Abstract
The matrilin-1 gene has the unique feature that it is expressed in chondrocytes in a developmental stage-specific manner. Previously, we found that the chicken matrilin-1 long promoter with or without the intronic enhancer and the short promoter with the intronic enhancer restricted the transgene expression to the columnar proliferative chondroblasts and prehypertrophic chondrocytes of growth-plate cartilage in transgenic mice. To study whether the short promoter shared by these transgenes harbours cartilage-specific control elements, we generated transgenic mice expressing the LacZ reporter gene under the control of the matrilin-1 promoter between −338 and +67. Histological analysis of the founder embryos demonstrated relatively weak transgene activity in the developing chondrocranium, axial and appendicular skeleton with highest level of expression in the columnar proliferating chondroblasts and prehypertrophic chondrocytes. Computer analysis of the matrilin-1 genes of amniotes revealed a highly conserved Pe1 (proximal promoter element 1) and two less-conserved sequence blocks in the distal promoter region. The inverted Sox motifs of the Pe1 element interacted with chondrogenic transcription factors Sox9, L-Sox5 and Sox6 in vitro and another factor bound to the spacer region. Point mutations in the Sox motifs or in the spacer region interfered with or altered the formation of nucleoprotein complexes in vitro and significantly decreased the reporter gene activity in transient expression assays in chondrocytes. In vivo occupancy of the Sox motifs in genomic footprinting in the expressing cell type, but not in fibroblasts, also supported the involvement of Pe1 in the tissue-specific regulation of the gene. Our results indicate that interaction of Pe1 with distal DNA elements is required for the high level, cartilage- and developmental stage-specific transgene expression.
Keywords: cartilage-specific gene regulation, conserved promoter element, in vivo footprinting, matrilin, Sox9-binding site, transgenic mice
Abbreviations: CEC, chicken embryo chondrocyte; CEF, chicken embryo fibroblast; DMS, dimethyl sulphate; Dpe, distal promoter element; DTT, dithiothreitol; EMSA, electrophoretic mobility-shift assay; GST, glutathione S-transferase; HDM, high-density mesenchyme; HMG, high-mobility group; LM-PCR, ligation-mediated PCR; NFI, nuclear factor I; Pe1, proximal promoter element 1; SI, silencer element I
INTRODUCTION
Vertebrate skeleton develops as a result of many morphogenetic and differentiation steps [1]. Most skeletal elements are formed by endochondral ossification through a cartilaginous template. Chondrocyte precursors originate from the lateral plate mesoderm, cranial neural crest and sclerotome. After migrating to their destiny and undergoing commitment, the mesenchymal cells first aggregate and form precartilaginous condensations in the developing embryo. Then the prechondrocytes differentiate into chondroblasts, which progress through several differentiation stages including proliferation, maturation to chondrocytes and hypertrophy. Finally, the mineralized hypertrophic cartilage is replaced by bone. These differentiation steps are accompanied by significant changes in the morphology and gene expression pattern of the cells [1–3]. Thus the prechondrocytes and chondroblasts turn off the mesenchymal marker genes and start to express the cartilage protein genes in a sequential order [2–5]. In accordance with the in vivo observations, activation of the genes for type II collagen, aggrecan and cartilage link protein takes place in the early proliferative stage (stage Ia), whereas the matrilin-1 gene is turned on only in the late proliferative stage (stage Ib) of in vitro chondrogenesis [2,6,7]. Recent advances shed light on the transcriptional control of the chondrocyte lineage [8,9], but our knowledge is still limited on the regulation of the sequential activation of cartilage protein genes during chondrogenesis.
The essential role of three Sox proteins was reported in chondrogenic differentiation and in the activation of cartilage protein genes [8,9]. Sox proteins carry a single HMG (high-mobility group) box DNA-binding domain highly similar to that of Sry, a mammalian testis-determining factor [10,11]. HMG box domains interact with the minor groove of the DNA helix and bend the DNA. They can also recognize four-way junction sequences [12]. Sox domains bind to the CA/TTTGA/TA/T motif with moderate affinity [9,11,13]. In addition, some of the Sox proteins (e.g. Sox9) have a transcription activation domain and thus work as typical transcription factors. Furthermore, Sox proteins playing key roles in development often interact with partner factors [11].
The Sox9, L-Sox5 and Sox6 genes are turned on in chondro-progenitor cells and have a high level of expression in chondrocytes and some other cell types [8,9]. In campomelic dysplasia, SOX9 haploinsufficiency leads to skeletal abnormalities and XY sex reversal [14,15]. The absence of mesenchymal condensation and endochondral bone formation as well as the lack of activation of cartilage protein genes in Sox9–/– transgenic mice identified Sox9 as a chondrogenic master transcription factor [16]. Double inactivation of L-Sox5 and Sox6 in transgenic mice also severely interfered with chondroblast differentiation, prevented the activation of the matrilin-1 gene and highly decreased the expression level of genes for type II collagen (Col2a1), aggrecan (Agc) and cartilage link protein (Crtl1) [17]. Furthermore, the interaction of the 48 bp Col2a1 enhancer element with Sox9 and L-Sox5/Sox6 in vitro indicated that Sox proteins could regulate the Col2a1 transcription [18].
Previously, we cloned the gene for chicken matrilin-1 [19], the first member of the matrilin family of multiadhesion proteins. Matrilins are expressed in a unique and partially overlapping pattern and function as oligomeric adaptor molecules in the extracellular matrix of skeletal and other tissues [20]. Matrilin-1 (previously called cartilage matrix protein, CMP) is highly abundant in certain forms of hyaline cartilage. It can covalently bind to aggrecan [21] and, through the vWFA domains, it can form both collagen-dependent and independent fibrillar extracellular networks [22]. Thus matrilin-1 may perform a bridging function between the two major macromolecular networks of cartilage. The matrilin-1 gene also serves as a marker gene for the late proliferative stage of chondrogenesis [6,7].
The major control regions of the chicken matrilin-1 gene were mapped previously [23–25]. In transient expression experiments, we found a chondrocyte-specific positive control region in the first intron [23]. We also showed that the promoter fragment between positions −1137 and +64 conferred tissue- and developmental stage-specific regulation to the reporter gene due to an interplay between two positive and two negative regions [24]. We characterized the TATA proximal SI (silencer element I), which functioned by binding NFI (nuclear factor I)-family proteins. Recently, we have also provided evidence in transgenic mice that the long promoter (between −2011 and +67) alone and the short promoter with the intronic fragment (between −338 and +1819) were equally capable of directing the differentiation stage-specific expression of the reporter gene in chondrocytes [25]. In congruence with the expression pattern of the endogenous matrilin-1 gene, activity of the transgenes was restricted to the columnar proliferating and prehypertrophic zones of the growth plate. However, the presence of both promoter upstream and intronic elements was necessary for the high-level transgene activity in all chondrogenic tissues and for the extraskeletal transgene expression pattern most closely resembling the chicken matrilin-1 gene [25]. Our results suggested that relatively weak cartilage-specific elements dispersed in the promoter and first intron regulate the chicken gene.
