Abstract
CaMCs (calcium-dependent mitochondrial carriers) represent a novel subfamily of metabolite carriers of mitochondria. The ATP-Mg/Pi co-transporter, functionally characterized more than 20 years ago, has been identified to be a CaMC member. There are three isoforms of the ATP-Mg/Pi carrier in mammals, SCaMC-1 (short CaMC-1), -2 and -3 (or APC-1, -3 and -2 respectively), corresponding to the genes SLC25A24, SLC25A25 and SLC25A23 respectively, as well as six N-terminal variants generated by alternative splicing for SCaMC-1 and -2 isoforms. In the present study, we describe four new variants of human SCaMC-3 generated by alternative splicing. The new mRNAs use the exon 9 3′-donor site and distinct 5′-acceptor sites from repetitive elements, in regions downstream of exon 10, the last exon in all SCaMCs. Transcripts lacking exon 10 (SCaMC-3b, -3b′, -3c and -3d) code for shortened proteins lacking the last transmembrane domain of 422, 456 and 435 amino acids, and were found in human tissues and HEK-293T cells. Mitochondrial targeting of overexpressed SCaMC-3 variants is incomplete. Surprisingly, the import impairment is overcome by removing the N-terminal extension of these proteins, suggesting that the hydrophilic N-terminal domain also participates in the mitochondrial import process, as shown for the CaMC members aralar and citrin [Roesch, Hynds, Varga, Tranebjaerg and Koehler (2004) Hum. Mol. Genet. 13, 2101–2111].
Keywords: Alu repeat, ATP-Mg/Pi carrier, calcium-dependent mitochondrial carrier (CaMC), mitochondrial import, spliced variant, transposable element
Abbreviations: AGC, aspartate/glutamate carrier; CaMC, calcium-dependent mitochondrial carrier; SCaMC, short CaMC; EST, expressed sequence tag; HEK-293T cell, human embryonic kidney 293T cell; LTR, long terminal repeat; MaLR, mammalian apparent LTR-retrotransposon; MC, mitochondrial carrier; MCF, MC family; NT, N-terminal; RT, reverse transcriptase
INTRODUCTION
Transport of metabolites across the inner mitochondrial membrane is performed by structurally related proteins belonging to the MCF (mitochondrial carrier family; reviewed in [1–3]). MCF proteins are made up of a 3-fold repeated module of 100 amino acids containing two transmembrane domains and the characteristic mitochondrial signature [4,5]. Their common features have been successfully used to identify a large number of MCF members in the last few years [6–12].
CaMCs (calcium-dependent mitochondrial carriers) are a subgroup of MCs (mitochondrial carriers) having a bipartite structure, a C-terminal-half homologous with MCF and a long N-terminal extension harbouring EF-hand calcium-binding motifs facing the intermembrane space [13–17]. Human CaMCs include the AGC (aspartate/glutamate carrier) isoforms, Aralar1 and citrin [16], and the SCaMCs (short CaMCs), which are present in three isoforms [17], and correspond to the isoforms of the ATP-Mg/Pi carrier [18]. This transporter was functionally identified 20 years ago [19] and plays a role in the net transport of adenine nucleotides across the inner mitochondrial membrane [20,21]. The transport activity of the AGCs is regulated by cytosolic calcium [16,22], and the ATP-Mg/Pi carrier is also stimulated by extramitochondrial calcium [23]. Therefore the CaMCs provide a new mechanism to transduce calcium signals to mitochondria.
The mechanisms of import of MC proteins are now beginning to be understood. After translocation across the outer mitochondrial membrane, MCs and import components utilize the TIM22 (where TIM stands for translocase of the inner membrane) import pathway in the inner mitochondrial membrane to reach their final destinations [24]. This pathway contains components in the inner mitochondrial membrane, as well as components in the intermembrane space, the small Tims, which act as chaperone-like molecules to guide the hydrophobic precursors [24]. Most MC precursors interact with the Tim9p–Tim10p complex of the intermembrane space [25]. However, the AGCs aralar and citrin, and Tim23p, an import protein of the inner mitochondrial mem-brane, interact with the Tim8p–Tim13p complex [24,26,27]. Both Tim23p and the AGCs have large N-terminal domains in the intermembrane space, which participate in the interaction with Tim8p–Tim13p [28,29]. Mutations in the locus for human Tim8p (DDP1/TIMM8a locus) cause the Mohr–Tranebjaerg syndrome (MTS/DFN-1, deafness/dystonia syndrome) and cause neuronal defects in the mitochondrial import of aralar. Impaired aralar activity, as a malate–aspartate NADH shuttle member, could contribute to the pathology of MTS [29].
Although splicing is a rare event in MCF [3], mammalian SCaMC-1 and -2 genes show a number of N-terminal variants due to alternative promoter use [17,18]. We have now found new C-terminal variants for the human SCaMC-3 isoform generated by alternative splicing. The new variants include repetitive transposable elements, Alu and a member of the MaLR (mammalian apparent LTR-retrotransposon) family of non-autonomous retrotransposons [30,31], inserted downstream of the last SCaMC-3 exon, making human SCaMCs one of the most complex subgroups of MCF genes. Subcellular and tissue distribution of the new SCaMC-3 variants have been investigated, and their functional significance discussed. Interestingly, the results indicate that the N-terminal extension of SCaMC-3 participates in mitochondrial import, suggesting that these carriers may utilize the Tim8p–Tim13p pathway in the intermembrane space for assembly in the inner mitochondrial membrane.
MATERIALS AND METHODS
Sequence search and analysis
Initial human SCaMC-3 sequence analysis was performed with the EST (expressed sequence tag) clone AI073878 and a previously reported human SCaMC-3 sequence (GenBank® accession no. AJ619988) using web-based programs of the National Center for Biotechnology Information available at http://www.ncbi.nlm.nih.gov. 3′-Variants were analysed by multiple sequence alignment performed using the ClustalW program of the European Bioinformatics Institute (http://www.ebi.ac.uk). Intron–exon boundaries were assigned by comparing cDNA sequences with the genomic BAC clones AC011539 and AC010503 deposited in GenBank® using BLAST programs. Analysis of repetitive sequences was performed in Repbase Uptake (http://www.girinst.org). Prediction of transmembrane regions and protein orientation were performed with the Tmpred program (http://www.ch.embnet.org).
