Abstract
The now-emerging mitochondrial DNA (mtDNA) population genomics provides information for reconstructing a well-resolved mtDNA phylogeny and for discerning the phylogenetic status of the subcontinentally specific haplogroups. Although several major East Asian mtDNA haplogroups have been identified in studies elsewhere, some of the most basal haplogroups, as well as numerous minor subhaplogroups, were not yet determined or fully characterized. To fill the lacunae, we selected 48 mtDNAs from >2,000 samples across China for complete sequencing that cover virtually all (sub)haplogroups discernible to date in East Asia. This East Asian mtDNA phylogeny can henceforth serve as a solid basis for phylogeographic analyses of mtDNAs, as well as for studies of mitochondrial diseases in East and Southeast Asia.
Recent progress in the analysis of complete or nearly complete mtDNA sequences has provided new insights into the origin and spread of modern humans and the phylogeny of the major African, European, Asian, and Native American mtDNA lineages (Ingman et al. 2000; Finnilä et al. 2001; Maca-Meyer et al. 2001; Torroni et al. 2001; Derbeneva et al. 2002; Herrnstadt et al. 2002; Mishmar et al. 2003). These studies differ in regard to sequencing technique (viz., Maca-Meyer et al. [2001] employed manual sequencing), inclusion of the control region (which was not disclosed by Herrnstadt et al. [2002]), and sampling scheme: mtDNAs either were chosen according to the language spoken by their bearers (Ingman et al. 2000), were randomly selected from a certain geographic range (Finnilä et al. 2001; Herrnstadt et al. 2002), or were preselected according to RFLP haplogroup status (Maca-Meyer et al. 2001; Torroni et al. 2001; Derbeneva et al. 2002; Mishmar et al. 2003). Prior to these systematic sequencing efforts, a number of single, nearly complete mtDNA sequences, mainly sampled from Europe and Japan, were published in the field of medical genetics. All available mtDNAs of Asian and Native American (as well as of Papuan and Australian) origin that were published before the year 2002 were summarized in a tree by Kivisild et al. (2002). On the basis of the information provided by additional screening of a particular fragment (10171–10659) and other sites of the coding region, Yao et al. (2002a) devised a classification tree of East Asian mtDNA haplogroups that highlights some diagnostic motifs from the control region and particular parts of the coding region. Can this tree stand the test from complete sequence data and fully reflect the relationships among the mtDNA lineages observed in East Asia? To answer these questions, a fully resolved phylogeny of complete mtDNA sequences covering all major East Asian haplogroups is indispensable.
In the present study, we selected 48 mtDNAs for complete sequencing from >2,000 samples across China that belong to different subhaplogroups and also include previously unclassified haplotypes (Yao et al. 2002a, 2002c, 2003a; Yao and Zhang 2002; authors’ unpublished data). With the exception of the mtDNA haplogroup M7a (prominent in Japan), which has four representatives in the compilation of Kivisild et al. (2002), each of the (sub)haplogroups in the classification tree of Yao et al. (2002a) is represented by at least 1 of these 48 mtDNAs. Several mtDNAs that were only roughly classified as B4*, F2*, D*, G*, R*, and M* in Yao et al. (2002a, 2003a) were also selected for sequencing to better understand their phylogenetic status.
The complete mtDNA sequences were amplified by use of 15 pairs of primers (available from the authors on request). After being purified on spin columns (Watson BioTechnologies), each of the 15 overlapping fragments was sequenced for both strands by use of the BigDye Terminator Cycle Sequence Kit (ABI Applied Biosystems) and was run on an ABI 377 and an ABI 3700 DNA sequencer (ABI Applied Biosystems). The primers used for sequencing are composed of the PCR primers and a set of 47 internal primers (available from the authors on request). The sequences were edited and aligned by use of the DNASTAR software, and the mutations were scored relative to the revised reference sequence (rCRS) (Andrews et al. 1999). The length variation of the A and C stretches in region 16180–16193 was disregarded in the analysis.