To gain further insight into the transcriptional control of the restricted tissue-specific expression of the matrilin-1 gene, we searched for conserved motifs in the promoters of various vertebrate species and addressed the question whether Sox proteins are involved in this regulation. In the present study, we report on the functional analysis of the chicken matrilin-1 short promoter in transgenic mice and identification of a highly conserved DNA element interacting with cartilage-specific Sox factors in vitro. In vivo occupancy of the identified factor-binding sites is also studied in genomic footprinting, and the putative role of the element in the transcriptional control of the gene is discussed.
EXPERIMENTAL
Sequence analysis
All sequences were obtained either from the ENSEMBL genome browser [26], or from the EMBL databank [27]. Sequence manipulations were performed using the programs of the EMBOSS package [28]. Multiple alignments were made by the DIALIGN2 program [29] and were further improved by hand. Conserved motifs were searched with the MEME motif discovery program [30].
Oligonucleotides and plasmid constructions
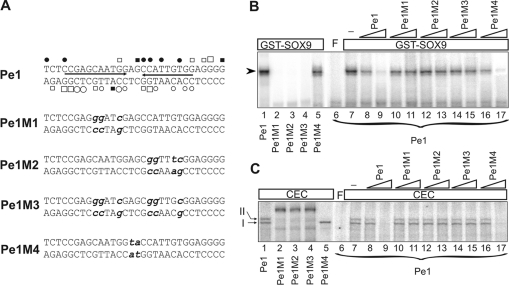
Double-stranded oligonucleotides were synthesized for Pe1 (promoter element 1): 5′-TCTCCGAGCAATGGAGCCATTGTGGAGGGG-3′; the consensus HMG box site: 5′-ACACTGAGAACAAAGCGCTCTCACAC-3′; and the consensus Sox element: 5′-GATCCGGACTAATAAACAATAAAGTCGACG-3′. Nucleotide sequences for the point mutant versions of Pe1 referred to as Pe1M1, Pe1M2, Pe1M3 and Pe1M4 are depicted in Figure 7(A).
Figure 7. Mutational analysis of Pe1 by EMSA.
(A) Nucleotide sequences of the wild-type and mutant versions of Pe1. The inverted repeats (arrows) harbouring the paired Sox motifs as well as protections and hyperreactivities are indicated as in Figure 5. Point mutations presented in bold lower case letters were introduced at nucleotides, which showed in vivo occupancy at the Sox motifs or in the spacer region. (B) Interaction of the wild-type (lane 1) and mutant versions (lanes 2–5) of radiolabelled Pe1 with 2.0 μg of purified GST–SOX9. Binding of radiolabelled Pe1 to GST–SOX9 was competed by 50- and 500-fold molar excess of unlabelled normal and mutant Pe1 as indicated on the top (lanes 8–17). (C) EMSA was performed to compare the CEC nucleoprotein complexes formed on the wild-type and mutant Pe1 elements (lanes 1–5). Formation of CEC nucleoprotein complexes was competed with 50- and 500-fold molar excess of unlabelled Pe1 and its mutant versions as indicated on the top (lanes 8–17). No competitor was added to lane 7. F, free probe.
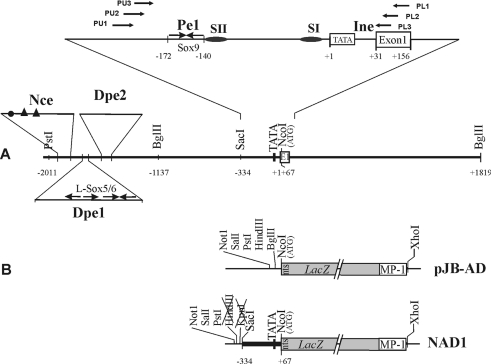
As reported in [23], all positions are given in bp from the first T of the TATA motif of the chicken matrilin-1 gene. Plasmid pJB-AD was made by ligating the NotI, SalI, PstI, HindIII, BglII and NcoI polylinker at the NotI and NcoI sites of vector pJB-HRV [25] upstream of the LacZ cassette bordered by a nuclear transport signal and the MP1 polyadenylation site. To construct NAD1, the KpnI (blunted)–NcoI fragment from a subclone carrying the matrilin-1 short promoter between −334 and +67 was inserted at the HindIII (blunted) and NcoI sites of pJB-AD.
The AvaI (blunted)–NcoI fragment harbouring the matrilin-1 minimal promoter between positions −15 to +67 was inserted into the SmaI and NcoI site of the pGL3-basic vector (Promega, Madison, WI, U.S.A.) to produce MpLuc. In construct 4×Pe1-MpLuc, four copies of the Pe1 element subcloned in the EcoRV site of pBluescript II SK(+) (Stratagene, La Jolla, CA, U.S.A.) was excised with SacI–HindIII (blunted) and ligated to the SacI–NheI (blunted) site of MpLuc. The SacI–NcoI fragment including the proximal promoter region of chicken matrilin-1 gene between −334 and +67 was ligated to the SacI and NcoI site of the pGL3-basic vector to produce the FO15Luc construct. FO15Luc derivatives ΔPe1M1 and ΔPe1M4 were made by PCR-based Quik Change® site-directed mutagenesis (Stratagene) according to the manufacturer's instructions using FO15Luc as a template and oligonucleotides Pe1M1 and Pe1M4 carrying point mutations in the Sox9 recognition site and in the spacer region of Pe1 respectively. Two synthetic oligonucleotide primers containing the desired mutation, each complementary to opposite strands of the vector, were extended during temperature cycling by Pyrococcus furiosus DNA polymerase (Fermentas AB, Vilnius, Lithuania). After the cycling, the products were treated with DpnI digestion to eliminate the methylated parental template. The nicked DNAs were then transformed into Escherichia coli and repaired. The BglII–NcoI matrilin-1 promoter fragment between −1137 and +67 was inserted into the BglII and NcoI sites of pGL3-basic to obtain pCMPLuc. AC8Luc carries the long PstI (blunted)–NcoI promoter fragment between positions −2011 and +67 at the SmaI (blunted) and NcoI sites of the same vector. The structure of all the constructs and the accuracy of the mutagenesis were verified by restriction mapping and nucleotide sequencing.