RNA isolation and RT (reverse transcriptase)–PCR analysis
The SCaMC-3 isoforms were confirmed by RT–PCR in HEK-293T cells (human embryonic kidney 293T cells). Total RNA was isolated by standard methods. The cDNA negative strand was synthesized from 3 μg of total RNA using the RT from avian myeloblastosis virus (Promega, Charbonnières, France) and oligo(dT) (Roche, Indianapolis, IN, U.S.A.) as primer. The PCR was performed using the following primers: sense, 9 5′-AGCAGTACAGCCACGACCG-3′ and antisense, 10a 5′-CCTGGACGTGACCCCCAAGG-3′ or 10b 5′-CCACATGGTCCTCTCCATAAG-3′. PCR fragments were cloned using pSTBlue1-blunt vector (Novagen, Madison, WI, U.S.A.) and sequenced. An aliquot of human cDNA multiple tissue library (Quick-clone cDNA; ClonTech, Palo Alto, CA, U.S.A.) was also PCR-amplified using the same primers. PCR products were transferred on to nylon membranes and hybridized under standard conditions with a 1.3 kb HindIII/KpnI fragment of SCaMC-3 cDNA containing common exon 9 sequences.
Construction of plasmids
A FLAG epitope was introduced at the C-terminus immediately preceding the termination codon of SCaMC-3a variants as described previously [17]. C-termini of SCaMC-3b and SCaMC-3c/3d were modified by similar procedures. C-terminal regions were amplified using the common sense primer, 9, and an oligonucleotide specific for the mentioned variants complementary to nucleotides preceding the stop codon. For PCRs, we used oligonucleotides 9 and 10b (described above) for SCaMC-3b, and 9 and 10Alu (5′-CTCTGGAGGCTGAGGCAGG-3′) for the SCaMC-3c/3d construct. The PCR fragments obtained were digested with KpnI, and then ligated with the 32-bp blunted-EcoRI sequence of FLAG epitope into KpnI/EcoRI-digested pBluescript vector (as described in [17]). Introduction of the correct FLAG sequence was confirmed by sequencing. Finally, these C-terminal FLAG-tagged KpnI–EcoRI fragments were fused with the common EcoRI–KpnI 5′-fragment obtained from AI073878 into pCMV5 vector digested with EcoRI, to obtain finally pCMV5-SCaMC-3bFLAG and pCMV5-SCaMC-3c/3dFLAG.
The expression vector for the ΔNT (where NT stands for N-terminal) truncated version of SCaMC-3a variant, pCMV5-ΔNT-SCaMC-3aFLAG (amino acids 151–468), in which the long N-terminal extension has been removed and is tagged with a FLAG epitope at the C-terminus, has been described previously [17]. To obtain the corresponding constructs for SCaMC-3b and -3c/d variants, pCMV5-ΔNT-SCaMC-3bFLAG (amino acids 151–422) and pCMV5-ΔNT-SCaMC-3c/3dFLAG (amino acids 151–435), specific C-terminal FLAG-tagged KpnI–EcoRI fragments for each variant were exchanged with its equivalent fragment into a pCMV5-ΔNT-SCaMC-3aFLAG vector.
Cell culture and transfection
HEK-293T and COS-7 cells were cultured in Dulbecco's modified Eagle's medium containing 5% (v/v) inactivated foetal bovine serum (Invitrogen) at 37 °C in a 7% CO2 atmosphere. For transfections, COS-7 and HEK-293T cells were grown in 6 cm dishes and transiently transfected using the Lipofectamine™ reagent (3 μl/μg of DNA; Invitrogen) according to the manufacturer's instructions. Reporter plasmid was used at the rate of 6 μg/dish. Cells were incubated with Lipofectamine™–DNA complexes for 6 h and then the media was replaced with fresh Dulbecco's modified Eagle's medium containing 5% (v/v) fetal bovine serum. After 16–36 h of incubation to allow expression, cells were harvested for Western-blot analysis. For immunofluorescence assays, cells were grown over coverslips, transfected and, after 36 h, fixed and processed for fluorescence microscopy.
Immunofluorescence analysis
For the study of mitochondrial location, living cells were incubated with 200 nM MitoTracker Red CMXRos (Molecular Probes, Eugene, OR, U.S.A.) for 30 min at 37 °C and rinsed in prewarmed PBS. MitoTracker-loaded cells were fixed in 2% (w/v) paraformaldehyde in PBS [room temperature (25 °C), 4 min] and 100% methanol (−20 °C, 3 min), washed and then used for immunofluorescence as described previously [13,17]. Incubation with the primary antibody anti-FLAG (M2 Sigma, 1:100) was performed at room temperature for 1 h. After washing with PBS, cells were incubated for 1 h with secondary antibody (FITC-conjugated anti-mouse rabbit IgG; Vector, Burlingame, CA, U.S.A.) at 1:200. Fluorescence microscopy was performed using an Axiovert epifluorescence microscope (Carl Zeiss, Cologne, Germany) at a nominal magnification of ×100. Images were taken in parallel and processed under identical conditions.
Subcellular fractionation and Western-blot analysis
HEK-293T cells were plated in 10 cm Petri dishes and transfected with 10 μg of expression plasmids; 36 h after transfection, cells were washed twice with PBS, scraped and harvested by centrifugation. Pelleted cells were homogenized in a dounce homogenizer and 0.8 ml of buffer A (250 mM sucrose, 20 mM Hepes, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol, pH 7.4, 1 mM iodoacetate and 1 mM PMSF). Homogenates were centrifuged at 500 g for 15 min and then the supernatants were centrifuged at the same speed. Mitochondria-enriched fractions were obtained by centrifugation at 9000 g for 15 min and resuspended in 100 μl of buffer A. Protein determination was performed by the Bradford method. The proteins were separated by SDS/PAGE (10% polyacrylamide) and the presence of transfected FLAG-tagged proteins was determined by Western blotting using anti-FLAG (1:1000; Sigma). Proteins were visualized by ECL® (Amersham Biosciences). Polyclonal serum against human SCaMC-1 was used at 1:5000 [17].
RESULTS
Identification of 3′-variants for the human SCaMC-3 gene
During the characterization of human SCaMC genes, we have detected in the databases two types of EST clones corresponding to the C-terminus of the human SCaMC-3 isoform. The first one, showing homology in all extensions with the coding 3′-ends of SCaMC-1 and -2 human isoforms, has been described previously (renamed here as SCaMC-3a) [17] and also termed APC2 [18]. The second one was made up of related ESTs (AA714326, AW975612, AW291265 and BQ188940) and cDNAs (BC001656 and NM_024103), which displayed divergent 3′-end sequences (Figure 1A). A multiple alignment of these sequences showed that, although all had a common 3′-end, some contained inserted sequences with variable lengths.
