asterisks. None of these amino acids is found within the regions assigned to functional domains (see below). It is evident that, while the overall sequences of the S.pombe and the human Translins share 36% identity and 54% similarity, the degrees of identity and similarity are higher in the C-terminal half of the proteins than in their N-terminal half. In the human Translin, the C-terminal region contains a putative nuclear export signal and a putative GTP binding site. Evidently, the nuclear export signal is highly conserved in the S.pombe Translin. The N-terminal half of the human and the mouse Translins include two basic regions. One basic region (amino acids 56–64), particularly the basic amino acids K60, R62 and H64, was shown by site-directed mutagenesis to be required for RNA-binding by the mouse Translin (15). Evidently, these basic amino acids have not been conserved in the S.pombe Translin. The second region (amino acids 86–97) also includes basic amino acids (R86, H88 and H90) that were found to be required for binding single-stranded DNA and RNA by the human and the mouse Translins (15,30). As shown in Figure 1, this region has been only partially conserved in the S.pombe Translin.
Figure 1.
Amino acid sequence alignment of the human and the S.pombe Translins. The sequences of the human Translin (Hs, Homo sapiens, GenBank™ gi: 12803111) and its S.pombe ortholog (Sp, S.pombe, GenBank™ gi: 19115469) were aligned using BLAST. Identical residues are highlighted in black. Similar residues are highlighted in gray. Previously identified motifs in the human Translin (also shared by the mouse Translin) are marked with boxes and are described in the Results. The asteriks indicate the amino acids in the human Translin that have been substituted with other amino acids in the mouse Translin.
The program ClustalW was used to align the amino acid sequences of the human, mouse and S.pombe TRAX proteins (data not shown). This alignment revealed that the sequence of 34 amino acids at the N-terminus of the human and the mouse TRAX proteins, which includes the bipartite nuclear targeting signal (amino acids 11–27, RKrkhdnfphnqRRegK) (13), is missing in the S.pombe TRAX protein (the two clusters of basic amino acids that constitute the signal are shown in boldface). However, we observed that the C-terminus of the S.pombe TRAX contains a putative bipartite nuclear targeting signal (amino acids 210–229, KRylnlevdtatppeeKRiR).
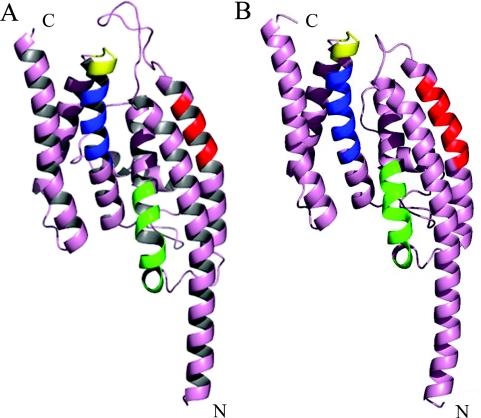
As noted in the Introduction, the structures of the human and the mouse Translins have been determined by X-ray crystallography and found to be very similar. We, therefore, built a sequence-dependent homology-based model of the S.pombe Translin, using the 3D-JIGSAW comparative modeling server (31) and the human Translin (PDB code 1J1J) as the structural template. A similar model based on the mouse Translin (PDB code 1KEY) using the 3D-PSSM protein fold recognition server (32) resulted in a highly similar structure (data not shown). The S.pombe Translin monomer model, shown in Figure 2A, predicts that the structure of this monomer is very similar to the structure of the human monomer, shown in Figure 2B, with an overall root-mean-square deviation of 0.91 Å over all superimposable α-carbons. The approximate positions of the Translin functional subdomains, based on the positions of these subdomains in the human and the mouse Translins, are also indicated on the S.pombe model. However, it should be noted that local structural differences within the putative RNA- and DNA-binding subdomains would not have been detected by this approach. Such changes may account for differences that we observed in the nucleic acids binding characteristics between the S.pombe and the human Translins (see below).
Figure 2.
Homology-based model of the S.pombe Translin monomer. (A) The S.pombe sequence was folded using the human Translin crystal structure (1J1J) as its template, by the 3D-JIGSAW comparative modeling server. Putative functional domains are identified by colored ribbons: red, RNA-binding domain; green, DNA/RNA-binding domain, blue, nuclear export signal; yellow, GTP binding site. (B) Visualization of one monomer of the human Translin structure (1J1J). The color coding is the same as that of (A). As denoted in Figure 1, the positions of the human functional domains are predicted to be as follows: RNA-binding domain: amino acids 56–64; RNA- and DNA-binding domain: amino acids 86–97; nuclear export signal: amino acids 147–157; GTP binding site: amino acids 159–163.
Evidence that the endogeneous tsn and trax genes are expressed in S.pombe
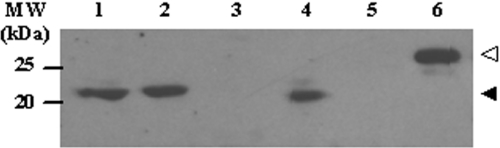
The S.pombe Translin and TRAX have been identified by the S.pombe genome sequencing project (10). This finding did not reveal whether the endogeneous genes encoding these proteins are actually expressed in S.pombe cells. To examine this question, we performed a western analysis of cell extracts prepared from wild-type S.pombe and from tsn− and trax− S.pombe strains (see below). The western blots were probed with a polyclonal anti-Translin antibody that we have prepared against recombinant S.pombe Translin, as described in Materials and Methods; Figure 3 shows these blots. It can be seen (lanes 1 and 2) that a band containing a protein of an apparent molecular weight of 23 kDa, which specifically interacted with the antibody, was found in the extracts prepared from a tsn+ wild-type haploid and from a tsn+ haploid member of a tetrad, which also contained a tsn− derivate (see below). A similar band was observed in an extract prepared from trax− cells (lane 4), but was absent from extracts of tsn− and tsn−,trax− haploids (lanes 3 and 5). These data indicated that this band contains Translin. Lane 6 shows a band containing recombinant Translin synthesized in E.coli, which was run in the same gel. As described below, the apparent molecular weight of the recombinant protein was 29 kDa. The discrepancy between the masses of the recombinant and the endogeneous Translins could be due to the presence of a histidine tag in the recombinant protein.
