Abstract
Hrs (hepatocyte growth factor-regulated tyrosine kinase substrate) and STAM (signal-transducing adaptor molecule) form a heterodimeric complex that associates with endosomal membranes and is tyrosine-phosphorylated in response to a variety of growth factors including EGF (epidermal growth factor), HGF (hepatocyte growth factor) and PDGF (platelet-derived growth factor). Phosphorylation of the Hrs–STAM complex requires receptor endocytosis. We show that an intact UIM (ubiquitin interaction motif) within Hrs is a conserved requirement for Hrs phosphorylation downstream of both EGF and HGF stimulations. Consistent with this, expression of a dominant-negative form of the E3 ubiquitin ligase, c-Cbl, inhibits EGF- and HGF-dependent Hrs phosphorylation. Despite this conservation, kinase inhibitor profiles using PP1 (4-amino-5-(4-methylphenyl)-7-(t-butyl)pyrazolo[3,4-d]pyrimidine) and SU6656 indicate that distinct non-receptor tyrosine kinases couple EGF, HGF and PDGF stimulation with the tyrosine phosphorylation of the Hrs–STAM complex. Crucially, analysis with phospho-specific antibodies indicates that these kinases generate a signal-specific, combinatorial phosphorylation profile of the Hrs–STAM complex, with the potential of diversifying tyrosine kinase receptor signalling through a common element.
Keywords: growth factor, Hrs, phosphorylation, signal transduction, signal-transducing adaptor molecule (STAM), ubiquitin interaction motif (UIM)
Abbreviations: EGF, epidermal growth factor; EGFR, EGF receptor; GFP, green fluorescent protein; GM-CSF, granulocyte/macrophage colony-stimulating factor; HA, haemagglutinin; HGF, hepatocyte growth factor; Hrs, hepatocyte growth factor-regulated tyrosine kinase substrate; IL-2, interleukin 2; MVB, multivesicular body; NP40, Nonidet P40; PDGF, platelet-derived growth factor; PDGFR, PDGF receptor; PKB, protein kinase B; PP1, 4-amino-5-(4-methylphenyl)-7-(t-butyl)pyrazolo[3,4-d]pyrimidine; STAM, signal-transducing adaptor molecule; UIM, ubiquitin interaction motif
INTRODUCTION
Activated tyrosine kinase receptors are removed from the cell surface by clathrin-coated vesicle-mediated endocytosis and delivered to a tubulovesicular sorting endosome [1]. Tyrosine kinase receptors such as EGFR [EGF (epidermal growth factor) receptor] and c-Met are ubiquitinated after stimulation through recruitment of the E3 ligase, c-Cbl [2,3]. Ubiquitinated receptors are recognized by a UIM (ubiquitin interaction motif)-containing protein, Hrs [HGF (hepatocyte growth factor)-regulated tyrosine kinase substrate], which is highly enriched in regions of the endosome that are characterized by a specialized clathrin coat [4,5]. Hrs is believed to couple the concentration of ubiquitinated receptors with recruitment of an extensive machinery for MVB (multivesicular body) formation [6]. This machinery comprises a set of multiprotein complexes [ESCRT (endosomal-associated complex required for transport) I, II and III], which participate in a vectorial chain of recruitment and association at the prevacuolar membrane necessary for MVB formation [7]. Once incorporated into these luminal vesicles, receptor signalling is terminated and the receptors are usually destined for lysosomal degradation.
Hrs was originally identified as a prominent target for tyrosine phosphorylation after HGF stimulation of the tyrosine kinase receptor c-Met [8]. Activation of other tyrosine kinase receptors and stimulation with cytokines such as IL-2 (interleukin 2) and GM-CSF (granulocyte/macrophage colony-stimulating factor) also result in phosphorylation of Hrs [9]. Both HGF- and EGF-dependent phosphorylation of Hrs requires coincident localization of activated receptor and Hrs at early endosomes, to which Hrs is targeted by interaction of its FYVE domain with PtdIns3P [10,11]. The channelling of such diverse signalling pathways into Hrs phosphorylation has suggested that it may play a conserved role in receptor sorting and/or signalling pathways. Enrichment of phospho-Hrs in the cytosol, despite the fact that phosphorylation occurs at the endosome led to the suggestion that this translocation may reflect a phosphorylation-dependent release mechanism that is important for the progression of receptor incorporation into MVBs [11,12]. However, results of a recent study have shown that Hrs contributes to sorting of a seven-transmembrane G-protein receptor in the absence of any detectable phosphorylation [13]. A role for Hrs phosphorylation in signalling relays is an alternative possibility. Hrs is constitutively associated with STAM (signal-transducing adaptor molecule) [9], which undergoes tyrosine phosphorylation in parallel with Hrs [11]. An SH3 (Src-homology 3) deletion mutant of STAM confers a dominant-negative effect on DNA synthesis mediated by IL-2 and GM-CSF [14].
We have previously confirmed the major EGF-dependent phosphorylation site on Hrs as Tyr334 [12], whilst systematic mass spectroscopic profiling of the EGF-signalling pathway has identified Tyr198 as a prominent phosphorylation site in STAM [15]. In the present study, we have compared the phosphorylation of the Hrs–STAM complex downstream of the acute activation of three tyrosine kinase receptors, which are all known to enter the lysosomal degradation pathway: EGFR, PDGFR [PDGF (platelet-derived growth factor) receptor] and c-Met. These three receptors activate many common signalling pathways, such as mitogen-activated protein kinase and PKB/Akt (where PKB stands for protein kinase B) pathways, yet have distinct physiological effects on cells. For example, in addition to cell growth, c-Met activation uniquely promotes motility and tubule formation of epithelial cells [16]. Our results demonstrate distinct combinations of phosphorylation sites associated with each stimulus, suggesting that Hrs–STAM phosphorylation may be one coding mechanism through which the respective signalling outputs are diversified.
