Abstract
Complexes formed between β2GPI (β2-glycoprotein I), a human plasma protein, and biological membranes are considered to be targets of macrophages and antiphospholipid autoantibodies involved in autoimmune diseases, such as antiphospholipid syndrome or systemic lupus erythematosus. The positively charged lysine-rich fifth domain of β2GPI facilitates its interaction with phospholipid membranes containing acidic phospholipids, which normally become exposed by apoptotic processes. In the present study, atomic force microscopy was applied to visualize the binding of β2GPI to a mixed phospholipid model membrane at physiological ionic strength. On supported lipid bilayers the formation of supramolecular assemblies of the protein with a height of approx. 3.3 nm was observed, suggesting a lateral agglomeration of β2GPI. Detailed analysis of kinetic constants using surface plasmon resonance revealed that the binding can be described by a two-state reaction model, i.e. a very fast interaction step, depending on the content of acidic phospholipids in the bilayer, and a second step with significantly lower kon and koff values. Taken together, our results suggest a biphasic interaction mechanism: a fast step of β2GPI binding to negatively charged lipids, mainly based on electrostatic interactions, and a slower phase of agglomeration of the protein on the bilayer surface accompanied by a protein-induced rigidification of the membrane, as revealed by electron paramagnetic resonance.
Keywords: antiphospholipid syndrome, apolipoprotein H, atomic force microscopy (AFM), β2-glycoprotein I, lupus, surface plasmon resonance (SPR)
Abbreviations: AFM, atomic force microscopy; aPLA, antiphospholipid autoantibody; G, Gauss; β2GPI, β2-glycoprotein I; 5-PCSL, palmitoyl-[2-(3-carboxypropyl)-4,4-dimethyl-2-tridecyl-3-oxazolidinyloxyl]-phosphatidylcholine; 5-PSSL, palmitoyl-[2-(3-carboxypropyl)-4,4-dimethyl-2-tridecyl-3-oxazolidinyloxyl]-phosphatidylserine; POPC, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine; POPS, 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine; RU, resonance units; SPR, surface plasmon resonance; SUV, small unilamellar vesicle
INTRODUCTION
β2-Glycoprotein I (β2GPI), also known as apolipoprotein H, is a soluble 54 kDa plasma glycoprotein, primarily synthesized in the liver. β2GPI circulates in human plasma at a concentration of about 0.2 mg/ml, of which approx. 40% is bound to lipoproteins [1]. The physiological function of this protein is not fully understood, but it is known that β2GPI is involved in a variety of physiologically relevant processes, such as the modulation of platelet-depending thrombosis [2,3], atherosclerosis [4], autoimmune diseases, such as antiphospholipid syndrome [5,6], systemic lupus erythematosus and recurrent fetal loss. Antiphospholipid syndrome is caused by the presence of antiphospholipid autoantibodies (aPLAs) that appear to interact with negatively charged phospholipids. Several studies demonstrated that these antibodies are not directed to the phospholipids themselves, but rather recognize epitopes of lipid-associated proteins [7,8]. Thus lipid-bound β2GPI is considered as target antigen for aPLAs [9,10]. Additionally, it was found that clustering of β2GPI increased its affinity for aPLAs [11], suggesting a possible mechanism for epitope formation by oligomerization [12]. There are also strong indications that β2GPI plays a critical role in the clearance of liposomes containing negatively charged lipids [13]. More recently, it was confirmed that β2GPI binds to membranes of apoptotic cells, acting as an immunological marker. The plasma membrane of living cells is structured asymmetrically regarding the phospholipid composition of the inner and outer leaflet. However, in the course of apoptosis negatively charged phosphatidylserine from the inner leaflet is gradually transferred to the outer leaflet. Thus bilayer asymmetry is lost and the membrane surface becomes negatively charged. β2GPI binds to these cells with high affinity and thereby significantly enhances immunological clearance of apoptotic cells by macrophages [14,15].
X-ray crystallography of β2GPI [16,17] revealed an elongated structure comprising five domains, four complement control protein domains and domain V, which is considered the membrane-binding domain. Domain V is highly conserved and carries a definite positive charge arising from 14 lysine residues (Figures 1A–1C). This structural feature is responsible for pronounced electrostatic interactions with negatively charged surfaces or particles [18,19]. Moreover, domain V has also been shown to interact with hydrophobic ligands [20]. The modular arrangement of the five domains implicates a very high structural flexibility depending on environmental conditions [21]. Moreover, variations in the spatial orientation and conformational changes upon binding of β2GPI to phospholipid membranes are observed [22–25].
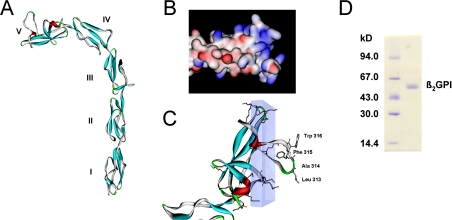
Figure 1. Schematic representations of β2GPI structure and SDS/PAGE.
(A) Crystal structure of β2GPI; domains are numbered from I to V. Crystallographic coordinates were taken from PDB (accession number 1C1Z) [16]. (B) Domain V in a surface potential representation (red, negative; blue, positive; white, uncharged). (C) Detailed structure of domain V. Schematic backbone with α-helix (red) and β-sheet motifs (blue). Residues of the hydrophobic loop and lysine residues are shown in a stick representation. The hydrophobic loop perpendicular to an area containing 11 out of the 14 lysine residues (transparent box) represents the putative phospholipid-binding site. Accelrys DS ViewerPro and POV-Ray were used. (D) Purity of β2GPI samples was verified by SDS/PAGE. The right-hand lane shows the β2GPI band with a molecular mass of approx. 54 kDa; left-hand lane, molecular-mass markers.
The present study entails the in vitro deposition of β2GPI on phospholipid membranes containing the negatively charged phospholipid phosphatidylserine at physiological ionic strength. AFM (atomic force microscopy) was applied to directly visualize the binding of the protein to the lipids and to characterize the morphology of the lipid–protein complexes. SPR (surface plasmon resonance) was used to investigate the binding kinetics and to establish a kinetic model to describe the interaction process. Additionally, EPR spectroscopic data revealed information about protein-induced membrane perturbations.
