Abstract
There is increasing evidence for strong genetic influences on athletic performance and for an evolutionary “trade-off” between performance traits for speed and endurance activities. We have recently demonstrated that the skeletal-muscle actin-binding protein α-actinin-3 is absent in 18% of healthy white individuals because of homozygosity for a common stop-codon polymorphism in the ACTN3 gene, R577X. α-Actinin-3 is specifically expressed in fast-twitch myofibers responsible for generating force at high velocity. The absence of a disease phenotype secondary to α-actinin-3 deficiency is likely due to compensation by the homologous protein, α-actinin-2. However, the high degree of evolutionary conservation of ACTN3 suggests function(s) independent of ACTN2. Here, we demonstrate highly significant associations between ACTN3 genotype and athletic performance. Both male and female elite sprint athletes have significantly higher frequencies of the 577R allele than do controls. This suggests that the presence of α-actinin-3 has a beneficial effect on the function of skeletal muscle in generating forceful contractions at high velocity, and provides an evolutionary advantage because of increased sprint performance. There is also a genotype effect in female sprint and endurance athletes, with higher than expected numbers of 577RX heterozygotes among sprint athletes and lower than expected numbers among endurance athletes. The lack of a similar effect in males suggests that the ACTN3 genotype affects athletic performance differently in males and females. The differential effects in sprint and endurance athletes suggests that the R577X polymorphism may have been maintained in the human population by balancing natural selection.
The α-actinins are a family of actin-binding proteins related to dystrophin. In humans, there are two genes encoding skeletal-muscle α-actinins: ACTN2 (MIM 102573), which is expressed in all fibers, and ACTN3 (MIM 102574), which is restricted to fast (type 2) fibers. The sarcomeric α-actinins are major components of the Z line, where they crosslink actin thin filaments; they likely perform a static function in maintaining the ordered myofibrillar array, as well as a regulatory function in coordinating myofiber contraction (Blanchard et al. 1989; Mills et al. 2001). We have recently demonstrated that α-actinin-3 deficiency is common in the general population and is due to homozygosity for a premature stop codon in ACTN3 (R577X) (North et al. 1999). It is likely that α-actinin-2 is able to “compensate” for the absence of α-actinin-3 in type 2 fibers, although there is no upregulation of α-actinin-2 levels in response to α-actinin-3 deficiency (authors' unpublished observations). However, there is strong evidence to suggest that ACTN3 has been maintained in the genome because of function(s) independent of ACTN2: ACTN3 sequence has remained highly conserved, in evolutionary terms, since its divergence from ACTN2 >300 million years ago; α-actinin-2 and α-actinin-3 are differentially expressed, spatially and temporally, during embryonic development; and ACTN2 expression does not completely overlap ACTN3 in mouse postnatal skeletal muscle (Mills et al. 2001). In addition, the frequency of the α-actinin-3–deficient genotype (577XX) varies from 25% in Asian populations to <1% in an African Bantu population; the frequency in Europeans is ∼18%. This raises the possibility that ACTN3 genotype confers differential fitness in humans, under certain environmental conditions. The force-generating capacity of type 2 muscle fibers at high velocity, the speed and tempo of movements, and the capacity of the individual to adapt to exercise training are all strongly genetically influenced (Rankinen et al. 2002). Thus, we hypothesized that ACTN3 genotype may be one of the factors that influence normal variation in muscle function. Since any effect on muscle function will be most readily observable at the extremes of human performance, we collaborated with the Australian Institute of Sport to study ACTN3 genotype frequencies in elite athletes.
We genotyped 436 unrelated white controls from three different sources (150 blood donors, 71 healthy children participating in an unrelated study, and 215 healthy adults participating in a talent-identification program with the Australian Institute of Sport), through use of the genotyping methodology described by Mills et al. (2001). Sex was known for 292 female controls and 134 male controls. We also genotyped 429 elite white athletes from 14 different sports. Athletes were defined as “elite” if they had represented Australia in their sport at the international level; 50 of the athletes had competed in Olympic Games. This study was approved by the institutional review boards of the Children’s Hospital at Westmead, the University of Sydney, and the Australian Institute of Sport.
