Abstract
The application of high-throughput techniques such as genomics, proteomics or transcriptomics means that vast amounts of heterogeneous data are now available in the public databases. Bioinformatics is responding to the challenge with new integrated management systems for data collection, validation and analysis. Multiple alignments of genomic and protein sequences provide an ideal environment for the integration of this mass of information. In the context of the sequence family, structural and functional data can be evaluated and propagated from known to unknown sequences. However, effective integration is being hindered by syntactic and semantic differences between the different data resources and the alignment techniques employed. One solution to this problem is the development of an ontology that systematically defines the terms used in a specific domain. Ontologies are used to share data from different resources, to automatically analyse information and to represent domain knowledge for non-experts. Here, we present MAO, a new ontology for multiple alignments of nucleic and protein sequences. MAO is designed to improve interoperation and data sharing between different alignment protocols for the construction of a high quality, reliable multiple alignment in order to facilitate knowledge extraction and the presentation of the most pertinent information to the biologist.
INTRODUCTION
The post-genomic era is presenting new challenges for bioinformatics. High-throughput genome sequencing and assembly techniques, together with new information resources, such as structural proteomics, interactomics, transcriptome data from microarray analyses, or light microscopy images of living cells have lead to a rapid increase in the amount of data available (1,2). As a result, there now exists a vast array of heterogeneous data resources distributed over different Internet sites that cover genomic, cellular, structure, phenotype and other types of biologically relevant information. A major challenge for bioinformaticians is the efficient integration of the experimental and predicted information with the vast number of applications that have been developed to manage and interpret this data into an integrated network, leading to improved cooperation and hopefully a more rapid pace of scientific discovery.
Multiple alignments of nucleic acid and protein sequences provide an ideal workbench for the integration and presentation of this mass of biological information (3,4). By placing the sequence in the context of the overall family, multiple alignments permit not only a horizontal analysis of the sequence along its length, but also a vertical view of its evolution. Since their introduction in the early seventies, multiple sequence alignments have been widely exploited in most aspects of molecular biology. They were originally used in evolutionary analyses to explore the phylogenetic relationships between organisms (5,6). More recently, new sequence database search methods have exploited multiple alignments to detect more and more distant homologues (7–9). Multiple sequence alignments have also led to a significant improvement of 3D fold recognition techniques and homology modelling techniques (10,11). Another important application is the functional characterization of nucleic acid and protein families, using either homology-based methods or mean ab initio predictions for a family of sequences. Furthermore, with the recent availability of programs to perform multiple structure alignments (12–14), it is now possible to analyse very distantly related proteins, whose sequence similarity is too low to be detected by sequence comparison methods. Of course, in the current era of complete genome sequences, it is now possible to perform comparative multiple sequence analysis at the genome level. Multiple alignment methods are responding to the challenges posed by these diverse applications, with current developments moving away from a single all-encompassing algorithm towards co-operative, knowledge-based systems, which exploit the new structural and functional data available (15–19). The success of these methods relies on the efficient integration of information from different databases and the close cooperation of the different alignment algorithms. Organization and analysis techniques are needed to ensure that the pertinent information can be extracted and presented to the biologist in a clear, user-friendly format.
The organization and merging of biological information from different domains, such as genetics, structural biology, protein chemistry or pharmacology, is currently hindered by syntactic differences in the file formats used by different applications and by semantic differences, such as naming conventions and terminology. The syntactic issue is now being addressed with the widespread adoption of standard file formats, such as the XML (eXtensible Markup Language) data exchange format. For example, the aim of the eFamily schema (http://www.efamily.org.uk/) is to allow different domain definitions and mappings to be exchanged between protein databases. However, if the data are to be truly understandable by multiple applications, semantic interoperability will also be necessary. Semantic ambiguities are ubiquitous, e.g. the same sequence may have different definitions, such as glycine-tRNA synthetase or glycine-tRNA ligase, in different sequence databases. The problem becomes more complex when natural language is used, e.g. for protein definitions. To resolve such semantic discrepancies, formal, structured vocabularies are now required, which constrain the use and interpretation of the terminology employed.