EXPERIMENTAL
Purification of β2GPI
β2GPI was purified from citrated human plasma by treatment with HClO4, followed by cation-exchange chromatography and heparin-affinity chromatography, as described previously [26]. Purity of the protein was confirmed by SDS/PAGE, as shown in Figure 1(D). Proteins were stored in aliquots at −20 °C for periods of less than 1 month and at −70 °C for longer periods.
Preparation of mixed phospholipid vesicles
POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) and POPS (1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine) (Avanti Polar Lipids, Alabaster, AL, U.S.A.) at a total concentration of 1 mg/ml lipids, at molar ratios of 100/0, 95/5, 90/10, 80/20, were dissolved in 1 ml of chloroform/methanol (4:1). The solvent was evaporated under vacuum and the lipids were swelled in 1 ml of Tris/HCl buffer (20 mM Tris/150 mM NaCl, pH 7.5). After freezing and thawing several times the suspension was extruded 10–15 times through polycarbonate membranes with a pore size of 100 nm.
AFM
A Macmode PicoSPM magnetically-driven dynamic force microscope (Molecular Imaging, Phoenix, AZ, U.S.A.) with a Molecular Imaging Macmode interface was used for AFM measurements. POPC and POPC/POPS vesicles were prepared as described above and fused to freshly cleaved mica in the AFM cell for 15 min. The single-supported bilayer formed was rinsed several times with Tris/HCl buffer with 3 mM Ca2+ before the addition of β2GPI to a final concentration of 5 μM. First topography images were recorded after 10 to 35 min in contact-mode using MicroLevers (Veeco, Santa Barbara, CA, U.S.A.) with a nominal spring constant of 0.01–0.03 N/m at a scanning speed of approx. 1 line/s. Calculations of β2GPI cluster heights within the images were performed using the PicoScan software from Molecular Imaging (variation expressed as S.E.M.).
SPR
Kinetic rates were measured using L1 sensor chips on a Biacore X device (Biacore AB, Uppsala, Sweden). The L1 chips comprise a carboxymethyl dextran hydrogel derivatized with lipophilic alkyl chain anchors to capture phospholipid vesicles. All measurements were carried out using Tris/HCl buffer with or without 3 mM Ca2+ as running and sample buffers at 25 °C. The sensor chips were washed several times with n-octyl-β-D-glucopyranoside (40 mM, 2 min) before injection of the vesicles (0.2 mg/ml) dispersed in buffer solution (60 min; flow rate, 2 μl/min). To remove residual multilayer structures and loosely bound vesicles, a short pulse of 10 mM NaOH was applied. Subsequently, BSA (1 mg/ml, 10 min) was added to block non-specific surface binding. The resulting bilayer linked to the chip surface was taken as a model membrane surface for studying β2GPI binding. Pure buffer solutions and the solution containing β2GPI were applied at a flow rate of 30 μl/min. In parallel, control experiments in the absence of negatively charged POPS (100 mol% POPC) were performed to evaluate unspecific binding of the protein. The control curves (pure POPC bilayer) were subtracted from the binding curves (bilayers containing POPS). In addition, β2GPI binding experiments were carried out at flow rates of 20 and 25 μl/min to test for a possible contribution of mass transport. As almost identical kinetic constants were obtained in these experiments (results not shown), the influence of mass transport can be ignored. All measurements were performed using Tris/HCl buffer containing 500 mM NaCl for desorption of β2GPI (2 min). Then the chips were regenerated by purging the flow cell with n-octyl-β-D-glucopyranoside (40 mM, 2 min) and SDS (0.5%, 2 min) to remove the lipid bilayer from the surface completely.
SPR data analysis
The sensorgrams for each protein–lipid bilayer interaction (L1 chip) were analysed by curve-fitting using numerical integration analysis [27]. The BIAevaluation software (version 4.1; Biacore AB, Uppsala, Sweden) offers various reaction models to perform complete kinetic analyses of the protein sensorgrams, from which the classical Langmuir 1:1 and the two-state binding model were applied (see the Results section). For each lipid composition, sensorgrams obtained at different protein concentrations were fitted collectively.
The following equation describes the two-state model:
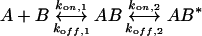 |
1 |
where A is the protein in solution and B is the lipid in the bilayer. AB represents the primary and AB* the secondary β2GPI–lipid complex bound to the membrane. The concentrations at time (t)=0 are: A=[β2GPI], B=Rmax, AB=0, AB*=0, where Rmax is the amount of available β2GPI-binding sites on the lipid bilayer (determined by the fit). The rate equations at t are:
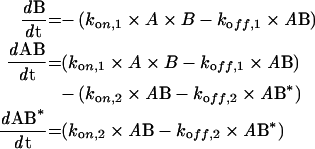 |
2 |
These equations were solved using the Biacore software. The equilibrium constant KD was calculated using the following equation:
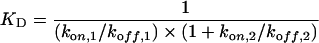 |
5 |
EPR
Phospholipid vesicles of POPC/POPS (80:20 molar ratio) were prepared as described above, with either 5-PCSL {palmitoyl-[2-(3-carboxypropyl)-4,4-dimethyl-2-tridecyl-3-oxazolidinyloxyl]-phosphatidylcholine} or 5-PSSL {palmitoyl-[2-(3-carboxypropyl)-4,4-dimethyl-2-tridecyl-3-oxazolidinyloxyl]-phosphatidylserine} at a molar ratio of spin label to total phospholipid of 1:100. The spin labels were synthesized according to a method described by Marsh et al. [28]. Briefly, the spin-labelled fatty acid 5-DSA [2-(3-carboxypropyl)-4,4-dimethyl-2-tridecyl-3-oxazolidinyloxyl; Sigma–Aldrich] was esterified with 1-palmitoyl-2-hydroxy-sn-glycero-3-phosphocholine and purified by silica gelcolumn chromatography. A part of the purified spin-labelled phosphatidylcholine (5-PCSL) was subject to phospholipase D-catalysed headgroup exchange to yield the spin-labelled phosphatidylserine (5-PSSL) derivative. The products were separated by preparative TLC on silica gel containing a fluorescence marker.
