To the Editor:
The Americas were the last continents to be settled by humans, yet many details of the earliest occupation remain poorly understood. Proposals for the date of first entry fall into two ranges, one suggesting a very early occupation ∼30,000–40,000 years before present (BP), and the other favoring dates ∼13,000 years BP, when the polar climate was again hospitable. We present Y-chromosomal data that support strongly the latter dates.
Recent activity in archeology has prompted a reinterpretation of the economy and culture of the earliest Americans (Dillehay 2000). Although this has revolutionized thinking about American prehistory, it has not pushed dates for the earliest human entry substantially backward in time. Indeed, the paucity of sites and skeletal material credibly dated to >14,000 years BP has been a consistent puzzle for those who would posit an extremely ancient history of human occupation in the Americas.
Only two Y-chromosome haplotypes seem to have reached the Americas from Asia before colonial contacts began in the 15th century. The most frequent haplotype—reaching a frequency of 100% in some populations—was identified by hypervariable markers (Pena et al. 1995) and was later confirmed by the discovery of two SNPs known as “M3” and “M45” (Underhill et al. 1996, 2000). A second haplotype, marked by a nucleotide substitution in the RPS4Y gene, was subsequently discovered at lower frequencies (Bergen et al. 1999), presumably reaching the Americas more recently. This pattern of limited haplotype diversity has also been observed in mtDNA (Torroni et al. 1994).
Putting a date on the earliest human entry into the continent has been hampered by the absence of a known mutation occurring before—but very close to—the time that the resulting haplotype entered the Americas. The M3/DYS199 polymorphism occurred after the first populations entered the Americas (Underhill et al. 1996), so knowing when it arose would not put an upper bound on the time of first colonization. Here, we describe a novel Y-chromosome SNP that can be reliably dated and that occurred before—but sufficiently close in time to—the initial human radiation into the Americas, so as to provide a meaningful upper bound on the time of entry.
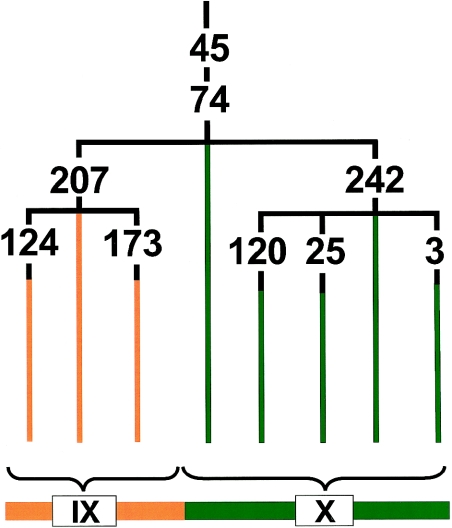
The polymorphism, which we call “M242” in the framework established by Underhill et al. (2000) (dbSNP accession number ss9805824), is a C→T transition residing in intron 1 (IVS-866) of the DBY gene. The M242-T allele is found only on chromosomes bearing the derived alleles at M45 and M74 (fig. 1). In addition, the derived allele (M242-T) is found on every chromosome bearing the M3 mutation, but not exclusively. Thus, as shown in figure 1, the M242 mutation arose after the M45/M74 mutations but before M3. This places it within a crucial gap that is very close in time to the entry of the first modern humans into the American continents. In the standard nomenclatural system of the Y Chromosome Consortium (2002), chromosomes bearing this polymorphism would be denoted as Q* in the absence of the Qa-defining mutation. The derived allele of M242 occurs at a frequency of 100% in indigenous Y chromosomes that do not carry the RPS4Y mutation (Bortolini et al. 2003 [in this issue]; P.U., unpublished data). It also occurs at a low but appreciable frequency (0%–17%; average ∼5%) in central Asian, Indian, and Siberian populations (table 1). On the basis of its widespread Eurasian distribution and its position within the Y-chromosome phylogeny (fig. 1), it is clear that the M242 mutation occurred before the first migration into the Americas. Thus, determining its age in Asian populations will establish the earliest time at which the first migrants could have reached the Americas.