In recent years, ontologies have been introduced in a number of areas for the management of biological knowledge (20). In computer science, an ontology is defined as a formal, structured representation of the knowledge in a particular domain (21). The most important aspect of an ontology is that it creates a shared understanding of a domain in a format that can be used by both humans and computers. Ontologies are thus used for automatic annotation of data, for the sharing of information from different resources and for the presentation of domain knowledge to researchers, and in particular to non-experts in the specific field. One of the most widely used bioinformatics ontologies is the Gene Ontology (GO) (22), which describes data about gene products. GO is composed of three separate hierarchical vocabularies, representing the function of a gene product, the process in which it plays a role and its cellular location. The GO is used for various tasks such as protein function inference and automated annotation (23–27). Numerous other ontologies have also been made publicly available, including developmental and anatomical ontologies, conditions for microarray experiments and phenotype attributes. Many of these ontologies are grouped together at the Open Biomedical Ontologies (OBO) website (http://obo.sourceforge.net). OBO is an umbrella web address for well-structured controlled vocabularies for shared use across different biological domains. One of the major goals of the OBO consortium is to provide a set of compatible ontologies, which can be used in combination in order to integrate individual data resources into a coherent whole. Although various ontologies have been developed for particular aspects of single sequences, such as gene structure (SO) (28), protein function (GO) or protein–protein interactions (MI) (29), they do not contain all the information required for analyses of gene families. Some work has also begun to develop standard data formats to represent RNA sequences and structures (30), and the RNA Ontology Consortium (ROC) (http://roc.bgsu.edu/) has been established to build a formal ontology. Recently, a protein family ontology has been developed (31) dedicated to protein family database creation and maintenance. However, this ontology does not cover multiple alignment concepts, such as column information or residue conservation.
We present here MAO, a new task-oriented ontology for data retrieval and exchange in the fields of DNA/RNA alignment, protein sequence and structure alignment. The ontology has been developed jointly by the members of the MAO work group, who intend to offer compatible multiple alignment tools and analysis results that commit to the MAO ontology. The purpose of MAO is to standardize descriptions of multiple sequence alignments in order to allow the different alignment construction and analysis methods to communicate with each other and also to allow the integration of structural or functional data with information about sequence family conservation and evolution. Similar to other ontologies, the MAO consists of a controlled vocabulary of terms or ‘concepts’ and a restricted set of relationships between the concepts. The MAO is organized as a complex hierarchy, known as a directed acyclic graph (DAG), where the nodes in the graph represent concepts and the branches joining the nodes represent relationships. Explicit text definitions are provided for all concepts, as well as unique identifiers for unambiguous access. The top-level concept is called the multiple_sequence_alignment, which may represent either nucleotide or protein sequences. Most of the basic features associated with multiple alignments are defined as MAO concepts, ranging from a single residue to sub-families of sequences. Attributes associated with the basic concepts allow the definition of more complex information, such as column conservation, residue or motif function, or 3D structural information. The MAO ontology has been implemented in the common shared syntax defined by OBO, using the open source Java software OBO-Edit (http://www.geneontology.org/). Wherever possible, cross-references are provided to related ontologies, such as the GO, SO, MI, Interpro (32) and the US National Center for Biotechnology Information (Bethesda, MD) organism classification. Thus, MAO permits the integration of diverse information in the context of the overall gene family, facilitating data cross-validation, complex analyses and knowledge extraction for presentation to the biologist in a user-friendly format.
MATERIALS AND METHODS
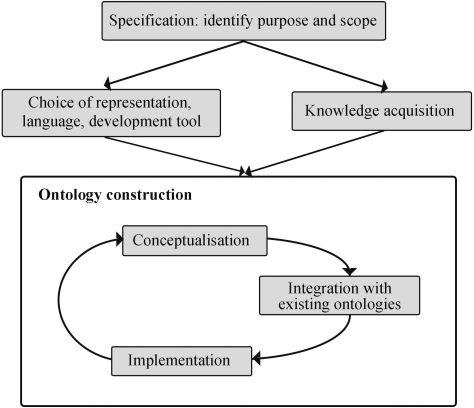
This section describes the framework for the development of MAO, including the design of the ontological model and the subsequent choices of representation and implementation tools. The development procedure shown in Figure 1 is based on the ontological building life cycle suggested by Stevens et al. (33). After the initial specification phase, the ontology is built using an iterative process designed to facilitate the maintenance and future evolution of the ontology, by allowing additional concepts to be incorporated when new knowledge becomes available in the domain. The individual steps in the life cycle are described in detail below.
Figure 1.
The MAO development life cycle.
Specification
The purpose of MAO is to facilitate the communication between the numerous methods for the construction, analysis and annotation of DNA/RNA and protein sequence alignments. The scope of the ontological concepts, therefore, ranges from a complete multiple alignment via subsets of sequences or individual sequences to single residues. In addition, structural or functional features associated with the sequences are defined, either as concepts within MAO or as cross-references to external resources, such as existing ontologies or public databases.