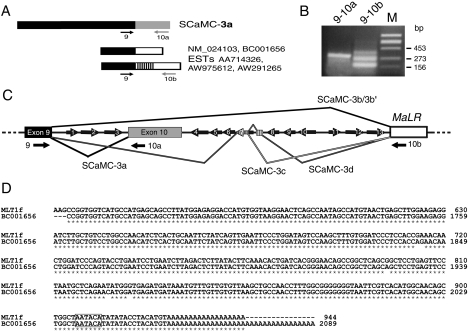
Figure 1. Human SCaMC-3 gene exhibits alternative spliced variants derived from 3′-flanking repetitive elements.
(A) Schematic representation of the different 3′ SCaMC-3 sequences found in databases. Black boxes indicate the common 5′-region, the novel 3′-end common to all SCaMC-3 variant analyses is indicated by an open box, and inserted sequences are indicated by stripes (accession numbers are shown). Numbered arrows indicate the relative position of selected specific primers used to amplify the different 3′-ends (described in the Materials and methods section). (B) RT–PCR analysis of SCaMC-3 3′-end variants. Reverse transcription was performed from total RNA of HEK-293T cells. The primer pairs used for each amplification are indicated at the top of each lane. The amplified products were verified by cloning and sequencing. The sizes of molecular-mass standards, HindIII-digested phage Φ29, are indicated. (C) Genomic organization of the 3′-region of the human SCaMC-3 gene. The genomic map was constructed from overlapping human BAC clones AC011539 and AC010503. The common exon 9 is shown as a black box. The grey box indicates the previously described SCaMC-3 last exon, exon 10, and the MaLR element is shown as an open box. Introns are shown as thick lines connecting these elements. Dark arrows represent intronic Alu elements (280 bp) and short arrows indicate truncated Alu sequences (120–150 bp), at their relative positions. The direction of the arrow shows the orientation of the inserted Alu element. Splicing events between upstream exon 9 and alternative 3′-exonized sequences are indicated by solid lines. (D) Alignment of the MLT1f consensus and that of novel SCaMC-3 3′-end nucleotide sequences. Only aligned regions with relevant homology are shown. Identical bases are indicated by asterisks. The MLT1f sequence is a fragment of a 944 bp-long consensus sequence deposited in RepBase Uptake [48]; nucleotide positions in the aligned MLT1f fragment are shown. The 2089 bp BC001656 nucleotide sequence comes from GenBank®. The polyadenylation signals are boxed.
BLAST searches revealed that most 3′-sequences matched genomic sequences located 3.6 kb downstream of the last SCaMC-3 exon and contained in BAC clones AC011539 and AC010503. Also we detected high homology to repetitive elements belonging to the MaLR family [30], suggesting that most of the 3′-alternative SCaMC-3 sequences were derived from an integrated MaLR element. An identical approach showed that nucleotides inserted between exon 9 and the MaLR element belong to human Alu repeat family. Interestingly, numerous Alu repeats, full-length or 5′-end-truncated, were detected around SCaMC-3 exon 10 (Figure 1C). There are 13 Alu elements, seven of them in opposite orientation to that of SCaMC-3 transcription. Antisense Alu elements contain several potential splice sites [32], and hence Alu-containing sequences could be incorporated into SCaMC-3 transcripts from genomic Alu elements flanking exon 10. Taken together, these observations suggested the existence of 3′-variants of SCaMC-3 generated by alternative splicing from transposable elements.
To confirm and characterize these novel spliced SCaMC-3 transcripts, we performed RT–PCR assays (see Figure 1A for primer location). We performed these assays using total RNA obtained from HEK-293T cells. As shown in Figure 1(B), a single band of the expected size, 311 bp, is obtained with a reverse SCaMC-3a-specific primer (10a; Figure 1B). However, when we used a reverse primer derived from divergent 3′-sequences, three major bands are observed (Figure 1B). The sequencing of these PCR products has confirmed the observations previously obtained from EST alignments. All fragments display a 5′-region coincident with SCaMC-3a, up to the end of exon 9, and 3′-divergent sequences. The smallest band corresponds to a doublet of 173 and 177 bp, named SCaMC-3b and -3b′ respectively, that contain new 3′-sequences homologous with that of a transposable MaLR element (see below, accession numbers AJ879080 and AJ879081). In SCaMC-3b′, an upstream 3′-acceptor site is used, resulting in the insertion of 4 nucleotides absent from SCaMC-3b. The sequence of the two longer co-amplified products, of 271 and 359 bp (denoted SCaMC-3c and -3d, accession numbers AJ879082 and AJ879083 respectively) matched well with that of SCaMC-3b, in both 5′- and 3′-ends, but included additional inserted sequences. The 271 bp fragment displayed a 98 bp insertion at the splice junction of SCaMC-3b between exon 9 and the divergent 3′-end, which matched sequences within the Alu element 7 (corresponding to nucleotides 2414–2591 of BAC AC010503; Figure 1B) that belongs to the Alu-S class. The longer PCR product, SCaMC-3d, contained the same 98 bp Alu repeat as SCaMC-3c, and additional 88 nucleotides corresponding to an intergenic sequence segment close to an Alu element (nucleotides 2137–2224 of BAC AC010503, Figure 1C).
Figure 1(C) shows a diagram of the splicing events involved in the generation of the four SCaMC-3 3′-variants. These transcripts are generated by alternative splicing mediated by the SCaMC-3 donor site at the 3′-end of exon 9 and four acceptor signals from the 3′-flanking repetitive elements. Inspection of flanking genomic sequences confirmed that all spliced transcripts contained conserved 5′-donor and 3′-acceptor sites. The 3′-region of SCaMC-3d mRNA was identical with sequences in BC001656 and NM_024103. Also we detected EST sequences matching the SCaMC-3b and -3b′ variants: CA436368, F35656 and F28491 for SCaMC-3b, and CB242135 and BQ016496, AW975612 and AA714326 for SCaMC-3b′.
When the 3′-end of BC001656, which contains the longest 3′-region including the poly(A)+ (polyadenylated) tail, was aligned with repetitive elements obtained from Repbase Uptake, we found 98% identity at nucleotide level with the 3′-half of MLT1f element, a member of the MaLR family (Figure 1D). The homologous region corresponds to the last 300 bp immediately upstream of poly(A) tails (Figure 1D), which resemble a single LTR (long terminal repeat). MaLR elements form a class of mobile genetic elements different from retroviruses, SINEs and LINEs, with structural similarities to the retrovirus [30]. Interestingly, most MaLRs seem to remain in the genome as solitary LTRs [30]. The MLT1f element is integrated in the positive orientation, and this could lead to a premature transcription termination due to the transcriptional termination site located within this element [33]. In fact, the 3′-ends of both MLT1f and BC001656 are exactly coincident, showing a proximal polyadenylation signal of AATACA with a single base mismatch from consensus (Figure 1D). All 3′-alternative transcripts probably use this polyadenylation signal derived from the integrated element, since we did not find any EST clone containing further 3′-sequences. In fact, all database sequences analysed contained a common 3′-end, indicating that the insertion of MLT1f leads to transcription ending in all messengers that contain it.