Figure 1.
Partial revision of the Y-chromosome phylogeny presented by Underhill et al. (2000), indicating the placement of M242 relative to other Y-chromosome polymorphisms in haplogroups IX and X, arising in central Asia and the Americas. As can be seen, M242 arises during a crucial gap between M45/M74—which both arose in Asia—and M3, which first arose in the Americas.
Table 1.
Frequency of M242 in 49 Populations Examined[Note]
| Population | N | Frequency of M242 |
| Svanetian | 25 | 0 |
| British | 25 | 0 |
| Kazbegi | 25 | 0 |
| Orkney | 26 | 0 |
| Ishkashimi | 25 | 0 |
| Tajik/Khojant | 22 | 0 |
| Kyrgyz | 52 | .02 |
| Lezgi | 12 | 0 |
| Kazak | 54 | .06 |
| Nenets | 54 | 0 |
| Sinte Romani | 15 | 0 |
| Pomor | 28 | 0 |
| Tehran | 24 | 0 |
| Russian/North | 49 | 0 |
| Kallar | 84 | .01 |
| Lebanon | 50 | 0 |
| Azeri | 21 | 0 |
| Ossetian | 17 | 0 |
| Shiraz | 12 | .08 |
| Bartangi | 30 | .13 |
| Kurd | 17 | 0 |
| Korean | 45 | .02 |
| Saami | 23 | 0 |
| Yagnobi | 31 | .03 |
| Sourashtran | 46 | 0 |
| Esfahan | 16 | .06 |
| Russian/Tashkent | 89 | 0 |
| Turkmen | 30 | .1 |
| Armenian | 47 | 0 |
| Dungan | 40 | .08 |
| Tajik/Dushanbe | 16 | 0 |
| Tuvinian | 42 | .17 |
| Crimean Tatar | 22 | 0 |
| Iranian/Samarkand | 53 | 0 |
| Uzbek/Kashkadarya | 19 | .05 |
| Shugnan | 44 | .11 |
| Uzbek/Bukhara | 58 | .02 |
| Uzbek/Surkhandarya | 68 | .04 |
| Yadhava | 129 | .03 |
| Kazan Tatar | 38 | .03 |
| Tajik/Samarkand | 40 | .05 |
| Uighur | 41 | .05 |
| Uzbek/Khorezm | 70 | .09 |
| Uzbek/Tashkent | 43 | .14 |
| Arab/Bukhara | 42 | .14 |
| Karakalpak | 44 | 0 |
| Uzbek/Fergana Valley | 63 | .05 |
| Uzbek/Samarkand | 45 | .07 |
| Mongolian | 24 | 0 |
Note.— Further details of samples and Y-chromosome typing are available in Wells et al. (2001).
We applied several approaches to dating this mutation in 69 Eurasian samples—out of a total 1,935 individuals tested—observed to have the M242-T allele (table 2 and Wells et al. 2001). These methods are based on the accumulation of variation at 15 microsatellite loci. One approach is a widely used, model-based technique (Su et al. 1999), which was corroborated independently by a technique less dependent on assumptions about a population’s demographic history (Stumpf and Goldstein 2001). Guided by the original description of the method (Su et al. 1999), the initial effective population size (Ne) for the Asian male population was assumed to fall between a low of 1,111 and a high of 1,500. If a Y-chromosome microsatellite mutation rate of 0.18% and a human generation time (g) of 25 years are used, this leads to an estimate of 9,200–9,700 years BP, when the average variance across all microsatellite loci is taken (0.589). Although a male generation time of 25 years is frequently assumed, there is now compelling evidence that the male generation time—at least within modern times—is closer to 32 or 35 years (Tremblay and Vezina 2000; Helgason et al. 2003). When a generation time of 35 years is applied, the age estimates range from 13,000 to 14,000 years BP (table 3). As can be seen from the table, the estimates of effective population size have a relatively minor effect on age estimates, whereas mutation rates have a somewhat greater impact. A solid consensus regarding an average Y-chromosome microsatellite mutation rate, for mostly the same loci used in the present study, is developing around a rate of 0.18%–0.20% per generation (Heyer et al. 1997; Kayser et al. 2000). The best estimates of the age of the M242 mutation when this method is used appear to center on the 14,000–15,000-year range. A significant drawback of this method is its lack of a clear approach for calculating standard errors.
