Abstract
Buccal cells were collected from 29 participants, by use of mouthwash rinses, and were split into equal aliquots, with one aliquot irradiated by electron-beam (E-beam) irradiation equivalent to the sterilizing dosage used by the U.S. Postal Service and the other left untreated. Aliquots were extracted and tested for DNA yields (e.g., TaqMan assay for quantifying human genomic DNA), genomic integrity, and amplification-based analysis of genetic variants (e.g., single-nucleotide polymorphisms [SNPs] and single tandem repeats [STRs]). Irradiated aliquots had lower median DNA yields (3.7 μg/aliquot) than untreated aliquots (7.6 μg/aliquot) (P<.0005) and were more likely to have smaller maximum DNA fragment size, on the basis of genomic integrity gels, than untreated aliquots (P<.0005). Irradiated aliquots showed poorer PCR amplification of a 989-bp β-globin target (97% for weak amplification and 3% for no amplification) than untreated aliquots (7% for weak amplification and 0% for no amplification) (P<.0005), but 536-bp and 268-bp β-globin targets were amplified from all aliquots. There was no detectable irradiation effect on SNP assays, but there was a significant trend for decreased detection of longer STRs (P=.01) in irradiated versus untreated aliquots. We conclude that E-beam irradiation reduced the yield and quality of buccal-cell specimens, and, although irradiated buccal-cell specimens may retain sufficient DNA integrity for some amplified analyses of many common genomic targets, assays that target longer DNA fragments (>989 bp) or require whole-genome amplification may be compromised.
Self-collected buccal-cell specimens are a relatively inexpensive, noninvasive collection method of genomic DNA for molecular epidemiologic studies (Lench et al. 1988; Lum et al. 1998; Walker et al. 1999; Garcia-Closas et al. 2001; Le Marchand et al. 2001). Specimens can be collected by a variety of methods, including a simple mouthwash rinse and expectoration into a collection vial that can be sent via mail. Several large U.S. epidemiologic studies—including The Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial (Gohagan et al. 2000) and Agricultural Health Study (Mage et al. 2000)—have chosen to use self-collected buccal cells to facilitate sample collection for genomic DNA. However, the use of electron-beam (E-beam) irradiation has been recently implemented by the U.S. Postal Service (USPS) to “sterilize” mail (via cleavage of microbe DNA) as a protective measure against terrorism with biological agents. (The USPS irradiation plan is currently limited to mail received at ZIP postal codes in Washington, DC, that begin 202–205. We are not aware of immediate plans to expand the irradiation treatment to other ZIP codes.) The potential effects of decontaminating radiation on DNA yields and quality from E-beam–exposed buccal cells sent through the U.S. Mail are largely unknown.
To address this issue, buccal-cell specimens were collected by expectoration of a mouthwash rinse (Scope [Procter & Gamble]) into a collection vial from 32 consenting volunteers, according to a National Cancer Institute (NCI) institutional review board–approved protocol (Garcia-Closas et al. 2001). Specimens were split into equal aliquots and were assigned unique identifiers. One aliquot remained untreated. The other aliquot for 29 pairs (3 were shipped but remained untreated, for use as shipping controls) was exposed to E-beam irradiation (Titan Scan Technologies) by use of a protocol that simulated potential exposure (⩾56 kilograys [KGy]) through the USPS (Hanson 2002). Aliquots were placed on a conveyor belt and exposed twice to 10 million electron–volt radiation, which resulted in a total dose in the range of 70–97 KGy. Specimens were returned to the NCI biorepository, cells were pelleted by centrifugation, and, after the supernatant was decanted, the cell pellet was stored at −70° C to −80° C until DNA extraction.