We have also examined the mechanism of Hrs–STAM phosphorylation. Previous work has shown a requirement for a functional endocytic pathway in order to achieve Hrs phosphorylation downstream of HGF and EGF stimulation [10,11]. Furthermore, an intact UIM domain within Hrs is required for EGF-dependent phosphorylation of Hrs [12]. In the present study, we have extended this analysis of the role of the UIM domain to HGF stimulation and in addition show that Cbl E3 ubiquitin ligase activity is required for growth factor-mediated Hrs phosphorylation, suggesting a common ubiquitin-dependent mechanism linking activated receptors to Hrs phosphorylation.
It has previously been suggested that a downstream kinase, such as c-Src, rather than EGFR kinase itself could be responsible for Hrs phosphorylation [17]. We have profiled the diverse phosphorylation events associated with distinct stimuli against the Src-specific tyrosine kinase inhibitor SU6656 [18] and the less-specific tyrosine kinase inhibitor PP1 (4-amino-5-(4-methylphenyl)-7-(t-butyl)pyrazolo[3,4-d]pyrimidine) [19]. We show that the differing phosphorylation outputs are due to the actions of distinct kinase activities, demonstrating that c-Src cannot be a universal intermediary between tyrosine kinase activation and Hrs–STAM phosphorylation.
The present study is one of the first detailed studies of a common node in phosphorylation-dependent signalling networks elicited by multiple growth factor stimulation for which the phosphorylation status of the substrate has been shown to differ. Our results suggest a cautionary approach in extrapolating the results of phospho-proteomic studies that pertain to individual growth factors.
EXPERIMENTAL
Cell culture, plasmids and transfections
HeLa cells and HER14 cells (NIH3T3 fibroblasts stably transfected with the human EGFR [20]) were cultured in 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) foetal calf serum and 1% non-essential amino acids. All tissue culture reagents were purchased from Invitrogen Life Technologies (Paisley, U.K.).
Plasmids comprising the open reading frame of Hrs and Leu269/S270A (Ser270→Ala) Hrs cloned into pEGFPC1 have been described previously [12]. A construct encoding a constitutively active Src (Y527F Src) was a gift from Dr S. Courtneidge (Van Andel Research Institute, Grand Rapids, MI, U.S.A.). Constructs encoding wild-type Cbl and 70Z-Cbl were gifts from Dr Y. Yarden (Weizmann Institute, Rehovet, Israel). HeLa cells were transfected with FuGENE 6 (Roche Diagnostics, Lewes, East Sussex, U.K.) or GeneJuice transfection reagent (Novagen, Merck Biosciences, Nottingham, U.K.) according to the manufacturer's instructions.
Antibodies and other reagents
Anti-Hrs and anti-phospho-Tyr334-Hrs (PY334) rabbit polyclonal antibodies have been described previously [12]. A rabbit polyclonal anti-Stam antibody was generated against the N-terminal peptide MPLFTANPFEQDVEC (Abcam, Cambridge, U.K.). Anti-phospho-Tyr198-Stam (PY198) antibody was generated against the peptide QHTETKSLpYPSSE (CovalAb, Cambridge, U.K.). Affinity-purified anti-GFP (where GFP stands for green fluorescent protein) sheep polyclonal antibody [21] was a gift from Dr F. Barr (Max Planck Institute, Martinsried, Germany). Other antibodies were obtained commercially as follows: anti-HA (where HA stands for haemagglutinin) rabbit polyclonal and mouse monoclonal antibodies (Covance Research Products, Princeton, NJ, U.S.A.), mouse monoclonal anti-EGFR R1, goat polyclonal anti-EGFR 1005, and anti-PDGFRβ rabbit polyclonal antibody (Santa Cruz Biotechnology, CA, U.S.A.), anti-phosphotyrosine monoclonal antibody PY20 (Transduction Laboratories, Lexington, KY, U.S.A.), Alexa Fluor 488- and 594-coupled secondary antibodies (Molecular Probes), horseradish peroxidase-coupled secondary antibodies and the mouse monoclonal anti-ubiquitin antibody P4G7 (Sigma). Protein A–agarose and Protein G–agarose were obtained from Sigma, purified mouse EGF was from Dr J. Smith (Liverpool, U.K.), purified human HGF was from Genentech (San Francisco, CA, U.S.A.) and PDGF-BB was from Calbiochem, Merck Biosciences (Nottingham, U.K.). PP1 was obtained from Alexis (San Diego, CA, U.S.A.) and SU6656 was from SUGEN (San Francisco, U.S.A.).
Stimulation of cells with growth factors
Cells were starved for 16 h in serum-free medium, preincubated with kinase inhibitors where indicated (PP1 at 2.5 μM for 15 min, SU6656 at 2 μM for 1 h) and stimulated with EGF (100 ng/ml), HGF (250 ng/ml) and PDGF (30 ng/ml) for 8 min at 37 °C. The cells were washed twice with ice-cold PBS and lysed for 10 min on ice in either NP40 (Nonidet P40) lysis buffer (25 mM Tris/HCl, pH 7.5, 100 mM NaCl, 0.5%, w/v, NP40 and 50 mM NaF) or NP40-RIPA buffer (10 mM Tris/HCl, pH 7.5, 100 mM NaCl, 1%, w/v, NP40, 0.1%, w/v, SDS, 1%, w/v, sodium deoxycholate and 50 mM NaF), supplemented in each case with mammalian protease inhibitor cocktail and phosphatase inhibitor cocktail II (Sigma). The lysates were precleared by centrifugation and immunoprecipitations were set up with antibodies and Protein A–agarose or Protein G–agarose for 4 h at 4 °C. For immunoprecipitation of ubiquitinated EGFR and PDGFR, 10 mM N-ethylmaleimide was added to the lysis buffer to inhibit protein deubiquitination. Immunoprecipitated proteins were analysed as described in [12]. Blots were routinely stripped and reprobed to ensure that equal amounts of protein had been loaded.