Table 2.
Microsatellite Genotypes for 69 M242-Bearing Chromosomes Identified in the Populations Listed in Table 1
|
No. of Repeats in |
|||||||||||||||||
| SampleIDa | Population | DYS425 | DYS426 | DYS388 | DYS389I | DYS389B | DYS434 | DYS437 | DYS435 | DYS436 | DYS390 | DYS391 | DYS392 | DYS393 | DYS19 | DYS439 | DYS438 |
| Ar41 | Arab (Bukhara) | 14 | 12 | 12 | 12 | 16 | 9 | 9 | 11 | 12 | 22 | 10 | 15 | 13 | 14 | 11 | 11 |
| Tat32 | Crimean Tatar | 10 | 12 | 12 | 12 | 16 | 9 | 8 | 11 | 12 | 21 | 10 | 15 | 13 | 13 | 11 | 11 |
| Tat36 | Crimean Tatar | 12 | 12 | 12 | 12 | 16 | 9 | 9 | 11 | 12 | 19 | 11 | 13 | 12 | 14 | 14 | 10 |
| Du46 | Dungan | 12 | 12 | 12 | 12 | 16 | 9 | 9 | 11 | 13 | 23 | 10 | 15 | 13 | 13 | 12 | 11 |
| Du40 | Dungan | 12 | 12 | 12 | 12 | 16 | … | 8 | 11 | 13 | 23 | 6 | 14 | 15 | 14 | 11 | 12 |
| Du71 | Dungan | 12 | 12 | 12 | 12 | 16 | … | 8 | 11 | 13 | 24 | 6 | 14 | 13 | 13 | 11 | 10 |
| Esf29 | Esfahan | 13 | 12 | 12 | 11 | 16 | … | 8 | 10 | 12 | 22 | 10 | 15 | 13 | 13 | 11 | 11 |
| 16Sm | Iranian (Samarkand) | 12 | 12 | 12 | 12 | 16 | 9 | 8 | 11 | 12 | 23 | 11 | 16 | 13 | 15 | 13 | 11 |
| Kz53 | Kazak | 12 | 12 | 12 | 11 | 17 | 9 | 7 | 11 | 12 | 23 | 10 | 14 | 13 | 13 | 13 | 12 |
| Kz138 | Kazak | 12 | 12 | 12 | 11 | 18 | 10 | 7 | 11 | 12 | 23 | 11 | 15 | 13 | 13 | 14 | 11 |
| Kz54 | Kazak | 12 | 11 | 12 | 11 | 17 | 9 | 8 | 11 | 12 | 22 | 10 | 14 | 14 | 13 | 12 | 11 |
| Kor35 | Korean | 11 | 12 | 12 | 12 | 16 | … | 9 | 11 | 13 | 25 | 10 | 15 | 13 | 13 | 12 | 11 |
| Kg63 | Kyrgyz | 12 | 12 | 12 | 12 | 17 | 9 | 8 | 12 | 12 | 23 | 10 | 14 | 14 | 13 | 11 | 11 |
| Pam35 | Pamiri | 12 | 12 | 12 | 11 | 17 | 9 | 8 | 12 | 12 | 25 | 10 | 14 | 13 | 14 | 13 | 11 |
| Pam66 | Pamiri | 10 | 12 | 12 | 11 | 16 | 9 | 9 | 11 | 12 | 22 | 11 | 15 | 12 | 13 | 11 | 11 |