Changes in motional dynamics and relative orientation of the spin-probes were examined by evaluation of the hyperfine splittings 2A∥ and 2A⊥ and calculation of the corresponding order parameters S (eqn 6). EPR spectra were recorded on an X-band ECS 106 spectrometer (Bruker, Rheinstetten, Germany) with modulation amplitude of 1.2 G (Gauss, where 10 G=1 mT) and a sweep width of the static field of 100 G. All spectra were recorded at room temperature. EPR spectra were evaluated using the Bruker WINEPR software package. Final values for 2A∥ and 2A⊥ were obtained by calculation of the arithmetic average from multiple measurements and error evaluation with a significance level of P<0.05.
The order parameter S was calculated using the following equations:
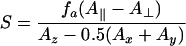 |
6 |
where fa is the polarity correction factor derived from EPR crystal data for Ax, Ay and Az [29]:
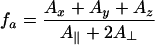 |
7 |
RESULTS
AFM reveals clustering of β2GPI proteins on to phospholipid bilayers
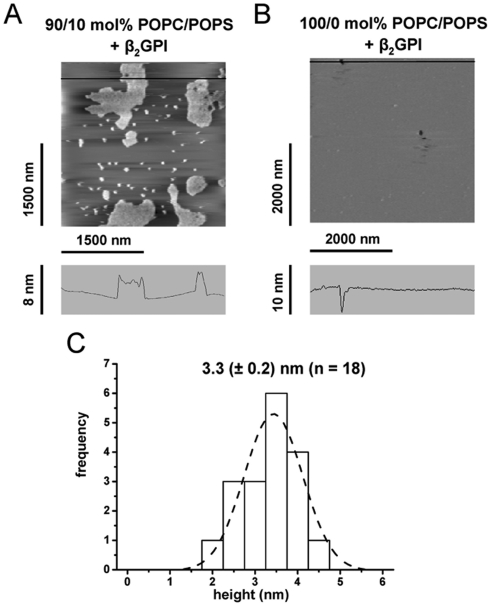
AFM experiments were carried out to visualize the binding of β2GPI to lipid bilayers. For this, vesicles containing pure POPC or a mixture of POPC/POPS were fused to freshly cleaved mica, which resulted in a planar-supported lipid bilayer. No phase separation between POPC and POPS was observed at a temperature of approx. 25 °C (Figure 2B). This finding is in good agreement with the POPC/POPS-phase diagram reported previously [30], which predicts a homogeneous fluid phase for the mixture of POPC and POPS within the molar concentrations used in our present study and for a temperature range between 10 and 37 °C. The bilayer remained stable after several washing cycles with Tris/HCl buffer and 3 mM Ca2+. Figure 2(A) shows an AFM image (POPC/POPS, 90/10 mol%) recorded 35 min after the injection of protein. Partially cohesive protein agglomerates could be resolved with a height of approx. 3 nm, as determined from the cross-section shown in Figure 2(A). A detailed statistical analysis revealed an average height of 3.3 (±0.2 nm) for the protein clusters (Figure 2C). In contrast, the lateral spread of the protein assemblies varied from a few nanometres up to 500 nm. Control experiments were performed by fusing vesicles containing 100 mol% POPC to mica. The appearance of only very few defect structures confirmed the formation of an almost complete bilayer (Figure 2B). No binding of β2GPI was observed to this layer (Figure 2B), demonstrating that the binding to the bilayer strongly depends on the presence of negatively charged lipids, such as POPS.
Figure 2. β2GPI bound to planar-supported lipid bilayers imaged by AFM.
(A) Binding of β2GPI to planar-supported lipid bilayers consisting of a mixture of 90 mol% POPC and 10 mol% POPS on mica. Images were obtained 30–35 min after injection of β2GPI (5 μM) into the AFM cell. The cross-section along the horizontal line is shown below. (B) Control experiment with pure POPC and protein concentration as stated above. The cross-section along the horizontal line shows one of a few defective structures. (C) Statistical height analysis of representative sites (n=18, 3 different experiments) upon binding of β2GPI. The calculated average height (±S.E.M.) of bound β2GPI agglomerates is 3.3±0.2 nm.
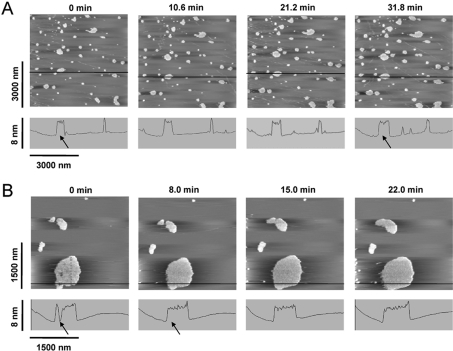
In order to gain more insight into the binding mechanism between β2GPI and lipids, time-resolved AFM experiments were carried out. For these experiments, bilayers comprising a mixture of POPC/POPS were prepared on mica. After 10 to 30 min following protein injection (final concentration, 5 μM), images were recorded every 7 to 11 min in the presence of excess protein in solution. Figure 3 shows typical examples of AFM images with both the formation of new clusters and the growth of pre-existing clusters, as it is expected for nucleation driven processes.
Figure 3. Time-resolved AFM imaging of β2GPI binding.
Time-resolved binding of β2GPI on to (A) 90/10 mol% and (B) 95/5 mol% POPC/POPS supported bilayers on mica. β2GPI was present at a total concentration of 5 μM in the AFM fluid cell. Note the increase in size of the β2GPI agglomerates, as indicated by the arrows in the cross-sections shown.
In Figure 3(A) (10 mol% POPS) AFM images recorded on a randomly chosen area (3000 nm×3000 nm) are presented, whereas the images in Figure 3(B) (5 mol% POPS) were obtained from a large pre-existing protein cluster. Both Figures show time-dependent formation of new protein ‘islands’ on the bilayer, as well as an increase in size of existing clusters. In some cases, gaps between protein agglomerates were filled up (Figure 3B). The height of the clusters, however, remained constant. No significant difference in the binding mechanism between 5 and 10 mol% POPS could be visualized by AFM.