Figure 3.
Western blot analysis of endogeneous Translin in S.pombe cell extracts. The western blot procedure was carried out, as described in Materials and Methods. 150 μg of total protein from wild-type or mutant S.pombe cells were loaded per lane. Recombinant S.pombe Translin served as a positive control. Polyclonal anti-S.pombe Translin antibodies were used to probe the membrane. Lane 1, wild-type strain (FY370); lane 2, tsn+ derivative of a heterozygous diploid tsn+/tsn−; lane 3, tsn− derivative of the heterozygous diploid tsn+/tsn−; lane 4, trax− mutant; lane 5, tsn−trax− mutant; lane 6, Recombinant S.pombe Translin. Closed arrowhead marks the position of endogenous Translin. Open arrowhead marks the position of recombinant Translin. Equal sample loading was confirmed by Coomassie blue staining. The mutant genotypes are described in the legend to Figure 4 and in the text.
The authenticity of the endogenous Translin was further confirmed by immunoprecipitation assays performed with the anti-Translin antibody, followed by SDS–PAGE of the immunoprecipitates and mass-spectrometry of the bands. Similar immunoprecipitation assays performed with an anti-TRAX antibody revealed that endogenous TRAX is also expressed in S.pombe cells (O. Laufman and H. Manor, unpublished data).
Evidence that tsn and trax are non-essential genes in S.pombe
In order to study the functions of the S.pombe Translin, we used a genetic approach, whereby we have undertaken the deletion of the tsn gene in S.pombe cells and the examination of the consequences. Since it was not known originally whether Translin was essential for S.pombe viability, we initially replaced one of the two Translin alleles in diploid S.pombe cells with the ura4 gene. This was carried out by homologous recombination with sequences flanking the tsn gene, as illustrated in Figure 4A. Next, we performed a tetrad analysis of haploid derivatives of the tsn+/tsn− heterozygotes. The results of this analysis are shown in Figure 4B. It can be seen that in all the tetrads the four haploid descendents were viable. To examine the genotypes of these haploids and of the parental strains, we performed a PCR analysis of DNA extracted from the cells, using the primers depicted in Figure 4A. Figure 4C shows an agarose gel profile of the PCR amplification products of DNA obtained from wild-type haploid tsn+ cells (lane 1), cells of a tsn+/tsn− heterozygote (lane 2) and haploid tsn− cells (lane 3). Clearly, the lengths of the amplified segments matched the lengths predicted by the diagram shown in Figure 4A. Additional PCR assays revealed that the larger colonies among the tetrads shown in Figure 4B contained tsn−ura4+ cells.
In a pilot gene deletion project carried out in S.pombe, Decottignies et al. (23) found that the S.pombe trax gene is non-essential. We obtained from them the trax− S.pombe strain that they have isolated by replacing the trax gene with the kanamycin gene, as indicated in Figure 4D. Using the primers depicted in Figure 4D, we have carried out a PCR analysis of DNA extracted from these cells and from wild-type cells (Figure 4E). It can be seen that the PCR assays yielded fragments of the expected lengths from the wild-type trax+ DNA (lane 1) and trax− DNA (lane 2). Next, we crossed our tsn− mutant with the trax− mutant and carried out a tetrad analysis of the haploid derivatives of this cross (data not shown). These haploids were also viable and included cells in which both the tsn and the trax genes have been deleted, as indicated by the PCR analyses shown in Figure 4C (lane 4) and Figure 4E (lane 3). The tsn−,trax+ and the tsn−,trax− genotypes were subsequently verified in the haploid strains described above by Southern blotting and direct sequencing. Detailed cell proliferation assays shown in the Supplementary Material confirmed that the deletions did not cause a reduction in the proliferation rates of the cells; rather, the growth of the tsn−,trax− double mutant was slightly stimulated, while a lesser stimulation observed in the proliferation of the tsn− mutant was within the experimental error. Thus, both tsn and trax are non-essential genes in S.pombe.
DNA-binding specificity of the S.pombe Translin
The S.pombe Translin ORF was cloned into an E.coli expression plasmid, such that it was fused in-frame downstream to a sequence encoding a histidine tag. The recombinant protein was expressed and purified, as described in the Supplementary Material, and used for biochemical studies described below.
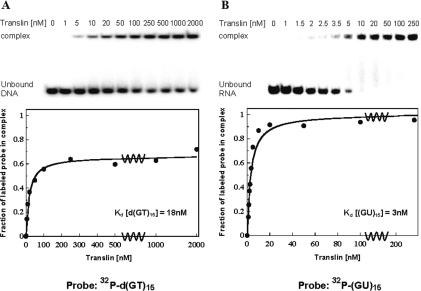
Preliminary studies indicated that, like the human Translin, the S.pombe Translin binds single-stranded d(GT)n repeats. Therefore, we carried out more detailed studies, in which we used the gel mobility shift technique to titrate the oligodeoxynucleotide d(GT)15 with the S.pombe Translin. In these assays, we incubated the 32P-labeled oligodeoxynucleotide d(GT)15 with increasing concentrations of the protein under conditions found to be optimal for binding of the DNA to the protein and ran the mixtures in a native polyacrylamide gel. Figure 5A (upper panel) shows a phosphorimage of this gel. A single retarded band containing the DNA–Translin complex is clearly visible, in addition to the band containing the unbound DNA. The intensity of the retarded band increased, and the intensity of the band containing the unbound DNA decreased, as the input concentration of the protein was increased. The lower panel in Figure 5A shows a plot of these data. It can be seen that an apparent plateau level was reached in this experiment, in which the maximal fraction of bound DNA was ∼0.70. The failure to completely saturate the DNA could be due to aggregation of the protein at the rather high concentrations required for reaching the apparent plateau level. The aggregation could effectively reduce the concentration of protein available for binding, such that a true plateau level could not be attained. Nevertheless, an apparent dissociation constant of 18 nM was estimated from these data, which is indicative of specific recognition and binding of single-stranded d(GT)n repeats by the S.pombe Translin.