Two different hierarchical relations are specified to describe the relationships between the various MAO concepts. First, specialization (is_a) relations are defined in which the child term is more restrictive than the parent term, e.g. amino_acid is_a residue. The is_a relationship implies inheritance, so that any attributes associated with the parent concept are inherited by its children. Second, partitive (part_of) relationships between concepts are also possible, e.g. residue is part_of sequence. Both the is_a and part_of relations imply irreflexivity (nothing is a part of itself), asymmetry (if atom is part_of nucleotide, then nucleotide is not part_of atom) and transitivity (if residue is part_of sequence, and sequence is part_of sub_alignment, then residue is part_of sub_alignment). Finally, two associative relationships is_name and is_attribute are specified in order to describe properties associated with particular concepts. For example, ‘sequence_name is_name of sequence’ is used to specify a user-defined name for a given sequence. Similarly, the relationship ‘column_conservation is_attribute of column’ is used to describe the level and type of conservation observed for a particular column in the multiple alignment. The range of allowed values for the attributes is not specified in MAO because attribute values are considered to be instances of attributes, which will be specific to the different applications that commit to the ontology.
Knowledge acquisition
The multiple alignment ontology was established in close collaboration with domain experts from both the DNA/RNA and protein communities, including experts in the fields of both primary sequence and 2D/3D structure comparisons. Each expert supplied a list of requirements for the types of data that should be represented in the ontology, as well as a list of potential cross-references to relevant external resources. Definitions were thus constructed from our known knowledge, from major textbooks and from colleagues. As knowledge in the field progresses, new concepts and new definitions will be added to the ontology, subject to agreement by the members of the MAO work group.
Representation
The ontological model described above, where concepts are organized in a hierarchical network, can be represented by a graph structure known as a DAG. In the DAG, the nodes of the graph represent concepts that are connected by directed edges representing the asymmetric relations between concepts. DAGs can be considered to be a generalization of trees in which child nodes (more specialized terms) may have multiple parents (less specialized terms) and multiple relationships to their parents. The DAG used in MAO has a single root node called multiple_sequence_alignment. All other nodes are connected to this root by one of the four relations described above, or by a chain of several hierarchical relations.
Conceptualization
This phase involves the identification of the key concepts, their properties and the relationships that hold between them. The ontology was built from the top-down, starting from the high-level multiple_sequence_alignment concept. Then, in an iterative process, more specific concepts are added to the more generic ones. Each concept was initially assigned a primary name, corresponding to the most generally accepted term in the field. Any alternative terminology is then defined as a synonym of the primary name. A number of conventions were systematically applied when naming concepts, in order to ensure coherence, and also to ensure that the terms are parsable by automatic programs or scripts. Thus, the concepts are all specified as singular entities, no plurals are allowed. In addition, the names contain no hyphens, black slashes or other characters that may have a special meaning in regular expression or programming language definitions. Compound terms, corresponding to short phrases, are systematically separated by underscore characters, rather than space characters. Lower case characters are used throughout to avoid potential clashes when using ontology tools that are not case sensitive. Finally, each concept has a unique identifier with the syntax RO: nnnnnnn, where RO specifies that the concept belongs to the MAO ontology and nnnnnnn is a unique integer within MAO.
In addition to the hierarchical relations, textual descriptions are also associated with each concept in the ontology. A number of rules were used for making a definition: (i) the definition should be positive, not negative; (ii) the definition should be free from words sharing the same root as the concept being defined and (iii) the definition should be as clear and concise as possible in order to convey the essence of the concept to the biologist or the software engineer.
Integration with existing ontologies
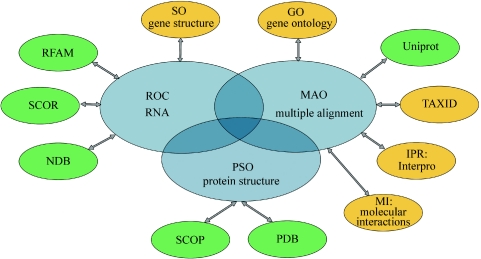
An important criterion in the design of MAO was the definition of the interface with other biological resources, in particular other related ontologies in OBO. Cross-references are provided to related ontologies, such as GO, SO, MI, Interpro and the NCBI organism classification, but the list of inter-relations will obviously grow as new domain ontologies are developed. Cross-references are also provided to a number of public databases, including the nucleic acid and protein sequence databases, such as GenBank (34) and UniProt (35), RNA databases, such as NDB (36), SCOR (37) and RFAM (38), and protein 3D structure databases, such as PDB (39) and SCOP (40).
Implementation
The ontology was constructed using the open source Java tool OBO-Edit. The tool provides a graphical interface to handle any vocabulary that has a DAG data structure. The OBO-Edit tool can export the resulting ontology in both the GO flat-file format and the newer OBO format, which is one of the formats supported by the OBO consortium. The MAO ontology is freely available in OBO format from the MAO website at http://bips.u-strasbg.fr/LBGI/MAO/mao.html or from the OBO site at http://obo.sourceforge.net.