Kinetic rate constants obtained by applying a two-state binding model
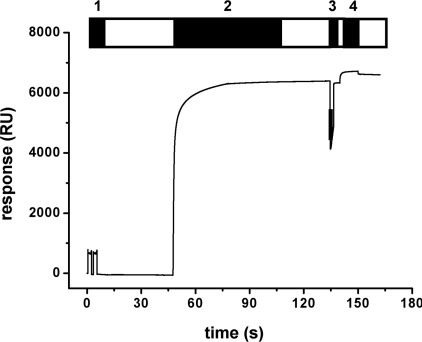
Biacore L1 chips were used for SPR measurements. Previously, it was shown that the liposomes fuse and form a lipid bilayer on the dextran matrix of this chip [31]. Thus, the L1 chip represents the system of choice to study the interaction of proteins, such as β2GPI, with lipid bilayers mimicking cellular membranes. The chip was treated as described in the Experimental section. Figure 4 shows a typical response curve for the loading procedure of the L1 chip. The rapid increase in the signal indicates the binding and fusion of the vesicles. After 1 h of incubation with vesicles a stable bilayer was formed on the chip, resulting in an SPR response in the range of 6000 RU (resonance units) for the POPC/POPS and 7000 RU for pure POPC. These values are comparable with those obtained in a previous study [32]. After injection of NaOH and BSA in order to remove multilayers and block non-specific binding sites, the surface was used to study the interaction between β2GPI and phospholipids.
Figure 4. Sensorgram of a typical SPR experiment.
A Biacore L1 chip was washed several times with detergent (step 1) before POPC/POPS (80/20 mol%) vesicles (step 2) were injected. After adsorption and fusion of vesicles a short pulse of NaOH (10 mM) was applied (step 3) to remove multilayers or loosely bound vesicles. For the blocking of non-specific binding sites, BSA (1 mg/ml) was added (step 4). The chips were subsequently used to study the interaction with β2GPI. Open bars indicate washing with running buffer.
The dependence of the binding kinetics on the content of negatively charged lipids was probed by variation of the molar ratio of POPS to POPC. Representative sensorgrams of the interaction of the protein with a bilayer consisting of 10 mol% POPS (90 mol% POPC) and 20 mol% POPS (80 mol% POPC) are shown in Figures 5(A) and 5(C) (continuous lines) respectively. A solution of β2GPI in the concentration range of 250 to 5000 nM for 10% POPS and 50 to 2500 nM for 20% POPS was applied from t=0 up to 2 min to record protein binding (left part of curves). Subsequently, a buffer solution was injected to initiate the dissociation phase (right part of curves). Binding experiments using bilayers comprising 5 mol% POPS (95 mol% POPC) showed low response (resonance signals lower than 100 RU) and were therefore not further investigated (results not shown).
Figure 5. Sensorgrams of β2GPI binding kinetics to POPC/POPS bilayers immobilized on a Biacore L1 chip.
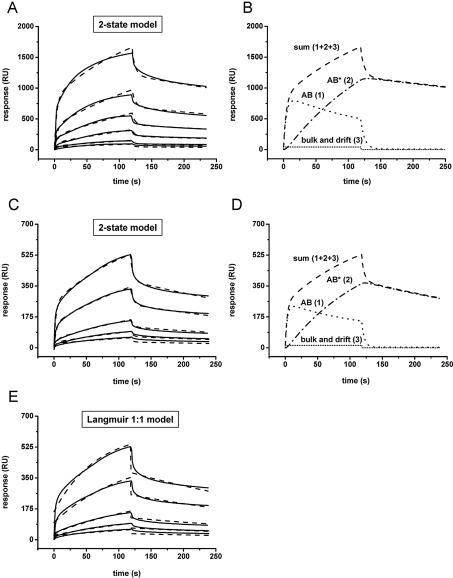
Control experiments with pure POPC vesicles were performed and curves were automatically subtracted. β2GPI was added at t=0 s and pure buffer at t=120 s. Binding curves (——) were measured on a bilayer comprising 80/20 mol% POPC/POPS (A) or 90/10 mol% POPC/POPS (C) with protein concentrations (bottom to top curve) of 50, 100, 250, 500, 1000 and 2500 nM (A) or 250, 500, 1000, 2500 and 5000 nM (C). Fits (–––) were obtained by applying the two-state model. (B, D) An analysis of the fits of the corresponding binding curves measured at the highest protein concentration [2500 and 5000 nM β2GPI, shown in (A) and (C)] (–––, sum of curves 1, 2 and 3) shows the formation of the initial lipid–protein complex (AB,·····, curve 1), the subsequent rearrangement to protein clusters (AB*, –·–·, curve 2) and the bulk and drift effect of the binding (········, curve 3). (E) Binding curves (——) from (C) (10 mol% POPS) fitted by the Langmuir 1:1 model (– – –). Note the significant deviation from raw data at the beginning and end of the protein injection.
The observed agglomeration of β2GPI on the lipid bilayers, as revealed by AFM experiments, strongly suggests a reaction mechanism comprising two steps: (i) initial binding of the protein to the lipids, and (ii) subsequent formation of protein clusters. Hence, a two-state reaction kinetic model was applied to the SPR data using the BiaEvaluation software (Figures 5A and 5C). Fits with good quality and low deviations from raw data were yielded (Figure 5, broken lines). Figures 5(B) and 5(D) show the contributions from the single components to the fit for binding curves measured at the highest protein concentrations from Figures 5(A) and 5(C) (2500 and 5000 nM for 20 and 10 mol% POPS respectively): (i) the formation of the initial lipid–protein complex (AB, curve 1), (ii) the subsequent rearrangement, leading to protein clusters (AB*, curve 2), and (iii) the bulk and drift effect of binding (curve 3). The much slower step of clustering of β2GPI (see curve 2) determines the speed of the overall reaction. For comparison, fits obtained by the classical Langmuir 1:1 binding model are shown in Figure 5(E). The fits differed significantly from the raw data, particularly at the beginning and end of the binding curves. Thus the Langmuir model cannot adequately describe the characteristics of the binding curves and was considered unsuitable as a model for the interaction of β2GPI with the phospholipids.