Given the localization of α-actinin-3 in fast skeletal-muscle fibers, we hypothesized that deficiency of α-actinin-3 would reduce performance in sprint/power events and would therefore be less frequent in elite sprint athletes. To test this hypothesis, we analyzed genotypes of a subset of 107 elite athletes (72 male and 35 female) in our cohort, classified a priori as specialist sprint/power athletes by one of the authors (J.P.G.) at the Australian Institute of Sport, blinded to genotyping results. This group comprised 46 track athletes competing in events of ⩽800 m, 42 swimmers competing in events ⩽200 m, 9 judo athletes, 7 short-distance track cyclists, and 3 speed skaters. For comparison, we analyzed a subset of 194 subjects (122 male and 72 female) classified independently as specialist endurance athletes, including 77 long-distance cyclists, 77 rowers, 18 swimmers competing over distances of ⩾400 m, 15 track athletes competing in events of ⩾5,000 m, and 7 cross-country skiers. Thirty-two sprint athletes (25 male and 7 female) and 18 endurance athletes (12 male and 6 female) had competed at the Olympic level. Because of the stringency of the classification criteria, 128 of our elite athletes could not be unambiguously assigned into either the sprint/power or endurance groups and were excluded from subsequent analyses.
To test for homogeneity of ACTN3 allele and genotype frequencies between athlete and control groups, we used the log-linear modeling approach described by Huttley and Wilson (2000), implemented in the statistical programming language R (version 1.6.2), through use of the package hwde (contributed by J. Maindonald; available from The R Project for Statistical Computing Web site). χ2 values were estimated using genotype numbers for comparisons between athletes and controls.
The genotypic profiles of the three control groups (150 blood donors, 71 healthy children, and 215 healthy adults) did not differ significantly from one another (χ2=0.19; P=.996) nor from a previously genotyped group of 107 white Europeans (Mills et al. 2001), suggesting that the genotype frequencies in our control cohort are representative of the broader white population. ACTN3 genotype frequencies did not vary significantly between male and female control subjects, and, overall, there was no significant deviation from Hardy-Weinberg (H-W) equilibrium.
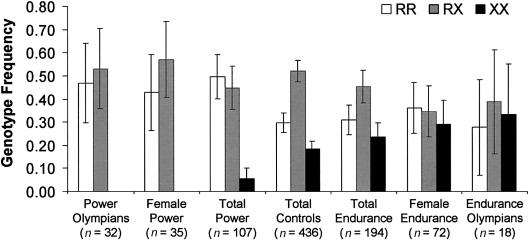
ACTN3 genotyping data from the control, sprint/power, and endurance groups are summarized in table 1 and figure 1. There were no significant allele or genotype frequency differences between the elite athlete group as a whole and the controls. However, when the athletes were divided into sprint/power and endurance groups and compared with controls, there was strong evidence of allele frequency variation (χ2[df=5]=23; P<.001). There were significant allele frequency differences between sprint athletes and controls for both males (χ2[df=1]=14.8; P<.001) and females (χ2[df=1]=7.2; P<.01). Sprint athletes had a lower frequency of the XX (α-actinin-3 null) genotype (6% vs. 18%), and no female elite sprint athletes or sprint Olympians were XX. The sprint athlete group also had a higher frequency of the RR genotype (50% vs. 30%) and a lower frequency of the heterozygous RX genotype (45% vs. 52%), compared with controls. Elite endurance athletes had a slightly higher frequency of the XX genotype (24%) than did controls (18%). Importantly, allele frequencies in sprint and endurance athletes deviated in opposite directions and differed significantly from each other in both males (χ2[df=1]=13.3; P<.001) and females (χ2[df=1]=5.8; P<.05). The differences between the two groups effectively “canceled each other out,” explaining the lack of association when the entire elite athlete cohort was compared with the control group.
Table 1.