| Kuz97 | Pamiri | 12 | 12 | 12 | 12 | 19 | 9 | 7 | 11 | 12 | 23 | 10 | 14 | 12 | 13 | 13 | 11 |
| Pam40 | Pamiri | 14 | 12 | 12 | 11 | 18 | 9 | 9 | 11 | 12 | 25 | 9 | 13 | 13 | 13 | 12 | 11 |
| Pam75 | Pamiri | 10 | 12 | 12 | 9 | 16 | … | 9 | 11 | 12 | 22 | 11 | 15 | 12 | 13 | 11 | 11 |
| Pam52 | Pamiri | 12 | 11 | 12 | 12 | 17 | 9 | 8 | 11 | 12 | 23 | 10 | 12 | 13 | 14 | 12 | 11 |
| Pam84 | Pamiri | 12 | 12 | 12 | 12 | 19 | 9 | 9 | 11 | 12 | 25 | 10 | 13 | 13 | 13 | 12 | 10 |
| Pam60 | Pamiri | 12 | 12 | 12 | 11 | 17 | 9 | 8 | 12 | 12 | 25 | 10 | 14 | 13 | 14 | 13 | 11 |
| Pam109 | Pamiri | 12 | 12 | 12 | 11 | 18 | 9 | 9 | 11 | 12 | 24 | 10 | 13 | 13 | 13 | 12 | 11 |
| Pam69 | Pamiri | 12 | 12 | 12 | 11 | 16 | 9 | 9 | 11 | 12 | 22 | 11 | 15 | 12 | 13 | 11 | 11 |
| PamlO2 | Pamiri | 12 | 12 | 12 | 11 | 17 | 9 | 9 | 11 | 12 | 22 | 11 | 13 | 13 | 14 | 12 | 10 |
| Pam104 | Pamiri | 13 | 12 | 12 | 11 | 16 | 9 | 9 | 11 | 12 | 22 | 11 | 15 | 12 | 13 | 11 | 11 |
| Pam105 | Pamiri | 13 | 12 | 12 | 11 | 16 | 9 | 9 | 11 | 12 | 22 | 11 | 15 | 12 | 13 | 11 | 11 |
| Rt7 | Russian (Tashkent) | 12 | 12 | 12 | 11 | 17 | 9 | 8 | 11 | 12 | 25 | 11 | 11 | 13 | 16 | 10 | 11 |
| Rt8 | Russian (Tashkent) | 12 | 12 | 12 | 11 | 17 | 9 | 8 | 11 | 12 | 25 | 10 | 11 | 13 | 16 | 10 | 11 |
| Rt10 | Russian (Tashkent) | 12 | 12 | 12 | 11 | 16 | 9 | 8 | 11 | 12 | 25 | 10 | 11 | 14 | 15 | 12 | 11 |
| Rt42 | Russian (Tashkent) | 12 | 12 | 12 | 11 | 17 | 9 | 8 | 11 | 12 | 25 | 11 | 11 | 13 | 15 | 12 | 11 |
| SR23 | Sinte Romani | 12 | 11 | 12 | 12 | 16 | … | 8 | 11 | 12 | 23 | 10 | 15 | 14 | 13 | 11 | 11 |
| Td1 | Tajik | 12 | 12 | 12 | 11 | 15 | 9 | 8 | 11 | 12 | 22 | 10 | 16 | 12 | 13 | 12 | 11 |
| Td36 | Tajik | 12 | 12 | 12 | 11 | 15 | 9 | 8 | 11 | 12 | 23 | 10 | 16 | 12 | 13 | 12 | 11 |
