Abstract
Using a combination of multiplex amplifiable probe hybridization and semiquantitative fluorescence in situ hybridization (SQ-FISH), we analyzed DNA copy number variation across chromosome band 8p23.1, a region that is frequently involved in chromosomal rearrangements. We show that a cluster of at least three antimicrobial β-defensin genes (DEFB4, DEFB103, and DEFB104) at 8p23.1 are polymorphic in copy number, with a repeat unit ⩾240 kb long. Individuals have 2–12 copies of this repeat per diploid genome. By segregation, microsatellite dosage, and SQ-FISH chromosomal signal intensity ratio analyses, we deduce that individual chromosomes can have one to eight copies of this repeat unit. Chromosomes with seven or eight copies of this repeat unit are identifiable by cytogenetic analysis as a previously described 8p23.1 euchromatic variant. Analysis of RNA from different individuals by semiquantitative reverse-transcriptase polymerase chain reaction shows a significant correlation between genomic copy number of DEFB4 and levels of its messenger RNA (mRNA) transcript. The peptides encoded by these genes are potent antimicrobial agents, especially effective against clinically important pathogens, such as Pseudomonas aeruginosa and Staphylococcus aureus, and DEFB4 has been shown to act as a cytokine linking the innate and adaptive immune responses. Therefore, a copy number polymorphism involving these genes, which is reflected in mRNA expression levels, is likely to have important consequences for immune system function.
Introduction
Defensin genes encode a family of small cationic antimicrobial peptides that form an important part of the innate immune system (Ganz 1999). The family is divided into three classes, α, β, and θ, according to the arrangement of disulfide bridges between cysteine residues. The α-defensins are strongly expressed in neutrophils and are also present in certain epithelia, such as the gut wall, endocervix, and vagina. They have been shown by both in vitro and in vivo methods to have powerful antimicrobial properties (Ghosh et al. 2002; Salzman et al. 2003) and contribute to the anti-HIV-1 properties of CD8 antiviral factor (Zhang et al. 2002). The β-defensins are expressed in a variety of epithelia, especially in the airways and epididymis, and have been shown to have broad antimicrobial properties (Schutte and McCray 2002). In particular, DEFB4 (MIM 602215) is effective against Escherichia coli and Pseudomonas aeruginosa at micromolar concentrations (Harder et al. 1997; Bals et al. 1998; Liu et al. 1998), and DEFB103 (MIM 606611) is extremely effective against Staphylococcus aureus (Harder et al. 2001). In addition to its antimicrobial properties, DEFB4 is expressed in leukocytes and acts as a chemokine for cells of the adaptive immune response (Yang et al. 1999; Biragyn et al. 2002). The only θ-defensin gene identified so far in humans is DEFT1, which appears to be an expressed pseudogene. The putative ortholog of DEFT1 in the rhesus monkey (Macacca mulatta) has strong anti-HIV properties (Cole et al. 2002). All α- and θ-defensins and most β-defensins occur in a cluster at 8p23.1, although recent in silico analysis has identified clusters of putative β-defensins at 20p13, 20q11.1, and 6p12 (Schutte et al. 2002). The evolutionary relationship between these clusters and the genes within them is not known.
Chromosome band 8p23.1 is known to be a frequent site of chromosomal rearrangements mediated by two olfactory repeat regions (ORRs) 5 cM apart. As many as one in four individuals from the normal population is a carrier of an inversion polymorphism between these two ORRs (Giglio et al. 2001, 2002). An apparent chromosomal duplication has been described, in this region, that is a euchromatic variant (EV) with no clinical phenotypic effect (Barber et al. 1998; O’Malley and Storto 1999).
To characterize the cytogenetic EV and to determine copy number variation at this locus, we used a combination of semiquantitative fluorescence in situ hybridization (SQ-FISH), for examination of relative signal ratios, and multiplex amplifiable probe hybridization (MAPH), for direct assay of the DNA copy number. We show that the EV is not a simple doubling of a chromosomal segment but is a high-copy-number allele of normal copy number variation involving the β-defensin gene cluster. Most individuals have 2–7 copies per diploid genome, whereas EV carriers have 9–12 copies. We also show that expression levels of DEFB4 are correlated with copy number, which suggests that this polymorphism may be an important component of genetic variation in susceptibility to infectious disease.
Material and Methods
DNA and RNA Extraction
Genomic DNA and total RNA were extracted from lymphoblastoid cells and whole peripheral blood, using standard techniques (Ausubel et al. 1997). All research samples from patients were collected under appropriate ethical committee approval.
MAPH and Analysis
MAPH is a DNA-based quantitative method for direct determination of DNA copy number and relies on the fact that amplifiable probes can be hybridized to genomic DNA fixed onto a nylon membrane, stringently washed, and then amplified so that the amount of amplified product is directly proportional to the copy number in the genomic DNA. Each amplifiable probe is a different length, so that the probes can be resolved by electrophoresis. All probes share primer-binding sites at each end, so that one pair of primers can amplify all probes simultaneously.
MAPH probes were generated by PCR amplification and cloning into pZero-2 vector (Invitrogen) and were sequenced to confirm identity. The probes spanning 8p23.1 were termed “A”–“N” (table 1 and fig. 1). The final probe set was a mixture of these probes with probes from a set used for screening subtelomeric regions for deletions and duplications (Hollox et al. 2002). These subtelomeric probes do not report common polymorphism in extensive testing (Hollox et al. 2002) and could therefore be used as reference probes for measurement of the relative dosage of the 8p23.1 probes.
Table 1.