Figure 5.
Gel mobility shift assays of the binding of Translin to the single-stranded oligodeoxynucleotide d(GT)15 and the single-stranded oligoribonucleotide (GU)15. Recombinant S.pombe Translin at the indicated concentrations was incubated with each of the two single-stranded 32P-labeled oligonucleotides. Complexes were analyzed by the gel mobility shift technique. The phosphorimager images of the gels are shown at the top of (A) and (B). Plots of the fraction of labeled oligonucleotide in the complex versus the total concentration of Translin in the binding reaction are shown at the bottom. (A) Binding of the S.pombe Translin to the oligodeoxynucleotide d(GT)15 at a concentration of 0.04 nM. (B) Binding of the S.pombe Translin to the oligoribonucleotide (GU)15 at a concentration of 0.03 nM. Kdis values were derived from these plots, as described previously (5).
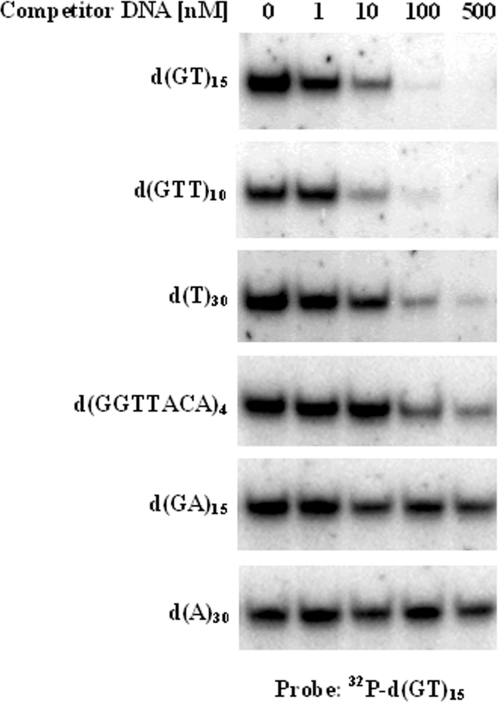
Figure 6 presents a series of competition assays that lent support to this conclusion. In these assays, the S.pombe Translin was incubated with 32P-labeled d(GT)15 in the presence of increasing concentrations of the indicated unlabeled oligodeoxynucleotides, and the mixtures were resolved by PAGE. It can be seen that the S.pombe Translin bound the oligodeoxynucleotide d(GTT)10 with an affinity that was comparable with the affinity with which it bound the oligodeoxynucleotide d(GT)15. Also, the affinity of the S.pombe Translin for the oligodeoxynucleotide d(T)30 was relatively high, while the affinity of the protein for the oligodeoxynucleotide d(GGTTACA)4, which contains four repeats of S.pombe telomeric DNA, was much lower. Evidently, the S.pombe Translin had an even lower affinity for the oligodeoxynucleotide d(GA)15 and did not bind the oligodeoxynucleotide d(A)30 at all. These results were corroborated by direct titration assays of the type shown in Figure 5A that were carried out with other samples of the same oligodeoxynucleotides, which had been labeled with 32P. Finally, similar gel shift assays revealed that, like the human Translin, the S.pombe Translin did not bind d(GT)n·d(AC)n repeats and other double-stranded DNA molecules (data not shown).
Figure 6.
Competition of unlabeled oligodeoxynucleotides with a 32P-labeled d(GT)15 probe for binding the S.pombe Translin. Recombinant S.pombe Translin at a concentration of 5 nM was incubated with 1 nM of the 32P-labeled d(GT)15 probe in the presence of increasing concentrations of the indicated unlabeled oligodeoxynucleotides. Gel mobility shift assays were performed, as described in Materials and Methods. Only the upper parts of the gels, which contain the retarded band, are shown.
Comparison of RNA binding by the S.pombe Translin with RNA binding by the human Translin
Figure 5B shows a titration of the oligoribonucleotide (GU)15 with the S.pombe Translin. This experiment was carried out by the same procedure used for titrating the oligodeoxynucleotide d(GT)15. It can be seen that the Kdis value for binding the oligoribonucleotide, 3 nM, was 6-fold lower than the Kdis value for binding the oligodeoxynucleotide (Figure 5A). It is also evident that at the relatively low concentrations required for reaching the plateau level in this experiment, the fraction of (GU)15 molecules that could be bound by Translin was close to 1.0. The very small dissociation constant of the complex S.pombe Translin–(GU)15 is an indication of specific binding of the protein to the GU repeats. The specificity of the binding to these GU repeats was also underscored by a much lower affinity that the S.pombe Translin was found to have for the oligoribonucleotide (U)24 (data not shown).