Frozen buccal-cell pellets were suspended in Tris-EDTA buffer, and genomic DNA was extracted using phenol-chloroform (Lum et al. 1998). Total-DNA yields were quantified by spectrophotometry at 260-nm and 280-nm wavelengths and the PicoGreen assay (Molecular Probes) (Hopwood et al. 1997). Human-specific genomic DNA was quantified using a TaqMan assay that amplifies a 119-bp region of the human DNA BRCA1 gene (Haque et al., in press). The integrity of genomic DNA was assessed using gel electrophoresis on 0.4% agarose gels stained with ethidium bromide. Electrophoretic migration of specimen DNA was compared with known molecular size markers (Ready-Load λ DNA/HindIII Fragments [Invitrogen]) and were rated for the maximum fragment size as “no visible DNA,” “<564 bp,” “<6,557 bp,” “<23,130 bp,” or “⩾23,130 bp” (Garcia-Closas et al. 2001). PCR amplifications of 268-bp, 536-bp, and 989-bp β-globin fragments were performed as described elsewhere, with 15–20 ng of extracted DNA per reaction (Greer et al. 1994; Lum et al. 1998; Garcia-Closas et al. 2001). Amplified products were separated on 4% agarose gels for the 268-bp and 536-bp fragments and 3% gels for the 989-bp fragment; a lane with a positive amplification control (K562 DNA [Promega]) and lanes with a 100-bp DNA ladder (Invitrogen) were run on each gel. Masked to the treatment status, results were evaluated independently by two raters as “negative” (no band observed), “weak positive” (a band was observed at the correct molecular size, but band intensity <25% of the K562 band), or “positive” (a band was observed at the correct molecular size, but band intensity ⩾25% of the K562 band). Discrepancies between “positive” and “weak positive” ratings were, by default, called “positive.” Discrepancies between “weak positive” and “negative” ratings were adjudicated by running the amplified product on a polyacrylamide gel and staining the gel with silver nitrate stain. A sample was called “weak positive” if the presence of a band at the correct molecular size was detected; otherwise, it was called “negative.” Differences in continuous measurements (DNA yield and purity [the ratio of the spectrophometric readings at 260 nm and 280 nm]) and categorical measurements (genomic integrity and PCR amplification) between irradiated and untreated aliquots were tested for significance (P<.05; two-sided) by use of the Wilcoxon matched-pair sign-rank test and the Pearson χ2 test, respectively.
Sixteen STRs and 10 SNPs were measured for each pair of aliquots, masked to the irradiation status. Specifically, 0.25 ng of extracted DNA were tested for the following 16 STR loci: D8S1179, D21S11, D7S820, CSF11PO, D3S1358, TH01, D13S317, S16S539, D2S1338, D19S433, vWA, TPOX, S18S51, D5S818, AMELI, and FGA. The AmpFLSTRR IdentifilerT PCR Amplification Kit (Applied Biosystems [ABI]) was used per manufacturer’s instructions and included the 13 core STR loci standardized under the Federal Bureau of Investigation’s Combined DNA Index System (ABI) (Buse et al. 2003). Differences between irradiated and untreated aliquots for detection of any single STR were evaluated using symmetry χ2 tests. A linear regression model was used to evaluate the effects of the predicted maximum size of the STRs, irradiation (yes or no), and their interaction on the failure rate of these measurements. Ten frequent and commonly tested National Cancer Institute cancer-related SNPs (BRCA1-02, BRCA2-01, MGMT-04, MPO-02, MTHFR-01, NAT1-20, NAT2-01, SOD2-01, XRCC1-01, and XRCC3-01) were assayed in duplicate, with 5 ng of extracted DNA by TaqMan (ABI). The duplicate results were combined and grouped as “measured” (“homozygous” or “heterozygous” for either test) or “unmeasured” (“undetermined” or “not amplified” for both tests), and differences between irradiated and untreated aliquots were tested for significance by use of symmetry χ2 tests.