A third kind of 3′-end SCaMC-3 sequences unrelated to those described above has also been observed. These sequences are found exclusively in two human EST clones, AI073878 and W24006, which exhibit a very short 3′-divergent end, 106 bp long, coincident with genomic sequences next to exon 9. Because they represented rare clones with an uncommonly short 3′-untranslated region sequence, and we did not find expressed sequences with these short 3′-ends, they have not been analysed further.
Tissue distribution of alternatively spliced SCaMC-3 transcripts
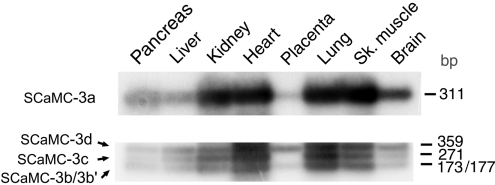
To examine whether the new 3′ SCaMC-3 transcripts naturally exist in human tissues and, to analyse their distribution pattern, we performed PCRs using cDNA libraries made from adult human tissues (see the Materials and methods for details). As shown in Figure 2, SCaMC-3a, -3b/-3b′, -3c and -3d isoforms coexist in all tissues tested. No gross differences were found in the distribution pattern of SCaMC-3a (Figure 2, upper panel) and the novel isoforms (Figure 2, bottom panel) in the different tissues. However, we observed changes in expression level among tissues; thus SCaMC-3a is present at higher levels in all tissues analysed than transcripts containing exonic sequences downstream of exon 10: SCaMC-3b/3b′, -3c or -3d. Moreover, although a 2.1 kb transcript could be assembled from databases using SCaMC-3b-specific sequences, we were unable to detect it by Northern-blot analysis in either human tissues [17] or HEK-293T cells (results not shown). It is likely that alternative transcripts display low expression levels, only detectable by PCR assays. It has been reported that Alu-containing exons are included in transcripts at lower frequencies than the alternatively spliced exons that do not contain an Alu sequence [34]. This also agrees with the low representation of sequences containing SCaMC-3c and -3d in the databases.
Figure 2. Expression analysis of SCaMC-3b 3′-end variants in human tissues.
Equivalent aliquots of a multiple human cDNA panel; pancreas, liver, kidney, heart, placenta, lung, skeletal (Sh.) muscle and brain (ClonTech) were used as templates. The upper panel shows the results obtained with primers specific for SCaMC-3a transcript, and the bottom one the PCR products co-amplified with primers 9 and 10b, corresponding to SCaMC-3b, -3b′, -3c and -3d transcripts. The sizes of amplified fragments are given in terms of bp. The products obtained were verified by sequencing. The results indicate a similar distribution pattern of SCaMC-3a and transcripts containing repetitive elements.
The new SCaMC-3 variants are C-terminal truncated variants
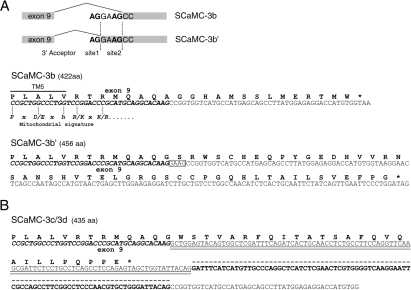
The predicted proteins for 3′-spliced SCaMC-3 variants display shorter C-terminal ends than that of the SCaMC-3a isoform (468 amino acids) [17]. Thus putative proteins of 422, 456 and 435 amino acids are encoded by SCaMC-3b, -3b′ and -3c/-3d transcripts respectively (Figure 3). In the SCaMC-3b transcript, the alternative 3′-sequence results in an insertion of 14 amino acids in-frame to the common 407 amino acids encoded by exons 1–9. This new C-terminus does not show homology, at the amino acid level, to that of SCaMC-3a (Figure 3A). The use of site 1 as splicing acceptor in SCaMC-3b′ generates a translation frame-shift, resulting in a C-terminal peptide unrelated to that of SCaMC-3b, of 50 amino acids in length (Figure 3A). Searches in databases did not reveal any protein with similarity to both novel C-terminal amino acid sequences. It is therefore likely that these sequences were generated from an usually untranslated MaLR insert.
Figure 3. Alternative 3′-SCaMC-3 transcripts encoding for C-terminal truncated variants.
(A) Upper panel: splicing acceptor sites involved in the generation of alternative SCaMC-3b and -3b′ transcripts. Genomic sequences derive from BAC clones mentioned in Figure 1. Alternative 3′-splice sites, site 1 and site 2, are shown. Lower panels: predicted amino acid sequences for the C-termini of SCaMC-3b and SCaMC-3b′ are shown. Exonic sequences from exon 9 are indicated in italic bold face and the sequence derived from MLT1f element by normal letters. Additional nucleotides in SCaMC-3b′ transcript are boxed. Predicted position of the TM-5 domain and equivalence with MC signature are shown. (B) The 3′-end sequence of SCaMC-3d transcript and its putative ORF, identical with SCaMC-3c, are shown at the top. Sequences from exon 9 and MLT1f element are represented as in (A). Alu-derived nucleotides are double-underlined and additional SCaMC-3d-specific sequences are indicated by the broken line.
On the other hand, the presence of an Alu element within a transcript results in premature translation termination since it contains numerous stop codons [32]. In the case of SCaMC-3c and -3d messengers, both result in the same polypeptide, because both use the same stop signal from their common Alu element (Figure 3B). The SCaMC-3c/-3d-specific segment encodes a short C-terminus of 28 amino acids, which does not show homology to any SCaMC-3 C-end, but shows a high degree of similarity to other proteins whose transcripts contain Alu-derived sequences.