Generation and analysis of transgenic mice
To generate transgenic mice, the purified NotI–XhoI fragment of NAD1 was microinjected into the male pronuclei of fertilized oocytes obtained from (C57BL/6×CBA)F1 intercrosses as described in [25]. Recipient females were killed on E15.5 (embryonic day 15.5), and the presence of the transgene in founder (G0) embryos was detected by PCR analysis and dot-blot hybridization of placental DNA. Positive G0 embryos were fixed and stained with X-Gal, photographed as whole mounts, and subjected for histological analysis as described previously [25]. All animal experiments were conducted according to the ethical standards of the Animal Health Care and Control Institute, Csongrád County, Hungary.
Cell culture
Preparation of CEC (chicken embryo chondrocyte) and CEF (chicken embryo fibroblast) cultures and HDM (high-density mesenchyme) cultures undergoing chondrogenesis in vitro was described in [6,24]. The mouse fibroblast cell line NIH3T3 was obtained from A.T.C.C. (Manassas, VA, U.S.A.) and maintained in Dulbecco's modified Eagle's medium and 10% (v/v) fetal calf serum (Sigma–Aldrich, MO, U.S.A.).
In vivo footprinting with DMS (dimethyl sulphate) and UV light
A total of 7.5×106–1×107 CEC and 5×106 CEF cells were treated with 0.2% DMS for 5 min at room temperature (22 °C) or irradiated with 2400 J/m2 UV light in vivo and subjected to genomic footprinting as described in [31]. Briefly, genomic DNA was isolated from the cultures treated in vivo (v) by incubation with 600 μg/ml proteinase K and compared with ‘naked’ DNA treated with DMS and UV light in vitro (t). DNA samples (30 μg of ‘v’ and ‘t’) were cleaved with 100 μl of 1 M piperidine at 88 °C for 30 min and amplified by LM-PCR (ligation-mediated PCR) [31] between positions −227 and +140 using gene-specific nested primers PU1, 5′-TGTTCCCATCCCAGATTCC-3′ and PU2, 5′-TCCCAGATTCCCCACATACCGC-3′ for the upper strand; PL1, 5′-GGAGGTGCCCCCCAGA-3′ and PL2, 5′-TGCCCCCCAGACTCCACAGCT-3′ for the lower strand (Figure 1A); and linker primers LP11, 5′-GAATTCAGATC-3′ and LP25, 5′-GCGGTGACCCGGGAGATCTGAATTC-3′. First, a gene-specific primer (PU1 or PL1) annealed to the cleaved and denatured genomic DNA was extended with Sequenase™ and the double-stranded linker was blunt-end ligated. Then the fragments were PCR-amplified using LP25 and PU2 or PL2. The PCR ladders were separated on sequencing gels with G+A and C+T sequence ladders, transferred on to nylon membranes and hybridized with 32P-labelled single-stranded PCR probes made with gene-specific primers PU3, 5′-GCGTGGCTGCGGGTCCCT-3′ or PL3, 5′-CCACAGCTCTGGAGGAGAAGCAG-3′ (Figure 1A).
Figure 1. Diagram of the promoter region of the chicken matrilin-1 gene analysed using in vivo and in vitro techniques.
(A) Schematic figure of the 5′-end of the gene depicts the location of conserved sequence blocks harbouring potential distal (Dpe1 and Dpe2) and proximal promoter elements (Pe1) and initiator element (Ine). Numbers indicate the positions relative to the TATA motif. In the expanded view of the proximal promoter region, horizontal arrows mark the location of primers used in genomic footprinting and oval symbols denote the SI and SII elements. Inverted arrows represent the inverted pairs of conserved Sox9 and L-Sox5/Sox6 motifs in Pe1 and Dpe1 respectively. Closed circle and closed triangles depict the putative Ikaros2- and ΔEF-binding sites in a potential neural crest-specific element (Nce) discussed previously [25]. (B) Schematic map of the vector pJB-AD and the fusion construct NAD1 carrying the short promoter of the chicken matrilin-1 gene, the LacZ cassette preceded by the SV40 nuclear transport signal (nts) and the MP-1 polyadenylation signal.
Preparation of cell extracts
Crude cell extracts were made from CEC, CEF and NIH3T3 cultures for EMSA (electrophoretic mobility-shift assay). Cells were lysed in 20 mM Hepes (pH 7.9), 350 mM NaCl, 0.5 mM EDTA, 0.5 mM EGTA, 2 mM DTT (dithiothreitol), 0.2 mM PMSF and 1% Nonidet P 40 on ice for 30 min. The chromatin was centrifuged for 5 min at 20800 g at 4 °C. The supernatants were supplemented with 10% (v/v) glycerol, aliquoted and stored at −80 °C.
Synthesis and purification of bacterially expressed GST (glutathione S-transferase)–SOX9
The GST–SOX9 vector for bacterial expression of the fusion of GST with full-length SOX9 and the empty control vector were kindly provided by P. Berta (University of Freiburg, Freiburg, Germany; [32]). The recombinant protein was produced in bacterial strain BL21-CodonPlus (DE3)-RIL after induction with 0.1 mM isopropyl-β-D-thiogalactopyranoside for 2 h at 37 °C. The cells were harvested, resuspended in 150 mM NaCl, 1 mM DTT, 5 mM EDTA, 25% (w/v) sucrose and 50 mM Tris (pH 7.5), and sonicated for 2 min at 4 °C. After centrifugation, the cleared lysate was applied on to glutathione–Sepharose™ 4B beads (Amersham Biosciences, Upsala, Sweden) and washed three times in 250 mM NaCl, 5 mM EDTA, 50 mM Tris (pH 7.6) and three times in 120 mM NaCl, 5 mM EDTA and 50 mM Tris (pH 7.6). Purified proteins were eluted from the matrix with the latter buffer supplemented with 10 mM GSH by incubating for 30 min at 4 °C. Yields were tested by SDS/PAGE.
EMSA and supershift experiments
Crude cell extracts were incubated in the presence of 100 ng of poly(dG-dC)·(dG-dC) and various unlabelled competitors with 10–20 fmol of end-labelled DNA probe, and loaded on prerun 5–6.6% polyacrylamide gel in 22.5 mM Tris/borate/0.25 mM EDTA (pH 8) as described in [24]. Supershift experiments were performed as described in [24] with antisera raised against SOX9, L-Sox5 and Sox6 [18] kindly provided by B. de Crombrugghe and V. Lefebvre (University of Texas, M.D. Anderson Cancer Center, Houston, TX, U.S.A.).