Applying eqns 2–5, as described in the Experimental section (Biacore Evaluation software), kinetic constants of the interaction of β2GPI with membranes containing 10 or 20 mol% POPS in the presence and absence of Ca2+ (3 mM) were calculated (Table 1). kon values of the first interaction step were significantly higher than those obtained for the second one (by a factor of approx. 106), which is also evident from the curves shown in Figures 5(B) and 5(D). Depending on the concentration of POPS within the lipid bilayers, significant differences in the kinetic constants kon, 1 and the KD values (Table 1) are observed. KD values decreased with increasing POPS content, whereas kon, 1 values increased. This result clearly demonstrates that the first reaction step in the interaction mechanism strongly depends on the content of negatively charged lipids within the lipid bilayer. The increase of the kon, 1 values reflects an enhanced affinity of the protein for membranes with higher content of POPS. This is in good agreement with the proposition that the primary mechanism of binding is based on electrostatic interactions. In the absence of Ca2+ the association of the protein occurred at a higher rate (increased kon, 1 value), which in turn results in a significant decrease of the KD value, confirming the role of Ca2+ as a modulator in the binding process. The lower affinity in the presence of Ca2+ is most likely due to the electrostatic shielding of the negatively charged POPS by bivalent cations.
Table 1. Kinetic constants of the interaction of β2GPI with POPC/POPS lipid bilayers at different phosphatidylserine contents and the effect of Ca2+ (two-state reaction model).
The results are expressed as the mean±S.E.M., n=3–5, P<0.01.
| Phosphatidylserine content (mol%) | [Ca2+] (mM) | kon, 1 (M−1·s−1×104) | koff, 1 (s−1×10−1) | kon, 2 (s−1×10−2) | koff, 2 (s−1×10−3) | KD (nM) |
|---|---|---|---|---|---|---|
| 10 | 3 | 1.67±0.23 | 3.35±1.13 | 3.93±2.28 | 1.80±0.04 | 1078±224 |
| 20 | 3 | 4.62±0.77 | 1.64±0.08 | 2.04±0.19 | 2.05±0.25 | 339±22 |
| 20 | 0 | 10.53±0.04 | 1.99±0.19 | 0.97±0.02 | 0.94±0.44 | 139±30 |
Kinetic constants calculated for the second reaction step (both kon, 2 and koff, 2) did not significantly differ from each other, suggesting that this step does not strongly depend on the content of negatively charged lipids in the membrane. The low koff, 2 rates determine the flat course of the final dissociation part of the sensorgrams (Figure 5).
Membrane phospholipids are subject to a motional restriction, as revealed by EPR
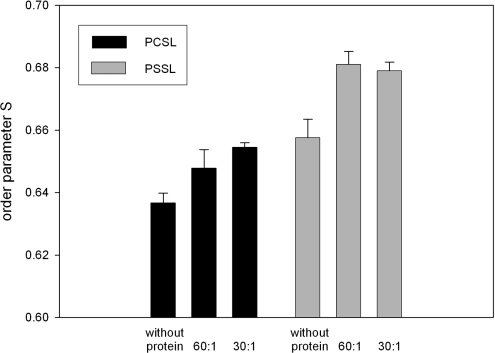
The evaluation of EPR spectra from SUVs (small unilamellar vesicles) containing either spin-labelled phosphatidylserine (5-PSSL) or phosphatidylcholine (5-PCSL) showed a significant increase in the outer hyperfine splittings (2A∥) and a slight decrease in the inner hyperfine splittings (2A⊥) upon addition of β2GPI. Table 2 shows the values for SUVs comprising either POPC/POPS/PSSL or POPC/POPS/PCSL with molar ratios of 80:20:1. Protein was added to these mixtures at a molar ratio (total lipid/protein) of either 60:1 or 30:1. The results indicated a motional restriction near the phospholipid headgroup region, obviously caused by β2GPI. There is no significant difference in the relative change of the rigidity of phospholipid acyl chains in vesicles containing either 5-PCSL or 5-PSSL. The absolute values of the order parameter S indicated a higher rigidity for POPS as compared with POPC. Increasing the protein concentration to a lipid/protein ratio of 30:1 resulted in no significant change compared with a ratio of 60:1 (Figure 6).
Table 2. Outer (2A∥) and inner hyperfine splitting (2A⊥) derived from ESR spectra measured for the interaction of β2GPI with POPC/POPS (80:20 molar ratio) vesicles containing either 5-PCSL or 5-PSSL as spin-labelled phospholipids.
The results are expressed as the mean±S.E.M., n=10, P<0.05.
| 5-PCSL | 5-PSSL | |||
|---|---|---|---|---|
| Ratio (lipid/protein) | 2A∥ (G) | 2A⊥ (G) | 2A∥ (G) | 2A⊥ (G) |
| Without protein | 51.24±0.11 | 18.28±0.09 | 51.59±0.45 | 17.79±0.07 |
| 60:1 | 51.93±0.08 | 18.20 ±0.03 | 52.96±0.17 | 17.58±0.06 |
| 30:1 | 52.13±0.40 | 18.07±0.11 | 52.95±0.29 | 17.63±0.08 |
Figure 6. Membrane perturbation determined by EPR.
Changes of the order parameter S of phospholipid vesicles comprising 20 mol% POPS and 80 mol% POPC as a function of protein/lipid ratio and the type of spin label used. n=10, P<0.05.
DISCUSSION
The present study was focused on the binding of β2GPI on to mixed lipid model membranes comprising the zwitterionic phosphatidylcholine and a smaller fraction of negatively charged phosphatidylserine in the fluid phase at physiological ionic strength. The model system mimics membranes of apoptotic cells that have lost lipid asymmetry with regards to the inner and outer leaflet, exposing negatively charged phospholipids. We found that β2GPI binds to such membranes with high affinity by forming protein agglomerates.
Surface topology of lipid-associated β2GPI
A single molecule of β2GPI exhibits an apparent length of 13.0 nm and an average diameter of 2.3 nm for the complement control protein domains, as revealed by the X-ray crystal structure [16]. For comparison, the average height of clusters obtained by AFM topography imaging is approx. 3.3 nm. This value roughly corresponds to the diameter of the protein and indicates a horizontal-like orientation of the molecules on the bilayer. The surface topography reveals densely packed protein clusters without ordered patterns. Over time, a two-dimensional growth of patches is observed with an increasing overall coverage of the surface. A further accretion of protein molecules perpendicular to the surface does not occur.