Immunofluorescence
Cells were washed twice with PBS and fixed either with methanol or with 3% (w/v) paraformaldehyde. For methanol fixation, cells were incubated with methanol at −20 °C for 5 min followed by drying for 8 min and rehydration in PBS. Alternatively, cells were processed as described in [12]. Cells were viewed using a Bio-Rad LaserSharp confocal microscope. Single confocal sections were taken at 260 nm steps and analysed with the accompanying software.
RESULTS
A common requirement for a UIM domain in Hrs for EGF- and HGF-mediated tyrosine phosphorylation
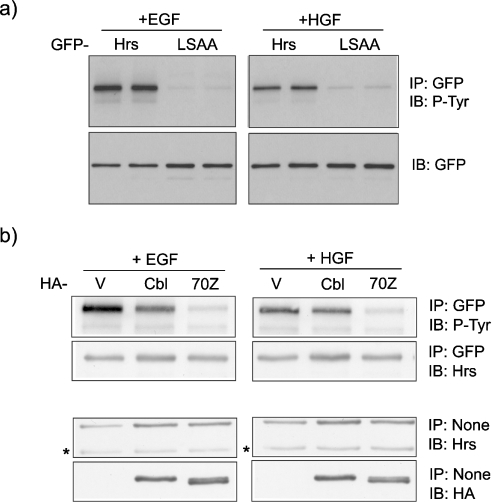
We have previously shown that Hrs phosphorylation in response to EGF is dependent on an intact UIM domain, which is proposed to mediate interaction with the ubiquitinated EGFR [12]. We now show that this requirement is conserved for phosphorylation of Hrs in response to HGF stimulation of the receptor tyrosine kinase c-Met (Figure 1a). GFP–Hrs or a mutant, containing two amino acid changes within the UIM domain (LSAA), which ablate ubiquitin binding activity [12], were expressed in HeLa cells. After stimulation of cells with EGF or HGF, the wild-type and mutant Hrs were immunoprecipitated and probed with an anti-phosphotyrosine antibody (PY20). Wild-type Hrs was phosphorylated in response to both growth factors, whereas this response was greatly reduced for the UIM domain mutant.
Figure 1. A common requirement for a UIM in Hrs for EGF- and HGF-mediated Hrs tyrosine phosphorylation.
(a) HeLa cells were transfected with GFP–Hrs or the Hrs–UIM mutant GFP–LSAA and stimulated 22 h post-transfection with EGF (100 ng/ml) or HGF (250 ng/ml) after a 16 h starvation period. GFP–Hrs was immunoprecipitated and tyrosine phosphorylation was assessed by immunoblotting with PY20 antibody (P-Tyr). Lysates prepared as described above were analysed by immunoblotting with anti-GFP antibody. (b) An oncogenic Cbl mutant interferes with Hrs phosphorylation. HeLa cells were transfected with GFP–Hrs and pCDNA3.1 control vector (V), HA-tagged c-Cbl or 70Z-Cbl and then stimulated 22 h post-transfection with EGF or HGF after a 16 h starvation period. GFP–Hrs was immunoprecipitated and samples were analysed by immunoblotting with P-Tyr and reprobed with anti-Hrs antibody. Lysate samples prepared as described above were analysed by immunoblotting with anti-Hrs or anti-HA antibody to assess relative expression levels (note that the band indicated by * corresponds to endogenous Hrs).
An oncogenic Cbl mutant interferes with Hrs phosphorylation
We have previously shown that Hrs phosphorylation in response to EGF and HGF is dependent on clathrin-mediated endocytosis and PtdIns3P-dependent localization of Hrs to endosomes [10,11]. Hrs has been proposed to recruit ubiquitinated receptors to the endosomal clathrin coat before their inclusion into the internal vesicles of the MVB [5,6,22]. If this interaction is necessary for Hrs phosphorylation, as suggested by the requirement for an intact UIM domain, then interference with receptor ubiquitination should also oppose Hrs phosphorylation.
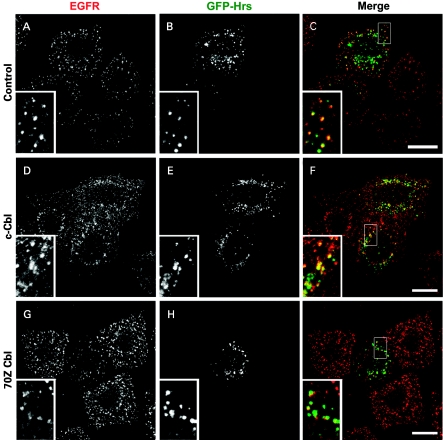
In common with several tyrosine kinases, EGFR and Met are both ubiquitinated by the E3-RING ligase, Cbl [2,3,23,24]. In order to interfere with receptor ubiquitination, we have co-expressed c-Cbl or a RING-deletion mutant 70Z-Cbl, which has been shown to lack E3-ligase activity [2] together with GFP–Hrs. Expression of 70Z-Cbl, but not c-Cbl, strongly suppressed both HGF- and EGF-mediated phosphorylation of Hrs (Figure 1b). Whilst the 70Z-Cbl mutant has previously been reported to reduce the rate of EGFR internalization [25], we clearly observe a high degree of coincident localization of GFP–Hrs and EGFR in 70Z-Cbl co-transfected cells after 8 min of EGF stimulation (Figure 2), and hence the moderate suppression of EGFR entry into Hrs-containing endosomes by 70Z-Cbl on its own is unlikely to cause the striking inhibition of GFP–Hrs phosphorylation observed in Figure 1(b).