| Tu22 | Turkmen | 12 | 12 | 12 | 11 | 15 | 9 | 8 | 11 | 12 | 23 | 10 | 16 | 13 | 13 | 13 | 11 |
| Tu25 | Turkmen | 12 | 12 | 12 | 11 | 15 | 9 | 8 | 11 | 12 | 23 | 10 | 16 | 13 | 13 | 12 | 11 |
| Tu29 | Turkmen | 12 | 12 | 12 | 11 | 15 | … | 8 | 10 | 12 | … | … | … | … | … | … | 11 |
| Tv47 | Tuvinian | 12 | 12 | 12 | 11 | 17 | 9 | 7 | 11 | 12 | 23 | 10 | 14 | 13 | 13 | 12 | 11 |
| Tv50 | Tuvinian | 12 | 12 | 12 | 11 | 18 | 9 | 7 | 11 | 12 | 24 | 10 | 14 | 13 | 13 | 12 | 11 |
| Tv-E165 | Tuvinian | 12 | 12 | 12 | 11 | 17 | 9 | 7 | 11 | 12 | 23 | 10 | 14 | 13 | 13 | 12 | 11 |
| Tv26 | Tuvinian | 12 | 12 | 12 | 11 | 18 | 9 | 7 | 11 | 12 | 23 | 10 | 14 | 13 | 13 | 12 | 11 |
| Tv29 | Tuvinian | 12 | 12 | 12 | 11 | 18 | 9 | 7 | 11 | 12 | 23 | 10 | 14 | 13 | 13 | 12 | 11 |
| Tv34 | Tuvinian | 12 | 12 | 12 | 11 | 17 | 9 | 7 | 11 | 12 | 23 | 10 | 14 | 13 | 13 | 12 | 11 |
| Tv8 | Tuvinian | 12 | 12 | 12 | 11 | 18 | … | 7 | 11 | 12 | … | … | … | … | … | … | 11 |
| Ui74 | Uighur | 12 | 12 | 12 | 12 | 17 | 9 | 8 | 11 | 12 | 23 | 11 | 16 | 13 | 13 | 12 | 11 |
| Ui69 | Uighur | 12 | 11 | 12 | 12 | 16 | 9 | 9 | 11 | 12 | 23 | 10 | 12 | 12 | 14 | 11 | 11 |
| Ui51 | Uighur | … | 11 | … | … | … | … | 8 | 11 | 12 | … | … | … | … | … | … | … |
| L1 | Uzbek (Bukhara) | 12 | 12 | 12 | 12 | 18 | 9 | 7 | 11 | 12 | 23 | 10 | 14 | 13 | 13 | 11 | 11 |
| Uz52 | Uzbek (Central) | 12 | 12 | 12 | 11 | 18 | 9 | 9 | 11 | 12 | 25 | 10 | 13 | 13 | 13 | 12 | 11 |
| Uz9 | Uzbek (Central) | 12 | 12 | 12 | 12 | 15 | 10 | 8 | 11 | 12 | 23 | 10 | 16 | 13 | 13 | 12 | 12 |
| 3F | Uzbek (Fergana) | 12 | 12 | 12 | 11 | 17 | 9 | 8 | 12 | 12 | 24 | 10 | 14 | 13 | 14 | 12 | 11 |
| 16F | Uzbek (Fergana) | 12 | 12 | 12 | 11 | 17 | 9 | 7 | 11 | 12 | 23 | 9 | 14 | 13 | 13 | 13 | 12 |
| 49F | Uzbek (Fergana) | 10 | 11 | 12 | 11 | 16 | 9 | 8 | 11 | 12 | 21 | 10 | 15 | 13 | 13 | 12 | 11 |