To avoid the five major types of errors observed in published mtDNA data (viz., base shifts, reference bias, phantom mutations, base misscoring, and artificial recombination), as classified by Bandelt et al. (2001), we took the following quality-control measures in the course of data generation and handling. First, every mtDNA was sequenced at least twice. Second, all mutations recorded in the phylogenetic tree (fig. 1) were confirmed by rechecking the sequence electropherograms or the original references. Third, all of the insertions and deletions (indels) in the samples, and some potentially misrecorded or seemingly recurrent polymorphisms compared with the reported data (Derbeneva et al. 2002; Herrnstadt et al. 2002; Kivisild et al. 2002; Mishmar et al. 2003)—such as site 4086 in samples GD7824 and GD7811, site 5108 in GD7812, sites 12358 and 12372 in GD7825, site 13928C in SD10313, site 8877 in EWK28, site 10658 in Miao271, site 10685 in SD10324; site 10801 in QD8147; and site 10810 in GD7809—were confirmed by independent amplification and sequencing. Moreover, the absence of mutations at sites 8450, 13827, 14180, 15217, and 15805 in GD7830; site 14340 in SD10324; site 4086 in QD8167; sites 5465, 9123, and 10238 in GD7812; and site 7933 in XJ8426 were also confirmed by at least two independent experiments. To test the quality of the complete mtDNA sequences obtained, all of the “private” polymorphisms in 15 of the 48 completely sequenced mtDNAs in the terminal branches of the phylogenetic tree (fig. 1) have been rechecked in another experiment together with some controls (authors’ unpublished data). As a result, we detected discrepancies with the partial information published earlier in Yao et al. (2002a) in three samples (QD8141 actually has the 16217 mutation; LN7595 has two “C” insertions at site 315 instead of three; and GD7842 carries the mutation at site 10586).
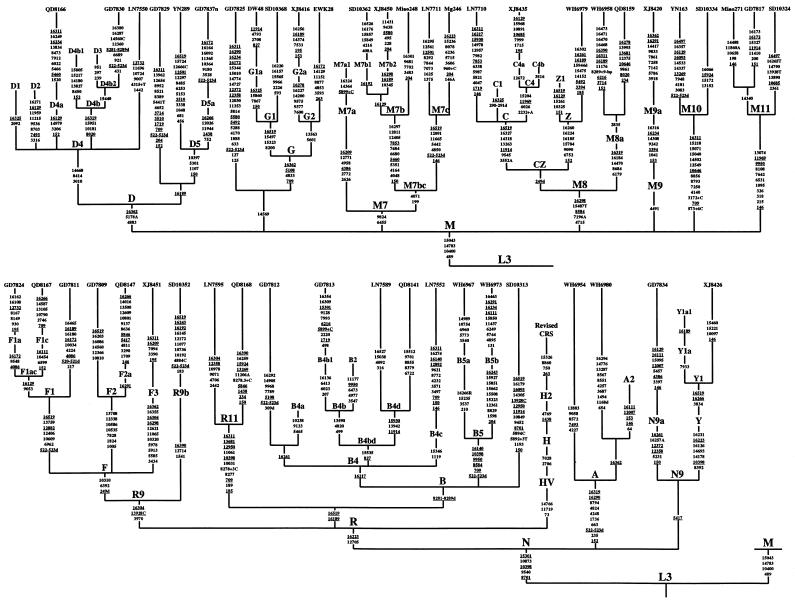
Figure 1.
Phylogenetic tree of 48 East Asian mtDNA lineages, which were sampled from various regional Han populations (Yao et al. 2002a, 2003a), except for those with prefixes DW (Daur, from Inner Mongolia), EWK (Ewenki, from Inner Mongolia), Mg (Mongolian, from Inner Mongolia), and Miao (Miaozu, from Hunan). This tree incorporates the information drawn from previous reports (Derbeneva et al. 2002; Herrnstadt et al. 2002; Kivisild et al. 2002, and references therein; Mishmar et al. 2003) by indicating the roots of several haplogroups from which those Asian sequences branch off. Also indicated are the Native American haplogroups A2, B2, C1, D1 (Bandelt et al., in press) and D2, D3 (Derbeneva et al. 2002), of which only C1 and D2 are found in Asia, too. The mutations (reconstructed most parsimoniously) on uninterrupted branches are listed (arbitrarily) in ascending order. Suffixes A, C, G, and T indicate transversion, “d” indicates a deletion, and a plus sign (+) indicates an insertion; the recurrent mutations are underlined. The reconstruction of the mutations at site 16519 and deletions of CA repeats around sites 522–523, which are known to be extremely variable, is tentative at best. Note that 2232+A, one of the specific mutations of haplogroup C4, was inadvertently scored as 2233+A in figure 1 of Kivisild et al. (2002). Further, the insertion defining haplogroup M7a1 is now scored as 5899+C.