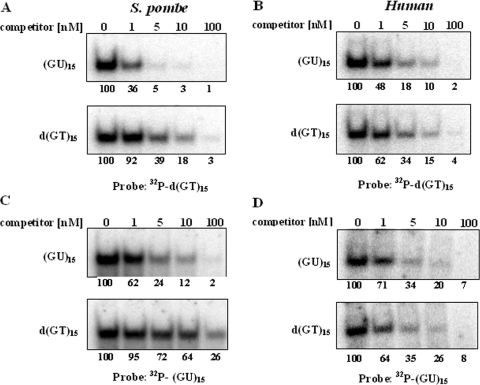
Figure 7A and C shows gel shift competition assays, which confirmed the conclusion that the S.pombe Translin has a much higher affinity for the oligoribonucleotide (GU)15 than for the corresponding oligodeoxynucleotide d(GT)15. In the experiment shown in Figure 7A, 32P-labeled d(GT)15 was incubated in the presence of various concentrations of either unlabeled d(GT)15 or unlabeled (GU)15. Clearly, (GU)15 was a much more efficient competitor than the homologous d(GT)15. The reciprocal experiment, in which 32P-labeled (GU)15 was used as a probe, is shown in Figure 7C. It can be seen that in this experiment too, (GU)15 was a much more efficient competitor than the homologous d(GT)15.
Figure 7.
Gel shift competition assays designed to determine the relative affinities of the human and the S.pombe Translins for the oligoribonucleotide (GU)15 and the oligodeoxynucleotide d(GT)15. Recombinant S.pombe Translin or human Translin was incubated with 1 nM of a 32P-d(GT)15 probe, or a 32P-(GU)15 probe, in the presence of increasing concentrations of the indicated unlabeled competitors. Gel mobility shift assays were performed, as described in Materials and Methods. Only the upper parts of the gels, which contain the retarded band, are shown. (A) S.pombe Translin, 32P-d(GT)15 probe. (B) Human Translin, 32P-d(GT)15 probe. (C) S.pombe Translin, 32P-(GU)15 probe. (D) Human Translin, 32P-(GU)15 probe. The numbers at the bottom of each part represent the percentage of bound probe as a function of the concentration of competitor, relative to the value obtained in the absence of competitor, which was designated as 100%.
So far, no direct comparison has been made between the affinities of either the human or the mouse Translin to specific single-stranded DNA sequences and homologous RNA sequences. Figure 7B and D shows gel shift competition assays of the type shown in Figure 7A and C, which were carried out with the human Translin. It can be seen that, unlike the assays performed with the S.pombe Translin, in the assays of the human Translin, the unlabeled oligonucleotides d(GT)15 and (GU)15 competed with the 32P-labeled d(GT)15 and (GU)15 probes with approximately equal efficiencies. Other experiments, in which direct titration assays of the binding of the human Translin to 32P-labeled (GU)15 and d(GT)15 were carried out, revealed that the Kdis value for binding the oligoribonucleotide was only twice smaller than the Kdis value for binding the oligodeoxynucleotide (data not shown).
The S.pombe Translin is an octamer
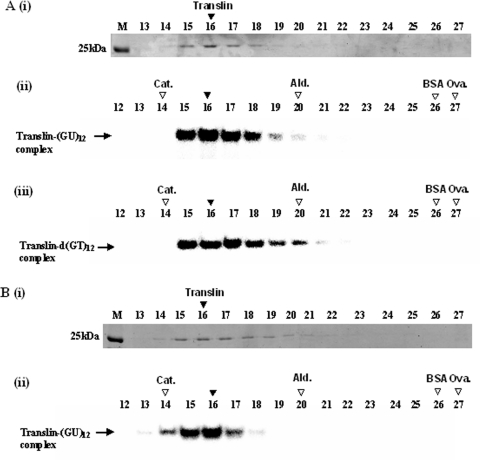
To determine the molecular weight of the native form of the S.pombe Translin, we centrifuged the protein in a glycerol gradient, as described in Materials and Methods. Figure 8A, (i), shows the sedimentation profile of the S.pombe Translin determined by SDS–PAGE of aliquots withdrawn from fractions collected from the gradient, followed by Coomassie blue staining of the gel. Figure 8A, (ii) and (iii), shows the sedimentation profiles of the RNA- and the DNA-binding forms of Translin. These profiles were determined by gel shift assays of other aliquots withdrawn from the same gradient fractions, using the 32P-labeled oligoribonucleotide (GU)12 and oligodeoxynucleotide d(GT)12 as probes. Figure 8B, (i) and (ii), shows the data obtained in a complementary experiment, in which the S.pombe Translin was first bound to the 32P-labeled oligoribonucleotide (GU)12 and was subsequently centrifuged in a similar glycerol gradient. Figure 8B, (i), shows the sedimentation profile of Translin in this gradient, determined by SDS–PAGE, followed by Coomassie blue staining. Figure 8B, (ii), shows the sedimentation profile of the preformed complexes between Translin and the 32P-labeled oligoribonucleotide (GU)12, determined by electrophoresis in a native polyacrylamide gel, followed by phosphorimaging. It can be seen that both the DNA- and the RNA-binding activities of Translin, as well as the preformed RNA–Translin complexes, cosedimented with the bulk of the protein. Using the sedimentation constants and molecular masses of the marker proteins shown in Figure 8, we estimated the sedimentation constant and the molecular mass of the native S.pombe Translin to be 9.9S and 217 kDa, respectively (27). These data indicated that both the DNA- and the RNA-binding forms of the S.pombe Translin are octamers, similar to the human Translin (3,33). Furthermore, the S.pombe Translin remained an octamer after binding the RNA probe.
Figure 8.