The resulting new peptides display relevant structural changes with respect to the long isoform, SCaMC-3a. The MC signature, PX(D/E)Xh(K/R)X(R/K)X20–30(D/E)GX4a(K/R)GRG (h is hydrophobic and a is aromatic) [5], corresponding to the third MC repeat is disrupted in all cases. The point of divergence between variants starts within the first half of MC signature located next to the TM5 end (see Figure 3). In the short SCaMC-3 variants, the characteristic-long hydrophilic segment connecting transmembrane α-helices TM5 and TM6 as well as the second half of the MC signature are substituted by the novel C-termini. The hydrophobic profile of SCaMC-3b and SCaMC-3b′ C-terminal extensions did not show any hydrophobic amino acid stretch that can form a typical α-helical domain equivalent to TM6. Thus, in these variants, the loss of TM6 might cause a change in the classical carrier topology in which the C-terminus faces the matrix side of the inner mitochondrial membrane rather than the intermembrane space. Analysis of SCaMC-3c/-3d C-terminal sequence indicated the possible formation of a hydrophobic transmembrane α-helical domain, a putative TM6, along with a shortening of the TM5–TM6 hydrophilic loop. These possible topological changes of the novel SCaMC-3 variants might also affect mitochondrial targeting and sorting of the precursor protein.
Subcellular location of SCaMC-3 variants
Most mitochondrial precursor proteins contain an N-terminal targeting presequence [24,35]. However, proteins of the inner membrane such as those from the MCF contain targeting and sorting information throughout the mature sequence [35]. In addition, it has been shown that the hydrophyilic N-terminal extension of the long CaMCs is not required for mitochondrial import of the AGCs Aralar1 and citrin [15,16,36], or the SCaMCs [17]. In order to determine whether the shortened SCaMC-3 C-terminal variants have any change in mitochondrial import, we have analysed the intracellular distribution of SCaMC-3 isoforms with the shortest C-terminus, SCaMC-3b and -3c/-3d. Initially, HEK-293 and COS-7 cells were transfected with FLAG-tagged versions of SCaMC-3b, -3c/-3d and -3a proteins (SCaMC-3a–FLAG, SCaMC-3b–FLAG and -3c/-3d–FLAG, all tags at the C-terminus) and analysed by immunofluorescence with an anti-FLAG antibody [17]. In each case, mitochondrial localization was verified by co-staining with MitoTracker Red CMXRos, a mitochondrial marker. In FLAG-positive COS-7 cells, we sometimes observed a clear punctuate staining of the SCaMC-3 isoforms, matching that of mitochondrial MitoTracker (see Figure 4A, panels I). However, in most cases, SCaMC-3 isoforms showed a diffuse staining pattern, together with a punctuate one. In these cases, the MitoTracker staining pattern often showed mitochondrial fragmentation and swelling of the mitochondrial network (Figure 4A, panels II). This appearance was also observed in HEK-293 cells (results not shown). Remarkably, overexpression of truncated SCaMC-3 variants (ΔNT-SCaMCs), which encompass the entire MC domain but lack the long N-ter-minal extensions of these proteins, showed the same Mitotracker staining pattern; a fragmented mitochondrial network, but no diffuse staining for SCaMC-3 (Figure 4B). Similar patterns were observed when these variants were transfected in HEK-293 cells. Fragmentation and swelling of the mitochondrial network are common observations with overexpressed mitochondrial proteins, which we have not pursued any further. However, the diffuse staining pattern of whole length SCaMC-3 variants was subsequently investigated after subcellular fractionation and Western-blot analysis of transfected cells.
Figure 4. Subcellular distribution of overexpressed SCaMC-3a, SCaMC-3b and SCaMC-3c/-3d variants.
(A) Immunofluorescence assays of COS-7 cells transiently transfected with FLAG-tagged SCaMC-3 variants. Cells were transfected with the indicated constructs, labelled with MitoTracker, processed using an anti-FLAG monoclonal antibody and visualized with FITC secondary antibody. Two different patterns (I and II) were detected. Identical fields labelled for MitoTracker and anti-FLAG for representative cells of each pattern are presented. All images are processed in parallel. Magnification, ×100. (B) Mitochondrial localization of SCaMC proteins with N-terminal deletions (ΔNT-SCaMCs). COS-7 cells were transfected with ΔNT-SCaMC-3–FLAG constructs (ΔNT-SCaMC-3a, ΔNT-SCaMC-3b and ΔNT-SCaMC-3c/-3d) coding for the MC homology domain tagged at its C-terminus with FLAG. The cells were labelled with MitoTracker and with anti-FLAG antibody as in (A). (C) Western-blot analysis of overexpressed SCaMC isoforms in mitochondrial-enriched extracts (MIT) and post-mitochondrial (Post-MIT) supernatants from HEK-293 cells. Protein (10 μg) of extracts from cells transiently transfected with the indicated constructs were analysed. Membranes were immunoblotted with anti-FLAG antibody and reblotted with polyclonal anti-SCaMC-1 to test for mitochondrial fractionation. Sizes of immunoreactive anti-FLAG bands are coincident with the expected ones. Positions of molecular-mass standards are indicated.
To this end, SCaMC-3 variants were transfected in HEK-293 cells, and mitochondrial-enriched and post-mitochondrial fractions were analysed by Western blotting with the anti-FLAG antibody. To correct for mitochondrial loading, we used an antibody against SCaMC-1, the SCaMC isoform most commonly represented in cell lines [17]. Surprisingly, all three overexpressed SCaMC-3 variants were detected both in mitochondrial and post-mitochondrial fractions, whereas control SCaMC-1 was only present in mitochondria (Figure 4C). These results suggest that the diffuse localization pattern of these variants (Figure 4A, panels II) corresponds to cytosolic non-imported SCaMC-3 precursor proteins. Transfection assays followed by immunofluorescence and Western-blot analysis have been performed several times with similar results. Thus both immunofluorescence and subcellular fraction analysis suggest that mitochondrial import of SCaMC-3 occurs unsuccessfully when proteins are overexpressed.
In contrast, the ΔNT forms of these variants were detected not in post-mitochondrial fractions, but only in mitochondria (Figure 4C). The occasional observation of immunoreactive material in post-mitochondrial fractions is probably due to carry over of mitochondrial proteins, as judged from the presence of SCaMC-1 in these fractions (Figure 4B). The subcellular localization of the ΔNT forms agrees with the clear mitochondrial pattern of these proteins with total lack of the diffuse localization observed for the full-length proteins (Figure 4B). Taken together, our results suggest that all full-length variants have difficulties with mitochondrial import, at least when overexpressed in cells, whereas these difficulties are no longer observed after the removal of the N-terminal extensions of the proteins. Our results suggest that mitochondrial import is unaffected by shortening and sequence changes of the C-termini and that unsatisfactory import observed for the whole length versions can be attributed to the N-terminal region common to all SCaMC-3.
DISCUSSION
In the present study, we report the characterization of new spliced variants for the human SCaMC-3 gene, an isoform of the ATP-Mg/Pi MC recently identified at the molecular level [17,18]. These variants are generated by alternative splicing promoted by interspersed repetitive elements, Alu sequences and an MaLR, located immediately downstream of the previously reported last SCaMC-3 exon (exon 10). To the best of our knowledge, this is the first time that sequences derived from transposable elements are found in MC transcripts. However, the functional relevance of these new variants is currently unknown.