RESULTS AND DISCUSSION
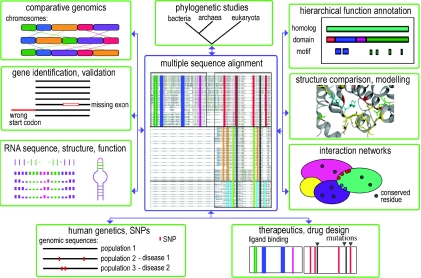
Multiple sequence alignments play a central role in a wide range of applications, including in-depth database searching, functional residue identification, structure prediction techniques and of course, evolutionary studies (Figure 2). Accurate multiple alignments, therefore, represent an ideal environment for the reliable integration, propagation and presentation of the most vital and relevant aspects of all the information associated with a sequence family.
Figure 2.
Examples of molecular biology applications (shown in green boxes) that rely on multiple sequence alignments. Conserved positions in the multiple sequence alignment (shown as coloured bars in the central figure) often correspond to functionally or structurally important sites, such as catalytic sites (as shown in structure comparison), interaction interfaces (in interaction networks) or secondary structure elements (as in RNA sequence, structure and function).
The MAO is a task ontology for the multiple alignment of DNA, RNA and protein sequences and 3D structures. MAO has been developed by a number of experts in the fields of RNA sequence alignment, protein family alignment and 3D structure comparisons and analyses. The ontology thus provides an objective, consensual specification of domain information that represents a consensual agreement on the concepts and relations that characterize the way knowledge in that domain is expressed. The MAO ontology has been registered at the OBO website, which provides an umbrella web address for well-structured controlled vocabularies for shared use across different biological domains. Acceptance on the OBO site implies that the ontology has been accepted as authoritative by the OBO group (41) and that the ontology meets a number of specific criteria defined by the community. In particular, only a single ontology should be specified for each domain or task, and new ontologies should be orthogonal to the other ontologies already hosted within OBO. An important issue in the development of the MAO was, therefore, to define the scope of the ontology and the relationships to other existing ontologies, in order to ensure orthogonality and to facilitate integration between the different domain ontologies. Figure 3 shows the main cross-references defined in MAO to external ontologies, particularly those covering RNA terms and protein 3D structure, as well as to the public sequence and structure databases.
Figure 3.
Major interactions between MAO and external resources. Ontologies developed in collaboration are shown in blue. Other ontologies are shown in yellow, while external databases are shown in green.
Design criteria
In theoretical terms, an ontology is generally described as ‘a formal representation of a domain of knowledge’. MAO uses a hierarchical model represented by a DAG, in which concepts are described by textual definitions and are linked by one of the four formal relations. Two different hierarchical relationships are defined, namely is_a and part_of. Characteristics are assigned to the concepts where appropriate using the associative relationships is_name and is_attribute, in order to permit the integration of more complex information, such as residue function or activity, sequence feature conservation or 3D structural location. The is_attribute relationship is also used to record the algorithm or program used for important alignment concepts, such as sub_alignment_construction_method or column_conservation_construction_method. This means that the results obtained by different alignment algorithms can be represented in the same framework for comparison and integration purposes.
Scope and structure
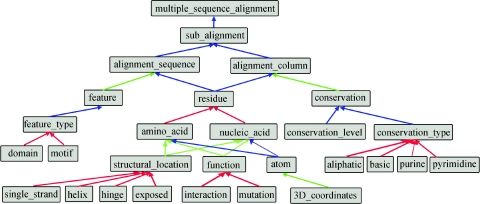
The use to which an ontology is put largely determines the content of the ontology (33). Thus, no ‘optimal’ ontology exists, but the quality of a particular ontology should be judged by its usefulness or suitability for a specific application. The MAO ontology covers the great majority of relevant concepts required when constructing or analysing multiple alignments of DNA, RNA or protein sequences, as shown in Figure 4. The top level multiple_sequence_alignment concept is divided into sub_alignments, defining a subset of sequences, which may be constructed by an automatic sequence clustering algorithm, or may be specified by some other factor, such as phylogenetic or functional criteria. Sub_alignments are then divided into alignment_sequences and alignment_columns. Alignment_sequences have various global attributes, such as function, taxonomy, sequence database cross-references, etc. In addition, sequence features can also be defined that represent a particular subsequence and may correspond to a domain, a transmembrane region, a signal peptide, a secondary structure element, etc. Alignment_columns can be characterized according to their conservation, described in terms of both the level and the type of conservation. In order to accommodate a wide range of conservation calculation methods, a large number of conservation attributes have been defined. Thus, the conservation level can be described by either a qualitative or a quantitative value, while the type of conservation might refer to either a single residue or a group of residues that share a similar feature, such as ‘small’, ‘negatively_charged’ or ‘hydrophilic’. Clearly, both columns and sequences should contain residues, but in addition the concept ‘gap’ is defined to represent insertions or deletions in the sequences. These gap positions are crucial for the multiple alignment definition and differentiate the alignment_sequence specified in MAO from the sequence concept in other ontologies, such as the SO or the protein family ontology (31). Residues are defined to be either amino acid or nucleotide. For amino acids, two main attributes exist. First, the structural location of the amino acid can be defined as exposed/buried, N/C-terminal, helix/strand/loop, etc. Second, the amino acid can be annotated by its functional activity, i.e. active site, binding site, post-transcriptional modification, mutation, etc. Attributes specific to RNA molecules, such as base pairs and ‘structural motifs’, are currently being defined in collaboration with the ROC and will be included in a future release of MAO. The lowest level concept is the atom that is a part_of both amino acids and nucleotides, and allows the integration of 3D structural information in the form of x, y, z atomic coordinates.