According to the X-ray crystal structure, domain V exhibits a hydrophobic loop that is considered to be involved in membrane interaction [18]. In addition, a total of 12 lysine residues are found in close proximity to the putative membrane-binding site (Figure 1C). Hence, long-range electrostatic interaction forces between positively charged lysine residues and negatively charged phospholipid headgroups are presumably responsible for the attraction of the protein molecules to the membrane, whereas hydrophobic short-range interactions will become effective as soon as the hydrophobic loop of the protein comes in close contact with the membrane. The calculated surface area around this hydrophobic loop is approx. 400 Å2. As membrane lipids are in the fluid phase, POPS molecules are likely to be attracted by the highly positive charge of domain V. Assuming a phospholipid headgroup area of approx. 65 Å2 [33], several POPS molecules could be accommodated directly beneath domain V. Given that the lateral diffusion coefficient of a single phospholipid molecule in the fluid membrane is approx. 2.5×10−12 m2/s [34], a migration of initially formed single protein–lipid complexes seems realistic. Hence, we hypothesize that β2GPI binding induces at least partial lipid demixing, resulting in lateral protein clustering accompanied by a locally limited segregation of lipids. However, diffusion of large clusters in the plane of the surface was not observed, most likely because the diffusion rate decreases with increasing cluster size.
Kinetics of two-state binding reaction
The two-state reaction model assumes two distinct reaction complexes, AB and AB*. β2GPI (A) binds to phospholipids (B) yielding a complex AB. Cluster formation leads to a complex AB*, which cannot dissociate into A+B. This model has already been successfully applied for the membrane binding of a toxin [35]. In this study [35], the two steps in the binding are explained by the mediation of two different regions within the toxin molecule. In the case of β2GPI, the first reaction step arises from the initial binding of the positively charged domain V to the negatively charged phospholipids. Most likely, a subsequent demixing of phospholipids promotes the formation of protein agglomerates AB* on the surface as already discussed above (reaction step 2).
The calculated KD values were in the range of 0.14–1.1 μM, depending on the POPS content and the presence of Ca2+. Willems et al. [36] obtained a KD of 0.17 μM for a 20 mol% containing POPC/POPS bilayer in the absence and 3.9 μM in the presence of Ca2+. These values are in good agreement with that obtained in our present study. The slight differences can be explained by two facts: (i) the use of a different kinetic model (Langmuir 1:1 model) that was not appropriate to our system, and (ii) different experimental conditions (e.g. buffers containing BSA and different NaCl concentrations). However, the presence of divalent cations, such as Ca2+, leads to an increase of observed KD values that suggests electrostatic shielding of negatively charged phospholipids by Ca2+ ions. This fact supports the idea that initial β2GPI binding is primarily mediated by electrostatic interactions between domain V and POPS.
SPR revealed the fast kinetics of association and dissociation with regard to membrane composition and bivalent cations. In contrast, AFM provided detailed insight into the topology of the protein lipid assemblies, as well as into the process of their formation. However, the most important difference between SPR and AFM experiments is the timescale of observation. SPR experiments are performed continuously and at very fast acquisition rates. Kinetic data are recorded within less than 2 min, whereas the timescale of AFM experiments extends to a period of approx. 50 min.
Influence of β2GPI binding on bilayer phospholipid organization
We have investigated the effects of β2GPI binding on membrane rigidity by EPR spectroscopy. We found that a rigidification of phospholipid side chains near the headgroup region occurs. This is reflected in an increase of the calculated order parameter value, not only for phosphatidylserine, but also for phosphatidylcholine spin-labelled phospholipids. As POPC is not directly involved into binding of β2GPI and no long-range electrostatic interaction between domain V and choline headgroups exists, the pronounced increase in rigidity of 5-PCSL could either be interpreted as being due to hydrophobic interactions between the hydrophobic C-terminal loop in domain V [20,37] and acyl chains, or by the fact that the hypothesized migration of POPS also affects the organization of POPC molecules.
β2GPI clusters as targets for macrophages and aPLAs
In aqueous solution β2GPI displays an inherent interdomain flexibility and might adopt a number of different conformations, depending on environmental conditions, such as ionic strength or membrane association [21]. This structural feature constitutes an important characteristic, which probably facilitates the exposure of cryptic epitopes. However, it is also conceivable that clustering of β2GPI is the crucial step for a contribution in: (i) the receptor-mediated uptake of apoptotic cells by macrophages [14,15], and (ii) the formation of target antigens for aPLAs in antiphospholipid syndrome. This notion is supported by recent results from Reddel et al. [12] who showed by ELISA techniques using modified plates that for the detection of anti-β2GPI autoantibodies a coating concentration threshold exists such that minimal or no binding occurred below a certain concentration of β2GPI, even though the antigen was easily detectable on the plate. On the basis of this finding, the authors speculated that two adjacent β2GPI are necessary for antibody binding. Moreover, Lutters et al. [10] showed that the formation of β2GPI dimers drastically enhanced the affinity of aPLAs to the protein. In keeping with these observations, the densely packed morphology of β2GPI on a negatively charged phospholipid bilayer as visualized in our present study, may expose structural features very similar to those found in dimeric forms and accordingly facilitates divalent antibody binding. Future AFM and SPR experiments with β2-GPI associated with aPLAs should further clarify the overall picture of the formation of antiphospholipid autoantibody complexes.
β2GPI promotes the recognition of anionic phospholipid vesicles to macrophages, which is mediated by a putative cell surface receptor on macrophages [38]. Consequently, a further goal should be the identification of a macrophage receptor responsible for specific recognition of β2GPI–phospholipid complexes. In summary, our results regarding the kinetics and the protein's membrane organization give further insight into mechanisms and molecular principles of protein functionality.
Acknowledgments
This work was supported by the Austrian Academy of Sciences (DOC 13/2001), the Austrian Science Fund (P-15387 to C.R., P-15295 to H.J.G., P-14549 to P.H. and P-16479 to R.P.) and the Austrian National Bank (NB 10352). The technical assistance of J. Kellner is gratefully appreciated.