Number and Frequency (%) of ACTN3 Genotypes and Frequency (%) of ACTN3 Alleles in Controls and Elite Sprint/Power and Endurance Athletes
|
No. (%) with Genotype |
AlleleFrequency(%) |
||||
| Group (n) | RR | RX | XX | R | X |
| Male: | |||||
| Controls (134) | 40 (30) | 73 (54) | 21 (16) | 57 | 43 |
| Sprint (72) | 38 (53) | 28 (39) | 6 (8) | 72 | 28 |
| Endurance (122) | 34 (28) | 63 (52) | 25 (20) | 54 | 46 |
| Female: | |||||
| Controls (292) | 88 (30) | 147 (50) | 57 (20) | 55 | 45 |
| Sprint (35) | 15 (43) | 20 (57) | 0 (0) | 71 | 29 |
| Endurance (72) | 26 (36) | 25 (35) | 21 (29) | 53 | 47 |
| Total: | |||||
| Controls (436) | 130 (30) | 226 (52) | 80 (18) | 56 | 44 |
| Sprint (107) | 53 (50) | 48 (45) | 6 (6) | 72 | 28 |
| Endurance (194) | 60 (31) | 88 (45) | 46 (24) | 54 | 46 |
Figure 1.
ACTN3 genotype frequency in controls, elite sprint/power athletes, and endurance athletes. Compared with healthy white controls, there is a marked reduction in the frequency of the ACTN3 577XX genotype (associated with α-actinin-3 deficiency) in elite white sprint athletes; remarkably, none of the female sprint athletes or sprint athletes who had competed at the Olympic level (25 males and 7 females) were α-actinin-3 deficient. Conversely, there is a trend toward an increase in the 577XX genotype in endurance athletes, although this association reaches statistical significance only in females. Error bars indicate 95% CIs.
Overall, there was also evidence of genotype variation that is not explained by allele frequency differences (χ2[df=5]=16.7; P<.01). This suggested variation in H-W disequilibrium coefficients among groups, despite there being no evidence for departure from H-W equilibrium overall. The effect was restricted to female sprint (χ2[df=1]=7.4; P<.01) and endurance (χ2[df=1]=6.0; P<.05) athletes, with more heterozygous female sprint athletes than expected at H-W equilibrium (20 vs. 15) and fewer than expected heterozygous female endurance athletes (25 vs. 36). The allele-frequency–independent genotype differences between female sprint and endurance athletes were highly significant (χ2[df=1]=13.8; P<.001). No effect was seen in males, suggesting that the effect of ACTN3 genotype on performance differs between males and females.
Our findings suggest that the ACTN3 577R allele provides an advantage for power and sprint activities. No female elite sprint athletes in our sample were α-actinin-3 deficient (compared with 8% of males). In males, the androgen hormone response to training is likely to make a significant contribution to improvements in performance, so that the relative effect of α-actinin-3 on muscle power may be reduced. Interestingly, all male Olympian power athletes in our cohort had at least one copy of the functional R allele of ACTN3 (associated with the presence of α-actinin-3 in skeletal muscle), suggesting that “every variable counts” at the highest levels of sporting competition. Although at least 73 genetic loci have been associated with fitness and performance phenotypes (Rankinen et al. 2002), ACTN3 is the first structural skeletal-muscle gene for which such an association has been demonstrated.
The functional basis for this advantage is likely related to the fact that α-actinin-3 is the predominant fast fiber isoform in both mouse and human (Mills et al. 2001) and may confer a greater capacity for the absorption or transmission of force at the Z line during rapid contraction. Approximately 45% of the variation in fiber type proportions is accounted for by genetic factors (Simoneau and Bouchard 1995). Sarcomeric α-actinins bind to the gluconeogenic enzyme fructose-1,6-bisphosphatase (Gizak et al. 2003), to the glycogen phosphorylase amorphin (Chowrashi et al. 2002), and to the calsarcins (Frey et al. 2000; Frey and Olson 2002), which interact with calcineurin, a signaling factor that plays a role in the specification of muscle fiber type (Serrano et al. 2001). Thus, α-actinin-3 may promote the formation of fast-twitch fibers or alter glucose metabolism in response to training. In addition, α-actinin-3 may be evolutionarily optimized for the minimization of damage caused by eccentric muscle contraction. The Z line in fast, glycolytic fibers is the structure most vulnerable to exercise-induced injury resulting in morphological damage and degradation of associated proteins, including the α-actinins (Friden and Lieber 2001). We are currently exploring the mechanism by which the presence or absence of α-actinin-3 alters muscle function—and, hence, athletic performance—through the generation and analysis of an Actn3 knockout mouse model.