Glycerol gradient centrifugation of the recombinant S.pombe Translin. (A) Sample of the recombinant S.pombe Translin was centrifuged in a 20–40% linear glycerol gradient, as described in Materials and Methods. Fractions of equal volumes were collected from the bottom of the gradient. Aliquots withdrawn from these fractions were analyzed by SDS–gel electrophoresis, followed by Commassie blue staining [A (i)]. Other aliquots withdrawn from the same fractions were analyzed by the gel mobility shift assay for binding the 32P-labeled oligoribonucleotide (GU)12 [A (ii)] or the 32P-labeled oligodeoxynucleotide d(GT)12 [A (iii)]. Another sample of the recombinant S.pombe Translin was preincubated with the 32P-labeled oligoribonucleotide (GU)12 and then centrifuged in another tube in the same run. Aliquots withdrawn from this tube were also analyzed by SDS–gel electrophoresis and Coomassie blue staining [B (i)]. Other aliquots were electrophoresed in a native polyacrylamide gel and the 32P-labeled bands were detected by phosphorimaging [B (ii)]. M indicates the gel lanes including the gel marker proteins. Horizontal arrows indicate the gel-shifted complexes generated by binding of Translin. Vertical filled arrowheads indicate the peaks of the Coomassie blue-stained Translin, and the peaks of the RNA- and DNA-binding activities. Vertical open arrowheads indicate the peaks of the centrifugation marker proteins: Cat., Catalase (232 kDa); Ald., Aldolase (158 kDa); BSA (66 kDa); Ova., Ovalbumin (43 kDa).
Evidence for specific association of the S.pombe Translin and TRAX
Figure 9 shows the results of yeast (S.cerevisiae) two-hybrid analysis of the interaction between the S.pombe Translin and TRAX in a selective medium lacking adenine. Identical results were obtained in a medium lacking histidine (data not shown). It can be seen that Translin specifically interacted with TRAX (D,E). Specific interactions also occurred between individual Translin subunits (C), but did not occur between individual TRAX molecules (F). The specific association of Translin and TRAX has also been confirmed by coimmunoprecipitation of the two proteins from S.pombe cell extracts, using the polyclonal antibodies that we have prepared against the S.pombe Translin and TRAX, as described in Materials and Methods (O. Laufman and H. Manor, unpublished data).
Figure 9.
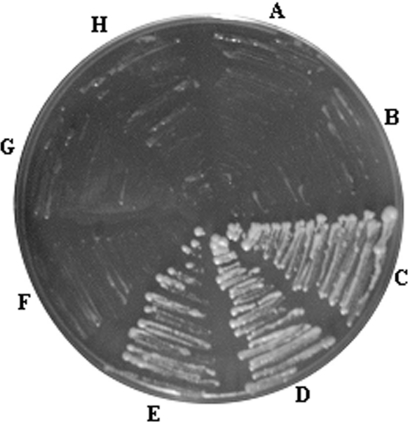
Analysis of the interaction between the S.pombe Translin and the S.pombe TRAX by the yeast two-hybrid technique. The complete ORFs of the S.pombe Translin and TRAX were subcloned in-frame into the pGAD-C (GAL4 activation domain) and pGBD-C (GAL4 binding domain) vectors (28). Pairs of binding domain (BD) and activation domain (AD) constructs were cotransfected into the S.cerevisiae strain Y1183. Co-transformants were assayed for the activation of the ADE2 reporter gene by streaking on SD medium lacking leucine, tryptophan and adenine, as described in Materials and Methods. A, AD and BD-Translin; B, AD-Translin and BD; C, AD-Translin and BD-Translin; D, AD-Translin and BD-TRAX; E, AD-TRAX and BD-Translin; F, AD-TRAX and BD-TRAX; G, AD-TRAX and BD; H, AD and BD-TRAX.
DISCUSSION
The S.pombe genes tsn and trax, encoding the Translin and TRAX homologs, have been identified by the whole genome sequencing project of the fission yeast (10). In this paper, we presented the first demonstration that the two genes are actually expressed in S.pombe (Figure 3), but are non-essential (Figure 4). It should be noted, in this connection, that the mouse, as well as the human, Translin and TRAX have been recently reported to be involved in the control of cell proliferation. Translin-null mice were generated and found to develop into mature fertile animals. However, at birth they were 10–30% smaller than their wild-type littermates and remained so until the age of 6–9 months (34). Mouse embryo fibroblasts derived from the tsn− mice also showed decreased proliferation (35). In line with these observations, overexpression of Translin in human embryo kidney cells accelerated the proliferation of these cells (36). TRAX was also reported to be essential for normal proliferation of human HeLa cells (37). In view of these data on the mammalian Translin and TRAX, we have carefully measured the rates of proliferation of cells of the S.pombe deletion mutants. We have detected a slight stimulation in the rate of proliferation of cells of the double mutant tsn−,trax− versus the wild-type cells and a lesser stimulation in the proliferation of tsn− cells that was within the experimental error (see Supplementary Material). Additional studies will be required to find out the reason for these apparent differences between the S.pombe and the mammalian Translin and TRAX proteins.
As shown in Figure 1, the S.pombe Translin shares only 36% identity and 54% similarity with the human and the mouse Translins. However, the model shown in Figure 2A indicates that its 3D structure most likely resembles the structure of the human and the mouse Translins. This is indeed quite apparent when compared with the monomer human Translin structure visualized in Figure 2B. In particular, the model predicts that, like the two mammalian Translins, the predominant secondary structure features in the S.pombe Translin are α helices. In addition, the sedimentation analysis presented in Figure 8 showed that the S.pombe Translin subunits, similar to the human Translin subunits, assemble into octamers, and that only the octamers bind DNA or RNA. Furthermore, the sedimentation analysis of complexes formed between the S.pombe Translin and the oligoribonucleotide (GU)12 revealed that the octameric configuration of the S.pombe Translin was preserved after binding the nucleic acid, as previously observed in studies of the human Translin (3,33). However, it has been reported that preformed mouse Translin–DNA complexes contain dimers of the protein (38).