MAPH Probes Used to Screen 8p23.1 for Copy Number Variation
| MAPH Probe (DDBJ/EMBL/GenBank Accession Number) | Location (Bases) |
| A (NM_001147) | ANGPT2 exon 9 (1716–1901) |
| B (AF287957) | Proximal to ANGPT2 (168026–168192) |
| C (Z45294) | FLJ11210 (41–174) |
| D (AF233439) | Between DEFB1 and DEFA4 (62370–62614) |
| E (AF238378) | DEFA3 exon 1 (41924–42309) |
| F (NM_04942) | DEFB4 exon 2 (117–237) |
| G (AC252830) | DEFB4 intron 1 (74061–74197) |
| H (G13705) | SPAG11 intron 2 (121–326) |
| I (AA687243) | Anonymous region (41–120) |
| J (AA226797) | Distal to MASL1 (121–211) |
| K (AA010611) | MASL1 intron 1 (50–241) |
| L (Z24258) | Next to D8S550 (296–380) |
| M (L34357) | GATA4 exon 6 (1415–1817) |
| N (AQ318792) | DLC1 intron 1 (21–238) |
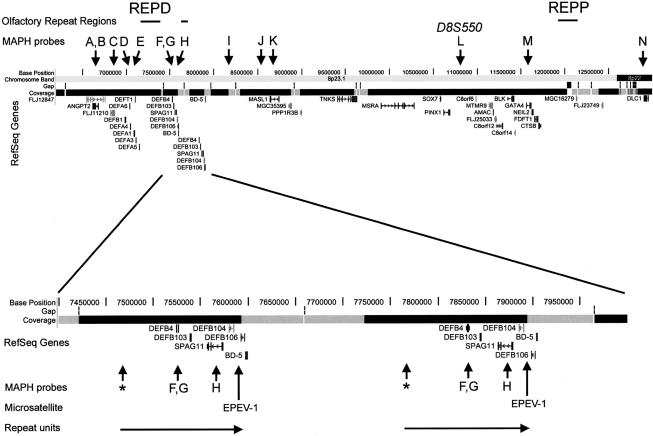
Figure 1.
MAPH probe locations on 8p23.1. Positions of the MAPH probes (A–N) used to screen 8p23.1 for copy number variation are shown by arrows, together with the ORRs named “REPP” and “REPD” by Giglio et al. (2001). The area identified as involved in the copy number variation is shown in detail and is represented twice in this genome assembly (November 2002 assembly; for details, see the University of California–Santa Cruz [UCSC] Genome Bioinformatics Web site). The probe marked with an asterisk (*) showed increased dosage in 8p23.1 EVs but maps to ORR sequences and was not included in the main MAPH probe set. RefSeq = reference sequence.
The full experimental details of MAPH have been published elsewhere (Armour et al. 2000), and updates are available at the Institute of Genetics, University of Nottingham, Web site (Multiplex Amplifiable Probe Hybridization). In brief, hybridization of immobilized genomic DNA with the 8p23.1 MAPH probe set was performed as described elsewhere, using other probe sets (Armour et al. 2000). After stringent washing, amplifiable probes that remained bound to the genomic DNA were released by incubation in a solution of 50 μl (containing 75 mM Tris-HCl [pH 8.8 at 25°C], 20 mM [NH4]2SO4, and 0.01% [v/v] Tween 20 [1 × PCR buffer IV; ABgene]) at 95°C for 5 min; 1 μl of this solution was used to seed a 20-μl PCR using a 5′ FAM-labeled PZA primer (5′-AGTAACGGCCGCCAGTGTGCTG-3′) and an unlabeled PZB primer (5′-CGAGCGGCCGCCAGTGTGATG-3′) and was amplified for 25 cycles, with each cycle being 95°C for 1 min, 60°C for 1 min, and 70°C for 1 min, followed by a final 20-min 72°C extension incubation. After ethanol precipitation of the product, 3 μl of formamide loading buffer/ROX-500 fluorescent marker was added to the pellet and was loaded on an ABI 377 fluorescent-gel-electrophoresis apparatus.
After electrophoretic separation of the 61 different probes in the probe set (14 probes mapping to 8p23.1 and 47 probes mapping to single-copy subtelomeric regions; see fig. 2), each probe was quantified by measuring the appropriate peak area by use of the Genescan software (Applied Biosystems). These values were normalized against the four nearest reference peaks within the sample. In the absence of normal copy number variation, these values are in an approximate Gaussian distribution around the mean, with the SD indicating the measurement error of the MAPH procedure (Hollox et al. 2002). SDs of ∼10% were observed, low enough to detect changes involving a large relative change in copy number, such as one copy instead of two, but not low enough to discriminate, for example, six copies from five copies. Multiple testing of each sample allowed us to increase the measurement precision and report 95% confidence limits on each copy number value (table 2). Multiple probes also allow an increase in precision of copy number measurement, although this assumes that each probe dosage reflects the same underlying dosage in the DNA. For this reason, table 2 reports separately the values of probes F–H.
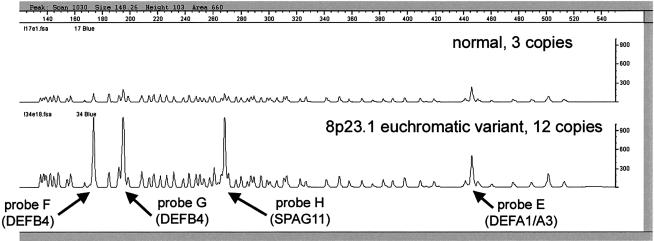
Figure 2.
MAPH analysis of normal control individual and 8p23.1 EV carrier. A MAPH chromatogram shows the analysis of an EV carrier (bottom), with 12 copies of the 8p23.1 repeat unit, and a control DNA sample (top), with 3 copies of the 8p23.1 repeat unit. The X-axis represents probe length (in bp), and the Y-axis represents relative fluorescence units. Each peak represents a different amplifiable probe separated by length. The three probes (F–H) measuring β-defensin–cluster copy number are indicated, together with a probe (E) mapping to DEFA1. Other peaks represent other probes, most of which (47 of the remaining 57) map to single-copy subtelomeric regions and have a copy number of two per diploid genome. The areas of the polymorphic probe peaks (F–H) in the control sample (three copies) are similar to, but slightly larger than, the subtelomeric probe peaks (two copies).