Figure 2. Co-localization of EGFR and GFP–Hrs in cells expressing c-Cbl and 70Z-Cbl mutant.
HeLa cells were seeded on to coverslips and transfected with GFP–Hrs and pCDNA3 (control), HA-tagged c-Cbl or 70Z-Cbl and then stimulated for 8 min with 100 ng/ml EGF 22 h post-transfection after a 16 h starvation period. The cells were fixed with 3% paraformaldehyde, permeabilized with Triton X-100 and stained with anti-EGFR (EGFR shown in red) followed by a secondary antibody coupled with Alexa Fluor 594. All panels show a single confocal section. Scale bars, 20 μm.
Src-kinase inhibitor profiles suggest distinct mechanisms of Hrs and STAM phosphorylation
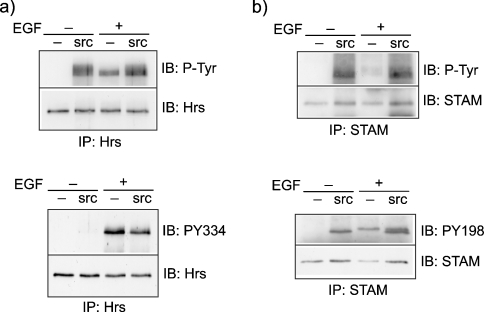
Hrs and STAM are both phosphorylated in response to a variety of growth signals including both receptor tyrosine kinase ligands and cytokines. A common denominator in these diverse signalling cascades is the activation of members of the Src-kinase family. A previous study has shown that Hrs can be phosphorylated by c-Src in vitro but is, nevertheless, phosphorylated in EGFR-transfected Src-Yes-Fyn-knockout cells [17]. We have previously characterized a phospho-specific Hrs antibody recognizing phospho-Tyr334, which is generated after EGF stimulation [12]. Whereas Hrs is clearly phosphorylated in non-stimulated cells transfected with constitutively active Src, as judged by blotting with an anti-phosphotyrosine antibody (PY20), Tyr334 is not a target for Src (Figure 3a). We have also generated a phospho-specific antibody against phospho-Tyr198 in STAM that has been identified by mass spectroscopic analysis of EGF-evoked phosphorylation of proteins [15]. Parallel analysis of the phosphorylation state of STAM in cells transfected with constitutively active Src (Y527F) versus EGF-stimulated cells shows that Tyr198 can be phosphorylated by Src in addition to other uncharacterized tyrosine residues (Figure 3b).
Figure 3. Comparison of tyrosine phosphorylation of Hrs and STAM in response to EGF and constitutively active Src.
HeLa cells were transfected with constitutively active Y527F-Src (src) or pCDNA3.1 (–), starved for 16 h in serum-free medium and then stimulated 22 h post-transfection for 8 min with EGF (100 ng/ml). (a) Lysates were subjected to immunoprecipitation with anti-Hrs antibody and analysed by immunoblotting (IB) with either PY20 antibody (P-Tyr) or anti-PY334-Hrs antibody. The blots were stripped and reprobed with anti-Hrs antibody. (b) Lysates were subjected to immunoprecipitation with anti-STAM antibody and analysed by immunoblotting with either P-Tyr or anti-PY198-STAM antibody. The blots were stripped and reprobed with anti-STAM antibody.
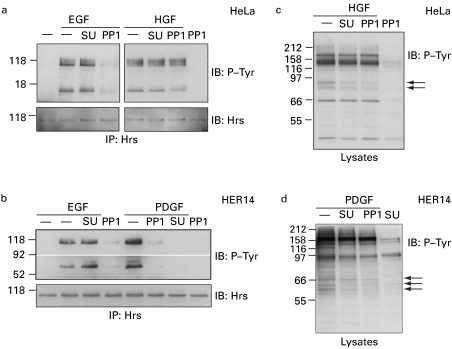
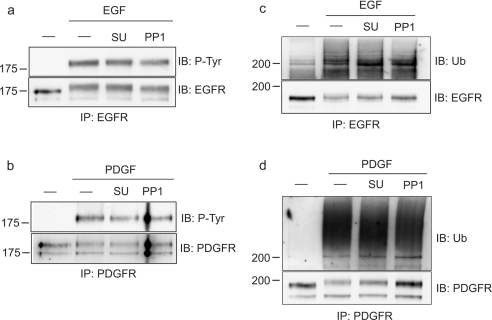
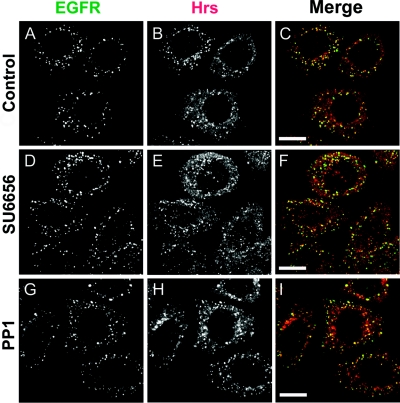
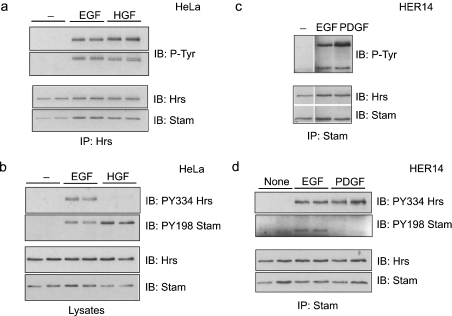
To analyse further the involvement of Src kinases in the phosphorylation of the Hrs–STAM complex in response to various signals, we made use of two distinct kinase inhibitors with high and moderate specificities for Src-kinase family members, SU6656 and PP1 [18,19]. First, we compared EGF- and HGF-mediated phosphorylation in HeLa cells and found that EGF, but not HGF-dependent phosphorylation of both Hrs and STAM, is sensitive to PP1, whereas neither was inhibited by SU6656 (Figure 4a). Secondly, we made use of HER14 cells, which are NIH3T3 cells that are stably transfected with EGFR [20], thus enabling us to compare directly EGF- with PDGF-mediated phosphorylation of Hrs and STAM, since PDGFR is not expressed in HeLa cells. In contrast with both EGF- and HGF-mediated phosphorylation of Hrs and STAM, we found that PDGF-mediated phosphorylation is inhibited by both Src-kinase inhibitors PP1 and SU6656, whereas the EGF-dependent phosphorylation showed an inhibitor profile identical with that observed in HeLa cells (Figure 4b and Table 1). As reported previously [18,19], both inhibitors generate discernible differences in the total cellular phospho-tyrosine profile (Figures 4c and 4d). Importantly, at the concentrations we have used, neither inhibitor significantly affects EGFR or PDGFR phosphorylation and ubiquitination (Figure 5), nor do they inhibit endocytosis of EGFR to an Hrs-positive endosomal compartment (Figure 6).