From an evolutionary point of view, the challenge is to explain the high frequency of the 577XX ACTN3 genotype, given the apparent power-performance advantage of the 577RR genotype. One possibility is that the power-performance effect of the 577RR genotype is only manifest in the extreme circumstances of athletic competition, outside the range of normal human activity, and is consequently of minimal evolutionary significance. In this model, the 577X allele could have been selectively neutral during human evolution and become established in the human population by random genetic drift. However, this explanation is difficult to reconcile with the high level of evolutionary conservation that we have previously demonstrated for ACTN3 (North et al. 1999; Mills et al. 2001).
It is also possible that the X allele is selectively neutral but has reached its current frequency because of positive selection on a beneficial polymorphism at a nearby locus (i.e., “genetic hitchhiking”) (Kaplan et al. 1989). This hypothetical variant would need to be in strong linkage disequilibrium with 577X to result in the strong association observed in our study; however, it would be unlikely to reside within the ACTN3 gene itself, since the 577X polymorphism results in deficiency of the α-actinin-3 protein. The distance over which “useful” linkage disequilibrium extends varies considerably between loci (Reich et al. 2001); however, a distance of 10 kb has been proposed as a rough average value on the basis of population modeling (Ardlie et al. 2002). The only identified gene other than ACTN3 in the 20-kb region centered on the R577X polymorphism is the CTSF gene (MIM 603539), which encodes the papain-like cysteine protease cathepsin F (Wang et al. 1998). The dbSNP Home Page identifies nine polymorphic sites within the CTSF gene, none of which alter the amino acid sequence of the encoded protein. Furthermore, the only characterized function of cathepsin F is related to antigen processing in macrophages (Shi et al. 2000), making it an unconvincing candidate for influencing athletic performance. Although we cannot completely rule out that variations in CTSF or in other, more distant genes have influenced our results, it is more likely that the R577X polymorphism is directly responsible for the observed association with elite athletic performance.
The evolutionary model most consistent with our results is one in which 577XX genotype has been acted on by positive natural selection. Our data demonstrate a trend toward a higher frequency of the XX genotype in endurance athletes, although the association reached statistical significance only in females; the frequency of the 577RX genotype was also lower in female elite endurance athletes than in female controls (35% vs. 50%). However, the effect may be stronger than is indicated by these results, since a specific allelic association may be difficult to detect in a heterogeneous cohort of mixed athletic disciplines. If the 577XX genotype enhances endurance performance as the 577R allele appears to enhance sprint ability, then the 577R and 577X alleles may be maintained in the population because they both confer selective advantages under different environmental conditions and are thus kept at high population frequencies by balancing selection. We are currently studying the frequency of alleles and the pattern of genetic polymorphisms flanking the R577X locus in different human populations, to determine the origin of the X allele and to identify the form and magnitude of any selective pressures that have acted on the R577X locus.
Distinct beneficial effects on sprint and endurance athletic performance by different genotypes at a single locus have also been observed in studies of the gene encoding angiotensin-converting enzyme (ACE). The ACE gene has two alleles, termed “I” and “D”; the I allele is associated with lower ACE activity in both serum and tissue (Rieder et al. 1999). An increased frequency of the ACE I allele has been observed in elite endurance athletes (Gayagay et al. 1998; Montgomery et al. 1998; Myerson et al. 1999; Nazarov et al. 2001). Conversely, an increased frequency of the ACE D allele has been associated with elite sprint performance (Myerson et al. 1999; Nazarov et al. 2001; Woods et al. 2001). The absence of a correlation between the ACE I/D polymorphism and performance in other studies of elite athletes (Taylor et al. 1999; Rankinen et al. 2000) may be explained by the failure of these studies to adequately restrict their subjects to well-defined categories according to area of specialist performance, so that the allelic association is “canceled out” (Nazarov et al. 2001).