| Kuz68 | Uzbek (Khorezm) | 12 | 12 | 12 | 11 | 17 | 9 | 9 | 11 | 12 | 24 | 10 | 13 | 13 | 14 | 12 | 11 |
| Kuz69 | Uzbek (Khorezm) | 12 | 12 | 12 | 11 | 17 | 9 | 9 | 11 | 12 | 24 | 10 | 13 | 13 | 14 | 12 | 11 |
| Kuz79 | Uzbek (Khorezm) | 12 | 12 | 12 | 11 | 15 | 9 | 8 | 11 | 12 | 23 | 10 | 17 | 13 | 13 | 13 | 11 |
| Kuz28 | Uzbek (Khorezm) | 11 | 12 | 12 | 12 | 16 | 8 | 8 | 12 | 12 | 23 | 10 | 14 | 13 | 14 | 12 | 11 |
| Kuz48 | Uzbek (Khorezm) | 13 | 12 | 12 | 11 | 16 | 9 | 9 | 11 | 12 | 22 | 10 | 15 | 13 | 13 | 11 | 11 |
| Uzs108 | Uzbek (Southern) | 13 | 12 | 12 | 11 | 16 | 9 | 9 | 11 | 12 | 22 | 9 | 13 | 13 | 13 | 12 | 11 |
| Uzs116 | Uzbek (Southern) | 12 | 11 | 12 | 11 | 17 | 9 | 9 | 11 | 12 | 23 | 10 | 12 | 13 | 14 | 11 | 11 |
| 25Sd | Uzbek (Surkhandarya) | 13 | 12 | 12 | 11 | 16 | 9 | 8 | 11 | 12 | 22 | 9 | 15 | 13 | 14 | 13 | 11 |
| 45Sd | Uzbek (Surkhandarya) | 12 | 12 | 12 | 9 | 17 | 9 | 9 | 12 | 12 | 24 | 12 | 14 | 13 | 15 | 12 | 11 |
| 46Sd | Uzbek (Surkhandarya) | 12 | 11 | 12 | 11 | 17 | 9 | 8 | 11 | 12 | 22 | 10 | 11 | 13 | 14 | 12 | 9 |
| 37T | Uzbek (Tashkent) | 12 | 12 | 12 | 12 | 15 | 9 | 8 | 11 | 12 | 23 | 10 | 16 | 13 | 13 | 12 | 11 |
| 40T | Uzbek (Tashkent) | 12 | 10 | 12 | 12 | 15 | 9 | 8 | 11 | 12 | 20 | 10 | 14 | 14 | 13 | 13 | 11 |
| 43T | Uzbek (Tashkent) | 12 | 11 | 12 | 11 | 17 | 9 | 9 | 11 | 12 | 24 | 11 | 13 | 12 | 14 | 11 | 12 |
| 6646 | Yadhava | 12 | 11 | 12 | 11 | 16 | … | 9 | 12 | 12 | 22 | 10 | 13 | 13 | 13 | 11 | 11 |
| 6653 | Yadhava | … | 12 | 12 | 12 | 16 | … | 9 | 11 | 12 | 22 | 10 | 16 | 13 | 13 | 11 | 11 |
| 6709 | Yadhava | 12 | 12 | 12 | 11 | 16 | … | 9 | 11 | 12 | 24 | 10 | 13 | 13 | 13 | 11 | … |
| YaglO | Yaghnob | 10 | 12 | 12 | 9 | 17 | 9 | 8 | 12 | 12 | 23 | 10 | 13 | 13 | 14 | 12 | 12 |
Sample IDs are from Wells et al. (2001).
Table 3.