Figure 4.
Graphical representation of part of the MAO ontology structure. Grey boxes represent concepts and coloured arrows represent relationships (red, is_a; blue, part_of; green, is_attribute). The major MAO concepts are described in the text.
Because many biological terms may be ambiguous, MAO concepts have associated textual definitions so that their precise meaning within the context of the ontology is clear to a human reader. Each concept in the ontology is defined as precisely and as succinctly as possible. Definitions are the basis for the relations between concepts, for semantic disambiguation and as such the foundation of an ontology and therefore indispensable. However, different experts can employ different terminologies for the same concepts and it is not the purpose of MAO to impose a particular terminology. Alternative terms for a concept are therefore defined as synonyms. In addition, each term in the ontology is assigned a unique ID that has two components: a two letter code RO that indicates the ontology namespace and a number. IDs can be used to link a biological database to the ontology. The user can query the database for data associated with a particular ID and use the logic of the rules in the ontology to ask further questions about the data. IDs can also be used to connect different databases directly.
Example applications
The vocabulary specified in MAO has been used to define an XML schema for annotated multiple alignments, in order to provide an unambiguous file format that is computer-friendly and easily readable. The XML schema has been incorporated in the BAliBASE benchmark database (version 3) (42) for the comparison and evaluation of multiple alignment algorithms. It has also been used in the Structural Proteomics in Europe (SPINE) project to generate HMTL format ‘identity cards’ for each potential protein target. These identity cards, containing the results of the automatic target identification and characterization process, are made accessible to all members of the SPINE consortium over the web.
Integrated gene family analysis
One of the most powerful features of the MAO ontology is that it provides a natural, intuitive link between a number of different ontologies in the domains of genomics and proteomics. Using the cross-references defined in MAO, diverse functional information from external data resources, such as active sites, mutation data and their associated phenotypes, etc. can be integrated, either for a single sequence or for a family of sequences. In the context of the overall family alignment, structural and functional data can be combined with information about the conservation of the family and the variability observed at different residue sites. As an example, Figure 5 shows a multiple alignment of the interleukin-1 (IL1) protein family. IL1 is a proinflammatory cytokine produced by activated macrophages and monocytes. It functions in the generation of systemic and local responses to infection, injury, and immunological challenges and is the primary cause of chronic and acute inflammation (43). The overall IL1 family alignment is divided into four sub_alignments, corresponding to two structurally distinct forms (IL1A and IL1B), one sub-family of IL1 homologues (I1Fx) and one sub-family containing IL1 receptor antagonist proteins (IL1X). The domain structure of the sequences was determined by cross-reference to the Interpro database, followed by propagation of the known domains in the conserved regions of the alignment. In particular, the interleukin domain was identified as a common attribute shared by all the sequences in the C-terminal half of the multiple alignment. However, IL1A and IL1B are both synthesized as larger precursors, with the N terminal ∼115 amino acids forming a propeptide that is cleaved off to release the active IL1. Sequence analysis of this propeptide region highlighted a number of conserved features in the IL1A sequences that were not present in the IL1B sub_alignment, including a continuous stretch of four columns of conserved basic amino acids (lysine or arginine) that corresponded exactly to the experimentally verified nuclear localization signal motif of human IL1A (44). These differentially conserved regions may be responsible for the functional disparities observed recently. In fact, it has been shown that IL1A produces apoptosis in malignant cell lines, whereas IL1B promotes invasiveness (45) and it has been suggested that within the nucleus, the IL1A propeptide may interact with elements of RNA processing affecting alternate splicing of genes involved in the regulation of apoptosis (44).