Figure 4. Differential effect of Src-kinase inhibitors on EGF-, HGF- and PDGF-stimulated phosphorylation of Hrs and STAM.
(a) HeLa cells or (b) HER14 cells were serum-starved for 16 h, preincubated either with 2 μM SU6656 (SU) or vehicle for 1 h or with 2.5 μM PP1 for 15 min, and stimulated with EGF (100 ng/ml), HGF (250 ng/ml) or PDGF (30 ng/ml) for 8 min at 37 °C or left unstimulated. Cell lysates were subjected to immunoprecipitation with anti-Hrs antibody and tyrosine phosphorylation was assessed by immunoblotting with PY20 antibody (P-Tyr, top panels). Note that the upper band corresponds to phospho-Hrs and the lower band to co-precipitated phospho-STAM. The blots were stripped and reprobed with anti-Hrs antibody (bottom panels). (c, d) Lysates, prepared from cells treated as in (a, b), were directly analysed by immunoblotting with P-Tyr antibody and they displayed altered tyrosine phosphorylation profiles (arrows).
Table 1. Distinct Src-kinase inhibitor profiles for Hrs–STAM phosphorylation in response to different growth factors.
| Phosphorylation of Hrs–STAM inhibited? | ||
|---|---|---|
| Growth factor | PP1 | SU6656 |
| EGF | Yes | No |
| HGF | No | No |
| PDGF | Yes | Yes |
Figure 5. Phosphorylation and ubiquitination of EGFR and PDGFRs in the presence of Src-kinase inhibitors.
HER14 cells were serum-starved for 16 h, preincubated either with 2 μM SU6656 or vehicle for 1 h or with 2.5 μM PP1 for 15 min and stimulated with (a, c) 100 ng/ml EGF or (b, d) 30 ng/ml PDGF, for 8 min at 37 °C. Cell lysates were subjected to immunoprecipitation with (a, c) anti-EGFR or (b, d) anti-PDGFR antibody. Phosphorylation and ubiquitination were assessed with PY20 antibody (P-Tyr) and anti-ubiquitin antibody respectively. The blots were reprobed with anti-EGFR or anti-PDGFR antibody (bottom panels).
Figure 6. Co-localization of internalized EGFR with Hrs in the presence of Src-kinase inhibitors.
HeLa cells were seeded on to coverslips, serum-starved for 16 h, preincubated either with 2 μM SU6656 or vehicle for 1 h or with 2.5 μM PP1 for 15 min, and stimulated with 100 ng/ml EGF for 8 min at 37 °C. The cells were fixed with methanol and stained with anti-EGFR (EGFR, shown in green) and anti-Hrs antibody (Hrs, shown in red) followed by secondary antibodies coupled with Alexa Fluor 488 and Alexa Fluor 594 respectively. All panels show a single confocal section. Scale bars, 20 μm.
Generation of distinct combinatorial tyrosine phosphorylation patterns in response to distinct signal inputs
Since Hrs–STAM phosphorylation displays different inhibitor profiles according to stimulus, we next asked whether the phosphorylation sites are conserved. To address this question, we made use of our phospho-specific Hrs and STAM antibodies (see above) and compared the total phospho-tyrosine signals in Hrs immunoprecipitates with specific phospho-Tyr334 and -Tyr198 signals in response to either EGF or HGF stimulation in HeLa cells. Whereas phosphorylation of STAM on Tyr198 can be observed in both EGF- and HGF-stimulated cells, Hrs is clearly phosphorylated on alternative tyrosine residues after HGF stimulation (Figures 7a and 7b). Turning to HER14 cells in order to analyse PDGF-mediated phosphorylation, PDGF, in common with EGF, elicits phosphorylation of Tyr334 in Hrs, but, in contrast with EGF, does not elicit a substantial signal for phospho-Tyr198 in STAM (Figures 7c and 7d). Thus each growth factor stimulus generates a distinct phosphorylation profile on the Hrs–STAM complex as summarized in Table 2.