References
- 1.Polz E., Kostner G. M. The binding of β2-glycoprotein-I to human serum lipoproteins: distribution among density fractions. FEBS Lett. 1979;102:183–186. doi: 10.1016/0014-5793(79)80955-2. [DOI] [PubMed] [Google Scholar]
- 2.Brighton T. A., Hogg P. J., Dai Y. P., Murray B. H., Chong B. H., Chesterman C. N. β2-Glycoprotein I in thrombosis: evidence for a role as a natural anticoagulant. Br. J. Haematol. 1996;93:185–194. doi: 10.1046/j.1365-2141.1996.446981.x. [DOI] [PubMed] [Google Scholar]
- 3.Shi T., Iverson G. M., Qi J. C., Cockerill K. A., Linnik M. D., Konecny P., Krilis S. A. β2-Glycoprotein I binds Factor XI and inhibits its activation by thrombin and Factor XIIa: loss of inhibition by clipped β2-glycoprotein I. Proc. Natl. Acad. Sci. U.S.A. 2004;101:3939–3944. doi: 10.1073/pnas.0400281101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harats D., George J. β2-glycoprotein I and atherosclerosis. Curr. Opin. Lipidol. 2001;12:543–546. doi: 10.1097/00041433-200110000-00010. [DOI] [PubMed] [Google Scholar]
- 5.McNeil H. P., Simpson R. J., Chesterman C. N., Krilis S. A. Anti-phospholipid antibodies are directed against a complex antigen that includes a lipid-binding inhibitor of coagulation: β2-glycoprotein I (apolipoprotein H) Proc. Natl. Acad. Sci. U.S.A. 1990;87:4120–4124. doi: 10.1073/pnas.87.11.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McNeil H. P., Chesterman C. N., Krilis S. A. Immunology and clinical importance of antiphospholipid antibodies. Adv. Immunol. 1991;49:193–280. doi: 10.1016/s0065-2776(08)60777-4. [DOI] [PubMed] [Google Scholar]
- 7.Galli M., Comfurius P., Maassen C., Hemker H. C., de Baets M. H., van Breda-Vriesman P. J., Barbui T., Zwaal R. F., Bevers E. M. Anticardiolipin antibodies (ACA) directed not to cardiolipin but to a plasma protein cofactor. Lancet. 1990;335:1544–1547. doi: 10.1016/0140-6736(90)91374-j. [DOI] [PubMed] [Google Scholar]
- 8.Roubey R. A., Eisenberg R. A., Harper M. F., Winfield J. B. ‘Anticardiolipin’ autoantibodies recognize β2-glycoprotein I in the absence of phospholipid. Importance of Ag density and bivalent binding. J. Immunol. 1995;154:954–960. [PubMed] [Google Scholar]
- 9.Lutters B. C., Derksen R. H., Tekelenburg W. L., Lenting P. J., Arnout J., de Groot P. G. Dimers of β2-glycoprotein I increase platelet deposition to collagen via interaction with phospholipids and the apolipoprotein E receptor 2′. J. Biol. Chem. 2003;278:33831–33838. doi: 10.1074/jbc.M212655200. [DOI] [PubMed] [Google Scholar]
- 10.Lutters B. C., Meijers J. C., Derksen R. H., Arnout J., de Groot P. G. Dimers of β2-glycoprotein I mimic the in vitro effects of β2-glycoprotein I–anti-β2-glycoprotein I antibody complexes. J. Biol. Chem. 2001;276:3060–3067. doi: 10.1074/jbc.M008224200. [DOI] [PubMed] [Google Scholar]
- 11.Sheng Y., Kandiah D. A., Krilis S. A. Anti-β2-glycoprotein I autoantibodies from patients with the ‘antiphospholipid’ syndrome bind to β2-glycoprotein I with low affinity: dimerization of β2-glycoprotein I induces a significant increase in anti-β2-glycoprotein I antibody affinity. J. Immunol. 1998;161:2038–2043. [PubMed] [Google Scholar]
- 12.Reddel S. W., Wang Y. X., Krilis S. A. Anti-β2-glycoprotein I autoantibodies require an antigen density threshold, consistent with divalent binding. Lupus. 2003;12:37–45. doi: 10.1191/0961203303lu280oa. [DOI] [PubMed] [Google Scholar]
- 13.Chonn A., Semple S. C., Cullis P. R. β2 glycoprotein I is a major protein associated with very rapidly cleared liposomes in vivo, suggesting a significant role in the immune clearance of ‘non-self’ particles. J. Biol. Chem. 1995;270:25845–25849. doi: 10.1074/jbc.270.43.25845. [DOI] [PubMed] [Google Scholar]
- 14.Balasubramanian K., Chandra J., Schroit A. J. Immune clearance of phosphatidylserine-expressing cells by phagocytes. The role of β2-glycoprotein I in macrophage recognition. J. Biol. Chem. 1997;272:31113–31117. doi: 10.1074/jbc.272.49.31113. [DOI] [PubMed] [Google Scholar]
- 15.Balasubramanian K., Schroit A. J. Characterization of phosphatidylserine-dependent β2-glycoprotein I macrophage interactions. Implications for apoptotic cell clearance by phagocytes. J. Biol. Chem. 1998;273:29272–29277. doi: 10.1074/jbc.273.44.29272. [DOI] [PubMed] [Google Scholar]
- 16.Schwarzenbacher R., Zeth K., Diederichs K., Gries A., Kostner G. M., Laggner P., Prassl R. Crystal structure of human β2-glycoprotein I: implications for phospholipid binding and the antiphospholipid syndrome. EMBO J. 1999;18:6228–6239. doi: 10.1093/emboj/18.22.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bouma B., de Groot P. G., van den Elsen J. M., Ravelli R. B., Schouten A., Simmelink M. J., Derksen R. H., Kroon J., Gros P. Adhesion mechanism of human β(2)-glycoprotein I to phospholipids based on its crystal structure. EMBO J. 1999;18:5166–5174. doi: 10.1093/emboj/18.19.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagihara Y., Hong D. P., Hoshino M., Enjyoji K., Kato H., Goto Y. Aggregation of β2-glycoprotein I induced by sodium lauryl sulfate and lysophospholipids. Biochemistry. 2002;41:1020–1026. doi: 10.1021/bi015693q. [DOI] [PubMed] [Google Scholar]