The now-available information provided by the complete mtDNA sequences analyzed in this study (GenBank accession numbers AY255133–AY255180) and those sequences reported by other labs (Derbeneva et al. 2002; Herrnstadt et al. 2002; Kivisild et al. 2002 and references therein; Mishmar et al. 2003) offers a solid basis for discerning the phylogenetic relationship of the mtDNA haplogroups (fig. 1). The definition of haplogroups D, M7, C, A, and N9a, as well as macrohaplogroups M, R, and N, is confirmed and remains unchanged (Kivisild et al. 2002). Our results also provide complementary information for the haplogroups that were defined only by control-region and/or partial coding-region information in Yao et al. (2002a, 2003a) and Kivisild et al. (2002): D5, G1, G2, M7b, M7c, M8, M8a, Z, M9a (originally named “M9” in Yao et al. [2002a]), M10, F1, F1a, F1c, F2, B4c, B5, B5a, and B5b are all further characterized by additional mutations (fig. 1). Some haplogroups are redefined here. We follow Bandelt et al. (in press) in broadening the definition of “G1” (Kivisild et al. 2002) by requiring only three coding-region mutations (8200, 15323, and 15497) for G1 status. Haplogroup R9 is now broadened by requiring only two characteristic coding-region mutations (3970 and 13928C). It embraces two haplogroups, F (first introduced as “R9” in Yao et al. [2002a]) and R9b (initially named “R10” in Yao and Zhang [2002]). Note that there are two equally parsimonious reconstructions for the evolution of 16304; here, we opt for the one that places a forward mutation at site 16304 on the way to haplogroup R9. This prompts yet another broadening of haplogroup F, which is now recognizable by an “A” deletion scored at site 249 and transitions at sites 6392 and 10310. Haplogroup F thus encompasses haplogroups F1, F2, and F3 (originally called “R9a” in Yao et al. [2002a]). The definition of “haplogroup Y” is also broadened, since the mtDNAs with motif 16126-16261-16311 lack the 3834 mutation (authors’ unpublished data). Furthermore, some mtDNA haplotypes that were previously not well classifiable relative to the employed classification tree (marked by a star after the corresponding haplogroup acronym in Yao et al. [2002a, 2003a]) can now be assigned to new (sub)haplogroups defined as follows. The three M* haplotypes, GD7817 (Yao et al. 2002a), SD10324 (Yao et al. 2003a), and Miao271 (authors’ unpublished data)—which share seven coding-region mutations (1095, 6531, 7642, 8108, 9950, 11969, and 13074) and four mutations in HVS-II (146, 215, 318, and 326)—form a new M branch, named “haplogroup M11.” It is then evident that the mtDNAs QD8130 and XJ8436 (Yao et al. 2002a) also belong to this haplogroup. The two R* haplotypes, LN7595 and QD8168, which bear a motif similar to that of B5 but do not show the 9-bp deletion in the COII/tRNALys intergenic region (Yao et al. 2002a), form a new branch that has eight characteristic mutations (709, 8277, 8278+3C, 10031, 10398, 11061, 12950, and 13681) in the coding region and four mutations (185, 189, 16189, and 16311) in the control region. This new haplogroup is designated as “R11.” The two B4* haplotypes, LN7589 and QD8141 (Yao et al. 2002a)—determined by three coding-region transitions at sites 11914, 13942, and 15930—form a new branch of B4 named “B4d,” which is a sister group to B4b. Then, the smallest haplogroup (designated as “B4bd”) that comprises both B4b and B4d is recognizable by two transitions (at sites 827 and 15535).
In total, >70 named nested haplogroups are discerned that can be regarded as sufficiently supported by the complete sequence data (fig. 1), and most of them can be recognized by specific mutations in both the coding and the control regions. As a result, the identification of haplogroup status in future East Asian mtDNA studies could be simplified, since it requires only a few coding-region mutations to be typed according to a preliminary prediction of haplogroup status based on control-region motifs. Furthermore, our phylogenetic strategy for sample selection employed here is much more efficient and effective than random sampling or use of nonphylogenetic criteria and thus can be widely used in other mtDNA phylogeographic studies.