Figure 5.
Knowledge-based sequence analysis of the IL1 protein family. Protein sequences were detected by a BlastP search using the query sequence IL1A_ HUMAN (outlined in red) and aligned using the PipeAlign web server (46). (a) Schematic view of the full length alignment. Alignment columns are coloured according to similar residue conservation (black, 100% conserved, dark blue/pink, 60% conserved; light blue/pink, 20% conserved; similar residue groups, DN; EQ; ST; KR; KYW; LIVM) using the GeneDoc program (http://www.psc.edu/biomed/genedoc). (b) Multiple alignment of IL1 propeptide in IL1A and IL1B subgroups, produced using the OrdAli program (L. Moulinier, manuscript in preparation). Alignment columns are coloured according to conservation; black, 100% conserved; grey, 80% similar residue groups (PAGST, DEQN, KRH, FYW, ILMV, C); red/blue, 100% conserved in sub_alignment IL1A/IL1B. Functional sites were experimentally verified for human IL1A and IL1B (black triangle above sequence: cleavage site). (c) Selected MAO concepts and associated instances, highlighting the differentially conserved features. Mutation predictions were obtained from SeattleSNPs (http://pga.mbt.washington.edu/).
Perspectives
An ontology provides the conceptual framework that is used to capture knowledge in a specific domain. The concepts in the ontology represent classes or sets of instances that exist in the real world, but the ontology itself should not contain any instances. This is roughly analogous to what is known as the schema for a relational database or XML document. The combination of an ontology with associated instances is known as a ‘knowledge base’. Work is now in progress to construct a MAO knowledge base of high quality, global multiple alignments that will cover most of the known protein fold space. Information as diverse as gene structure, protein 3D structure/function or specific residue interactions will be combined together with taxonomic and evolutionary information to produce a detailed description of a protein family. An important part of this development will be the analysis and cross-validation of this mass of heterogeneous information, the presentation of the pertinent information in a user-friendly, graphical interface and the easy accessibility of these annotated alignments. The potential applications for such a knowledge base are numerous, but will include such fields as the definition of characteristic motifs for specific protein folds, or the automatic annotation of the ever-increasing number of hypothetical proteins being produced by the high-throughput genome sequencing projects.
Acknowledgments
The authors are grateful to many colleagues for their invaluable contributions to the ontology. E.W. thanks the Institut Universitaire de France for support. J.D.T., D.M. and O.P. were supported by institute funds from the Institut National de la Santé et de la Recherche Médicale, the Centre National de la Recherche Scientifique, the Hôpital Universitaire de Strasbourg, the Fond National de la Science (GENOPOLE) and the SPINE project (E.C. contract number QLG2-CT-2002-00988). Funding to pay the Open Access publication charges for this article was provided by the Centre National de la Recherche Scientifique.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kitano H. Computational systems biology. Nature. 2002;420:206–210. doi: 10.1038/nature01254. [DOI] [PubMed] [Google Scholar]
- 2.Tong W. Analyzing the biology on the system level. Genomics Proteomics Bioinformatics. 2004;2:6–14. doi: 10.1016/S1672-0229(04)02002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woese C.R., Pace N.R. The RNA World. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. Probing RNA structure, function and history by comparative analysis. [Google Scholar]
- 4.Lecompte O., Thompson J.D., Plewniak F., Thierry J., Poch O. Multiple alignment of complete sequences (MACS) in the post-genomic era. Gene. 2001;270:17–30. doi: 10.1016/s0378-1119(01)00461-9. [DOI] [PubMed] [Google Scholar]
- 5.Woese C.R., Fox G.E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl Acad. Sci. USA. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fox G.E., Stackebrandt E., Hespell R.B., Gibson J., Maniloff J., Dyer T.A., Wolfe R.S., Balch W.E., Tanner R.S., Magrum L.J., et al. The phylogeny of prokaryotes. Science. 1980;209:457–463. doi: 10.1126/science.6771870. [DOI] [PubMed] [Google Scholar]