- 19.Del Papa N., Sheng Y. H., Raschi E., Kandiah D. A., Tincani A., Khamashta M. A., Atsumi T., Hughes G. R., Ichikawa K., Koike T., et al. Human β2-glycoprotein I binds to endothelial cells through a cluster of lysine residues that are critical for anionic phospholipid binding and offers epitopes for anti-β2-glycoprotein I antibodies. J. Immunol. 1998;160:5572–5578. [PubMed] [Google Scholar]
- 20.Hong D. P., Hagihara Y., Kato H., Goto Y. Flexible loop of β2-glycoprotein I domain V specifically interacts with hydrophobic ligands. Biochemistry. 2001;40:8092–8100. doi: 10.1021/bi010196v. [DOI] [PubMed] [Google Scholar]
- 21.Hammel M., Kriechbaum M., Gries A., Kostner G. M., Laggner P., Prassl R. Solution structure of human and bovine β2-glycoprotein I revealed by small-angle X-ray scattering. J. Mol. Biol. 2002;321:85–97. doi: 10.1016/s0022-2836(02)00621-6. [DOI] [PubMed] [Google Scholar]
- 22.Wang F., Xia X. F., Sui S. F. Human apolipoprotein H may have various orientations when attached to lipid layer. Biophys. J. 2002;83:985–993. doi: 10.1016/S0006-3495(02)75224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J., Sykes B. D., Ryan R. O. Structural basis for the conformational adaptability of apolipophorin III, a helix-bundle exchangeable apolipoprotein. Proc. Natl. Acad. Sci. U.S.A. 2002;99:1188–1193. doi: 10.1073/pnas.032565999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang S. X., Cai G. P., Sui S. F. The insertion of human apolipoprotein H into phospholipid membranes: a monolayer study. Biochem. J. 1998;335:225–232. doi: 10.1042/bj3350225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang S. X., Sun Y. T., Sui S. F. Membrane-induced conformational change in human apolipoprotein H. Biochem. J. 2000;348:103–106. [PMC free article] [PubMed] [Google Scholar]
- 26.Gries A., Nimpf J., Wurm H., Kostner G. M., Kenner T. Characterization of isoelectric subspecies of asialo-β2-glycoprotein I. Biochem. J. 1989;260:531–534. doi: 10.1042/bj2600531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morton T. A., Myszka D. G., Chaiken I. M. Interpreting complex binding kinetics from optical biosensors: a comparison of analysis by linearization, the integrated rate equation, and numerical integration. Anal. Biochem. 1995;227:176–185. doi: 10.1006/abio.1995.1268. [DOI] [PubMed] [Google Scholar]
- 28.Marsh D., Watts A., Pates R. D., Uhl R., Knowles P. F., Esmann M. ESR spin-label studies of lipid–protein interactions in membranes. Biophys. J. 1982;37:265–274. doi: 10.1016/S0006-3495(82)84675-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hubbell W. L., McConnell H. M. Molecular motion in spin-labeled phospholipids and membranes. J. Am. Chem. Soc. 1971;93:314–326. doi: 10.1021/ja00731a005. [DOI] [PubMed] [Google Scholar]
- 30.Silvius J. R., McMillen D. A., Saley N. D., Jost P. C., Griffith O. H. Competition between cholesterol and phosphatidylcholine for the hydrophobic surface of sarcoplasmic reticulum Ca2+-ATPase. Biochemistry. 1984;23:538–547. doi: 10.1021/bi00298a022. [DOI] [PubMed] [Google Scholar]
- 31.Erb E. M., Chen X., Allen S., Roberts C. J., Tendler S. J., Davies M. C., Forsen S. Characterization of the surfaces generated by liposome binding to the modified dextran matrix of a surface plasmon resonance sensor chip. Anal. Biochem. 2000;280:29–35. doi: 10.1006/abio.1999.4469. [DOI] [PubMed] [Google Scholar]
- 32.Baird C. L., Courtenay E. S., Myszka D. G. Surface plasmon resonance characterization of drug/liposome interactions. Anal. Biochem. 2002;310:93–99. doi: 10.1016/s0003-2697(02)00278-6. [DOI] [PubMed] [Google Scholar]
- 33.May S., Harries D., Ben-Shaul A. Lipid demixing and protein–protein interactions in the adsorption of charged proteins on mixed membranes. Biophys. J. 2000;79:1747–1760. doi: 10.1016/S0006-3495(00)76427-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benes M., Billy D., Hermens W. T., Hof M. Muscovite (mica) allows the characterisation of supported bilayers by ellipsometry and confocal fluorescence correlation spectroscopy. Biol. Chem. 2002;383:337–341. doi: 10.1515/BC.2002.037. [DOI] [PubMed] [Google Scholar]
- 35.Hong Q., Gutierrez-Aguirre I., Barlic A., Malovrh P., Kristan K., Podlesek Z., Macek P., Turk D., Gonzalez-Manas J. M., Lakey J. H., Anderluh G. Two-step membrane binding by Equinatoxin II, a pore-forming toxin from the sea anemone, involves an exposed aromatic cluster and a flexible helix. J. Biol. Chem. 2002;277:41916–41924. doi: 10.1074/jbc.M204625200. [DOI] [PubMed] [Google Scholar]
- 36.Willems G. M., Janssen M. P., Pelsers M. M., Comfurius P., Galli M., Zwaal R. F., Bevers E. M. Role of divalency in the high-affinity binding of anticardiolipin antibody-β2-glycoprotein I complexes to lipid membranes. Biochemistry. 1996;35:13833–13842. doi: 10.1021/bi960657q. [DOI] [PubMed] [Google Scholar]
- 37.Hammel M., Schwarzenbacher R., Gries A., Kostner G. M., Laggner P., Prassl R. Mechanism of the interaction of β2-glycoprotein I with negatively charged phospholipid membranes. Biochemistry. 2001;40:14173–14181. doi: 10.1021/bi0114372. [DOI] [PubMed] [Google Scholar]
- 38.Thiagarajan P., Le A., Benedict C. R. β2-Glycoprotein I promotes the binding of anionic phospholipid vesicles by macrophages. Arterioscler. Thromb. Vasc. Biol. 1999;19:2807–2811. doi: 10.1161/01.atv.19.11.2807. [DOI] [PubMed] [Google Scholar]