All 29 irradiated aliquots underwent a color change from green to light brown, with a concomitant increase in temperature, as did unused bottles of Scope (fig. 1); the three aliquots that were shipped but not irradiated did not change color. Median DNA yields per aliquot were significantly less for irradiated aliquots than for untreated aliquots, as measured by spectrophotometry (5.9 μg vs. 12.3 μg; P=.002), PicoGreen assay (3.9 μg vs. 8.9 μg; P=.01), and TaqMan assay (3.7 μg vs. 7.6 μg; P<.0005) (table 1). (N.B.: The expected median yield for the whole specimen is twice that of an aliquot [e.g., 15.2 μg/specimen, on the basis of the TaqMan assay] and is similar to the median yield [16.6 μg/specimen] observed elsewhere from participants in a breast cancer case-control study [Garcia-Closas et al. 2001].) Irradiated aliquots had a slightly greater DNA purity, as measured by the spectrophotometric ratio of 260:280 nm, than untreated aliquots (untreated=1.67 vs. irradiated=1.74; P=.07).
Figure 1.
Color of Scope before (left) and after (right) sterilizing E-beam irradiation
Table 1.
Summary of DNA Yields, DNA Integrity Measurement, and Outcomes of PCR Amplification of β-Globin Fragments of Lengths 989 bp, 536 bp, and 268 bp
|
Results for Paired Aliquots(n=29) |
|||
| Method | Untreated | Irradiated | Pa |
| DNA yields, medianb: | |||
| Spectrophotometric (260 nm) | 12.3 μg | 5.9 μg | .002 |
| PicoGreen | 8.9 μg | 3.9 μg | .01 |
| TaqMan | 7.6 μg | 3.7 μg | <.0005 |
| Genomic integrity (maximum size)c: | |||
| ⩾23,130 bp | 93.1% | .0% | |
| <23,130 bp | 6.9% | 31.0% | |
| <6,557 bp | .0% | 65.5% | |
| <564 bp | .0% | 3.5% | <.0005 |
| PCR Amplification of a 989-bp β-globin: | |||
| Positive | 93% | 0% | |
| Weak positive | 7% | 97% | |
| Negative | 0% | 3% | <.0005 |
| PCR Amplification of a 536-bp β-globin: | |||
| Positive | 100% | 100% | |
| Weak positive | 0% | 0% | |
| Negative | 0% | 0% | 1.0 |
| PCR Amplification of a 268-bp β-globin: | |||
| Positive | 100% | 100% | |
| Weak positive | 0% | 0% | |
| Negative | 0% | 0% | 1.0 |
Wilcoxon matched-pair sign-rank test (continuous) and Pearson χ2 test (categorical).
Yield per aliquot. Each aliquot represents one-half of the whole mouthwash specimen.
Electrophoretic migration of specimen DNA was compared with known molecular size markers (Ready-Load λ DNA/HindIII Fragments [Invitrogen]) and rated for the maximum fragment size as “no visible DNA,” “<564 bp,” “<6,557 bp,” “<23,130 bp,” or “⩾23,130 bp.”
The irradiated aliquots were more likely to have a smaller maximum size of DNA fragments on integrity gels than were untreated aliquots (P<.0005) (table 1); irradiated aliquots (93.1%) were also more likely than untreated aliquots (3.5%) to have genomic DNA that did not migrate into the gel (P<.0005) and exhibited weaker or no PCR amplification of the 989-bp β-globin DNA fragment (0% positive, 97% weak positive, and 3% negative), compared with the irradiated aliquots (93% positive, 7% weak positive, and 0% negative) (P<.0005) (fig. 2; table 1). All aliquots, irradiated or untreated, were positive for amplification of 268-bp and 536-bp β-globin DNA fragments. (The amplification of the 536-bp fragment appeared to be more efficient than for the other fragments, which resulted in ratings of some amplified products as “strong positive” by the test lab. “Positive” and “strong positive” ratings were conservatively combined. However, untreated aliquots were more likely to be called “strong positive” than irradiated aliquots [P<.0005].)
Figure 2.
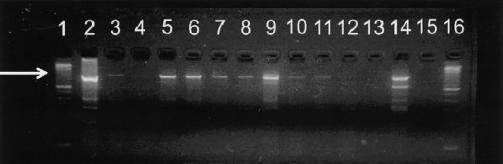
Results of 989-bp β-globin fragment PCR amplification of DNA extracted from aliquots of buccal cells. The arrow indicates the 989-bp fragment. Lanes 3–12 and 14 (3,4,8, and 10–12 are E-beam irradiated, and 5–7, 9, and 14 are untreated) are test specimens, lanes 13 and 15 are blank controls, lane 2 is a positive amplification control (K562 cell DNA), and lanes 1 and 16 are molecular size standards. Lanes 5, 6, 9, and 14 were rated as “positive,” lanes 3, 4, 7, 8, 10, and 11 were rated as “weak positive,” and lanes 12, 13, and 15 were rated as “negative.”