The conservation of the overall structure of the S.pombe Translin versus the human Translin could account for the similarity in their modes of binding to nucleic acids, namely their ability to bind single-stranded DNA and RNA, and their inability to bind double-stranded DNA. On the other hand, the low degree of conservation of the putative single-stranded DNA and RNA-binding motifs in the S.pombe Translin (Figure 1) suggests that the local 3D structures of the nucleic acid binding subdomains were altered in the yeast protein. Such differences could account for the alteration of the DNA recognition profile of the fission yeast Translin versus the human Translin. In particular, we have shown that even though the S.pombe Translin specifically binds the oligodeoxynucleotide d(GT)15, it has a 9-fold larger Kdis value for this oligodeoxynucleotide than the Kdis value found for the human Translin. Second, unlike the human Translin, the S.pombe Translin has a relatively high affinity for d(T)n repeats. Third, although the human Translin has a high affinity for single-stranded human telomeric repeats (5), the S.pombe Translin very poorly binds single-stranded S.pombe telomeric repeats (Figure 6).
As described in the Results, we compared the binding of the human Translin to the oligodeoxynucleotide d(GT)15 versus the oligoribonucleotide (GU)15. We found that it had approximately equal affinities for these DNA and RNA repeats. In contrast, a similar comparison made with the S.pombe Translin revealed that the yeast protein had a much higher affinity for the oligoribonucleotide (GU)15 than for the oligodeoxynucleotide d(GT)15 (Figures 5 and 7). This difference between the human and the S.pombe Translins is also indicative of local alterations in the 3D structures of the nucleic acid binding subdomains in the yeast Translin octamer. Since the human Translin has approximately equal affinities for homologous single-stranded DNA and RNA sequences, it might perform functions that depend on its sequence-specific association with either DNA or RNA. Furthermore, the virtual identity of the human and the mouse Translins indicates that they are engaged in similar activities in human and mouse cells, respectively. On the other hand, our observation that the S.pombe Translin has much higher affinities for RNA sequences than for homologous DNA sequences suggests that the S.pombe Translin could be primarily involved in functions related to its RNA-binding characteristics.
Among the functions proposed for the human and the mouse Translins on account of their RNA-binding capability are control of translation of specific mRNAs (19,21) and regulation of mRNA sorting and transport (20,39). In particular, it has been suggested that complexes consisting of Translin and associated proteins carry specific mRNAs and proteins from the nucleus into the cytoplasm, and that proteins associated with the Translin nuclear export signal are involved in this intracellular traffic (40) [The latter suggestion appears plausible in view of experiments, which indicated that the mouse Translin nuclear export signal is functional (15)]. Interestingly, these functions were proposed to be mediated through binding of Translin to regions including GU clusters and other G-rich sequences in mRNAs (39,41). Our demonstration that the S.pombe Translin strongly binds (GU)n repeats (Figures 5 and 7), and the observation that the nuclear export signal has been highly conserved in the S.pombe Translin (Figure 1), suggests that the S.pombe Translin may perform similar functions in the fission yeast cells.
As noted earlier, there are homologous trax genes in the various species that also contain tsn orthologs. This co-evolution of the two genes suggests that Translin and TRAX are functionally related. We have shown in this study that the S.pombe Translin specifically interacts with the S.pombe TRAX, as was previously observed in studies of the human and the mouse Translins (13,15,42). This observation supports the notion that in S.pombe cells, as well as in human and mouse cells, the two proteins are engaged as a complex in a common metabolic pathway.
It is also possible that either TRAX or Translin has additional functions, which they perform independently in association with other proteins. One possibility was indicated by a report that the mouse TRAX (but not Translin) interacts with two centrosomal proteins and a Golgi-associated protein (43). These findings suggested that complexes including TRAX and these proteins might be engaged in biochemical activities related to the activities that these proteins are known to perform.
Our observation that neither the tsn gene nor the trax gene is essential for S.pombe cell growth and division suggests that S.pombe cells might contain a backup system that can efficiently replace Translin and TRAX in providing the functions required for cell proliferation. Such a backup system may also exist in mammalian cells, and in cells of the budding yeast S.cerevisiae that do not contain Translin and TRAX orthologs. We are currently using the S.pombe Translin- and TRAX-null strains for further genetic and biochemical studies. The combined genetic and biochemical approach, which was initiated by the work reported here, should further clarify the biological functions of Translin and TRAX.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