Transient expression assay
CEC, CEF and HDM cultures were transfected with the calcium phosphate co-precipitation method as described in [24]. Cells were transfected 24 h after plating with 5 μg of each luciferase construct. To test for transfection efficiency, 0.5 μg of pCAT®3 control vector (Promega) was included as an internal control, and an empty vector was added to give a total DNA amount of 10 μg. Parallel plates were also transfected with pGL3-control vector (Promega). The transfected cells were harvested 48 h later by centrifugation and the cell pellet was lysed by two cycles of freezing and thawing in 200 μl of 50 mM Tris/HCl, 2 mM DTT, 0.5 mM EDTA, 0.5 mM EGTA and 1% Nonidet P40. Luciferase activity was measured from the supernatant in a Luminoscan Ascent (ThermoLabsystem 2.6) using luciferin substrate (Promega) [33]. Relative luciferase activities were expressed in percentage of that of FO15Luc taken as 100%. All transfections were performed in duplicates with at least two different DNA preparations. Data are presented as means±S.E.M.
RESULTS
Identification of conserved sequence blocks in the promoters of amniote matrilin-1 genes
To identify potential control elements in the regulatory region of the matrilin-1 gene, we searched for conserved sequence blocks in various vertebrate species using the DIALIGN2 program [29]. The location of the most-conserved DNA segments is shown in Figure 1(A). Within the 2–3 kb promoter region tested, only one putative element exhibited very strong sequence conservation between mammals and chicken. This proximal promoter element, herein referred to as Pe1, is located 100–200 bp upstream of TATA in amniotes and includes a pair of inverted motifs highly similar to the AGAACAATGG motif, which was shown to be the preferred binding site of Sox9 in vitro [13] (Figure 2). The inverted Sox motifs are separated by two nucleotides within Pe1. Apart from this, only two distal DNA segments, Dpe1 (distal promoter element 1) and Dpe2, show a certain degree of sequence similarity between mammals and chicken (Figures 1A and 2). Dpe1 harbours motifs sharing 6/8 bp identity with the preferred recognition sequence for L-Sox5/Sox6 [13]. Dpe2 includes the fully conserved GACACAGAGAA motif, which does not match with any consensus motif of known transcription factors of the TRANSFAC® database (http://www.gene-regulation.com/pub/databases.htm1#transfac). The degree of sequence similarity around the TATA motif was relatively weak when compared with other eukaryotic promoters. Apart from the TATA box, only the TGTGCCG/A motif was found uniformly in the initiator element (Ine) of various amniote species tested (Figure 2). This motif harbours the first transcription start site of the chicken gene.
Figure 2. Conserved sequence blocks in the matrilin-l promoter region of amniotes.
In the alignment made by the DIALIGN2 program [29], the most conserved regions between the human, mouse, rat, dog and chicken matrilin-1 promoter sequences are shown. The human, mouse and rat matrilin-1 promoter sequence data are from the ENSEMBL database [26], the dog sequence is from the EMBL database (accession no. AACN010934065). Positions are given relative to the TATA box. Asterisks mark the fully conserved nucleotides. Interrupted horizontal arrows above and below the consensus sequence of Dpe1 and Pe1 indicate putative Sox motifs marking those nucleotides that are identical with the consensus L-Sox5/Sox6 and Sox9 recognition sequences respectively shown in a separate box.
Functional analysis of the short matrilin-1 promoter in transgenic mice
The TR70, VAM1 and VAM2 transgenes exhibiting zonal expression in the growth plate [25] shared the short matrilin-1 promoter. To study whether the short promoter harbouring the conserved Pe1 element can significantly contribute alone to the transcriptional regulation of the gene, we fused the chicken matrilin-1 promoter between –334 and +67 to the LacZ reporter gene. The fusion construct NAD1 (Figure 1B) was microinjected into the pronuclei of fertilized mouse eggs, and the founder embryos were killed at E15.5 to study the β-galactosidase activity by whole-mount X-Gal staining (Figure 3) and histological techniques (Figure 4). Altogether 12 positive founder embryos expressing the transgene at detectable level by histology were analysed in detail. Each of these embryos exhibited LacZ expression of a similar pattern in all the developing cartilaginous elements of the chondrocranium, appendicular and axial skeleton (Figures 3 and 4). The level of LacZ expression was somewhat weaker than that in the killed VAM2 embryos carrying intronic sequences in addition to the same short promoter as harboured by NAD1 [25]. X-Gal staining showed zonal differences in the transgene expression pattern in the cartilage growth plates of developing long bones and vertebral bodies (Figure 4), albeit the zonal differences were less pronounced than it was demonstrated for the TR70 and VAM2 transgenes carrying longer promoter and intronic regions respectively [25]. In agreement with the expression pattern of the endogenous matrilin-1 gene, the level of NAD1 transgene expression was generally highest in the columnar proliferating chondroblasts and prehypertrophic chondrocytes, whereas a lower level of expression was seen in the zones of epiphyseal and resting (also called source [34]) chondroblasts and upper hypertrophic chondrocytes (panels A–F). The site of integration only slightly influenced the level and pattern of expression in cartilage. Similar to previous observations [25], not all cells exhibited LacZ activity even in the zones of expression (panels D, E, I and Q). Differing from the TR70, VAM1 and VAM2 transgenes, however, no decrease was observed in the β-galactosidase activity in proximal as compared with distal structures during the progression of endochondral bone formation. For example, the NAD1 transgene activity in the cranial vertebral bodies was comparable with that in the caudal vertebral bodies of the same embryo (compare panels G and H). This suggests that the short promoter lacks control elements repressing the transgene expression in proximal structures.
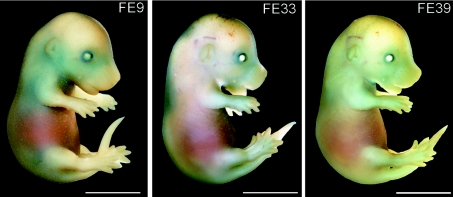
Figure 3. Transgene expression pattern in NAD1 embryos.
Transgenic founder mice carrying the NAD1 transgene were killed at E15.5. Whole-mount X-Gal staining reveals relatively weak and diffuse LacZ expression in the developing chondrocranium, axial and appendicular skeleton. FE, G0 founder embryos. Scale bar, 2 mm.