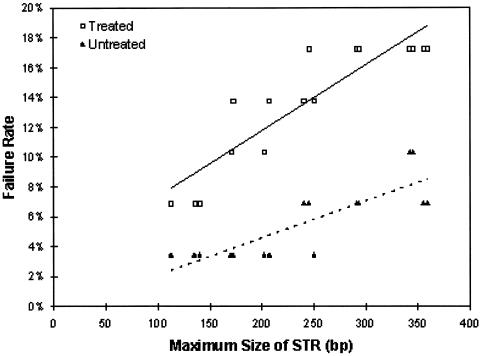
Measurement failure was nonsignificantly more likely to occur in irradiated aliquots than untreated aliquots for each STR that was considered individually (fig. 3). In a linear regression model of failure rates versus size of test STR (range 112–359 bp) and irradiation status, fragment size (P<.0005), irradiation status (P=.08), and the interaction of fragment size and irradiation status (P=.01) were associated with increased measurement failure. There were no significant differences in detection of SNPs from irradiated and untreated aliquots.
Figure 3.
The effect of E-beam irradiation on the failure rate of detection of STRs as a function of size of the STR for irradiated (□) versus untreated (▴) aliquots. Fragment size (P<.0005), irradiation status (P=.08), and the interaction of fragment size and irradiation status (P=.01) were associated with increased measurement failure.
In our study using paired aliquots, one irradiated by E-beam irradiation and the other untreated, we demonstrated that there was a significant decrease in DNA yield following irradiation. A decrease in the yield for extracted DNA in irradiated aliquots may be due, in part, to DNA cross-linking and subsequent degradation following strand damage. This is consistent with an observed DNA fraction that did not migrate in the genomic integrity gels in the irradiated aliquots only (data not shown). There was also evidence (e.g., reduced amplification of the 989-bp β-globin fragment, shorter maximum DNA fragment size, and reduced efficiency for longer STRs) that the irradiation caused cleavage of DNA strands. Whether DNA cross-linking and cleavage are the result of direct absorption of E-beam energy by DNA or the generation of heat by the E-beam treatment remains an unanswered question.
Additional studies are needed to evaluate the impact of E-beam irradiation on other biospecimens. A recent study of E-beam–irradiated dried blood spots made from umbilical cord blood found that β-globin fragments (300–1300 bp)—but not whole genomes—were amplifiable (Dr. Joanne Mei, Centers for Disease Control and Prevention, Department of Health and Human Services; personal communication).
We conclude that E-beam irradiation reduces the yields and quality of DNA extracted from buccal-cell collections. Although there might be sufficient DNA integrity for small fragment amplifications for at least some genetic analyses, we infer from our data that amplification of longer DNA targets and whole genomes may be compromised. One limitation to our study is that we did not incorporate a quantitative PCR method to more accurately measure losses in amplifiable DNA, and, thus, we may have underestimated the impact of the irradiation on genomic DNA. We emphasize that this was a pilot study to measure the effects of E-beam irradiation on buccal-cell genomic DNA, with a controlled exposure. Further studies of buccal-cell specimens sent through the U.S. Mail are now planned to evaluate the true E-beam exposure and the impact those exposures on the genomic DNA. The change in mouthwash color, which could occur prior to its use during the shipping of the mouthwash to participants if E-beam sterilization becomes more widely used, could negatively impact subject compliance because of the color change and possible changes in taste. The toxicity of the irradiated mouthwash is also uncertain. These issues warrant consideration for future implementation of this collection method.