For comparison with previous approaches and published data, we also estimate the ages of the major haplogroups, on the basis of our collection of 48 lineages. We adopt the mutation rate of one base substitution (i.e., one mutation other than indel) in the coding region per 5,140 years (Mishmar et al. 2003), which was calibrated on the basis of an assumed human-chimp split of 6.5 million years ago. This yields the following ages of those branches of the tree of figure 1 that carry more than five sampled lineages: 50.8±6.6 thousand years (ky) for B, 57.4±8.2 ky for D, 60.0±9.2 ky for F, 65.4±10.3 ky for R9, 62.3±6.3 ky for R, 64.6±6.8 ky for N, and 69.3±5.4 ky for M, where age ±SD is calculated as in Saillard et al. (2000). The ages of the three macrohaplogroups M, N, and R are thus only slightly larger than those calculated by Mishmar et al. (2003). Since there is no positive evidence yet that the East Asian haplogroups would share any mutations with the West Eurasian or South Asian haplogroups other than those defining M, N, and R (Kivisild et al. 2002), it seems that the first modern humans carried exactly the root haplotypes of the three Eurasian macrohaplogroups into Southeast Asia. The founder age for the three root haplotypes that are based on the set of 48 coding-region sequences is then estimated as 65.4±3.8 ky. Incidentally, this value nearly equals the corresponding age of 66.0 ky, on the basis of the heuristic rate of one transition within 16090–16365 per 20,180 years (but see Saillard et al. [2000] for a critical view on the calibration of this rate). Thus, we do not see the necessity yet that “conjectures about the timing of human migrations may need to be reassessed” (Mishmar et al. 2003).
The detailed phylogeny of complete mtDNA sequences is particularly important for the study of mtDNA-related diseases. It allows us to allocate each identified mutation to a certain branch of the mtDNA phylogeny, so that pathogenic and/or disease-associated mutations can be clearly distinguished from haplogroup-specific mutations. For example, our previous analysis of the 5178A polymorphism, which is a basal mutation specific to haplogroup D, in different age samples, showed no evidence for association between this mutation and longevity (contra Tanaka et al. [1998]). This highlights the importance of examining pathogenic mtDNA mutations from a phylogenetic point of view (Rocha et al. 1999; Yao et al. 2002b). Although the mtDNA phylogenetic background does not seem to make any contribution to the phenotypic presentation of the pathogenic mutation 3243 in patients with either MELAS syndrome or a wide array of disease phenotypes (Torroni et al. 2003), the observed association between certain mtDNA haplogroup(s) and either longevity (De Benedictis et al. 1999; Niemi et al. 2003), Leber hereditary optic neuropathy (LHON [Brown et al. 1997; Torroni et al. 1997]), or Parkinson disease (van der Walt et al. 2003) strongly suggests that this phylogenetic approach should be more widely used in mtDNA-related medical genetics. Moreover, tracing mtDNA mutations along phylogenetic pathways is helpful in pinpointing potential oversights and artificial recombination (e.g., as shown in Yao et al. [2003b] and Yao and Zhang [2003]). The B5b sequence of a patient suffering from LHON and cardiomyopathy recently reported by Mimaki et al. (2003) evidently missed a batch of mutations relative to the rCRS (73, 204, 263, 1438, 8281–8289del, 8584, 10398, 15223, 16140, and 16189).
The mutational pattern can also be studied in detail with a large complete mtDNA phylogeny at hand. For instance, transversions A→G or T→G are apparently rather rare in the coding region (cf. Herrnstadt et al. [2003]). The only shared transversions to G in the Eurasian mtDNA tree reported to date by more than one lab seem to be 961G in haplogroup H, 12083G in haplogroup I, and 12738G in haplogroup K1 (Ingman et al. 2000; Finnilä et al. 2001; Maca-Meyer et al. 2001; Herrnstadt et al. 2002). Further transversions to G found in lineages from haplogroups J, T, W, and X by Mishmar et al. (2003) may thus be problematic, at least 14974G (Herrnstadt et al. 2003). On the other hand, indels in the coding region seem to occur at an absolute frequency comparable with that of transversions but might be missed occasionally, owing to conservative reading of ambiguous sequencer outputs. For example, only a single private coding-region indel (15944d in an African haplogroup L1c lineage) can be scored in the 53 complete mtDNA sequences of Ingman et al. (2000) (contrast this to nine private indels and five shared ones detected in our 48 complete mtDNA sequences); moreover, their single haplogroup F sequence (closely related to the lineage XJ8440 of Yao et al. [2002a]) misses the 249 deletion. We agree with Herrnstadt et al. (2003) that the solution to the problem of mtDNA databases containing errors “is further effort, both at the front end (the sequencing process itself) and at the back end (increased quality control), of mtDNA database construction.”