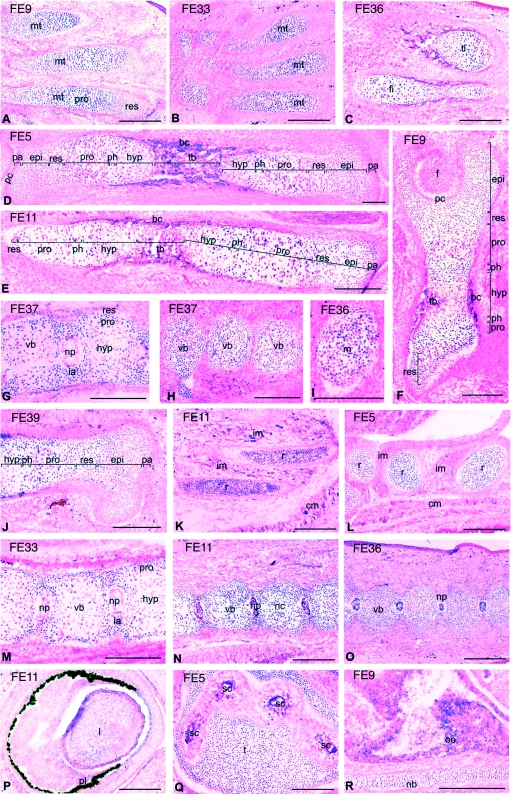
Figure 4. Histological analysis of the NAD1 transgene expression.
Sagittal cryosections were made from E15.5 G0 founder embryos (FE, G0 founder embryos) whole-mount stained with X-Gal. The sections were counterstained with eosin. LacZ expression is seen in the developing metatarsals (mt) (A, B), tibia (ti) and fibula (fi) (C), femur (D) of the hind limbs, humerus (E) and iliac bone (F). The intensity of X-Gal staining is relatively the highest in the zones of columnar proliferating chondroblasts (pro) and prehypertrophic chondrocytes (ph), and weaker in the epiphyseal chondroblasts (epi), resting (res) and hypertrophic (hyp) zones of growth plate cartilage (A–F, J). X-Gal staining is visible in the proliferating chondroblasts and prehypertrophic chondrocytes of cranial (G), lumbal (M) and caudal (H, N, O) vertebral bodies (vb), in the inner annulus (ia) and nucleus pulposus (np) of developing intervertebral discs (G, M–O). The level of LacZ expression is similar in the proliferating chondroblasts of cranial (G) and caudal (H) vertebral bodies within the same embryo. Expression can also be seen in the cartilaginous primordia of Meckel's cartilage (m) (I), temporal (t) (Q) and nasal bones (nb; R), in the longitudinal (K) and cross (L) sections of the developing ribs (r) and in the eye (P). Ectopic expression is occasionally observed in non-chondrogenic tissues, e.g. the olfactory epithelium (oe) (R), epithelial cells of the semicircular canals (sc) of inner ear (Q), in the intervertebral (im) and cutaneous muscles (cm) (K, L) and in the osteoblasts of trabecular bone (tb) (D–F). bc, bone collar; f, head of femur; pa, periarticular cartilage; pc, perichondrium. Scale bar, 200 μm (A–H, J–R); 100 μm (I).
X-Gal staining of similar intensity was also observed in the inner annulus chondroblasts and nucleus pulposus of intervertebral discs, in the eye and collecting tubules of the developing kidney in most of the NAD1 transgenic embryos (panels G, M–P; and results not shown). Apart from this, ectopic LacZ expression of very faint to moderate intensity and of variable pattern was noticed in other tissues of some of the embryos, such as in osteoblasts, olfactory epithelium and semicircular canals of inner ear (panels D, Q and R). Occasionally, staining was also seen in skeletal muscle, heart muscle or lung of certain embryos, or in certain elements of the central and peripheral nervous system (panels K, L; and results not shown). The variable ectopic expression pattern in non-chondrogenic tissues rather suggests the influence of the integration site than the intrinsic activity of the promoter elements.
From these results, we conclude that the short promoter harbours a cartilage-specific control element, which is preferably active in certain developmental stages and in distinct zones of the growth plate. However, it is alone not sufficient to direct a high-level transgene activity fully restricted to certain chondrocyte differentiation stages and completely independent of the site of integration.
Cartilage-specific occupancy of the conserved Pe1 element by nuclear proteins in vivo
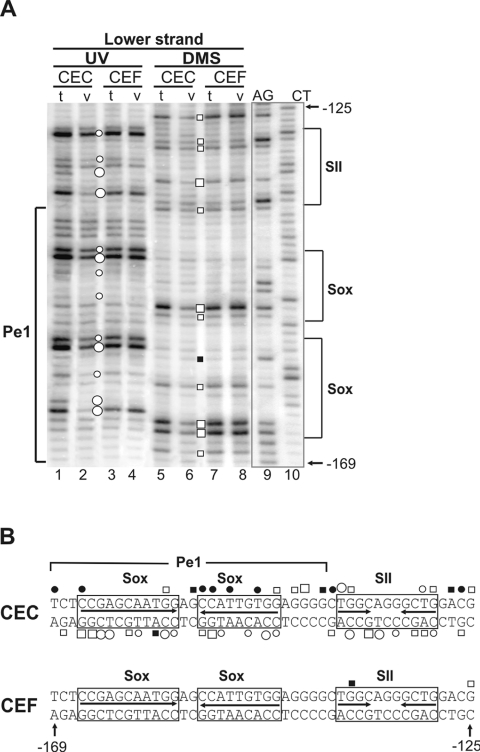
To detect whether transcription factors are bound to the putative recognition sequences within the proximal promoter region in vivo, we used the sensitive genomic footprinting strategy in combination with LM-PCR [31]. A series of primers was designed to cover 360 bp around the TATA motif (Figure 1A). The matrilin-1 expressing CEC cultures in comparison with the non-expressing CEF cultures were subjected to in vivo analysis. Genomic DNA was treated with DMS or UV light to modify G residues at N-7 position or produce (6–4) photoproducts at TC and CC dinucleotides respectively. These modifications are sensitive to bound proteins, therefore the areas of protein–DNA interactions appear as footprints on LM-PCR genomic sequencing ladders [31]. LM-PCR analysis of naked CEC and CEF DNA treated with the same reagents in vitro served as a reference. Differences in the modification patterns between the in vivo and in vitro treated samples, apparent as hyporeactivities (protections) or hyperreactivities, indicated in vivo DNA–protein contacts at specific sequences.
Sets of independent experiments revealed cartilage-specific binding of transcription factors to the short promoter (Figure 5). We focused on the region where sequence conservation during evolution was obvious, that is on the vicinity of the Pe1 element between –169 and –125. Results of DMS and UV footprinting obtained on the upper and lower strands of DNA (Figure 5A; and results not shown) are summarized in Figure 5(B). Apart from Pe1, the first region also covered the SII element identified previously [24]. In chondrocytes, occupancy was observed at the NFI contact points of the SII element (Figure 5A, lanes 1, 2, 5 and 6). The conserved Pe1 between –169 and –139 was also clearly protected by bound transcription factors at certain G, CC and CT nucleotides of the inverted Sox motifs, whereas hypersensitivity was seen on the opposite strand. Furthermore, hyporeactivity indicated factor binding at G residues of an Sp1-like motif between the Pe1 and SII elements (Figure 5) and also at G residues of several Sp1-like elements located between SII and the TATA motif (results not shown). These results demonstrate in vivo occupancy of potential recognition sequences for Sox9 and certain ubiquitous factors as well within Pe1 and its vicinity in chondrocytes (Figure 5B). As opposed to this, the complete absence of footprints in the short promoter in fibroblasts in repeated experiments indicated no factors bound to their recognition sequences in the non-expressing cell type (Figure 5A, lanes 3, 4, 7 and 8, Figure 5B; and results not shown). This suggests that regulation at the chromatin level can be involved in the activation of the gene in chondrocytes.