- 7.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eddy S.R. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 9.Karplus K., Barrett C., Hughey R. Hidden Markov models for detecting remote protein homologies. Bioinformatics. 1999;14:846–856. doi: 10.1093/bioinformatics/14.10.846. [DOI] [PubMed] [Google Scholar]
- 10.Michel F., Costa M., Massire C., Westhof E. Modeling RNA tertiary structure from patterns of sequence variation. Methods Enzymol. 2000;317:491–510. doi: 10.1016/s0076-6879(00)17031-4. [DOI] [PubMed] [Google Scholar]
- 11.Cozetto D., Tramontano A. Relationship between multiple sequence alignments and quality of protein comparative models. Proteins. 2005;58:151–157. doi: 10.1002/prot.20284. [DOI] [PubMed] [Google Scholar]
- 12.Guda C., Lu S., Scheeff E.D., Bourne P.E., Shindyalov I.N. CE-MC: a multiple protein structure alignment server. Nucleic Acids Res. 2004;32:W100–W103. doi: 10.1093/nar/gkh464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dror O., Benyamini H., Nussinov R., Wolfson H.J. Multiple structural alignment by secondary structures: algorithm and applications. Protein Sci. 2003;12:2492–2507. doi: 10.1110/ps.03200603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye Y., Godzik A. Multiple flexible structure alignment using partial order graphs. Bioinformatics. 2005;21:2362–2369. doi: 10.1093/bioinformatics/bti353. [DOI] [PubMed] [Google Scholar]
- 15.Simossis V.A., Heringa J. Integrating protein secondary structure prediction and multiple sequence alignment. Curr. Protein Pept. Sci. 2004;5:249–266. doi: 10.2174/1389203043379675. [DOI] [PubMed] [Google Scholar]
- 16.O'Sullivan O., Suhre K., Abergel C., Higgins D.G., Notredame C. 3DCoffee: combining protein sequences and structures within multiple sequence alignments. J. Mol. Biol. 2004;340:385–395. doi: 10.1016/j.jmb.2004.04.058. [DOI] [PubMed] [Google Scholar]
- 17.Ren T., Veeramalai M., Choon Tan A., Gilbert D. MSAT: a multiple sequence alignment tool based on TOPS. Appl. Bioinformatics. 2004;3:149–158. doi: 10.2165/00822942-200403020-00009. [DOI] [PubMed] [Google Scholar]
- 18.Johnson J.M., Mason K., Moallemi C., Xi H., Somaroo S., Huang E.S. Protein family annotation in a multiple alignment viewer. Bioinformatics. 2003;19:544–545. doi: 10.1093/bioinformatics/btg021. [DOI] [PubMed] [Google Scholar]
- 19.Jossinet F., Westhof E. Sequence to Structure (S2S): display, manipulate and interconnect RNA data from sequence to structure. Bioinformatics. 2005;21:3320–3321. doi: 10.1093/bioinformatics/bti504. [DOI] [PubMed] [Google Scholar]
- 20.Bard J.B., Rhee S.Y. Ontologies in biology: design, applications and future challenges. Nature Rev. Genet. 2004;5:213–222. doi: 10.1038/nrg1295. [DOI] [PubMed] [Google Scholar]
- 21.Gruber T.R. Formal Ontology in Conceptual Analysis and Knowledge Representation. Deventer, The Netherlands: Kluwer Academic Publishers; 1993. Toward principles for the design of ontologies used for knowledge sharing. [Google Scholar]
- 22.Gene Ontology Consortium. Creating the gene ontology resource: design and implementation. Genome Res. 2001;11:1425–1433. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Camon E., Barrell D., Lee V., Dimmer E., Apweiler R. The Gene Ontology Annotation (GOA) Database—an integrated resource of GO annotations to the UniProt Knowledgebase. In Silico Biol. 2004;4:5–6. [PubMed] [Google Scholar]
- 24.Hennig S., Groth D., Lehrach H. Automated Gene Ontology annotation for anonymous sequence data. Nucleic Acids Res. 2003;31:3712–3715. doi: 10.1093/nar/gkg582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen L.J., Gupta R., Staerfeldt H.H., Brunak S. Prediction of human protein function according to Gene Ontology categories. Bioinformatics. 2003;19:635–642. doi: 10.1093/bioinformatics/btg036. [DOI] [PubMed] [Google Scholar]
- 26.Chalmel F., Lardenois A., Thompson J.D., Muller J., Sahel J.A., Leveillard T., Poch O. GOAnno: GO annotation based on multiple alignment. Bioinformatics. 2005;21:2095–2096. doi: 10.1093/bioinformatics/bti252. [DOI] [PubMed] [Google Scholar]
- 27.Ponomarenko J.V., Bourne P.E., Shindyalov I.N. Assigning new GO annotations to protein data bank sequences by combining structure and sequence homology. Proteins. 2005;58:55–65. doi: 10.1002/prot.20355. [DOI] [PubMed] [Google Scholar]