In short, the phylogenetic tree of East Asian mtDNAs obtained in the present study covers all of the major haplogroups in the region and testifies to the phylogenetic status of the newly identified haplogroups (Kivisild et al. 2002; Yao et al. 2002a, 2003a; authors’ unpublished data) that were formerly defined on the basis of control-region and/or only partial coding-region information. This tree, then, can serve as a basis for haplogroup inferences in future studies of East Asian populations and for distinguishing pathogenic mutations from rare polymorphisms in mtDNA medical genetics.
Acknowledgments
We thank Shi-Fang Wu for technical assistance. We also thank Dr. Vincent Macaulay for helpful comments on the manuscript. This study was supported by grants from Chinese Academy of Sciences (KSCX2-SW-2010), Natural Sciences Foundation of China, and Natural Sciences Foundation of Yunnan Province.
Electronic-Database Information
Accession numbers and URL for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for the mtDNA complete sequence data [accession numbers AY255133–AY255180])
References
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N (1999) Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23:147 [DOI] [PubMed] [Google Scholar]
- Bandelt H-J, Herrnstadt C, Yao Y-G, Kong Q-P, Kivisild T, Rengo C, Scozzari R, Richards M, Villems R, Macaulay V, Howell N, Torroni A, Zhang Y-P. Identification of Native American founder mtDNAs through the analysis of complete mtDNA sequences: some caveats. Ann Hum Genet (in press) [DOI] [PubMed] [Google Scholar]
- Bandelt H-J, Lahermo P, Richards M, Macaulay V (2001) Detecting errors in mtDNA data by phylogenetic analysis. Int J Legal Med 115:64–69 [DOI] [PubMed] [Google Scholar]
- Brown MD, Sun F, Wallace DC (1997) Clustering of Caucasian Leber hereditary optic neuropathy patients containing the 11778 or 14484 mutations on an mtDNA lineage. Am J Hum Genet 60:381–387 [PMC free article] [PubMed] [Google Scholar]
- De Benedictis G, Rose G, Carrieri G, De Luca M, Falcone E, Passarino G, Bonafé M, Monti D, Baggio G, Bertolini S, Mari D, Mattace R, Franceschi C (1999) Mitochondrial DNA inherited variants are associated with successful aging and longevity in humans. FASEB J 13:1532–1536 [DOI] [PubMed] [Google Scholar]
- Derbeneva OA, Sukernik RI, Volodko NV, Hosseini SH, Lott MT, Wallace DC (2002) Analysis of mitochondrial DNA diversity in the Aleuts of the Commander Islands and its implications for the genetic history of Beringia. Am J Hum Genet 71:415–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnilä S, Lehtonen MS, Majamaa K (2001) Phylogenetic network for European mtDNA. Am J Hum Genet 68:1475–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrnstadt C, Elson JL, Fahy E, Preston G, Turnbull DM, Anderson C, Ghosh SS, Olefsky JM, Beal MF, Davis RE, Howell N (2002) Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am J Hum Genet 70:1152–1171 (erratum 71:448–449) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrnstadt C, Preston G, Howell N (2003) Errors, phantom and otherwise, in human mtDNA sequences. Am J Hum Genet 72:1585–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingman M, Kaessmann H, Pääbo S, Gyllensten U (2000) Mitochondrial genome variation and the origin of modern humans. Nature 408:708–713 [DOI] [PubMed] [Google Scholar]
- Kivisild T, Tolk H-V, Parik J, Wang Y, Papiha SS, Bandelt H-J, Villems R (2002) The emerging limbs and twigs of the East Asian mtDNA tree. Mol Biol Evol 19:1737–1751 [DOI] [PubMed] [Google Scholar]
- Maca-Meyer N, González AM, Larruga JM, Flores C, Cabrera VC (2001) Major genomic mitochondrial lineages delineate early human expansions. BMC Genetics 2:13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimaki M, Ikota A, Sato A, Komaki H, Akanuma J, Nonaka I, Goto Y (2003) A double mutation (G11778A and G12192A) in mitochondrial DNA associated with Leber’s hereditary optic neuropathy and cardiomyopathy. J Hum Genet 48:47–50 [DOI] [PubMed] [Google Scholar]