Estimated Ages of M242 for a Range of Male Generation Times, Y-Chromosome Microsatellite Mutation Rates, and Effective Population Sizes, Calculated Using the Method of Su et al. (1999)
|
Estimated Age(years) |
||||||||
|
Ne=1,111 |
Ne=1,500 |
Ne=3,000 |
Ne=10,000 |
|||||
| MutationRate | g=25 | g=35 | g=25 | g=35 | g=25 | g=35 | g=25 | g=35 |
| .10% | 20,975 | 29,365 | 18,700 | 26,180 | 16,375 | 22,925 | 15,175 | 21,245 |
| .18% | 9,675 | 13,545 | 9,225 | 12,915 | 8,650 | 12,110 | 8,300 | 11,620 |
| .20% | 8,550 | 11,970 | 8,175 | 11,445 | 7,750 | 10,850 | 7,450 | 10,430 |
| .30% | 5,400 | 7,560 | 5,250 | 7,350 | 5075 | 7,105 | 4,950 | 6,930 |
Although this method is now well established for a range of demographic scenarios, we wanted to compare these estimates with those from an entirely different method—one that is insensitive to assumptions of Ne and population growth rate (Stumpf and Goldstein 2001). This approach relies on an inference of the ancestral microsatellite haplotype, which we identified as the haplotype with the most frequent allele at each locus. Again using a mutation rate of 0.18% per generation, we estimate the age of the M242 mutation to be 15,000 years BP, averaged over each of the 15 loci (table 4). The SD for this average estimate is only 1,700 years, which definitively precludes an arrival time of ⩾30,000 years BP. Table 4 shows the results obtained when a variety of mutation rates and generation times are used, and here, too, our best estimate of the mutation’s age is ∼18,000 years BP. This establishes a fairly solid upper bound on the time of first entry into the Americas that seems to preclude a time of entry >20,000 years BP, and our best guess would be closer to 15,000–18,000 years BP. Since our estimate derives solely from the ancestral Asian populations, it is unaffected by changes of diversity occurring after European conquest of the Americas. Even demographic changes that occurred in central Asia and Siberia after Russians established greater sway in the region are likely to have had a minor effect, because we have sampled haplotype diversity over such a huge geographic area. Furthermore, microsatellite diversity on the M242-bearing haplotype in several Native American populations (Bortolini et al. 2003 [in this issue]) is nearly the same as in our Asian sample, substantiating our result and suggesting that the M242-T haplotype entered the Americas very soon after it arose.
Table 4.
Estimated Age of M242 for a Range of Male Generation Times and Y-Chromosome Microsatellite Mutation Rates, Calculated Using the Method of Stumpf and Goldstein (2001)
| MutationRate | EstimatedAge(generations) | Estimated Age ± SDwhen g=25(years) | Estimated Agewhen g=35(years) |
| .10% | 1,068 | 26,700 ± 3,075 | 37,380 |
| .18% | 593 | 14,825 ± 1,708 | 20,755 |
| .20% | 534 | 13,350 ± 1,537 | 18,690 |
| .30% | 356 | 8,900 ± 1,025 | 12,460 |
This discovery, which indicates a rather more recent entry into the Americas than suggested by previous genetic studies (Cavalli-Sforza et al. 1994; Torroni et al. 1994), places the DNA evidence more in line with archeological data, which is characterized by a clear dearth of sites credibly dated beyond 14,000 years BP. Our results do not contradict earlier studies of mtDNA (Torroni et al. 1994) and the autosomes (Cavalli-Sforza et al. 1994), whose standard errors were large and whose authors noted several reasons to expect their dates to overestimate the timing of the first human arrivals to the Americas. In addition, a more recent time of entry into the continent makes the proposal of the Amerind language family more plausible; or, conversely—given the rapidity of linguistic change—the existence of a unified Amerind family would itself imply a fairly recent settling of the Americas, as we have suggested here.