The insertion of mobile elements into a mature mRNA is not unusual [32,37]. This may cause genetic damage, but insertion may also contribute to protein variability and versatility of the genome without compromising its integrity [31,32,34,37]. In fact, this may be the case for SCaMC-3. In all the cell samples tested, there were 3′-variants of the SCaMC-3 gene generated by alternative splicing, in addition to mRNA without the repetitive element in the coding region. Furthermore, SCaMC-3 mRNAs containing repeat-derived sequences are under-represented. We were unable to detect them by Northern-blot analysis, and low levels of expression are found by PCR amplification, in agreement with Sorek et al. [34] who reported that the vast majority of Alu-containing exons that are alternatively spliced have a small retention rate and are only found in approx. one-fifth of all mRNA transcripts. The incorporation of retroviral-derived elements in transcripts as alternative-spliced sequences has already been reported [38,39]. As in SCaMC-3 variants, in FLT4 and Leptin receptor genes, retroviral integration occurs into the 3′-end, leading to the formation of alternative products in the same tissues as the full-length species. Furthermore, although insertion of these LTR-containing sequences results in premature transcriptional termination, their integration in the 3′-end may cause less damage than internal insertion, and be maintained if harmless [39]. In conclusion, the effects of transposable sequences within SCaMC-3 transcripts can be viewed as essentially neutral.
Mitochondrial targeting and assembly of SCaMC3 variants
In the MC proteins, all three modules (I, II and III) are important for mitochondrial targeting, and all are involved in translocation across the TOM (translocase of the outer membrane) complex and insertion in the inner mitochondrial membrane [40]. Partial or total deletion of any of these modules partially disrupts correct mitochondrial targeting of the protein [40]. Thus it was surprising that the SCaMC-3 C-terminal-shortened variants that lack the characteristic TM6 of module III did not show any relevant change with respect to SCaMC-3a mitochondrial localization. The intracellular distribution of FLAG-tagged overexpressed short SCaMC-3 variants, SCaMC-3c/-3d and -3b, is essentially the same as that of the longer SCaMC-3a variant that contains all six TM spanners. In all cases, MitoTracker signals co-localized with anti-FLAG staining, thus demonstrating that these short variants can still be targeted to mitochondria. Other MCs (human UCP-3 and MRS3/4) [41,42] also have splicing variants encoding proteins lacking one or more TM segments. MRS3/4, a 177-amino-acid form that contains only the last three TM spanners (TM4–TM6) is effectively targeted to the mitochondria [42,43]. Whether the short SCaMC-3 variants are correctly assembled and functionally active is still an open question. Indeed, recent structural work has shown that the adenine nucleotide translocase and possibly all MCs form a helical bundle with 3-fold symmetry [2,44]. These SCaMC-3 variants lacking TM6 in module III, would have difficulties in forming a stable bundle, and thus, a functional transporter.
Another surprising observation was the presence of a significant extramitochondrial localization for all FLAG-tagged SCaMC-3 overexpressed proteins. This altered distribution is not due to alteration of the mitochondrial membrane potential, Δψmit, because Mitotracker staining remains unaffected. It is also not due to a simple overexpression artifact, as it is not observed for the ΔNT deletion mutants of these carriers when similarly over-expressed. Since only full-length proteins but not their N-terminal truncated versions have extramitochondrial localizations, this suggests that the wild-type proteins are inefficiently imported in mitochondria, and only a fraction of the precursor proteins are able to mature in the organelle. Thus overexpression of these proteins unmasks this inefficiency by making the extramitochondrial products visible. In fact, other membrane proteins, such as the cystic fibrosis transmembrane conductance regulator, have difficulties in folding/maturation [45,46] that are exposed through the use of mutants.
If the full-length SCaMC proteins have difficulties in mitochondrial targeting, why don't they exist for the N-terminal truncated SCaMC proteins? We have found a possible explanation for this question in the recent findings of Koehler's group [29]. For their translocation to mitochondrial structures, most inner-membrane proteins use a specialized transport route known as the carrier pathway that recognizes internal targeting signals [24,35,47]. During transport across the intermembrane space, inner membrane proteins may interact with two complexes composed of distinct small Tim proteins: the Tim9p–Tim10p complex that binds to hydrophobic segments of the typical MC precursor [25], and on the other hand, the Tim8p–Tim13p complex that binds to the hydrophilic N-terminus of Tim23p protein (one component of TIM23 translocon) as well as to AGC isoforms [27,29]. The pathway chosen seems to be determined by the presence of the N-terminal extension; thus import of aralar1 variants containing most of the N-terminal extension was decreased by 80% in mitochondria that lacked the Tim8p–Tim13p complex [29].
It is likely that this alternative import pathway is also used by SCaMC proteins, as suggested by Roesch et al. [29]. In this case, the inefficient import observed when full-length SCaMC3 proteins were overexpressed may be due to a low efficacy for binding/translocation through the Tim8p–Tim13p pathway, a process depending on the N-terminus. When this is removed, translocation of the N-truncated versions, performed by the Tim9p–Tim10p complex, goes on unimpaired. Since this inefficient import is specific for SCaMC-3, and has not been observed for any other SCaMC or AGC isoform [13,15,17], the results open up the possibility that the differences between the N-terminal extensions of SCaMC-3 and other isoforms may affect their interaction with the translocation pathway.
In conclusion, our results suggest that the SCaMC-3 isoform of ATP-Mg/Pi carriers might use the Tim8p–Tim13p complex for mitochondrial import, as found for the ACGs aralar and citrin. The finding of new C-terminal variants for SCaMC-3 adds further complexity to the SCaMC family of ATP-Mg/Pi transporters of mitochondria. Mitochondrial localization is unimpaired in variants lacking TM6. Whether these variants are correctly assembled and functional in mitochondria is still unknown, but they seem to be present, albeit at low levels, in all cells. Further studies are required to address these questions.
Acknowledgments
I am grateful to J. Satrústegui, in whose laboratory this work was carried out, for her constant help and also for critical comments and improvements of the manuscript. I thank C. Galián for her collaboration in initial experiments and I. Ocaña for technical support. This work was supported by grants from the Spanish Ministerio de Ciencia y Tecnología (PM1998-021 and BMC2002-02072), Ministerio de Sanidad (FIS 01/0395) and Química Farmaceútica Bayer. The Centro de Biología Molecular Severo Ochoa is the recipient of institutional aid from the Ramón Areces Foundation.
