Abstract
A great deal of effort has been devoted to measuring the rates of different types of nucleotide substitutions. Mutation rates are known to depend on factors such as methylation status and nearest-neighbor nucleotide effects. However, until recently, in eukaryotes, the rates have not been considered to be strand specific. In a recent analysis of mammalian lineages, Green et al. (2003) uncovered an asymmetry in the frequencies of substitutions on the coding and noncoding strands of genes and showed that this resulted in a nucleotide-content asymmetry within most genes. The authors argue that this bias may be caused by the mammalian transcription-coupled repair in germ cells, but they did not demonstrate an association with germ-cell gene expression. In this work, I analyze nucleotide contents in genes with known expression patterns and levels and provide evidence that the observed asymmetry in mutation rates is, in fact, caused by transcription. The results also imply that germline transcription may occur in a large percentage, 71%–91%, of all human genes.
Numerous efforts have been undertaken to determine the rates of mutation that occurred during evolution and the more recent history of individual species. Some early studies (Gojobori et al. 1982; Bulmer 1986) demonstrated that mutation rates are specific to each type of nucleotide substitution. It is important to note that the rates of forward and back mutations are generally not equal; in mammals, the C:G→T:A base-pair transition is usually the most frequently occurring substitution, and its frequency is significantly higher than that of the reverse T:A→C:G transition. Thus, accumulation of neutral substitutions results in a generally GC-poor composition of mammalian genomes. We now also recognize that mutation rates are usually species specific and depend on numerous additional factors, such as nearest neighbor nucleotide effects (Hess et al. 1994; Krawczak et al. 1998) and base modification (Bulmer 1986). The most notable example is the process of deamination of methylated cytosines—usually present in CpG dinucleotides—which results in a significant CpG underrepresentation in mammalian genomes.
Mutations are usually detected as base-pair substitutions, but the underlying mutational mechanisms—which, in most cases, are still poorly understood—act on single bases. A base may be altered by DNA-damaging processes (such as, for example, deamination) or misincorporated during replication. If the damaged base is correctly repaired by DNA-repair systems, or if the mismatched base is correctly resolved by mismatch repair, no mutation is recovered. However, if the mutation escapes repair, an incorrect base is later synthesized on the complementary DNA strand, and the mutation is detected as a base-pair difference from the ancestral sequence. It is important to note that, since mutation acts on single bases, it is possible for mutation rates to be “strand specific.” In fact, in some simple organisms, it has been demonstrated that the rates may vary with respect to the polarity of transcription and replication (Tanaka and Ozawa 1994; Beletskii and Bhagwat 1996; Lobry 1996; Kano-Sueoka et al. 1999).
If the mutation rates of two complementary DNA strands were identical, over a sufficiently long interval the frequencies of complementary bases should approach equality (i.e., A=T and C=G). This is known as Chargaff’s second parity rule (PR2) (Chargaff 1951). Deviations from PR2 in simple organisms have been described elsewhere. Such asymmetries may result from the processes of transcription and replication, both of which distinguish between complementary DNA strands. During transcription, the antisense strand is thought to be stabilized by the transcription machinery, whereas the sense strand is exposed and prone to undergoing mutational processes, such as deamination. Hence, in enterobacteria, the rate of deamination of cytosine, which results in C→T transitions, is increased on sense-DNA strands (Beletskii and Bhagwat 1996). During replication, the lagging and leading DNA strands are subject to different frequencies of base misincorporation, and replication-associated biases may result in an excess of G over C and T over A on the leading strands. Such biases exist in most genomes that possess single origins of replication: bacteria, many viruses, mitochondria, and chloroplasts (reviewed by Frank and Lobry [1999]). In eukaryotic genomes, where multiple origins of replication exist, replication-associated biases are generally not observed. In the proximity of known origins of replication, such effects have been suggested (Wu and Maeda 1987) but have not been confirmed, in general (Bulmer 1991). Similarly, transcription-associated biases have not been suggested until recently (Green et al. 2003).