Table 2.
MAPH, Microsatellite, SQ-FISH, and Cytogenetic Analyses of a Subset of Individuals[Note]
|
MAPH Analysisa |
Microsatellite EPEV-1 Analysis |
SQ-FISH Analysisb |
||||||||||
| Sample | nd | SPAG11(probe H) | DEFB4a(probe G) | DEFB4b(probe F) | Other 8pe | β-Defensin–ClusterCopy Number | Genotype | ApproximateDosage Ratio | BAC 51D11 Signal Intensity Ratio | YAC HTY3020 Signal Intensity Ratio | β-Defensin–ClusterAlleles | G-BandCytogeneticAnalysisof 8p23.1 EVc |
| Family 1: | ||||||||||||
| I:2 | 4 | ND | 12 ± 1.6 | 10 ± .54 | 2.0 ± .19 | 11 | 169,182,184,188 | 1:2:7:1 | ND | ND | … | Yes |
| II:3 | 2 | ND | 10 ± 1.2 | 9.1 ± .59 | 2.0 ± .49 | 9 | 169,182,184 | 2:1:6 | ND | ND | … | Yes |
| II:5 | 3 | 3.4 ± .18 | 4.0 ± .21 | 3.0 ± .18 | 1.7 ± .11 | 4 | 169,184,188 | 1:2:1 | ND | ND | … | No |
| II:8 | 4 | ND | 8.9 ± .41 | 8.8 ± .61 | 1.9 ± .30 | 9 | 169,182,184 | 2:1:6 | 1:3.9 (2.4–6.3) | 1:7.6f (5.5–10) | 1, 8 | Yes |
| II:11 | 2 | ND | 12 ± 2.8 | 10.3 ± 1.7 | 2.2 ± .39 | 11 | 169,182,184 | 4:2:5 | ND | ND | … | Yes |
| Family 2: | ||||||||||||
| I:1 | 2 | 8.8 ± .01 | 11 ± 0.27 | 8.1 ± .55 | 2.3 ± .19 | 9 | 182,184,186,188 | 3:4:1:1 | ND | 1:2.8 (1.9–4.1) | 2, 7 | Yes |
| I:2 | 4 | 4.5 ± .23 | 4.3 ± .31 | 4.4 ± .38 | 2.1 ± .26 | 4 | 169,182,184,190 | 1:1:1:1 | ND | 1:1.3 (1.1–1.7) | 2, 2 | No |
| II:2 | 4 | 9.3 ± .66 | 9.4 ± .89 | 8.6 ± .35 | 2.2 ± .49 | 9 | 169,182,184,186 | 1:4:3:1 | 1:2.4 (1.6–3.5) | 1:3.2 (2.1–4.7) | 2, 7 | Yes |
| III:3 | 4 | 9.1 ± 1.8 | 9.8 ± .74 | 10 ± 1.1 | 2.1 ± .13 | 9/10 | 169,182,184,186 | 1:3:3:2 | 1:3.0 (2.1–4.4) | 1:3.5f (2.8–5.2) | 2, 7 | Yes |
| Family 3: | ||||||||||||
| I:1 | 5 | 4.4 ± .42 | 4.2 ± .36 | 4.4 ± .29 | 2.1 ± .10 | 4 | 182,184,188,190 | 1:1:1:1 | 1:1.6 (1.3–2.0) | 1:1.3f (1.1–1.5) | 2, 2 | No |
| I:2 | 3 | ND | 14 ± 3.4 | 12 ± .16 | 2.3 ± .78 | 12 | 169,182,184,186,188 | 1:1:8:1:1 | 1:2.1 (1.9–2.3) | 1:1.9f (1.5–2.4) | 4, 8 | Yes |
| II:1 | 3 | 12 ± 1.2 | 11 ± .65 | 13 ± .29 | 2.1 ± .89 | 12 | 169,182,184,188 | 1:2:8:1 | 1:2.8 (2.2–3.7) | 1:2.3f (1.5–3.4) | 4, 8 | Yes |
| Unrelated individuals: | ||||||||||||
| J1 | 5 | 2.0 ± .096 | 2.0 ± .14 | 2.0 ± .11 | 2.0 ± .15 | 2 | 169 | … | ||||
| N005 | 5 | 3.0 ± .28 | 2.5 ± .33 | 3.1 ± .44 | 1.9 ± .28 | 3 | 169, 186 | 1:2 | ||||
| N008 | 5 | 3.4 ± .42 | 3.2 ± .27 | 3.3 ± .25 | 1.8 ± .10 | 3 | 182,186,188 | 1:1:1 | ||||
| N015g | 5 | 4.6 ± .25 | 3.9 ± .13 | 5.4 ± .48 | 1.9 ± .35 | 5 | 169,184,186,188 | 2:1:1:1 | ||||
| N025 | 3 | 6.5 ± .85 | 7.3 ± .85 | 7.1 ± .85 | 2.0 ± .13 | 7 | 169,182,184,186 | 2:1:2:2 | ||||
| N029 | 3 | 5.1 ± .34 | 5.6 ± .19 | 6.2 ± .34 | 2.0 ± .36 | 6 | 169,184,186, 190 | 1:3:1:1 | ||||
| Chimpanzeeh | 1 | 4.1 | ND | 4.2 | 2.1 ± 1.0 | 4 | 169 | … | ||||
| Gorillah | 1 | 1.4 | ND | 2.5 | 1.9 ± 1.2 | 2 | 186 | … | ||||
Note.— ND = not determined.
Dosage per diploid genome ± 95% confidence limits, with J1 or N005 as the standard reference.
Mean ratio (95% confidence limits).
Yes = presence of an 8p23.1 EV chromosome; No = normal chromosome 8.