References
- 1.Walker J. E., Runswick M. J. The mitochondrial transport protein superfamily. J. Bioenerg. Biomembr. 1993;4:35–46. doi: 10.1007/BF01108401. [DOI] [PubMed] [Google Scholar]
- 2.Kunji E. R. The role and structure of mitochondrial carriers. FEBS Lett. 2004;564:239–244. doi: 10.1016/S0014-5793(04)00242-X. [DOI] [PubMed] [Google Scholar]
- 3.Palmieri F. The mitochondrial transporter family (SLC25): physiological and pathological implications. Pflugers Arch. 2004;447:689–709. doi: 10.1007/s00424-003-1099-7. [DOI] [PubMed] [Google Scholar]
- 4.Aquila H., Link T. A., Klingenberg M. The uncoupling protein from brown fat mitochondria is related to the mitochondrial ADP/ATP carrier. Analysis of sequence homologies and of folding of the protein in the membrane. EMBO J. 1985;4:2369–2376. doi: 10.1002/j.1460-2075.1985.tb03941.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Indiveri C., Iacobazzi V., Giangregorio N., Palmieri F. The mitochondrial carnitine carrier protein: cDNA cloning, primary structure and comparison with other mitochondrial transport proteins. Biochem. J. 1997;321:713–719. doi: 10.1042/bj3210713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.el Moualij B., Duyckaerts C., Lamotte-Brasseur J., Sluse F. E. Phylogenetic classification of the mitochondrial carrier family of Saccharomyces cerevisiae. Yeast. 1997;13:573–581. doi: 10.1002/(SICI)1097-0061(199705)13:6<573::AID-YEA107>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 7.Nelson D. R., Felix C. M., Swanson J. M. Highly conserved charge-pair networks in the mitochondrial carrier family. J. Mol. Biol. 1998;277:285–308. doi: 10.1006/jmbi.1997.1594. [DOI] [PubMed] [Google Scholar]
- 8.Dyall S. D., Koehler C. M., Delgadillo-Correa M. G., Bradley P. J., Plumper E., Leuenberger D., Turck C. W., Johnson P. J. Presence of a member of the mitochondrial carrier family in hydrogenosomes: conservation of membrane-targeting pathways between hydrogenosomes and mitochondria. Mol. Cell. Biol. 2000;20:2488–2497. doi: 10.1128/mcb.20.7.2488-2497.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dolce V., Fiermonte G., Runswick M. J., Palmieri F., Walker J. E. The human mitochondrial deoxynucleotide carrier and its role in the toxicity of nucleoside antivirals. Proc. Natl. Acad. Sci. U.S.A. 2001;98:2284–2288. doi: 10.1073/pnas.031430998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prohl C., Pelzer W., Diekert K., Kmita H., Bedekovics T., Kispal G., Lill R. The yeast mitochondrial carrier Leu5p and its human homologue Graves’ disease protein are required for accumulation of coenzyme A in the matrix. Mol. Cell. Biol. 2001;21:1089–1097. doi: 10.1128/MCB.21.4.1089-1097.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marobbio C. M., Vozza A., Harding M., Bisaccia F., Palmieri F., Walker J. E. Identification and reconstitution of the yeast mitochondrial transporter for thiamine pyrophosphate. EMBO J. 2002;21:5653–5661. doi: 10.1093/emboj/cdf583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Millar A. H., Heazlewood J. L. Genomic and proteomic analysis of mitochondrial carrier proteins in Arabidopsis. Plant Physiol. 2003;131:443–453. doi: 10.1104/pp.009985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.del Arco A., Satrústegui J. Molecular cloning of Aralar, a new member of the mitochondrial carrier superfamily that binds calcium and is present in human muscle and brain. J. Biol. Chem. 1998;273:23327–23334. doi: 10.1074/jbc.273.36.23327. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi K., Sinasac D. S., Iijima M., Boright A. P., Begum L., Lee J. R., Yasuda T., Ikeda S., Hirano R., Terazono H., et al. The gene mutated in adult-onset type II citrullinaemia encodes a putative mitochondrial carrier protein. Nat. Genet. 1999;22:159–163. doi: 10.1038/9667. [DOI] [PubMed] [Google Scholar]
- 15.del Arco A., Agudo M., Satrústegui J. Characterization of a second member of the subfamily of calcium-binding mitochondrial carriers expressed in human non-excitable tissues. Biochem. J. 2000;345:725–732. doi: 10.1042/0264-6021:3450725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmieri L., Pardo B., Lasorsa F. M., del Arco A., Kobayashi K., Iijima M., Runswick M. J., Walker J. E., Saheki T., Satrústegui J., et al. Citrin and aralar1 are Ca(2+)-stimulated aspartate/glutamate transporters in mitochondria. EMBO J. 2001;20:5060–5069. doi: 10.1093/emboj/20.18.5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.del Arco A., Satrústegui J. Identification of a novel human subfamily of mitochondrial carriers with calcium-binding domains. J. Biol. Chem. 2004;279:24701–24713. doi: 10.1074/jbc.M401417200. [DOI] [PubMed] [Google Scholar]
- 18.Fiermonte G., De Leonardis F., Todisco S., Palmieri L., Lasorsa F. M., Palmieri F. Identification of the mitochondrial ATP-Mg/Pi transporter. Bacterial expression, reconstitution, functional characterization, and tissue distribution. J. Biol. Chem. 2004;279:30722–30730. doi: 10.1074/jbc.M400445200. [DOI] [PubMed] [Google Scholar]
- 19.Austin J., Aprille J. R. Carboxyatractyloside-insensitive influx and efflux of adenine nucleotides in rat liver mitochondria. J. Biol. Chem. 1984;259:154–160. [PubMed] [Google Scholar]
- 20.Aprille J. R. Regulation of the mitochondrial adenine nucleotide pool size in liver: mechanism and metabolic role. FASEB J. 1988;2:2547–2556. doi: 10.1096/fasebj.2.10.3290024. [DOI] [PubMed] [Google Scholar]
- 21.Joyal J. L., Aprille J. R. The ATP-Mg/Pi carrier of rat liver mitochondria catalyzes a divalent electroneutral exchange. J. Biol. Chem. 1992;267:19198–19203. [PubMed] [Google Scholar]
- 22.Lasorsa F. M., Pinton P., Palmieri L., Fiermonte G., Rizzuto R., Palmieri F. Recombinant expression of the Ca(2+)-sensitive aspartate/glutamate carrier increases mitochondrial ATP production in agonist-stimulated Chinese hamster ovary cells. J. Biol. Chem. 2003;278:38686–38692. doi: 10.1074/jbc.M304988200. [DOI] [PubMed] [Google Scholar]