In this work, I investigate the bias, measured as B=[(G+T)-(A+C)]/(A+C+G+T), in all known human genes (a total of 13,870 unique genes, from the RefSeq track of the UCSC genome annotation database, Hg14 November 2002 [Kent et al. 2002]). In the absence of mutational asymmetry, this bias is expected to equal zero (Sueoka 1995). Within each gene, the average bias was determined from all noncoding intronic sequences, excluding the first and last 50 bp of each intron, since those regions are most likely to contain control elements and evolve in a nonneutral fashion (Majewski and Ott 2002). It is important to exclude potentially functional regions, since I am interested in investigating sequences that evolve neutrally and are under no selective pressure. To exclude repetitive elements that might have inserted recently and would not reflect long-term evolutionary patterns, I used the repeat-masked version of the genome. However, similar results are obtained with the unmasked sequence.
Transcription-associated, neutral asymmetric patterns should be heritable only in genes that are expressed in the germline. Hence, I expect such genes to have higher average biases than tissue-specific genes. In addition, if the asymmetric substitutions occur during transcription, highly expressed genes should have larger deviations from symmetry than genes with low average expression levels.
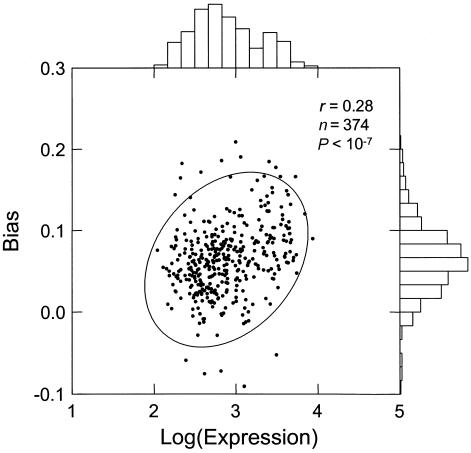
I used the HuGE Index Database (Haverty et al. 2002) to categorize genes by tissue specificity and expression levels. I used the subset of ubiquitously expressed housekeeping genes (n=374) to represent genes that are very likely to be transcribed in the germline. Although exact expression levels of those genes throughout their germline history are not known, I assume that their average expression over 19 different tissue types should be representative of their mean expression in the germline. Using the average values minimizes tissue-specific variations. The values were log-transformed to ensure a better approximation to normality. Within the housekeeping set, I demonstrate a highly significant Pearson correlation between expression intensity and the mutational bias (B) (r=0.28; P<10-7). This result, shown in figure 1, is also significant under the nonparametric Spearman rank correlation test (r=0.29; P<10-7). The correlation is even stronger for the subset of genes with lowest variability (as measured by the coefficient of variation) across tissues; that is, genes for which tissue-averaged expression should be a particularly good estimate of germline activity. For the set of the 30 least-variable genes, Spearman correlation increases to r=0.46.
Figure 1.
Pearson correlation between mean expression levels of housekeeping genes (Hsiao et al. 2001) and intronic mutational bias. The sample consists of 374 genes that are common to both the RefSeq and the HuGE databases and have appreciable intronic sequences (>100 total bp). In case of alternatively spliced genes, only the longest transcript was used. A 95% concentration ellipse is shown. The histogram plots illustrate that the logarithmic transformation provides a good approximation of a normal distribution of expression levels.
I also investigated the individual components of the overall compositional bias: the GC skew, which can be measured as (G-C)/(G+C), and the AT skew, measured as (T-A)/(A+T). Within the housekeeping-gene sample, both the GC skew (Pearson r=0.25; P=7.10-7) and the AT skew (r=0.17; P=8.10-4) are significantly correlated with the expression level.
As a control, I used a set of brain-specific genes: 431 genes that are highly expressed in the brain but not in any other tissue in the database (Hsiao et al. 2001). Although some of those genes may still be expressed in germ cells, there should be no correlation between brain-specific expression levels and B. In fact, no such correlation exists (Spearman r=0.02; P=.53).
Having established the correlation between gene expression and strand asymmetry, it is of interest to compare average biases across different groups of genes (table 1). Within the entire human genome (13,870 genes), B=4.3%. Within the brain-specific sample, the bias is close to this genomewide average (B=4.0%). However, within the housekeeping-gene sample, the mean bias is significantly elevated (B=6.4%; χ21=1,699; P<10−16). Similarly, in an independently assessed sample of 675 oocyte-expressed genes (Stanton and Green 2001), the bias is also significantly elevated (B=5.8%; χ21=3,092; P<10-16), which demonstrates that genes that are known to be expressed in germ cells have a higher average bias. The bias remains elevated (B=5.7%) even after removing all known housekeeping genes from the oocyte sample. Finally, there also exist variations in mean bias levels across chromosomes, with the largest bias (B=6.2%) observed on the Y chromosome, which is consistent with the involvement of Y-linked genes in reproductive functions.