Number of independent MAPH tests.
Excluding DEFA1.
Using YAC 820b4.
Heterogeneity of repeat unit (P<.01, by t test).
The large CIs seen in the gorilla and chimpanzee samples are due to dosage differences in reference subtelomeric probes as compared with humans.
Pulsed-Field Gel Electrophoresis
Peripheral blood leukocytes were taken from individual N025 (table 2), and DNA was prepared in agarose blocks, using standard methods (Ausubel et al. 1997). Restriction-endonuclease digestion, pulsed-field gel electrophoresis using a CHEFIII gel apparatus (BioRad), and blotting onto an uncharged nylon membrane (Osmonics) were performed using standard procedures (Ausubel et al. 1997).
FISH Analysis
Conventional FISH and SQ-FISH were performed in a similar way to previous work (Barber et al. 1999). In brief, images were captured, enhanced, and analyzed using a Photometrics 200 cooled charge-coupled–device camera and a Macintosh PowerPC equipped with Powergene MacProbe software (Perceptive Scientific Instruments). For SQ-FISH, fluorescent images were normalized to remove background, and the intensity ratios (EV chromosome:normal chromosome) of the individual signals on each homolog were measured in a series of 10–12 metaphase chromosome spreads. Standard parametric methods were then used on the natural-logarithm ratios to obtain mean, SD, and SEM. The 95% CIs were obtained from the antilog of the value of t(n − 1) × SEM, where t is the appropriate two-tailed t-test parameter and n is the number of paired observations.
Microsatellite Analysis
Genotypes for D8S1140 and D8S550 for CEPH families 1331 and 1332 were downloaded from the CEPH database (for details, see the Fondation Jean Dausset–CEPH Web site). To genotype microsatellite EPEV-1, we amplified 20 ng of genomic DNA by using the primers 5′-GGCAGTATTCCAGGATACGG-3′ (MSAT3F, fluorescently labeled at the 5′ end) and 5′-GAACAATTAGATATCCCTATGC-3′ (MSAT3R) for 25 cycles; 1.5 μl of this PCR product was then mixed with formamide loading buffer and was run on an ABI 377 with a ROX-500 marker. Peak areas were estimated after correction for PCR stutter, which was always <10% of the main peak area. Approximate allelic dosage ratios were calculated with prior knowledge of MAPH dosage results, by dividing the area under each allelic peak by the average per copy and rounding to the nearest integer.
Duplex Semiquantitative RT-PCR Analysis
One microgram of total RNA from lymphoblastoid cells was reverse transcribed using 5 U of Reverse-iT Reverse Transcriptase blend (ABgene) and 500 ng of an anchored oligo-dT primer (5′-TTTTTTTTTTTTTVN-3′) in a final volume of 20 μl. Five microliters of cDNA was used to seed a duplex PCR with 1.25 U of Taq polymerase, a pair of primers for the gene of interest, and a pair for TBP, a control housekeeping gene (Murphy et al. 1990). After 32 cycles of 94°C for 30 s, 60°C for 1 min, and 72°C for 1 min, a 10-μl aliquot was run on a 1.5% agarose gel, and each band was quantified using ArrayPro software (Media Cybernetics). To identify any genomic contamination, we designed all primers used for RT-PCR such that they spanned at least one intron in the genomic sequence, and a negative control for the reverse-transcriptase reaction was included in each PCR experiment. Each experiment was repeated three times, to obtain the SEM, and repeat analysis using different amounts of cDNA showed that the PCR had not reached saturation under these conditions. The log10 of the mean ratio ± SEM between the two bands was calculated. All RT-PCR analysis was performed blind to copy number.
Results
We analyzed locus copy number by use of MAPH (Armour et al. 2000) across 8p23.1 (fig. 1) in families with a cytogenetically visible EV of 8p23.1 (Barber et al. 1998) (figs. 2 and 3) and in 90 unrelated control individuals. Two probes spanning the DEFB4 β-defensin locus showed reproducibly polymorphic signals correlated between probes (r2=0.8, between probes F and G; see fig. 1). Further analysis showed the same variation at the SPAG11 gene (MIM 606560), 47 kb away (probe H), indicating that three probes reported concordant copy number variation of the same segment. Analysis of a microsatellite (EPEV-1 [DDBJ/EMBL/GenBank accession number BK001119]) in this region in CEPH families 1331 and 1332 demonstrated one to two alleles per haplotype and linkage to neighboring markers D8S550 and D8S1140 (data not shown). No copy number variation was found with any of the other 8p23.1 probes, with the exceptions of probe E, which detected the known independent copy number polymorphism of DEFA1 (Mars et al. 1995 and data not shown), and probe J, for which a putative copy number polymorphism was detected in 2 of 90 individuals.
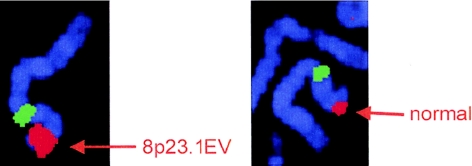
Figure 3.
SQ-FISH of 8p23.1 EV (left) and normal (right) homologs in individual II:1 of family 3, using 8p23.1 BAC 51D11 (red) and D8Z2 centromere probe (green).
The data are consistent with a minimum copy number of one SPAG11/DEFB4 gene per chromosome, for several reasons. First, the absolute strength of the three MAPH probes in individuals with the lowest copy number is very similar to that of the subtelomeric probes (two copies per genome) (Hollox et al. 2002) acting as reference loci in this probe set (e.g., see fig. 2). Second, when this model is used, the number of microsatellite alleles never exceeds the copy number (table 2). Third, the model also fits closely with the observed distribution of signals in the 90 control individuals; most have ∼1.5–2 times the signal found in the individuals with the lowest copy number, suggesting three and four as the most frequent copy numbers (from 35/90 and 38/90 samples, respectively). Under the assumption of a lowest copy number of two, other samples can be normalized to a standard reference sample, to determine copy number and to show that the unrelated control individuals have two to seven copies (table 2). Blind MAPH analysis of the three probes showing normal copy number variation in three EV-carrying families indicated that EV carriers had total copy numbers of 9–12 (figs. 2–4).