The authors thank Dr A. Decottignies for sending them the S.pombe trax− strain, Dr Y. Kassir for the gift of the S.cerevisiae two-hybrid host strain Y1183 and Dr A. Cohen for sending them an S.pombe cDNA library. The authors also thank Drs A. Cohen, J. Cooper and Y. Kassir for helpful advice. This work was supported by the Israel Science Foundation (Grants No. 373/99 and No. 378/03). Funding to pay the Open Access publication charges for this article was provided by the Israel Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Aoki K., Nakahara K., Ikegawa C., Seto M., Takahashi T., Minowada J., Strominger J.L., Maziarz R.T., Kasai M. Nuclear proteins binding to a novel target sequence within the recombination hotspot regions of Bcl-2 and the immunoglobulin DH gene family. Oncogene. 1994;9:1109–1115. [PubMed] [Google Scholar]
- 2.Aoki K., Suzuki K., Sugano T., Tasaka T., Nakahara K., Kuge O., Omori A., Kasai M. A novel gene, Translin, encodes a recombination hotspot binding protein associated with chromosomal translocations. Nature Genet. 1995;10:167–174. doi: 10.1038/ng0695-167. [DOI] [PubMed] [Google Scholar]
- 3.VanLoock M.S., Yu X., Kasai M., Egelman E.H. Electron microscopic studies of the translin octameric ring. J. Struct. Biol. 2001;135:58–66. doi: 10.1006/jsbi.2001.4383. [DOI] [PubMed] [Google Scholar]
- 4.Aharoni A., Baran N., Manor H. Characterization of a multisubunit human protein which selectively binds single stranded d(GA)n and d(GT)n sequence repeats in DNA. Nucleic Acids Res. 1993;21:5221–5228. doi: 10.1093/nar/21.22.5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacob E., Pucshansky L., Zeruya E., Baran N., Manor H. The human protein translin specifically binds single-stranded microsatellite repeats, d(GT)n, and G-strand telomeric repeats d(TTAGGG)n: a study of the binding parameters. J. Mol. Biol. 2004;344:939–950. doi: 10.1016/j.jmb.2004.09.095. [DOI] [PubMed] [Google Scholar]
- 6.Cohen S., Jacob E., Manor H. Effects of single-stranded DNA binding proteins on primer extension by telomerase. Biochim. Biophys. Acta. 2004;1679:129–140. doi: 10.1016/j.bbaexp.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Han J.R., Yiu G.K., Hecht N.B. Testis/brain RNA-binding protein attaches translationally repressed and transported mRNAs to microtubules. Proc. Natl Acad. Sci. USA. 1995;92:9550–9554. doi: 10.1073/pnas.92.21.9550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aoki K., Inazawa J., Takahashi T., Nakahara K., Kasai M. Genomic structure and chromosomal localization of the gene encoding translin, a recombination hotspot binding protein. Genomics. 1997;43:237–241. doi: 10.1006/geno.1997.4796. [DOI] [PubMed] [Google Scholar]
- 9.Castro A., Peter M., Magnaghi-Jaulin L., Vigneron S., Loyaux D., Lorca T., Labbe J.C. Part of Xenopus translin is localized in the centrosomes during mitosis. Biochem. Biophys. Res. Commun. 2000;276:515–523. doi: 10.1006/bbrc.2000.3482. [DOI] [PubMed] [Google Scholar]
- 10.Wood V., Gwilliam R., Rajandream M.A., Lyne M., Lyne R., Stewart A., Sgouros J., Peat N., Hayles J., Baker S., et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- 11.Pascal J.M., Hart P.J., Hecht N.B., Robertus J.D. Crystal structure of TB-RBP, a novel RNA-binding and regulating protein. J. Mol. Biol. 2002;319:1049–1057. doi: 10.1016/S0022-2836(02)00364-9. [DOI] [PubMed] [Google Scholar]
- 12.Sugiura I., Sasaki C., Hasegawa T., Kohno T., Sugio S., Moriyama H., Kasai M., Matsuzaki T. Structure of human translin at 2.2 angstrom resolution. Acta Crystallogr. D Biol. Crystallogr. 2004;60:674–679. doi: 10.1107/S0907444904002549. [DOI] [PubMed] [Google Scholar]
- 13.Aoki K., Ishida R., Kasai M. Isolation and characterization of a cDNA encoding a Translin-like protein, TRAX. FEBS Lett. 1997;401:109–112. doi: 10.1016/s0014-5793(96)01444-5. [DOI] [PubMed] [Google Scholar]
- 14.Cho Y.S., Chennathukuzhi V.M., Handel M.A., Eppig J., Hecht N.B. The relative levels of translin-associated factor X (TRAX) and testis brain RNA-binding protein determine their nucleocytoplasmic distribution in male germ cells. J. Biol. Chem. 2004;279:31514–31523. doi: 10.1074/jbc.M401442200. [DOI] [PubMed] [Google Scholar]
- 15.Chennathukuzhi V.M., Kurihara Y., Bray J.D., Hecht N.B. Trax (translin-associated factor X), a primarily cytoplasmic protein, inhibits the binding of TB-RBP (translin) to RNA. J. Biol. Chem. 2001;276:13256–13263. doi: 10.1074/jbc.M009707200. [DOI] [PubMed] [Google Scholar]
- 16.Han J.R., Gu W., Hecht N.B. Testis-brain RNA-binding protein, a testicular translational regulatory RNA-binding protein, is present in the brain and binds to the 3′ untranslated regions of transported brain mRNAs. Biol. Reprod. 1995;53:707–717. doi: 10.1095/biolreprod53.3.707. [DOI] [PubMed] [Google Scholar]
- 17.Wu X.Q., Hecht N.B. Mouse testis brain ribonucleic acid-binding protein/translin colocalizes with microtubules and is immunoprecipitated with messenger ribonucleic acids encoding myelin basic protein, alpha calmodulin kinase II, and protamines 1 and 2. Biol. Reprod. 2000;62:720–725. doi: 10.1095/biolreprod62.3.720. [DOI] [PubMed] [Google Scholar]
- 18.Finkenstadt P.M., Kang W.S., Jeon M., Taira E., Tang W.Z., Baraban J.M. Somatodendritic localization of Translin, a component of the Translin/Trax RNA binding complex. J. Neurochem. 2000;75:1754–1762. doi: 10.1046/j.1471-4159.2000.0751754.x. [DOI] [PubMed] [Google Scholar]
- 19.Morales C.R., Lefrancois S., Chennathukuzhi V., El-Alfy M., Wu X.Q., Yang J.X., Gerton G.L., Hecht N.B. A TB-RBP and Ter ATPase complex accompanies specific mRNAs from nuclei through the nuclear pores and into intercellular bridges in mouse male germ cells. Dev. Biol. 2002;246:480–494. doi: 10.1006/dbio.2002.0679. [DOI] [PubMed] [Google Scholar]
- 20.Chennathukuzhi V., Morales C.R., El-Alfy M., Hecht N.B. The kinesin KIF17b and RNA-binding protein TB-RBP transport specific cAMP-responsive element modulator-regulated mRNAs in male germ cells. Proc. Natl Acad. Sci. USA. 2003;100:15566–15571. doi: 10.1073/pnas.2536695100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J.X., Chennathukuzhi V., Miki K., O'Brien D.A., Hecht N.B. Mouse testis brain RNA-binding protein/translin selectively binds to the messenger RNA of the fibrous sheath protein glyceraldehyde 3-phosphate dehydrogenase-S and suppresses its translation in vitro. Biol. Reprod. 2003;68:853–859. doi: 10.1095/biolreprod.102.008631. [DOI] [PubMed] [Google Scholar]