Figure 5. Cartilage-specific occupancy of the Pe1 and SII elements in genomic footprinting.
(A) Footprints are shown between positions –169 and –125 on the lower strand of the DNA. AG and CT are Maxam–Gilbert control sequences. DNA from CEC and CEF cultures treated in vivo (v) with DMS (open and closed boxes) or UV light (open and closed circles) is compared with the in vitro (t) DNA samples treated with these reagents after isolation from CEC and CEF. Differences in the modification patterns between ‘v’ and ‘t’ treatments, visible as hyper-reactivities (large and small closed circles or closed boxes) or protections (large and small open circles or open boxes), indicate in vivo DNA–protein contacts at specific sequences. (B) Summary of the in vivo footprinting data is shown on both strands. The previously identified NFI-binding site in the SII element and the inverted repeat harbouring the putative paired Sox-binding sites are boxed.
Sox9, L-Sox5 and Sox6 interact with the Pe1 element in vitro
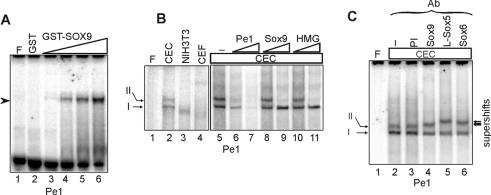
The inverted repeats within the conserved Pe1 element, which exhibited cartilage-specific protection in genomic footprinting, share 7/10 bp identity with the consensus Sox9-binding site (Figure 2). To confirm that Sox9 can indeed bind to Pe1 in the DNA-binding assay in vitro, bacterially expressed and purified GST–SOX9 fusion protein was incubated with the element. As demonstrated in Figure 6(A), the recombinant SOX9 formed a nucleoprotein complex efficiently on the element (lanes 3–6). When EMSA was performed with nuclear proteins from CEC, NIH3T3 and CEF cultures, we noticed tissue-specific differences in the pattern and behaviour of complexes (Figure 6B, lanes 1–4). Two specific DNA–protein complexes absent from fibroblasts were formed with CEC nuclear proteins (lane 2). Competition EMSA revealed that double-stranded oligonucleotides harbouring consensus Sox or HMG box motifs interfered only with the formation of the slowly migrating complex II (lanes 8–11). The same complex was supershifted with antibodies specifically recognizing Sox9, L-Sox5 and Sox6 (Figure 6C), indicating that each of the cartilage-specific Sox transcription factors participated in the formation of complex II on Pe1 in chondrocytes. On the other hand, the observation that complex I was neither competed nor supershifted with Sox-specific oligonucleotides or antibodies respectively, indicated that the element also interacted with other transcription factors in the expressing cell type. Together, these results provide sufficient evidence to conclude that the chondrocyte-specific in vivo footprints at the inverted Sox motifs of Pe1 might be due to the binding of Sox9, L-Sox5 and Sox6 proteins. In addition, the Pe1 element can also form a complex with an unrelated chondrocyte nuclear protein.
Figure 6. Analysis of the interaction of the Pe1 element with Sox proteins in vitro.
(A) 32P-labelled Pe1 was incubated with 2.0 μg of GST alone (lane 2) and increasing amounts (0.25, 0.5, 1.0 and 2.0 μg) of purified GST–SOX9 (lanes 3–6). No protein was added to lane F (free probe). (B) Radiolabelled Pe1 was incubated with 3 μg of nuclear proteins extracted from CEC, CEF and NIH3T3 cells (lanes 2–4). The binding of chondrocyte nuclear proteins was competed with 50- and 500-fold molar excesses of Pe1, consensus Sox9 and HMG elements (lanes 6–11). No competitor was added to lane 5. (C) Supershift experiment was performed without antiserum (lane 2) and with 1 μl each of preimmune antiserum (PI) (lane 3) and antibodies specifically recognizing Sox9, L-Sox5 and Sox6 (lanes 4–6). The arrows point to the supershifted complexes.
Mutations in either of the paired Sox motifs of Pe1 interfere with protein binding in EMSA
To examine further the Pe1 element, point mutations were introduced into either one (Pe1M1 and Pe1M2) or both (Pe1M3) of the inverted nonameric Sox motifs, or the spacer region between them (Pe1M4) (Figure 7A). These oligonucleotides were used as probes in EMSA with purified recombinant SOX9 and CEC nuclear proteins. Mutants Pe1M1, Pe1M2 and Pe1M3 did not bind to GST–SOX9 (Figure 7B, lanes 1–4) and did not compete for binding of Pe1 to either purified recombinant SOX9 or CEC nuclear proteins (Figures 7B and 7C, lanes 6–15). In other words, point mutations in either of the paired Sox motifs prevented the interaction between GST–SOX9 and Pe1, indicating that both nonameric Sox motifs within the pair were essential for recognition by SOX9. Consistent with earlier observations [35], these results clearly demonstrate the importance of paired Sox sites in cartilage-specific gene regulation. Mutations M1, M2 and M3 also equally interfered with the formation of CEC nucleoprotein complexes I and II, but yielded a very slowly migrating complex (Figure 7C, lanes 2–4). Since competition with a Sox-binding site or wild-type Pe1 element did not diminish the latter complex (results not shown), we concluded that the mutagenesis possibly created a binding site for an unknown nuclear factor synthesized in chondrocytes. However, the unknown nuclear factor did not recognize the wild-type element. The observation that mutations in either one of the inverted Sox motifs disrupted not only the Sox-specific complexes II, but complex I as well suggests that the formation of the two complexes is not completely independent. This gives rise to two possibilities. One explanation can be that the mutations also disrupted the overlapping binding site for a currently unidentified factor forming complex I. The alternative, and more likely explanation can be that binding of Sox9 homodimers to the paired sites can cause bending of the DNA, and thereby may promote an otherwise very weak interaction between Pe1 and the unidentified factor. Thus abolition of Sox9-binding to Pe1 may prevent the binding of the unidentified factor as well.