Electronic-Database Information
The accession number and URL for data presented herein are as follows:
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/ (for the M242 polymorphism [accession number ss9805824])
References
- Bergen AW, Wang CY, Tsai J, Jefferson K, Dey C, Smith KD, Park SC, Tsai SJ, Goldman D (1999) An Asian-Native American paternal lineage identified by RPS4Y resequencing and by microsatellite haplotyping. Ann Hum Genet 63:63–80 [DOI] [PubMed] [Google Scholar]
- Bortolini M-C, Salzano FM, Thomas MG, Stuart S, Nasanen SPK, Bau CHD, Hutz MH, Layrisse Z, Petzl-Erler ML, Tsuneto LT, Hill K, Hurtado AM, Castro-de-Guerra D, Torres MM, Groot H, Michalski R, Nymadawa P, Bedoya G, Bradman N, Labuda D, Ruiz-Linares A (2003) Y-chromosome evidence for differing ancient demographic histories in the Americas. Am J Hum Genet 73:524–539 (in this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A (1994) The history and geography of human genes. Princeton University Press, Princeton [Google Scholar]
- Dillehay TD (2000) The settlement of the Americas: a new prehistory. Basic Books, New York [Google Scholar]
- Helgason A, Hrafnkelsson B, Gulcher J, Ward R, Stefansson K (2003) A populationwide coalescent analysis of Icelandic matrilineal and patrilineal genealogies: evidence for a faster evolutionary rate of mtDNA lineages than Y chromosomes. Am J Hum Genet 72:1370–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer E, Puymirat J, Dieltjes P, Bakker E, de Knijff P (1997) Estimating Y chromosome specific microsatellite mutation frequencies using deep rooting pedigrees. Hum Mol Genet 6:799–803 [DOI] [PubMed] [Google Scholar]
- Kayser M, Roewer L, Hedman M, Henke L, Henke J, Brauer S, Kruger C, Krawczak M, Nagy M, Dobosz T, Szibor R, de Knijff P, Stoneking M, Sajantila A (2000) Characteristics and frequency of germline mutations at microsatellite loci from the human Y chromosome, as revealed by direct observation in father/son pairs. Am J Hum Genet 66:1580–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena SDJ, Santos FR, Bianchi NO, Bravi CM, Carnese FR, Rothhammer F, Gerelsaikhan T, Munkhtuja B, Oyunsuren T (1995) A major founder Y-chromosome haplotype in Amerindians. Nat Genet 11:15 [DOI] [PubMed] [Google Scholar]
- Stumpf MP, Goldstein DB (2001) Genealogical and evolutionary inference with the human Y chromosome. Science 291:1738–1742 [DOI] [PubMed] [Google Scholar]
- Su B, Xiao J, Underhill P, Deka R, Zhang W, Akey J, Huang W, Shen D, Lu D, Luo J, Chu J, Tan J, Shen P, Davis R, Cavalli-Sforza L, Chakraborty R, Xiong M, Du R, Oefner P, Chen Z, Jin L (1999) Y-chromosome evidence for a northward migration of modern humans into Eastern Asia during the last Ice Age. Am J Hum Genet 65:1718–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Neel JV, Barrantes R, Schurr TG, Wallace DC (1994) Mitochondrial DNA “clock” for the Amerinds and its implications for timing their entry into North America. Proc Natl Acad Sci USA 91:1158–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay M, Vezina H (2000) New estimates of intergenerational time intervals for the calculation of age and origins of mutations. Am J Hum Genet 66:651–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Jin L, Zemans R, Oefner PJ, Cavalli-Sforza LL (1996) A pre-Columbian Y chromosome-specific transition and its implications for human evolutionary history. Proc Natl Acad Sci USA 93:196–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, Kauffman E, Bonne-Tamir B, Bertranpetit J, Francalacci P, Ibrahim M, Jenkins T, Kidd JR, Mehdi SQ, Seielstad MT, Wells RS, Piazza A, Davis RW, Feldman MW, Cavalli-Sforza LL, Oefner PJ (2000) Y chromosome sequence variation and the history of human populations. Nat Genet 26:358–361 [DOI] [PubMed] [Google Scholar]
- Wells RS, Yuldasheva N, Ruzibakiev R, Underhill PA, Evseeva I, Blue-Smith J, Jin L, et al (2001) The Eurasian heartland: a continental perspective on Y-chromosome diversity. Proc Natl Acad Sci USA 98:10244–10249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Y Chromosome Consortium (2002) A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res 12:339–348 [DOI] [PMC free article] [PubMed] [Google Scholar]