- Mishmar D, Ruiz-Pesini E, Golik P, Macaulay V, Clark AG, Hosseini S, Brandon M, Easley K, Chen E, Brown MD, Sukernik RI, Olckers A, Wallace DC (2003) Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci USA 100:171–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemi AK, Hervonen A, Hurme M, Karhunen PJ, Jylha M, Majamaa K (2003) Mitochondrial DNA polymorphisms associated with longevity in a Finnish population. Hum Genet 112:29–33 [DOI] [PubMed] [Google Scholar]
- Rocha H, Flores C, Campos Y, Arenas J, Vilarinho L, Santorelli FM, Torroni A (1999) About the “pathological” role of the mtDNA T3308C mutation…. Am J Hum Genet 65:1457–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saillard J, Forster P, Lynnerup N, Bandelt H-J, Nørby S (2000) mtDNA variation among Greenland Eskimos: the edge of the Beringian expansion. Am J Hum Genet 67:718–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Gong JS, Zhang J, Yoneda M, Yagi K (1998) Mitochondrial genotype associated with longevity. Lancet 351:185–186 [DOI] [PubMed] [Google Scholar]
- Torroni A, Campos Y, Rengo C, Sellitto D, Achilli A, Magri C, Semino O, Garcia A, Jara P, Arenas J, Scozzari R (2003) Mitochondrial DNA haplogroups do not play a role in the variable phenotypic presentation of the A3243G mutation. Am J Hum Genet 72:1005–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Petrozzi M, D’Urbano L, Sellitto D, Zeviani M, Carrara F, Carducci C, Leuzzi V, Carelli V, Barboni P, De Negri A, Scozzari R (1997) Haplotype and phylogenetic analyses suggest that one European-specific mtDNA background plays a role in the expression of Leber hereditary optic neuropathy by increasing the penetrance of the primary mutations 11778 and 14484. Am J Hum Genet 60:1107–1121 [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Rengo C, Guida V, Cruciani F, Sellitto D, Coppa A, Luna Calderon F, Simionati B, Valle G, Richards M, Macaulay V, Scozzari R (2001) Do the four clades of the mtDNA haplogroup L2 evolve at different rates? Am J Hum Genet 69:1348–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Walt JM, Nicodemus KK, Martin ER, Scott WK, Nance MA, Watts RL, Hubble JP, et al (2003) Mitochondrial polymorphisms significantly reduce the risk of Parkinson disease. Am J Hum Genet 72:804–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y-G, Kong Q-P, Bandelt H-J, Kivisild T, Zhang Y-P (2002a) Phylogeographic differentiation of mitochondrial DNA in Han Chinese. Am J Hum Genet 70:635–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y-G, Kong Q-P, Man X-Y, Bandelt H-J, Zhang Y-P (2003a) Reconstructing the evolutionary history of China: a caveat about inferences drawn from ancient DNA. Mol Biol Evol 20:214–219 [DOI] [PubMed] [Google Scholar]
- Yao Y-G, Kong Q-P, Zhang Y-P (2002b) Mitochondrial DNA 5178A polymorphism and longevity. Hum Genet 111:462–463 [DOI] [PubMed] [Google Scholar]
- Yao Y-G, Macaulay V, Kivisild T, Zhang Y-P, Bandelt H-J (2003b) To trust or not to trust an idiosyncratic mitochondrial data set. Am J Hum Genet 72:1341–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y-G, Nie L, Harpending H, Fu Y-X, Yuan Z-G, Zhang Y-P (2002c) Genetic relationship of Chinese ethnic populations revealed by mtDNA sequence diversity. Am J Phys Anthropol 118:63–76 [DOI] [PubMed] [Google Scholar]
- Yao Y-G, Zhang Y-P (2002) Phylogeographic analysis of mtDNA variation in four ethnic populations from Yunnan Province: new data and a reappraisal. J Hum Genet 47:311–318 [DOI] [PubMed] [Google Scholar]
- ——— (2003) Pitfalls in the analysis of ancient human mtDNA. Chinese Sci Bull 48:826–830 [Google Scholar]