Pe1M4 was able to compete with the wild-type element for binding to recombinant SOX9 or CEC nuclear proteins (Figures 7B and 7C, lanes 16 and 17). However, the spacer mutation reduced the binding efficiency of Pe1M4 to GST–SOX9 (Figure 7B, lane 5). Contrary to Pe1, Pe1M4 formed only a single complex of altered mobility with CEC nuclear proteins in repeated experiments (Figure 7C, lane 5). This indicates that although the spacer mutation did not abolish the interaction of Pe1 with GST–SOX9, it modified the formation of CEC multiprotein complexes on the element. The dramatic effect of point mutations supports the conclusion that Pe1 can interact with Sox9 in vitro at the nonameric palindrome and suggests the binding of an unknown factor to the spacer region.
Functional analysis of the Pe1 element in transient expression studies
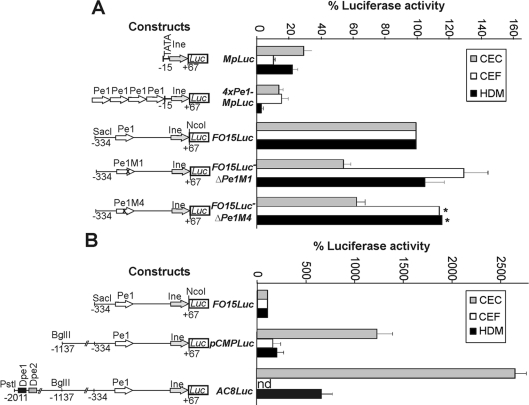
To confirm that the highly conserved Pe1 element significantly contributes to the transcriptional activity of the short promoter, wild-type and mutant versions of the promoter between positions −334 and +67 were fused to the luciferase reporter gene, and the promoter activity was measured in transient expression assays (Figure 8A). In agreement with the results in transgenic mice [25], the minimal promoter between −15 and +67 (MpLuc) had a very low activity in expressing and non-expressing cell types. Extension of the sequence up to −334 in the 5′-direction in the construct FO15Luc elevated the activity of the short promoter by 3.4-fold in chondrocytes. Activation was also observed in CEF and HDM cultures, possibly due to the binding of ubiquitous factors (Sp1, NFI, etc.) to the putative motifs of the short promoter in transient expression assays. Introducing mutations M1 into one of the paired Sox9 motifs of the Pe1 element by in situ mutagenesis in ΔPe1M1 decreased the luciferase activity by almost 2-fold in chondrocytes. The same mutation did not decrease but rather slightly increased the promoter activity in CEF and HDM cultures, thus demonstrating the tissue- and developmental stage-specific effect of mutation in the Sox-binding sites of Pe1. Interestingly, Pe1M4 containing mutations in the spacer region between the intact Sox motifs decreased the short promoter activity in derivative ΔPe1M4 to 60% in chondrocytes, but not in CEF and HDM cultures (Figure 8A). In agreement with the altered complex formation with chondrocyte nuclear proteins in EMSA (Figure 7C), this suggests that Pe1 probably interacts with another protein in chondrocytes that is also critical for the activity of the element. Based on the functional assays, we conclude that Pe1 is likely to contribute significantly to the short promoter activity by binding to cartilage-specific Sox proteins through the inverted motifs and to a currently unidentified factor in the spacer region.
Figure 8. Functional analysis of Pe1 in transient expression assays.
Luciferase reporter constructs harbouring the minimal or the proximal promoter region of the matrilin-1 gene (A) or promoter fragments of increasing length (B) between positions indicated are shown on the left. Construct 4×Pe1-MpLuc carries four tandem copies of Pe1 upstream of the minimal promoter. Point mutations Pe1M1 and Pe1M4 were introduced into Pe1 of FO15Luc in derivatives ΔPe1M1 and ΔPe1M4. Luciferase activities of constructs are expressed in percentage of that of the FO15Luc set at 100%. Values represent the averages±S.E.M. of 5–10 independent transfection experiments in CEC, CEF and HDM cultures. Asterisks indicate the values shown as averages of three independent experiments.
When longer promoter fragments including the putative Dpe1 and Dpe2 elements were tested in transient expression assays, we observed 12.4- and 26.9-fold enhancement of the reporter gene activity of constructs pCMPLuc and AC8Luc respectively in CEC cultures (Figure 8B). In contrast, the upstream promoter elements of pCMPLuc resulted in only 1.8- and 2.6-fold increase of the luciferase activity in CEF and HDM cultures. This indicates that upstream promoter elements can highly increase the tissue- and stage-specific activity of the matrilin-1 promoter.
The Pe1 element is found within 100–200 bp upstream of the TATA motif in various amniotes (Figure 2). To test whether this conserved location is important for the function, four copies of the element were placed upstream of the minimal promoter in construct 4×Pe1-MpLuc. However, this further decreased (CEC and HDM) or did not alter significantly (CEF) the luciferase activity as compared with the minimal promoter construct MpLuc (Figure 8A), suggesting that the Pe1 element cannot drive the promoter in such a close proximity to the TATA motif or it may work co-operatively with adjacent sequences binding to other factors.
DISCUSSION
Organization of chondrocytes into distinct zones of the growth plate plays an important role in the formation and longitudinal growth of long bones. Disturbance of the zonal distribution leads to severe developmental problems [1,3]. Identification of regulatory mechanisms driving the gene expression to specific zones would be very useful. The unique feature of the matrilin-1 gene among cartilage-specific genes is the characteristic expression pattern restricted to distinct zones of the growth plate in vivo or developmental stages in tissue cultures. Recent analysis of the major regulatory regions of the chicken matrilin-1 gene in transgenic mice revealed that the long promoter alone or in combination with the intronic enhancer as well as the short promoter with the intronic enhancer restricted the transgene expression to the columnar proliferative chondroblasts and prehypertrophic chondrocytes [25]. As all these transgenes shared the short promoter between −338 and +67, we asked (1) whether the proximal promoter region itself harbours cartilage-specific control elements and (2) whether the same elements are able to exert developmental-stage specificity to the promoter as well.