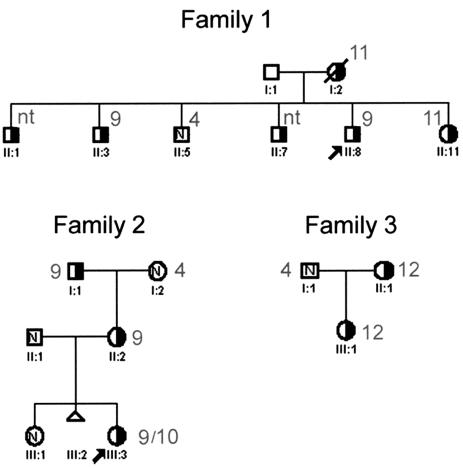
Figure 4.
MAPH analysis of 8p23.1 copy number in three families. Pedigrees of families 1–3 are annotated with MAPH-determined copy number (see also table 2). Half-blackened symbols indicate EV carriers. N = chromosomally normal; nt = not tested.
Analysis of allelic diversity and dosage at the EPEV-1 microsatellite can begin to reveal the evolution of the different-copy-number alleles (table 2). The EV chromosomes in family 2 have an expanded cassette of two microsatellite alleles showing increased dosage (182, 184, and 186 in the ratio 3:3:1, giving a seven-copy chromosome), which implies that the high-copy-number allele was formed recently by triplication of two repeats carrying the 182 and 184 alleles. In contrast, family 1 carries an EV chromosome that has been generated by expansion of one or more repeat units carrying the 184 allele. Because the EV chromosomes in different families carry different microsatellite alleles, we infer that they had independent origins (table 2). In the 16 different CEPH chromosomes analyzed, which each had one or two EPEV-1 alleles, nine different alleles were identified, showing that the underlying paralogy has existed long enough to allow microsatellite divergence between paralogs. Indeed, analysis of chimpanzee (Pan troglodytes) and gorilla (Gorilla gorilla) DNA shows that initial duplication of this β-defensin cluster may have occurred before the chimpanzee-human divergence, 6 million years ago (table 2).
The 120 kb between the EPEV-1 microsatellite and the farthest MAPH probe showing variation (shown by an asterisk in fig. 1 and not included in the main probe set) includes the β-defensins DEFB104 and DEFB103, as well as DEFB4 and SPAG11, which all map to clone SCb-295j18 (DDBJ/EMBL/GenBank accession number AF252830). This clone has been identified as “duplicated” by in silico analysis of whole-genome shotgun sequence (Bailey et al. 2002). Clones that share paralogous regions can be identified in GenBank, and two of these were included in the UCSC November 2002 assembly of the human genome (see the UCSC Genome Bioinformatics Web site). Measuring the distance between two paralogous SPAG11 genes in this assembly gives a repeat size of 306 kb. However, because of the interchromosomal paralogs of the adjacent ORR and the possible presence of sequence derived from inversion alleles, in silico assembly and identification of the repeat-unit boundaries has many potential problems. Pulsed-field gel electrophoresis analysis of SfiI-digested genomic DNA from an individual with seven copies of the repeat shows a single band of ∼240 kb after hybridization with a SPAG11 or DEFB4 probe, indicating that this is a minimum repeat size (data not shown). Therefore, a cluster of β-defensins occurs on this large repeat unit showing copy number variation. SQ-FISH of EV carriers demonstrated increased signal from BAC 51D11 (Trask et al. 1998), which hybridizes to a subset of ORRs that includes those in 8p. This shows that the adjacent ORR is involved in the EV and, probably, in the normal copy number variation. Further SQ-FISH analyses using YACs containing DEFB4 sequence show specific hybridization to 8p23.1 and increased signal on EV chromosomes (table 2).
Signal intensity ratios from SQ-FISH, calculated by comparison of signals on the two 8p23.1 homologs in each of ⩾10 cells, can be combined with total-copy-number measurements by MAPH to estimate the copy number on individual chromosomes. For family 2, SQ-FISH analysis of EV carriers (with a total copy number of nine) shows signal intensity ratios consistent with two copies on the normal chromosome and seven copies on the EV chromosome (table 2). For families 1 and 3, the SQ-FISH results are less straightforward. It is likely that heterogeneity between repeat-unit copies results in reduced hybridization of BAC 51D11 and YAC 820b4 to the EV chromosome in family 3, and reduced hybridization of BAC 51D11 may also be the cause of the discrepancy in SQ-FISH results between the BAC and the YAC in family 1. Paralogous repeats involving ORRs are thought to be a complex patchwork of sequence (Trask et al. 1998), yet there are no significant differences in MAPH results for each individual between probes F–H, except for individual N015 (P<.05, by t test, corrected for multiple observations), which demonstrates that any small-scale heterogeneity that affects MAPH is uncommon and, thus, that repeat units share similar sequence, at least across the β-defensin cluster.