Figure 7. Generation of distinct combinatorial tyrosine phosphorylation patterns in response to distinct signal inputs.
(a, b) HeLa cells were starved for 16 h in serum-free medium and stimulated for 8 min with EGF (100 ng/ml) or HGF (250 ng/ml). (a) Lysates were subjected to immunoprecipitation with anti-Hrs antibody and tyrosine phosphorylation was assessed by immunoblotting with (P-Tyr; top panels). Blots were stripped and reprobed with anti-Hrs or anti-Stam antibody (bottom panels). (b) Lysates were analysed by immunoblotting with anti-PY334-Hrs or anti-PY198-STAM antibody and blots were stripped and reprobed as in (a). (c, d) HER14 cells were serum-starved for 16 h and either not stimulated or stimulated for 8 min with EGF (100 ng/ml) or PDGF (30 ng/ml). (c) Lysates were subjected to immunoprecipitation with anti-STAM antibody and immunoblotted with P-Tyr (top panels). Blots were reprobed with anti-Hrs or anti-STAM antibody (bottom panels). (d) Lysates were subjected to immunoprecipitation with anti-STAM antibody and subjected to immunoblotting with anti-PY334-Hrs or anti-PY198-STAM antibody. Blots were stripped and reprobed as in (a).
Table 2. Differential tyrosine phosphorylation of the Hrs–STAM complex in response to different growth factors.
| Phosphorylation? | ||||
|---|---|---|---|---|
| Protein phosphorylated | Residue | EGF | HGF | PDGF |
| Hrs | Tyr334 | Yes | No | Yes |
| STAM | Tyr198 | Yes | Yes | No |
DISCUSSION
Tyrosine kinase receptors channel signals through shared pathways but generate distinct biological outputs. This may be due to cell-type restriction of responses or to the specific combination of signals produced by receptor activation [26,27]. It has recently become well appreciated that receptor trafficking dynamics can influence the signal output [28,29]. Phosphorylation of the endosome-restricted substrate, Hrs, is a clear-cut example of this principle. Our previous results have suggested that coincident endosomal localization of the receptor (EGFR or Met) and Hrs, as well as correct sorting of that receptor within the endosome, are a prerequisite for Hrs phosphorylation [10,11]. Hrs is a key player in the sorting mechanism through recognition of ubiquitinated receptors by its UIM domain. Overexpression of Hrs has a dominant-negative effect on EGFR sorting to MVBs by inhibiting internal vesicle formation [12]. This inhibitory effect of Hrs overexpression does not require phosphorylation of Tyr334 and is mediated by the UIM domain, which effectively couples receptor sorting to internal vesicle formation. Results of the present study suggest that Hrs phosphorylation may be contingent upon direct interaction between ubiquitinated receptors and Hrs, since disruption of the UIM domain inhibits phosphorylation downstream of either HGF or EGF.
The corollary to UIM domain mutation experiments is the inhibition of receptor ubiquitination, which can be effected by expression of dominant-negative mutants of the E3 ligase, c-Cbl [23]. Our finding that the 70Z-mutated form of c-Cbl inhibits Hrs phosphorylation is consistent with the above interpretation. However, we should acknowledge the caveat that there is some reduction in receptor endocytosis after the expression of this mutant [25], although this does not appear to be as acute.
If a requirement for direct receptor interaction is conserved between HGF- and EGF-dependent phosphorylation of Hrs, the simplest model is that the receptor tyrosine kinase activity is directly responsible for Hrs phosphorylation. Although this may yet be true in the specific case of Met receptor, it cannot be true for EGFR or PDGFR. This is shown by our finding that pharmacological treatment with particular tyrosine kinase inhibitors can diminish growth factor-dependent Hrs phosphorylation without affecting the receptor tyrosine kinase activity (Figures 4 and 5). Our results also show that there is no universal tyrosine kinase adapter between tyrosine kinase receptors and Hrs. In fact, the inhibitor profile indicates that distinct kinases mediate Hrs phosphorylation downstream of all the three specific signals. For PDGF stimulation, the relevant kinase could conceivably be an Src-family member since it is inhibited by both PP1 and SU6656 at concentrations for which each has optimal specificity (but see below). PP1 has previously been reported to inhibit PDGF receptor tyrosine phosphorylation [30]. We do not observe a significant effect of PP1 on PDGFR phosphorylation in our cells with the short incubation times that we used. The fact that the more specific inhibitor SU6656 equally inhibits Hrs phosphorylation makes it unlikely that this inhibition is due to an upstream effect on PDGFR. c-Src cannot mediate either HGF- or EGF-dependent Hrs phosphorylation since this is either insensitive to both inhibitors (HGF) or the more specific SU6656 inhibitor (EGF). Note that, at the concentrations of inhibitors we have used, we do not see any defect in receptor trafficking or ubiquitination. This is in contrast with previous studies, which have used an order of magnitude higher concentrations of PP1, which is capable of inhibiting EGFR ubiquitination [31].
Although rigorous comparison of stimulus-dependent phospho-proteomes is in its infancy, one might expect that highly conserved elements would also show conservation of phosphorylation profiles. This is certainly true in some cases where phosphorylation provides a very specific switch, which governs enzyme activity (e.g. PKB). Alternatively, phosphorylation could also be used as a means of diversifying signals by providing distinct recruitment sites for downstream signalling elements to shared adapter proteins.