Acknowledgments
The authors gratefully acknowledge help from Carla Chorley and Kathi Shea at BBI Biotech and Drs. Jeff Struewling, Arthur Schatzkin, and Juan Alguacil at the National Cancer Institute.
Electronic-Database Information
Accession numbers and the URL for data presented herein are as follows:
- National Cancer Institute, http://snp500cancer.nci.nih.gov/home.cfm (for BRCA1-02 [dbSNP id: rs799917], BRCA2-01 [dbSNP id: rs144848], MGMT-04 [dbSNP id: rs1803965], MPO-02 [dbSNP id: rs2333227], MTFHR-01 [dbSNP id: rs1801131], NAT1-20 [dbSNP id: rs4986993], NAT2-01 [dbSNP id: rs1208], SOD2-01 [dbSNP id: rs1799725], XRCC1-01 [dbSNP id: rs25487], and XRCC3-01 [dbSNP id: rs861539])
References
- Buse EL, Putinier JC, Hong MM, Yap AE, Hartmann JM (2003) Performance evaluation of two multiplexes used in fluorescent short tandem repeat DNA analysis. J Forensic Sci 48:348–357 [PubMed] [Google Scholar]
- Garcia-Closas M, Egan KM, Abruzzo J, Newcomb PA, Titus-Ernstoff L, Franklin T, Bender PK, Beck JC, Le Marchand L, Lum A, Alavanja M, Hayes RB, Rutter J, Buetow K, Brinton LA, Rothman N (2001) Collection of genomic DNA from adults in epidemiological studies by buccal cytobrush and mouthwash. Cancer Epidemiol Biomarkers Prev 10:687–696 [PubMed] [Google Scholar]
- Gohagan JK, Prorok PC, Hayes RB, Kramer BS, Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial Project Team (2000) The Prostate, Lung, Colorectal and Ovarian (PLCO) cancer screening trial of the National Cancer Institute: history, organization, and status. Control Clin Trials 21:S251–S272 [DOI] [PubMed] [Google Scholar]
- Greer CE, Wheeler CM, Manos MM (1994) Sample preparation and PCR amplification from paraffin-embedded tissues. PCR Methods Appl 3:S113–S122 [DOI] [PubMed] [Google Scholar]
- Hanson DJ (2002) Zapping the mail. Chemical and Engineering News 80:30–32 [Google Scholar]
- Haque KA, Pfeiffer RM, Beerman MB, Struewing JP, Chanock SJ, Bergen AW. Accuracy and precision of high-throughput DNA quantification methods. BMC Biotechnol (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood A, Oldroyd N, Fellows S, Ward R, Owen SA, Sullivan K (1997) Rapid quantification of DNA samples extracted from buccal scrapes prior to DNA profiling. Biotechniques 23:18–20 [DOI] [PubMed] [Google Scholar]
- Le Marchand L, Lum-Jones A, Saltzman B, Visaya V, Nomura AMY, Kolonel LN (2001) Feasibility of collecting buccal DNA by mail in a cohort study. Cancer Epidemiol Biomarkers Prev 10:701–703 [PubMed] [Google Scholar]
- Lench N, Stanier P, Williamson R (1988) Simple non-invasive method to obtain DNA for gene analysis. Lancet 1:1356–1358 [DOI] [PubMed] [Google Scholar]
- Lum A, Le Marchand L (1998) A simple mouthwash method for obtaining genomic DNA in molecular epidemiological studies. Cancer Epidemiol Biomarkers Prev 7:719–724 [PubMed] [Google Scholar]
- Mage DT, Alavanja MC, Sandler DP, McDonnell CJ, Kross B, Rowland A, Blair A (2000) A model for predicting the frequency of high pesticide exposure events in the Agricultural Health Study. Environ Res 83:67–71 [DOI] [PubMed] [Google Scholar]
- Walker AH, Najarian D, Rebbeck TR, Jaffe JM, Kanetsky PA, Rebbeck TR (1999) Collection of genomic DNA by buccal swabs for polymerase chain reaction-based biomarker assays. Environ Health Perspect 107:517–520 [DOI] [PMC free article] [PubMed] [Google Scholar]