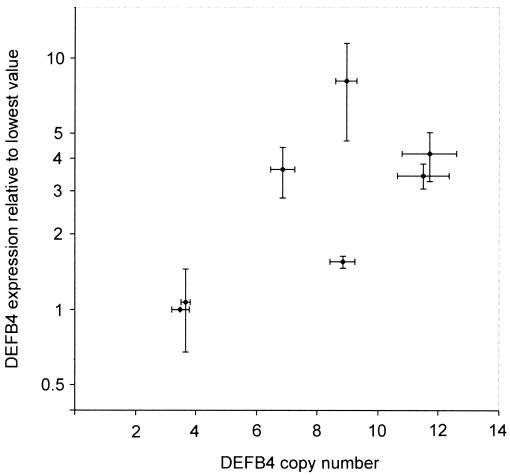
Microarray data show that expression levels of DEFB4 (previously known as “DEFB2”; see the GNF Gene Expression Atlas Web site) in whole blood are high (Su et al. 2002), and, given its chemokine function in addition to its antimicrobial properties, most if not all of this expression is likely to be in cells involved in the immune response (Yang et al. 1999). Therefore, we used lymphoblastoid cell lines, which are derived from peripheral blood lymphocytes, as a model for initial experiments measuring mRNA levels of the DEFB4 cytokine in different individuals. Total RNA was extracted from seven lymphoblastoid cell lines derived from individuals of the different families (table 2). As predicted, of the four genes (SPAG11, DEFB4, DEFB103, and DEFB104) known to map within the repeat unit, only DEFB4 was expressed in lymphoblastoid cells. Analysis of DEFB4 expression by semiquantitative duplex RT-PCR (Harder et al. 1997) shows that DEFB4 mRNA expression is significantly correlated with DEFB4 copy number, and variation in copy number accounts for 50% of the observed variation in expression (fig. 5).
Figure 5.
DEFB4 transcript levels versus β-defensin–cluster DNA copy number for a series of seven lymphoblastoid cell lines. DEFB4 transcript was measured in triplicate by using duplex RT-PCR with TBP as a control transcript. Mean ratio (transformed to log10) ± SEM of DEFB4:TBP intensity ratios is shown on the Y-axis, and MAPH copy number is shown (mean of five tests ± SEM) on the X-axis. There is a significant correlation between DEFB4 transcript levels and β-defensin–cluster copy number (r2=0.5; P<.05). A similar pattern of variation in DEFB4 expression level was also obtained, using G3PD as a control transcript (data not shown).
Discussion
Our results have shown that the molecular basis for the 8p23.1 EV is an extreme of normal copy number variation involving a cluster of β-defensins. These results provide a molecular basis for distinguishing the 8p23.1 EVs that are consistent with a normal phenotype (Barber et al. 1998) from other duplications of distal 8p, which may have phenotypic consequences (Pehlivan et al. 1999; Engelen et al. 2000; Fan et al. 2001; Kennedy et al. 2001; Harada et al. 2002; Tsai et al. 2002). Aberrant recombination between the ORRs in 8p23.1 is believed to underlie other clinically important chromosome rearrangements of 8p (Giglio et al. 2001, 2002), and microsatellite analysis of CEPH families will help to determine whether copy number predisposes to or is itself the product of aberrant recombination.
We have shown that MAPH, like other methods for directly assaying DNA copy number, is suitable for the analysis and genotyping of copy number polymorphisms, an often overlooked source of variation in the human genome (Siniscalco et al. 2000). In light of the apparently recent burst of segmental duplications (Bailey et al. 2001, 2002; Liu et al. 2003; Locke et al. 2003), these are likely to be frequent in the human genome, and recent reports support this hypothesis (Pramanik and Li 2002; Robledo et al. 2002; Yu et al. 2002).
The copy number variation at 8p23.1 involves a repeat unit of unprecedented size containing several genes and must be appreciated when interpreting SNP data from this region. An attempt at characterization of SNP variation has already been made (Jurevic et al. 2002), and distinguishing allelic polymorphisms from differences between paralogs is impossible without knowledge of the copy number. The combination of paralog differences and polymorphisms can produce an extremely diverse repertoire of gene variants, and careful characterization of this variation will provide the data for important clinical association studies. DEFB4 has been suggested as a modifier locus for cystic fibrosis (CF [MIM 219700]) (Bals et al. 1998; Singh et al. 1998) because of its efficacy against P. aeruginosa, which is a major cause of morbidity in patients with CF. β-Defensins have also been implicated in the etiology of inflammatory bowel disease and skin diseases such as eczema and psoriasis (Schutte and McCray 2002).
Defensins have evolved by duplication followed by adaptive evolution (Hughes 1999; Morrison et al. 2003). There is also evidence that certain defensin genes maintain general antimicrobial activity yet acquire high efficacy against certain species or take on other more diverse functions (e.g., signaling or sperm maturation). A model of gene evolution has been proposed in which one gene acquires multiple functions and then duplicates, with each paralog undergoing rapid adaptive evolution to specialize in its own functional niche (Hughes 1994). Discovery of polymorphic copy number variation of β-defensins suggests an ongoing “birth-and-death” process of gene-family evolution (Gu and Nei 1999), possibly driven by epidemic infections in human evolutionary history.
Acknowledgments
We thank all the family members involved in this project; Amanda Collins, Sheila Lane, and Jackie Langdon, for DNA samples; Hiroaki Shizuya and Barbara Trask, for BAC 51D11; Paul Strike, for statistical advice; and Ingrid Davies, for technical assistance. This work was supported by Wellcome Trust grant 060578 (to J.A.L.A.).