- 23.Nosek M. T., Dransfield D. T., Aprille J. R. Calcium stimulates ATP-Mg/Pi carrier activity in rat liver mitochondria. J. Biol. Chem. 1990;265:8444–8450. [PubMed] [Google Scholar]
- 24.Koehler C. M. New developments in mitochondrial assembly. Annu. Rev. Cell Dev. Biol. 2004;20:309–335. doi: 10.1146/annurev.cellbio.20.010403.105057. [DOI] [PubMed] [Google Scholar]
- 25.Curran S. P., Leuenberger D., Oppliger W., Koehler C. M. The Tim9p-Tim10p complex binds to the transmembrane domains of the ADP/ATP carrier. EMBO J. 2002;21:942–953. doi: 10.1093/emboj/21.5.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rothbauer U., Hofmann S., Muhlenbein N., Paschen S. A., Gerbitz K. D., Neupert W., Brunner M., Bauer M. F. Role of the deafness dystonia peptide 1 (DDP1) in import of human Tim23 into the inner membrane of mitochondria. J. Biol. Chem. 2001;276:37327–37334. doi: 10.1074/jbc.M105313200. [DOI] [PubMed] [Google Scholar]
- 27.Curran S. P., Leuenberger D., Schmidt E., Koehler C. M. The role of the Tim8p-Tim13p complex in a conserved import pathway for mitochondrial polytopic inner membrane proteins. J. Cell Biol. 2002;158:1017–1027. doi: 10.1083/jcb.200205124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paschen S. A., Rothbauer U., Kaldi K., Bauer M. F., Neupert W., Brunner M. The role of the TIM8-13 complex in the import of Tim23 into mitochondria. EMBO J. 2000;19:6392–6400. doi: 10.1093/emboj/19.23.6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roesch K., Hynds P. J., Varga R., Tranebjaerg L., Koehler C. M. The calcium-binding aspartate/glutamate carriers, citrin and aralar1, are new substrates for the DDP1/TIMM8a-TIMM13 complex. Hum. Mol. Genet. 2004;13:2101–2111. doi: 10.1093/hmg/ddh217. [DOI] [PubMed] [Google Scholar]
- 30.Smit A. F. Identification of a new, abundant superfamily of mammalian LTR-transposons. Nucleic Acids Res. 1993;21:1863–1872. doi: 10.1093/nar/21.8.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peaston A. E., Evsikov A. V., Graber J. H., de Vries W. N., Holbrook A. E., Solter D., Knowles B. B. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev. Cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 32.Makalowski W., Mitchell G. A., Labuda D. Alu sequences in the coding regions of mRNA: a source of protein variability. Trends Genet. 1994;10:188–193. doi: 10.1016/0168-9525(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 33.Smit A. F. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- 34.Sorek R., Ast G., Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12:1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfanner N., Wiedemann N. Mitochondrial protein import: two membranes, three translocases. Curr. Opin. Cell Biol. 2002;14:400–411. doi: 10.1016/s0955-0674(02)00355-1. [DOI] [PubMed] [Google Scholar]
- 36.Iijima M., Jalil A., Begum L., Yasuda T., Yamaguchi N., Xian Li M., Kawada N., Endou H., Kobayashi K., Saheki T. Pathogenesis of adult-onset type II citrullinemia caused by deficiency of citrin, a mitochondrial solute carrier protein: tissue and subcellular localization of citrin. Adv. Enzyme Regul. 2001;41:325–342. doi: 10.1016/s0065-2571(00)00022-4. [DOI] [PubMed] [Google Scholar]
- 37.Deininger P. L., Batzer M. A. Mammalian retroelements. Genome Res. 2002;12:1455–1465. doi: 10.1101/gr.282402. [DOI] [PubMed] [Google Scholar]
- 38.Kapitonov V. V., Jurka J. The long terminal repeat of an endogenous retrovirus induces alternative splicing and encodes an additional carboxy-terminal sequence in the human leptin receptor. J. Mol. Evol. 1999;48:248–251. doi: 10.1007/pl00013153. [DOI] [PubMed] [Google Scholar]
- 39.Hughes D. C. Alternative splicing of the human VEGFGR-3/FLT4 gene as a consequence of an integrated human endogenous retrovirus. J. Mol. Evol. 2001;53:77–79. doi: 10.1007/s002390010195. [DOI] [PubMed] [Google Scholar]
- 40.Wiedmann N., Pfanner N., Ryan M. T. The three modules of ADP/ATP carrier cooperate in receptor recruitment and translocation into mitochondria. EMBO J. 2001;20:951–960. doi: 10.1093/emboj/20.5.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gong D. W., He Y., Reitman M. L. Genomic organization and regulation by dietary fat of the uncoupling protein 3 and 2 genes. Biochem. Biophys. Res. Commun. 1999;256:27–32. doi: 10.1006/bbrc.1999.0239. [DOI] [PubMed] [Google Scholar]
- 42.Li F. Y., Nikali K., Gregan J., Leibiger I., Leibiger B., Schweyen R., Larsson C., Suomalainen A. Characterization of a novel human putative mitochondrial transporter homologous to the yeast mitochondrial RNA splicing proteins 3 and 4. FEBS Lett. 2001;494:79–84. doi: 10.1016/s0014-5793(01)02319-5. [DOI] [PubMed] [Google Scholar]
- 43.Li F. Y., Leibiger B., Leibiger I., Larsson C. Characterization of a putative murine mitochondrial transporter homology of hMRS3/4. Mamm. Genome. 2002;13:20–23. doi: 10.1007/s00335-001-2094-y. [DOI] [PubMed] [Google Scholar]
- 44.Pebay-Peyroula E., Dahout-Gonzalez C., Kahn R., Trezeguet V., Lauquin G. J., Brandolin G. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature (London) 2003;426:39–44. doi: 10.1038/nature02056. [DOI] [PubMed] [Google Scholar]
- 45.Ward C. L., Kopito R. R. Intracellular turnover of cystic fibrosis transmembrane conductance regulator. Inefficient processing and rapid degradation of wild-type and mutant proteins. J. Biol. Chem. 1994;269:25710–25718. [PubMed] [Google Scholar]
- 46.Johnston J. A., Ward C. L., Kopito R. R. Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rehling P., Brandner K., Pfanner N. Mitochondrial import and the twin-pore translocase. Nat. Rev. Mol. Cell. Biol. 2004;5:519–530. doi: 10.1038/nrm1426. [DOI] [PubMed] [Google Scholar]
- 48.Jurka J. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 2000;16:418–420. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]