Table 1.
Average Mutational Biases within Functional Divisions of Human Genes
| Sample | No. of Genes | B | Compared with Meana |
| RefSeq | 13,870 | .043 | = |
| Housekeeping | 374 | .064 | > |
| Oocyte | 675 (545)b | .058 (.057)b | > |
| Y chromosome | 54 | .062 | > |
| Brain | 431 | .040 | < |
All differences are highly significant under a χ2 test of homogeneity that compares G+T and A+C counts within each sample with those within the entire RefSeq gene complement.
The values in parentheses refer to a subset of the oocyte-expressed genes from which all known housekeeping genes have been removed.
Conversely, investigating the bias within individual genes may allow us to identify the genes that are significantly expressed in germ cells, since such genes should all have elevated biases. In fact, within the housekeeping-gene sample, 94% of genes have biases B>0. Within the oocyte-expressed sample, this value is even higher (97%). One may expect that this leads to a bimodal distribution of individual B values within the entire gene complement, genes absent in the germline with biases clustered around B=0, and germline-expressed genes distributed with a positive mean. However, if we examine all known genes with total nonrepetitive intronic lengths >10,000 nucleotides (n=7,054, selected for length to increase statistical power), 91% have positive biases; in 83%, the asymmetry is significant at the P<.05 level (χ2 test for deviation from G+T=A+C); and in 71%, it is significant at the Bonferroni-corrected P<7.1×10-6 level. This genomewide result extends the chromosome 22 analysis of Green et al. (2003) and suggests that a very large proportion, certainly >71% and possibly as high as 91%, of all genes are transcribed in reproductive cells, which reflects the great complexity of processes required for reproduction and early development. However, it should be noted that transcriptional activity does not necessarily imply functionality; some of the genes may not be essential for germline function but may still be transcribed at low, residual levels (Chelly et al. 1989).
The above analysis implies that mutational asymmetry in human genes is caused by transcription. In bacteria, such biases may be caused by deamination of cytosine. A similar mechanism has been proposed to explain some features of codon bias in human genes (Duret 2002) and may be applicable here. However, Green et al. (2003) argue that, in mammals, the bias is more likely to be caused by the action of transcription-coupled repair (TCR). The results presented here are consistent with both hypotheses. Increased levels of transcription should result in longer periods of separation for the two DNA strands, which may lead to increased deamination of cytosine. On the other hand, Leadon and Lawrence (1991) have also shown that the efficiency of TCR is dependent on the intensity of transcription. Thus, TCR may also cause the covariation of mutational asymmetry and intensity of transcription. Also, the fact that both the GC skew and AT skew are correlated with transcription does not offer a conclusive argument in favor of either mechanism. Whereas TCR may alter various mutation rates and hence affect both the GC and AT skews, deamination of cytosine would directly cause the GC skew by depleting the cytosine pool, but it would also result in the AT skew by converting cytosines into thymines. Although this work is not able to distinguish between the two hypotheses, the study of Green et al. (2003) provides two strong arguments in favor of TCR: in primates, the C→T transition rate is not elevated on the sense-DNA strand, and the overall frequency of neutral mutations does not vary between transcribed and nontranscribed sequences. Since both of the above effects would be expected under the deamination hypothesis, the action of TCR remains the preferable explanation for the observed mutational asymmetry.
Whereas experimental verification of the above hypothesis should be a welcome development in the future, the analysis presented here provides the strongest evidence to date that, in the human genome, transcription is the cause of strand-specific mutational asymmetry. The knowledge of such issues is crucial to the understanding of mutational processes that occur in the human genome.
Acknowledgments
I thank Dr. Loydie Majewska, Dr. Lilia Iakoucheva, Mark Levenstien, Dr. Phil Green, and an anonymous reviewer for helpful comments on the manuscript. This work was supported by grant HG00008 from the National Human Genome Research Institute, Bethesda.