Footnotes
Nucleotide sequence data reported herein are available in the DDBJ/EMBL/GenBank databases; for details, see the Electronic-Database Information section of this article.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Fondation Jean Dausset–CEPH, http://www.cephb.fr/ (for marker database)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for EPEV-1 [accession number BK001119], SCb-295j18 [accession number AF252830], and MAPH probes A [accession number NM_001147], B [accession number AF287957], C [accession number Z45294], D [accession number AF233439], E [accession number AF238378], F [accession number AC252830], G [accession number NM_04942], H [accession number G13705], I [accession number AA687243], J [accession number AA226797], K [accession number AA010611], L [accession number Z24258], M [accession number L34357], and N [accession number AQ318792])
- GNF Gene Expression Atlas, http://expression.gnf.org/cgi-bin/index.cgi
- Multiplex Amplifiable Probe Hybridization, http://www.nottingham.ac.uk/~pdzjala/maph/maph.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for DEFB4, DEFB103, SPAG11, and CF)
- UCSC Genome Bioinformatics, http://genome.cse.ucsc.edu/
References
- Armour JA, Sismani C, Patsalis PC, Cross G (2000) Measurement of locus copy number by hybridisation with amplifiable probes. Nucleic Acids Res 28:605–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (eds) (1997) Short protocols in molecular biology. Wiley, New York [Google Scholar]
- Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE (2002) Recent segmental duplications in the human genome. Science 297:1003–1007 [DOI] [PubMed] [Google Scholar]
- Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE (2001) Segmental duplications: organization and impact within the current Human Genome Project assembly. Genome Res 11:1005–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bals R, Wang X, Wu Z, Freeman T, Bafna V, Zasloff M, Wilson JM (1998) Human β-defensin 2 is a salt-sensitive peptide antibiotic expressed in human lung. J Clin Invest 102:874–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber JC, Joyce CA, Collinson MN, Nicholson JC, Willatt LR, Dyson HM, Bateman MS, Green AJ, Yates JR, Dennis NR (1998) Duplication of 8p23.1: a cytogenetic anomaly with no established clinical significance. J Med Genet 35:491–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber JC, Reed CJ, Dahoun SP, Joyce CA (1999) Amplification of a pseudogene cassette underlies euchromatic variation of 16p at the cytogenetic level. Hum Genet 104:211–218 [DOI] [PubMed] [Google Scholar]
- Biragyn A, Ruffini PA, Leifer CA, Klyushnenkova E, Shakhov A, Chertov O, Shirakawa AK, Farber JM, Segal DM, Oppenheim JJ, Kwak LW (2002) Toll-like receptor 4–dependent activation of dendritic cells by β-defensin 2. Science 298:1025–1029 [DOI] [PubMed] [Google Scholar]
- Cole AM, Hong T, Boo LM, Nguyen T, Zhao C, Bristol G, Zack JA, Waring AJ, Yang OO, Lehrer RI (2002) Retrocyclin: a primate peptide that protects cells from infection by T- and M-tropic strains of HIV-1. Proc Natl Acad Sci USA 99:1813–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelen JJ, Moog U, Evers JL, Dassen H, Albrechts JC, Hamers AJ (2000) Duplication of chromosome region 8p23.1→p23.3: a benign variant? Am J Med Genet 91:18–21 [PubMed] [Google Scholar]
- Fan YS, Siu VM, Jung JH, Farrell SA, Côté GB (2001) Direct duplication of 8p21.3→p23.1: a cytogenetic anomaly associated with developmental delay without consistent clinical features. Am J Med Genet 103:231–234 [DOI] [PubMed] [Google Scholar]
- Ganz T (1999) Defensins and host defense. Science 286:420–421 [DOI] [PubMed] [Google Scholar]
- Ghosh D, Porter E, Shen B, Lee SK, Wilk D, Drazba J, Yadav SP, Crabb JW, Ganz T, Bevins CL (2002) Paneth cell trypsin is the processing enzyme for human defensin-5. Nat Immunol 3:583–590 [DOI] [PubMed] [Google Scholar]
- Giglio S, Broman KW, Matsumoto N, Calvari V, Gimelli G, Neumann T, Ohashi H, Voullaire L, Larizza D, Giorda R, Weber JL, Ledbetter DH, Zuffardi O (2001) Olfactory receptor–gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements. Am J Hum Genet 68:874–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglio S, Calvari V, Gregato G, Gimelli G, Camanini S, Giorda R, Ragusa A, Guerneri S, Selicorni A, Stumm M, Tonnies H, Ventura M, Zollino M, Neri G, Barber J, Wieczorek D, Rocchi M, Zuffardi O (2002) Heterozygous submicroscopic inversions involving olfactory receptor–gene clusters mediate the recurrent t(4;8)(p16;p23) translocation. Am J Hum Genet 71:276–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Nei M (1999) Locus specificity of polymorphic alleles and evolution by a birth-and-death process in mammalian MHC genes. Mol Biol Evol 16:147–156 [DOI] [PubMed] [Google Scholar]
- Harada N, Takano J, Kondoh T, Ohashi H, Hasegawa T, Sugawara H, Ida T, Yoshiura K, Ohta T, Kishino T, Kajii T, Niikawa N, Matsumoto N (2002) Duplication of 8p23.2: a benign cytogenetic variant? Am J Med Genet 111:285–288 [DOI] [PubMed] [Google Scholar]
- Harder J, Bartels J, Christophers E, Schröder JM (1997) A peptide antibiotic from human skin. Nature 387:861 [DOI] [PubMed] [Google Scholar]
- ——— (2001) Isolation and characterization of human β-defensin-3, a novel human inducible peptide antibiotic. J Biol Chem 276:5707–5713 [DOI] [PubMed] [Google Scholar]
- Hollox EJ, Atia T, Cross G, Parkin T, Armour JAL (2002) High-throughput screening of human subtelomeric DNA for copy number changes using multiplex amplifiable probe hybridisation (MAPH). J Med Genet 39:790–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL (1994) The evolution of functionally novel proteins after gene duplication. Proc R Soc Lond B Biol Sci 256:119–124 [DOI] [PubMed] [Google Scholar]