We have shown that stimulus-specific tyrosine kinases mediate Hrs phosphorylation. Would this necessarily lead to an altered phosphorylation profile of the Hrs–STAM complex? For EGF stimulation, we and others have characterized Tyr334 on Hrs and Tyr198 on STAM as principal sites for phosphorylation [12,15]. We have raised phospho-specific antibodies that specifically recognize these modifications. Application of these antibodies to the examination of stimulus-dependent phosphorylation revealed that each stimulus produces a unique combination of phosphotyrosines on the Hrs–STAM complex, although individual modifications may be shared. For example, both EGF stimulation and HGF stimulation result in STAM phosphorylation at Tyr198, whereas HGF promotes the phosphorylation of an as yet unidentified tyrosine distinct from Tyr334 on Hrs (summarized in Table 2). Tyr334 is, however, phosphorylated in response to PDGF. Although inhibitor studies were suggestive that c-Src might be the relevant kinase downstream of PDGF, we do not find any Tyr334 phosphorylation when we express a constitutively active form of Src, although it efficiently phosphorylates Hrs at (an) alternative site(s). This suggests that the relevant kinase may be an alternative member of the Src family.
Although we have not directly addressed the specific cellular role of Hrs–STAM phosphorylation in the present study, our results clearly show that it cannot be a general requirement for Hrs-dependent receptor sorting. We favour the idea that it provides a way to couple the sorting of specific receptors with other downstream functions that the complex may engage in. The fact that a significant pool of the phosphorylated form can be found in the cytosol [12], as well as the wealth of interactions detected by two-hybrid screening in various model studies, suggests that Hrs–STAM function is not confined to receptor sorting at the endosome.
It is of great interest to understand how distinct biological outcomes are determined by the activation of specific tyrosine kinase receptors, despite the apparent engagement of common signalling pathways. The differential phosphorylation profile of the Hrs–STAM complex, described in this case study, provides a potential novel mechanism for diversifying signals through a common node in tyrosine kinase receptor signalling pathways. It will be a challenge for future phospho-proteomic studies to determine how widely this principal has been adopted.
Acknowledgments
We thank the Wellcome Trust for support and S. Courtneidge (Van Andel Research Institute, Grand Rapids, MI, U.S.A.) for helpful discussions.
References
- 1.Clague M. J. Molecular aspects of the endocytic pathway. Biochem. J. 1998;336:271–282. doi: 10.1042/bj3360271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levkowitz G., Waterman H., Zamir E., Kam Z., Oved S., Langdon W. Y., Beguinot L., Geiger B., Yarden Y. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998;12:3663–3674. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peschard P., Fournier T. M., Lamorte L., Naujokas M. A., Band H., Langdon W. Y., Park M. Mutation of the c-Cbl TKB domain binding site on the Met receptor tyrosine kinase converts it into a transforming protein. Mol. Cell. 2001;8:995–1004. doi: 10.1016/s1097-2765(01)00378-1. [DOI] [PubMed] [Google Scholar]
- 4.Raiborg C., Bache K. G., Gillooly D. J., Madshus I. H., Stang E., Stenmark H. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat. Cell Biol. 2002;4:394–398. doi: 10.1038/ncb791. [DOI] [PubMed] [Google Scholar]
- 5.Sachse M., Urbé S., Oorschot V., Strous G. J., Klumperman J. Bilayered clathrin coats on endosomal vacuoles are involved in protein sorting toward lysosomes. Mol. Biol. Cell. 2002;13:1313–1328. doi: 10.1091/mbc.01-10-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clague M. J., Urbe S. Hrs function: viruses provide the clue. Trends Cell Biol. 2003;13:603–606. doi: 10.1016/j.tcb.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Katzmann D. J., Odorizzi G., Emr S. D. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- 8.Komada M., Kitamura N. Growth-factor-induced tyrosine phosphorylation of hrs, a novel 115-kilodalton protein with a structurally conserved putative zinc finger domain. Mol. Cell. Biol. 1995;15:6213–6221. doi: 10.1128/mcb.15.11.6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asao H., Sasaki Y., Arita T., Tanaka N., Endo K., Kasai H., Takeshita T., Endo Y., Fujita T., Sugamura K. Hrs is associated with STAM, a signal-transducing adaptor molecule. J. Biol. Chem. 1997;272:32785–32791. doi: 10.1074/jbc.272.52.32785. [DOI] [PubMed] [Google Scholar]
- 10.Hammond D. E., Carter S., McCullough J., Urbe S., Vande Woude G., Clague M. J. Endosomal dynamics of met determine signaling output. Mol. Biol. Cell. 2003;14:1346–1354. doi: 10.1091/mbc.E02-09-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Urbé S., Mills I. G., Stenmark H., Kitamura N., Clague M. J. Endosomal localization and receptor dynamics determine tyrosine phosphorylation of hepatocyte growth factor-regulated tyrosine kinase substrate. Mol. Cell. Biol. 2000;20:7685–7692. doi: 10.1128/mcb.20.20.7685-7692.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Urbé S., Sachse M., Row P. E., Preisinger C., Barr F. A., Strous G., Klumperman J., Clague M. J. The UIM domain of Hrs couples receptor sorting to vesicle formation. J. Cell Sci. 2003;116:4169–4179. doi: 10.1242/jcs.00723. [DOI] [PubMed] [Google Scholar]