It is likely that there is a “trade-off” between sprint and endurance traits that imposes important constraints on the evolution of physical performance in humans and other vertebrates (Garland et al. 1990). This hypothesis is supported by recent data from world-class decathletes, which demonstrated that performance in the 100-m sprint, shot put, long jump, and 110-m hurdles (which rely on explosive power and fast fatigue-susceptible muscle fibers) is negatively correlated with performance in the 1,500-m race (which requires endurance and fatigue-resistant slow fiber activity) (Van Damme et al. 2002). This suggests that an individual is inherently predisposed toward specialist performance in one area (sprint/power vs. endurance). In humans, this appears to have been achieved, in part, through the maintenance of genetic variation by balancing natural selection. The result is that there are genetic differences among individuals, such as we have demonstrated for the ACTN3 locus, that may be useful predictors of athletic performance at the elite level.
Acknowledgments
We are grateful to J. Maindonald, for assistance in implementing the hwde package in R, and to Professor Ron Trent and Dr. Bing Yu, for provision of athlete DNA samples.
Electronic-Database Information
The URLs for data presented herein are as follows:
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/index.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for ACTN2, ACTN3, and CTSF)
- R Project for Statistical Computing, The, http://www.r-project.org/ (for the hwde package in R)
References
- Ardlie KG, Kruglyak L, Seielstad M (2002) Patterns of linkage disequilibrium in the human genome. Nat Rev Genet 3:299–309 [DOI] [PubMed] [Google Scholar]
- Blanchard A, Ohanian V, Critchley D (1989) The structure and function of α-actinin. J Muscle Res Cell Motil 10:280–289 [DOI] [PubMed] [Google Scholar]
- Chowrashi P, Mittal B, Sanger JM, Sanger JW (2002) Amorphin is phosphorylase; phosphorylase is an alpha-actinin-binding protein. Cell Motil Cytoskeleton 53:125–135 [DOI] [PubMed] [Google Scholar]
- Frey N, Olson EN (2002) Calsarcin-3, a novel skeletal muscle-specific member of the calsarcin family, interacts with multiple Z-disc proteins. J Biol Chem 277:13998–14004 [DOI] [PubMed] [Google Scholar]
- Frey N, Richardson JA, Olson EN (2000) Calsarcins, a novel family of sarcomeric calcineurin-binding proteins. Proc Natl Acad Sci USA 97:14632–14637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friden J, Lieber RL (2001) Eccentric exercise-induced injuries to contractile and cytoskeletal muscle fiber components. Acta Physiol Scand 171:321–326 [DOI] [PubMed] [Google Scholar]
- Garland T Jr, Bennett AF, Daniels CB (1990) Heritability of locomotor performance and its correlates in a natural population. Experientia 46:530–533 [Google Scholar]
- Gayagay G, Yu B, Hambly B, Boston T, Hahn A, Celermajer DS, Trent RJ (1998) Elite endurance athletes and the ACE I allele—the role of genes in athletic performance. Hum Genet 103:48–50 [DOI] [PubMed] [Google Scholar]
- Gizak A, Rakus D, Dzugaj A (2003) Immunohistochemical localization of human fructose-1,6-bisphosphatase in subcellular structures of myocytes. Histol Histopathol 18:135–142 [DOI] [PubMed] [Google Scholar]
- Huttley GA, Wilson SR (2000) Testing for concordant equilibria between population samples. Genetics 156:2127–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan NL, Hudson RR, Langley CH (1989) The “hitchhiking effect” revisited. Genetics 123:887–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills M, Yang N, Weinberger R, vander Woude DL, Beggs AH, Easteal S, North K (2001) Differential expression of the actin-binding proteins, α-actinin-2 and -3, in different species: implications for the evolution of functional redundancy. Hum Mol Genet 10:1335–1346 [DOI] [PubMed] [Google Scholar]