- ——— (1999) Evolutionary diversification of the mammalian defensins. Cell Mol Life Sci 56:94–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurevic RJ, Chrisman P, Mancl L, Livingston R, Dale BA (2002) Single-nucleotide polymorphisms and haplotype analysis in beta-defensin genes in different ethnic populations. Genet Test 6:261–269 [DOI] [PubMed] [Google Scholar]
- Kennedy SJ, Teebi AS, Adatia I, Teshima I (2001) Inherited duplication, dup (8) (p23.1p23.1) pat, in a father and daughter with congenital heart defects. Am J Med Genet 104:79–80 [DOI] [PubMed] [Google Scholar]
- Liu G, Zhao S, Bailey JA, Sahinalp SC, Alkan C, Tuzun E, Green ED, Eichler EE (2003) Analysis of primate genomic variation reveals a repeat-driven expansion of the human genome. Genome Res 13:358–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Wang L, Jia HP, Zhao C, Heng HH, Schutte BC, McCray PB Jr, Ganz T (1998) Structure and mapping of the human β-defensin HBD-2 gene and its expression at sites of inflammation. Gene 222:237–244 [DOI] [PubMed] [Google Scholar]
- Locke DP, Segraves R, Carbone L, Archidiacono N, Albertson DG, Pinkel D, Eichler EE (2003) Large-scale variation among human and great ape genomes determined by array comparative genomic hybridization. Genome Res 13:347–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mars WM, Patmasiriwat P, Maity T, Huff V, Weil MM, Saunders GF (1995) Inheritance of unequal numbers of the genes encoding the human neutrophil defensins HP-1 and HP-3. J Biol Chem 270:30371–30376 [DOI] [PubMed] [Google Scholar]
- Morrison GM, Semple CA, Kilanowski FM, Hill RE, Dorin JR (2003) Signal sequence conservation and mature peptide divergence within subgroups of the murine β-defensin gene family. Mol Biol Evol 20:460–470 [DOI] [PubMed] [Google Scholar]
- Murphy LD, Herzog CE, Rudick JB, Fojo AT, Bates SE (1990) Use of the polymerase chain reaction in the quantitation of mdr-1 gene expression. Biochemistry 29:10351–10356 [DOI] [PubMed] [Google Scholar]
- O’Malley DP, Storto PD (1999) Confirmation of the chromosome 8p23.1 euchromatic duplication as a variant with no clinical manifestations. Prenat Diagn 19:183–184 [PubMed] [Google Scholar]
- Pehlivan T, Pober BR, Brueckner M, Garrett S, Slaugh R, Van Rheeden R, Wilson DB, Watson MS, Hing AV (1999) GATA4 haploinsufficiency in patients with interstitial deletion of chromosome region 8p23.1 and congenital heart disease. Am J Med Genet 83:201–206 [PubMed] [Google Scholar]
- Pramanik S, Li H (2002) Direct detection of insertion/deletion polymorphisms in an autosomal region by analyzing high-density markers in individual spermatozoa. Am J Hum Genet 71:1342–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robledo R, Orru S, Sidoti A, Muresu R, Esposito D, Grimaldi MC, Carcassi C, Rinaldi A, Bernini L, Contu L, Romani M, Roe B, Siniscalco M (2002) A 9.1-kb gap in the genome reference map is shown to be a stable deletion/insertion polymorphism of ancestral origin. Genomics 80:585–592 [DOI] [PubMed] [Google Scholar]
- Salzman NH, Ghosh D, Huttner KM, Paterson Y, Bevins CL (2003) Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature 422:522–526 [DOI] [PubMed] [Google Scholar]
- Schutte BC, McCray PB Jr (2002) β-Defensins in lung host defense. Annu Rev Physiol 64:709–748 [DOI] [PubMed] [Google Scholar]
- Schutte BC, Mitros JP, Bartlett JA, Walters JD, Jia HP, Welsh MJ, Casavant TL, McCray PB Jr (2002) Discovery of five conserved β-defensin gene clusters using a computational search strategy. Proc Natl Acad Sci USA 99:2129–2133 (erratum 99:14611) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh PK, Jia HP, Wiles K, Hesselberth J, Liu L, Conway BA, Greenberg EP, Valore EV, Welsh MJ, Ganz T, Tack BF, McCray PB Jr (1998) Production of β-defensins by human airway epithelia. Proc Natl Acad Sci USA 95:14961–14966 (erratum 96:2569 [1999]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniscalco M, Robledo R, Orru S, Contu L, Yadav P, Ren Q, Lai H, Roe B (2000) A plea to search for deletion polymorphism through genome scans in populations. Trends Genet 16:435–437 [DOI] [PubMed] [Google Scholar]
- Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, Patapoutian A, Hampton GM, Schultz PG, Hogenesch JB (2002) Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci USA 99:4465–4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trask BJ, Friedman C, Martin-Gallardo A, Rowen L, Akinbami C, Blankenship J, Collins C, Giorgi D, Iadonato S, Johnson F, Kuo WL, Massa H, Morrish T, Naylor S, Nguyen OT, Rouquier S, Smith T, Wong DJ, Youngblom J, van den Engh G (1998) Members of the olfactory receptor gene family are contained in large blocks of DNA duplicated polymorphically near the ends of human chromosomes. Hum Mol Genet 7:13–26 [DOI] [PubMed] [Google Scholar]
- Tsai CH, Graw SL, McGavran L (2002) 8p23 Duplication reconsidered: is it a true euchromatic variant with no clinical manifestation? J Med Genet 39:769–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Chertov O, Bykovskaia SN, Chen Q, Buffo MJ, Shogan J, Anderson M, Schröder JM, Wang JM, Howard OMZ, Oppenheim JJ (1999) β-Defensins: linking innate and adaptive immunity through dendritic and T cell CCR6. Science 286:525–528 [DOI] [PubMed] [Google Scholar]
- Yu CE, Dawson G, Munson J, D’Souza I, Osterling J, Estes A, Leutenegger AL, Flodman P, Smith M, Raskind WH, Spence MA, McMahon W, Wijsman EM, Schellenberg GD (2002) Presence of large deletions in kindreds with autism. Am J Hum Genet 71:100–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yu W, He T, Yu J, Caffrey RE, Dalmasso EA, Fu S, Pham T, Mei J, Ho JJ, Zhang W, Lopez P, Ho DD (2002) Contribution of human α-defensin 1, 2, and 3 to the anti–HIV-1 activity of CD8 antiviral factor. Science 298:995–1000 [DOI] [PubMed] [Google Scholar]