- 22.Moreno S., Klar A., Nurse P. Molecular genetic-analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 23.Decottignies A., Sanchez-Perez I., Nurse P. Schizosaccharomyces pombe essential genes: a pilot study. Genome Res. 2003;13:399–406. doi: 10.1101/gr.636103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suga M., Hatakeyama T. High efficiency transformation of Schizosaccharomyces pombe pretreated with thiol compounds by electroporation. Yeast. 2001;18:1015–1021. doi: 10.1002/yea.753. [DOI] [PubMed] [Google Scholar]
- 25.Hoffman C.S., Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning, A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Martin R.G., Ames B.N. A method for determining the sedimentation behavior of enzymes: application to protein mixtures. J. Biol. Chem. 1961;236:1372–1379. [PubMed] [Google Scholar]
- 28.James P., Halladay J., Craig E.A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harlow E., Lane D.P. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 30.Aoki K., Suzuki K., Ishida R., Kasai M. The DNA binding activity of Translin is mediated by a basic region in the ring-shaped structure conserved in evolution. FEBS Lett. 1999;443:363–366. doi: 10.1016/s0014-5793(99)00010-1. [DOI] [PubMed] [Google Scholar]
- 31.Bates P.A., Kelley L.A., MacCallum R.M., Sternberg M.J.E. Enhancement of protein modeling by human intervention in applying the automatic programs 3D-JIGSAW and 3D-PSSM. Proteins. 2001;(Suppl. 5):39–46. doi: 10.1002/prot.1168. [DOI] [PubMed] [Google Scholar]
- 32.Kelley L.A., MacCallum R.M., Sternberg M.J.E. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 2000;299:499–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- 33.Lee S.P., Fuior E., Lewis M.S., Han M.K. Analytical ultracentrifugation studies of translin: analysis of protein–DNA interactions using a single-stranded fluorogenic oligonucleotide. Biochemistry. 2001;40:14081–14088. doi: 10.1021/bi010302t. [DOI] [PubMed] [Google Scholar]
- 34.Chennathukuzhi V., Stein J.M., Abel T., Donlon S., Yang S.C., Miller J.P., Allman D.M., Simmons R.A., Hecht N.B. Mice deficient for testis-brain RNA-binding protein exhibit a coordinate loss of TRAX, reduced fertility, altered gene expression in the brain, and behavioral changes. Mol. Cell. Biol. 2003;23:6419–6434. doi: 10.1128/MCB.23.18.6419-6434.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang S.C., Cho Y.S., Chennathukuzhi V.M., Underkoffler L.A., Loomes K., Hecht N.B. Translin-associated factor X is post-transcriptionally regulated by its partner protein TB-RBP, and both are essential for normal cell proliferation. J. Biol. Chem. 2004;279:12605–12614. doi: 10.1074/jbc.M313133200. [DOI] [PubMed] [Google Scholar]
- 36.Ishida R., Okado H., Sato H., Shionoiri C., Aoki K., Kasai M. A role for the octameric ring protein, Translin, in mitotic cell division. FEBS Lett. 2002;525:105–110. doi: 10.1016/s0014-5793(02)03095-8. [DOI] [PubMed] [Google Scholar]
- 37.Yang S.C., Hecht N.B. Translin associated protein X is essential for cellular proliferation. FEBS Lett. 2004;576:221–225. doi: 10.1016/j.febslet.2004.08.082. [DOI] [PubMed] [Google Scholar]
- 38.Wu X.Q., Xu L.H., Hecht N.B. Dimerization of the testis brain RNA-binding protein (translin) is mediated through its C-terminus and is required for DNA- and RNA-binding. Nucleic Acids Res. 1998;26:1675–1680. doi: 10.1093/nar/26.7.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohashi S., Kobayashi S., Omori K., Ohara T., Omae A., Muramatsu T., Li Y.M., Anzai K. The single-stranded DNA- and RNA-binding proteins pur alpha and pur beta link BC1 RNA to microtubules through binding to the dendrite-targeting RNA motifs. J. Neurochem. 2000;75:1781–1790. doi: 10.1046/j.1471-4159.2000.0751781.x. [DOI] [PubMed] [Google Scholar]
- 40.Hecht N.B. Intracellular and intercellular transport of many germ cell mRNAs is mediated by the DNA- and RNA-Binding protein, testis-brain RNA-binding protein (TB-RBP) Mol. Reprod. Dev. 2000;56:252–253. doi: 10.1002/(SICI)1098-2795(200006)56:2+<252::AID-MRD8>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 41.Li Z., Baraban J.M. High affinity binding of the Translin/Trax complex to RNA does not require the presence of Y or H elements. Brain Res Mol Brain Res. 2004;120:123–129. doi: 10.1016/j.molbrainres.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 42.Finkenstadt P.M., Jeon M., Baraban J.M. Trax is a component of the Translin-containing RNA binding complex. J. Neurochem. 2002;83:202–210. doi: 10.1046/j.1471-4159.2002.01158.x. [DOI] [PubMed] [Google Scholar]
- 43.Bray J.D., Chennathukuzhi V.M., Hecht N.B. Identification and characterization of cDNAs encoding four novel proteins that interact with translin associated factor-X. Genomics. 2002;79:799–808. doi: 10.1006/geno.2002.6779. [DOI] [PubMed] [Google Scholar]
- 44.Ayoub N., Goldshmidt I., Lyakhovetsky R., Cohen A. A fission yeast repression element cooperates with centromere-like sequences and defines a mat silent domain boundary. Genetics. 2000;156:983–994. doi: 10.1093/genetics/156.3.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ayoub N., Goldshmidt I., Cohen A. Position effect variegation at the mating-type locus of fission yeast: a cis-acting element inhibits covariegated expression of genes in the silent and expressed domains. Genetics. 1999;152:495–508. doi: 10.1093/genetics/152.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.