- Montgomery H, Marshall R, Hemingway H, Myerson S, Clarkson P, Dollery C, Hayward M, Holliman D, Jubb M, World M, Thomas E, Brynes A, Saeed N, Barnard M, Bell J, Prasad K, Rayson M, Talmud P, Humphries S (1998) Human gene for physical performance. Nature 393:221–222 [DOI] [PubMed] [Google Scholar]
- Myerson S, Hemingway H, Budget R, Martin J, Humphries S, Montgomery H (1999) Human angiotensin I-converting enzyme gene and endurance performance. J Appl Physiol 87:1313–1316 [DOI] [PubMed] [Google Scholar]
- Nazarov IB, Woods DR, Montgomery HE, Shneider OV, Kazakov VI, Tomilin NV, Rogozkin VA (2001) The angiotensin converting enzyme I/D polymorphism in Russian athletes. Eur J Hum Genet 9:797–801 [DOI] [PubMed] [Google Scholar]
- North KN, Yang N, Wattanasirichaigoon D, Mills M, Easteal S, Beggs AH (1999) A common nonsense mutation results in α-actinin-3 deficiency in the general population. Nat Genet 21:353–354 [DOI] [PubMed] [Google Scholar]
- Rankinen T, Perusse L, Raurama R, Rivera MA, Wolfarth B, Bouchard C (2002) The human gene map for performance and health-related fitness phenotypes: the 2001 update. Med Sci Sports Exerc 34:1219–1233 [DOI] [PubMed] [Google Scholar]
- Rankinen T, Wolfarth B, Simoneau JA, Maier-Lenz D, Rauramaa R, Rivera MA, Boulay MR, Chagnon YC, Perusse L, Keul J, Bouchard C (2000) No association between the angiotensin-converting enzyme ID polymorphism and elite endurance athlete status. J Appl Physiol 88:1571–1575 [DOI] [PubMed] [Google Scholar]
- Reich DE, Cargill M, Bolk S, Ireland J, Sabeti PC, Richter DJ, Layery T, Kouyoumjian R, Farhadian SF, Ward R, Lander ES (2001) Linkage disequilibrium in the human genome. Nature 411:199–204 [DOI] [PubMed] [Google Scholar]
- Rieder MJ, Taylor SL, Clark AG, Nickerson DA (1999) Sequence variation in the human angiotensin converting enzyme. Nat Genet 22:59–62 [DOI] [PubMed] [Google Scholar]
- Serrano AL, Murgia M, Pallafacchina G, Calabria E, Coniglio P, Lomo T, Schiaffino S (2001) Calcineurin controls nerve activity-dependent specification of slow skeletal muscle fibers but not muscle growth. Proc Natl Acad Sci USA 98:13108–13113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi GP, Bryant RAR, Riese R, Verhelst S, Driessen C, Li Z, Bromme D, Ploegh HL, Chapman HA (2000) Role for cathepsin F in invariant chain processing and major histocompatibility complex class II peptide loading by macrophages. J Exp Med 191:1177–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoneau JA, Bouchard C (1995) Genetic determinism of fiber type proportion in human skeletal muscle. FASEB J 9:1091–1095 [DOI] [PubMed] [Google Scholar]
- Taylor RR, Mamotte CDS, Fallon K, van Bockxmeer FM (1999) Elite athletes and the gene for angiotensin-converting enzyme. J Appl Physiol 87:1035–1037 [DOI] [PubMed] [Google Scholar]
- Van Damme R, Wilson RS, Vanhooydonck B, Aerts P (2002) Performance constraints in decathletes. Nature 415:755–756 [DOI] [PubMed] [Google Scholar]
- Wang B, Shi GP, Yao PM, Li Z, Chapman HA, Bromme D (1998) Human cathepsin F: molecular cloning, functional expression, tissue localization, and enzymatic characterization. J Biol Chem 273:32000–32008 [DOI] [PubMed] [Google Scholar]
- Woods D, Hickman M, Jamshidi Y, Brull D, Vassiliou V, Jones A, Humphries S, Montgomery H (2001) Elite swimmers and the D allele of the ACE I/D polymorphism. Hum Genet 108:230–232 [DOI] [PubMed] [Google Scholar]