- 28.Eilbeck K., Lewis S.E., Mungall C.J., Yandell M., Stein L., Durbin R., Ashburner M. The Sequence Ontology: a tool for the unification of genome annotations. Genome Biol. 2005;6:R44. doi: 10.1186/gb-2005-6-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hermjakob H., Montecchi-Palazzi L., Bader G., Wojcik J., Salwinski L., Ceol A., Moore S., Orchard S., Sarkans U., von Mering C., et al. The HUPO PSI's molecular interaction format—a community standard for the representation of protein interaction data. Nat. Biotechnol. 2004;22:177–183. doi: 10.1038/nbt926. [DOI] [PubMed] [Google Scholar]
- 30.Waugh A., Gendron P., Altman R., Brown J.W., Case D., Gautheret D., Harvey S.C., Leontis N., Westbrook J., Westhof E., Zuker M., Major F. RNAML: a standard syntax for exchanging RNA information. RNA. 2002;8:707–717. doi: 10.1017/s1355838202028017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolstencroft K., McEntire R., Stevens R., Tabernero L., Brass A. Constructing ontology-driven protein family databases. Bioinformatics. 2005;21:1685–1692. doi: 10.1093/bioinformatics/bti158. [DOI] [PubMed] [Google Scholar]
- 32.Mulder N.J., Apweiler R., Attwood T.K., Bairoch A., Bateman A., Binns D., Bradley P., Bork P., Bucher P., Cerutti L., et al. InterPro, progress and status in 2005. Nucleic Acids Res. 2005;33:D201–D205. doi: 10.1093/nar/gki106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stevens R., Goble C.A., Bechhofer S. Ontology-based knowledge representation for bioinformatics. Brief Bioinform. 2000;1:398–414. doi: 10.1093/bib/1.4.398. [DOI] [PubMed] [Google Scholar]
- 34.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank. Nucleic Acids Res. 2005;33:D34–D38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bairoch A., Apweiler R., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berman H.M., Westbrook J., Feng Z., Iype L., Schneider B., Zardecki C. The Nucleic Acid Database. Acta Crystallogr. D Biol. Crystallogr. 2002;58:889–898. doi: 10.1107/s0907444902003487. [DOI] [PubMed] [Google Scholar]
- 37.Klosterman P.S., Tamura M., Holbrook S.R., Brenner S.E. SCOR: a Structural Classification of RNA database. Nucleic Acids Res. 2002;30:392–394. doi: 10.1093/nar/30.1.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Griffiths-Jones S., Moxon S., Marshall M., Khanna A., Eddy S.R., Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andreeva A., Howorth D., Brenner S.E., Hubbard T.J., Chothia C., Murzin A.G. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Westbrook J., Feng Z., Jain S., Bhat T.N., Thanki N., Ravichandran V., Gilliland G.L., Bluhm W., Weissig H., Greer D.S., Bourne P.E., Berman H.M. The Protein Data Bank: unifying the archive. Nucleic Acids Res. 2002;30:245–248. doi: 10.1093/nar/30.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bard J., Rhee S.Y., Ashburner M. An ontology for cell types. Genome Biol. 2005;6:R21. doi: 10.1186/gb-2005-6-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson J.D., Koehl P., Ripp R., Poch O. BAliBASE 3.0: latest developments of the multiple sequence alignment benchmark. Proteins. 2005 doi: 10.1002/prot.20527. in press. [DOI] [PubMed] [Google Scholar]
- 43.Dinarello C.A. Interleukin-1, interleukin-1 receptors and interleukin-1 receptor antagonist. Int. Rev. Immunol. 1998;16:457–499. doi: 10.3109/08830189809043005. [DOI] [PubMed] [Google Scholar]
- 44.Pollock A.S., Turck J., Lovett D.H. The prodomain of interleukin 1alpha interacts with elements of the RNA processing apparatus and induces apoptosis in malignant cells. FASEB J. 2003;17:203–213. doi: 10.1096/fj.02-0602com. [DOI] [PubMed] [Google Scholar]
- 45.Song X., Voronov E., Dvorkin T., Fima E., Cagnano E., Benharroch D., Shendler Y., Bjorkdahl O., Segal S., Dinarello C.A., Apte R.N. Differential effects of IL-1 alpha and IL-1 beta on tumorigenicity patterns and invasiveness. J. Immunol. 2003;171:6448–6456. doi: 10.4049/jimmunol.171.12.6448. [DOI] [PubMed] [Google Scholar]
- 46.Plewniak F., Bianchett L., Brelivet Y., Carles A., Chalmel F., Lecompte O., Mochel T., Moulinier L., Muller A., Muller J., et al. PipeAlign: a new toolkit for protein family analysis. Nucleic Acids Res. 2003;31:3829–3832. doi: 10.1093/nar/gkg518. [DOI] [PMC free article] [PubMed] [Google Scholar]