- 13.Marchese A., Raiborg C., Santini F., Keen J. H., Stenmark H., Benovic J. L. The E3 ubiquitin ligase AIP4 mediates ubiquitination and sorting of the G protein-coupled receptor CXCR4. Dev. Cell. 2003;5:709–722. doi: 10.1016/s1534-5807(03)00321-6. [DOI] [PubMed] [Google Scholar]
- 14.Takeshita T., Arita T., Higuchi M., Asao H., Endo K., Kuroda H., Tanaka N., Murata K., Ishii N., Sugamura K. STAM, signal transducing adaptor molecule, is associated with Janus kinases and involved in signaling for cell growth and c-myc induction. Immunity. 1997;6:449–457. doi: 10.1016/s1074-7613(00)80288-5. [DOI] [PubMed] [Google Scholar]
- 15.Steen H., Kuster B., Fernandez M., Pandey A., Mann M. Tyrosine phosphorylation mapping of the epidermal growth factor receptor signaling pathway. J. Biol. Chem. 2002;277:1031–1039. doi: 10.1074/jbc.M109992200. [DOI] [PubMed] [Google Scholar]
- 16.Birchmeier C., Birchmeier W., Gherardi E., Vande Woude G. F. Met, metastisis, motility and more. Nat. Rev. Mol. Cell Biol. 2003;4:915–925. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- 17.Bache K. G., Raiborg C., Mehlum A., Madshus I. H., Stenmark H. Phosphorylation of Hrs downstream of the epidermal growth factor receptor. Eur. J. Biochem. 2002;269:3881–3887. doi: 10.1046/j.1432-1033.2002.03046.x. [DOI] [PubMed] [Google Scholar]
- 18.Blake R. A., Broome M. A., Liu X., Wu J., Gishizky M., Sun L., Courtneidge S. A. SU6656, a selective src family kinase inhibitor, used to probe growth factor signaling. Mol. Cell. Biol. 2000;20:9018–9027. doi: 10.1128/mcb.20.23.9018-9027.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanke J. H., Gardner J. P., Dow R. L., Changelian P. S., Brissette W. H., Weringer E. J., Pollok B. A., Connelly P. A. Discovery of a novel, potent, and Src family-selective tyrosine kinase inhibitor. Study of Lck- and FynT-dependent T cell activation. J. Biol. Chem. 1996;271:695–701. doi: 10.1074/jbc.271.2.695. [DOI] [PubMed] [Google Scholar]
- 20.Honegger A. M., Szapary D., Schmidt A., Lyall R., Van Obberghen E., Dull T. J., Ullrich A., Schlessinger J. A mutant epidermal growth factor receptor with defective protein tyrosine kinase is unable to stimulate proto-oncogene expression and DNA synthesis. Mol. Cell. Biol. 1987;7:4568–4571. doi: 10.1128/mcb.7.12.4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barr F. A., Preisinger C., Kopajtich R., Korner R. Golgi matrix proteins interact with p24 cargo receptors and aid their efficient retention in the Golgi apparatus. J. Cell Biol. 2001;155:885–891. doi: 10.1083/jcb.200108102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clague M. J. Membrane transport: a coat for ubiquitin. Curr. Biol. 2002;12:R529–R531. doi: 10.1016/s0960-9822(02)01030-8. [DOI] [PubMed] [Google Scholar]
- 23.Thien C. B. F., Langdon W. Y. Cbl: many adaptations to regulate protein tyrosine kinases. Nat. Rev. 2001;2:294–307. doi: 10.1038/35067100. [DOI] [PubMed] [Google Scholar]
- 24.Peschard P., Park M. Escape from Cbl-mediated downregulation: a recurrent theme for oncogenic deregulation of receptor tyrosine kinases. Cancer Cell. 2003;3:519–523. doi: 10.1016/s1535-6108(03)00136-3. [DOI] [PubMed] [Google Scholar]
- 25.Jiang X., Sorkin A. Epidermal growth factor receptor internalization through clathrin-coated pits requires Cbl RING finger and proline-rich domains but not receptor polyubiquitylation. Traffic. 2003;4:529–543. doi: 10.1034/j.1600-0854.2003.t01-1-00109.x. [DOI] [PubMed] [Google Scholar]
- 26.Hunter T. Signaling – 2000 and beyond. Cell (Cambridge, Mass.) 2000;100:113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 27.Tan P. B., Kim S. K. Signaling specificity: the RTK/RAS/MAP kinase pathway in metazoans. Trends Genet. 1999;15:145–149. doi: 10.1016/s0168-9525(99)01694-7. [DOI] [PubMed] [Google Scholar]
- 28.Sorkin A., von Zastrow M. Signal transduction and endocytosis; close encounters of many kinds. Nat. Rev. Mol. Cell Biol. 2002;3:600–614. doi: 10.1038/nrm883. [DOI] [PubMed] [Google Scholar]
- 29.Clague M. J., Urbe S. The interface of receptor trafficking and signalling. J. Cell Sci. 2001;114:3075–3081. doi: 10.1242/jcs.114.17.3075. [DOI] [PubMed] [Google Scholar]
- 30.Waltenberger J., Uecker A., Kroll J., Frank H., Mayr U., Bjorge J. D., Fujita D., Gazit A., Hombach V., Levitzki A., et al. A dual inhibitor of platelet-derived growth factor beta-receptor and Src kinase activity potently interferes with motogenic and mitogenic responses to PDGF in vascular smooth muscle cells. A novel candidate for prevention of vascular remodeling. Circ. Res. 1999;85:12–22. doi: 10.1161/01.res.85.1.12. [DOI] [PubMed] [Google Scholar]
- 31.Kassenbrock C. K., Hunter S., Garl P., Johnson G. L., Anderson S. M. Inhibition of Src family kinases blocks epidermal growth factor (EGF)-induced activation of Akt, phosphorylation of c-Cbl, and ubiquitination of the EGF receptor. J. Biol. Chem. 2002;277:24967–24975. doi: 10.1074/jbc.M201026200. [DOI] [PubMed] [Google Scholar]