Electronic-Database Information
URLs for data presented herein are as follows:
- HuGE Index, http://www.hugeindex.org/
- UCSC Genome Bioinformatics, http://genome.cse.ucsc.edu/
References
- Beletskii A, Bhagwat AS (1996) Transcription-induced mutations: increase in C to T mutations in the nontranscribed strand during transcription in Escherichia coli. Proc Natl Acad Sci USA 93:13919–13924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulmer M (1986) Neighboring base effects on substitution rates in pseudogenes. Mol Biol Evol 3:322–329 [DOI] [PubMed] [Google Scholar]
- ——— (1991) Strand symmetry of mutation rates in the β-globin region. J Mol Evol 33:305–310 [DOI] [PubMed] [Google Scholar]
- Chargaff E (1951) Structure and function of nucleic acids as cell constituents. Fed Proc 10:654–659 [PubMed] [Google Scholar]
- Chelly J, Concordet JP, Kaplan JC, Kahn A (1989) Illegitimate transcription: transcription of any gene in any cell type. Proc Natl Acad Sci USA 86:2617–2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L (2002) Evolution of synonymous codon usage in metazoans. Curr Opin Genet Dev 12:640–649 [DOI] [PubMed] [Google Scholar]
- Frank AC, Lobry JR (1999) Asymmetric substitution patterns: a review of possible underlying mutational or selective mechanisms. Gene 238:65–77 [DOI] [PubMed] [Google Scholar]
- Gojobori T, Li WH, Graur D (1982) Patterns of nucleotide substitution in pseudogenes and functional genes. J Mol Evol 18:360–369 [DOI] [PubMed] [Google Scholar]
- Green P, Ewing B, Miller W, Thomas PJ, Green ED, NISC Comparative Sequencing Program (2003) Transcription-associated mutational asymmetry in mammalian evolution. Nat Genet 33:514–517 [DOI] [PubMed] [Google Scholar]
- Haverty PM, Weng Z, Best NL, Auerbach KR, Hsiao LL, Jensen RV, Gullans SR (2002) HugeIndex: a database with visualization tools for high-density oligonucleotide array data from normal human tissues. Nucleic Acids Res 30:214–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess ST, Blake JD, Blake RD (1994) Wide variations in neighbor-dependent substitution rates. J Mol Biol 236:1022–1033 [DOI] [PubMed] [Google Scholar]
- Hsiao LL, Dangond F, Yoshida T, Hong R, Jensen RV, Misra J, Dillon W, Lee KF, Clark KE, Haverty P, Weng Z, Mutter GL, Frosch MP, Macdonald ME, Milford EL, Crum CP, Bueno R, Pratt RE, Mahadevappa M, Warrington JA, Stephanopoulos G, Gullans SR (2001) A compendium of gene expression in normal human tissues. Physiol Genomics 7:97–104 [DOI] [PubMed] [Google Scholar]
- Kano-Sueoka T, Lobry JR, Sueoka N (1999) Intra-strand biases in bacteriophage T4 genome. Gene 238:59–64 [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler AD (2002) The human genome browser at UCSC. Genome Res 12:996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczak M, Ball EV, Cooper DN (1998) Neighboring-nucleotide effects on the rates of germ-line single-base-pair substitution in human genes. Am J Hum Genet 63:474–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leadon SA, Lawrence DA (1991) Preferential repair of DNA damage on the transcribed strand of the human metallothionein genes requires RNA polymerase II. Mutat Res 255:67–78 [DOI] [PubMed] [Google Scholar]
- Lobry JR (1996) Asymmetric substitution patterns in the two DNA strands of bacteria. Mol Biol Evol 13:660–665 [DOI] [PubMed] [Google Scholar]
- Majewski J, Ott J (2002) Distribution and characterization of regulatory elements in the human genome. Genome Res 12:1827–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton JL, Green DP (2001) A set of 840 mouse oocyte genes with well-matched human homologues. Mol Hum Reprod 7:521–543 [DOI] [PubMed] [Google Scholar]
- Sueoka N (1995) Intrastrand parity rules of DNA base composition and usage biases of synonymous codons. J Mol Evol 40:318–325 [DOI] [PubMed] [Google Scholar]
- Tanaka M, Ozawa T (1994) Strand asymmetry in human mitochondrial DNA mutations. Genomics 22:327–335 [DOI] [PubMed] [Google Scholar]
- Wu CI, Maeda N (1987) Inequality in mutation rates of the two strands of DNA. Nature 327:169–170 [DOI] [PubMed] [Google Scholar]