To address these questions, in the present study, we characterized the short promoter of the chicken matrilin-1 gene in transgenic mice and demonstrated cartilage-specific interaction of transcription factors with the conserved Pe1 element containing inverted Sox motifs using in vitro assays and in vivo footprinting. We provided evidence that each of the 12 founder mice tested histologically exhibited similar zonal differences in the transgene expression in the developing cartilaginous elements of the chondrocranium, appendicular and axial skeleton. This implies that the proximal promoter region indeed harbours DNA elements, which can direct the developmental stage-specific expression of the transgene in chondrocytes. The X-Gal staining, however, showed a relatively low transgene activity, which was often accompanied by weak or hardly detectable ectopic expression of variable patterns in other tissues, indicating that the DNA elements carried by the short promoter were alone not sufficient to drive a high-level transgene expression, fully restricted to chondrocytes independent of the site of integration. Our results suggest that the tissue- and the developmental stage-specific control mechanisms are at least partly connected to one another. Comparison of the reporter gene activity between FO15Luc and AC8Luc and between the NAD1 (present study), TR70 and VAM1 transgenes [25] revealed that distal promoter and intronic elements highly enhanced the promoter activity and greatly increased the zonal differences in the transgene expression. The lack of significant differences in the NAD1 transgene expression between proximal and distal structures supports the assumption that DNA elements, which restricted the LacZ expression of the TR70, VAM1 and VAM2 transgenes to distal structures [25], are also located outside the proximal promoter region.
Results published in the present study are consistent with our previous observations [6,23–25]. Results presented here line up with previous reports of others [8,9,16–18] demonstrating the essential role of Sox9, L-Sox5 and Sox6 in chondrogenesis and cartilage-specific gene regulation. Binding of Sox9 to the enhancer elements of the Col2a1, Col11a2, CD-RAP and aggrecan genes has been well documented using in vitro DNA–protein interaction assays [18,35–37]. However, differing from the regulatory mechanisms reported for other cartilage protein genes, the present study describes for the first time the significance of a proximal promoter element recognized by cartilage-specific Sox transcription factors in the tissue- and stage-specific transcriptional control of the matrilin-1 gene.
Pe1 elements harbouring inverted Sox motifs were found in similar position in the proximal promoter region of the various amniote matrilin-1 genes (Figure 2). Although we found the matrilin-1 gene in the fugu and zebrafish genome, we could not identify their first exons, and thus the potential promoter regions. The observation that, contrary to the sequence divergence in other parts of the matrilin-1 regulatory regions in the various species, Pe1 remained strongly conserved under evolutionary pressure between chicken and mammals implies that the element performs a very important function in the transcriptional regulation of the matrilin-1 gene in amniotes. Phylogenetic conservation of important regulatory sequences has also been reported in other systems, leading to a similar conclusion. For example, Sox2 enhancers were functionally identified within extragenic sequence blocks clearly conserved between chicken and mammals [38]. From the 25 conserved sequence blocks, however, only two occurred in the fish genome as well, but similarly to our findings reported in the present study, the conserved regulatory elements were hidden within longer stretches of sequence similarity, when only mammalian species were compared. Species-specific variations in the occurrence of conserved regulatory Sox2 sequence blocks were related to distinct spatio-temporal differences in the gene expression between vertebrate species [39,40]. These and our findings support the conclusion that sequence conservations between chicken and mammals are reliable indications of important regulatory regions within a genetic locus.
Tissue-specific control elements have been identified in the promoter of certain eukaryotic genes, for example between 15 and 200 bp upstream of the TATA box of liver- or osteoblast-specific genes [41–43]. Furthermore, it has also been reported that SOX9 interacting with the partner transcription factor SF-1 recognizes a conserved DNA element approx. 100 bp upstream of the TATA box of mammalian anti-Müllerian hormone genes [32]. Even though inverted Sox motifs are known to play an essential role in the function of chondrocyte-specific enhancers of the Col2a1 and Col11a2 genes, conserved blocks similar to Pe1 could be found neither in their proximal promoter elements nor within the 3 kb region of the putative promoters of these genes in human, mouse and rat by computer search using the programs DIALIGN2 and MEME [29,30]. In accordance with this observation, the proximal promoter region of neither the Col2a1 and Col11a1 nor the Agc and the CDRAP genes were reported to mediate cartilage-specific regulation [44–46]. Interestingly, consistent with its conserved position in amniotes, the Pe1 element seems to function at a certain distance from the TATA box. In this respect, it also clearly differs from the cartilage-specific enhancer elements of other cartilage-protein genes [18,35–37,44–47]. These findings imply that Pe1 is a unique control element of the matrilin-1 gene not shared by other cartilage protein genes.
In our experiments, GST–Sox9 formed only a single complex with Pe1. Even though Sox9 is not capable of forming homodimers in solution [18,48,49], consistent with observations from other laboratories [35,50], we found that it could bind only to intact inverted pairs of Sox motifs (Figure 7B), thus supporting the conclusion that Sox9 dimerization might have occurred upon DNA binding. As opposed to this, Sox9 was reported to bind as a monomer to cis elements involved in sex determination [50].
Transient expression studies confirmed that Pe1 significantly contributed to the moderate activity of the short promoter in chondrocytes. Based on the data, we hypothesize that the element does not drive the high cartilage-specific expression of the promoter as an enhancer, but may rather act by modulating the promoter activity and mediating the effect of distal promoter and intronic enhancer elements. We also hypothesize that distal promoter and intronic elements may also function by forming multi-protein complexes by interacting with Pe1. Although Pe1 can bind to Sox9, L-Sox5 and Sox6 in vitro, it may show a preference for Sox9 in vivo, as the inverted Sox motifs carried by Pe1 are more similar to the preferred binding sites of Sox9, than to those of L-Sox5/Sox6. On the other hand, the conserved Dpe1 element carries motifs more similar to the preferred binding sites of L-Sox5/Sox6. Binding of Dpe1 to these factors and the interaction with nucleoprotein complexes formed on Pe1 may be necessary for the high tissue- and developmental stage-specific activity of the 2011 bp promoter in AC8Luc and TR70 transgenic mice [25]. Thus the Sox9 and L-Sox5/Sox6-binding sites may be separated over a large distance in the regulatory region of the matrilin-1 gene. The importance of L-Sox5/Sox6 binding is supported by the observation that Sox9 alone is not sufficient for the activation of the matrilin-1 gene in the absence of Sox5 and Sox6 proteins [17].
By bending the DNA, HMG box proteins are known to promote the binding of other transcription factors to the DNA. Lining up with these observations, our results suggest that, in addition to Sox proteins, other transcription factors may also be involved in the activity of the short matrilin-1 promoter. Sox9 is known to interact with a number of partner factors, including SF-1 and LcMaf [11,32,51]. One explanation for the conserved position of the Pe1 element can be that it may function by bending the DNA and promoting the interaction between the components of the polymerase II transcription machinery and ubiquitous factors bound to the proximal promoter elements. This assumption is supported by the observation that the short promoter includes several putative binding sites for ubiquitous transcription factors, such as Sp1 and NFI, and based on our genomic footprinting studies, these motifs were also occupied by transcription factors bound in vivo in chondrocytes (Figure 5; and results not shown). Furthermore, Sox9 may interact with different partner factors during subsequent steps of chondrogenesis, thereby contributing to the developmental stage-specific activity of the matrilin-1 gene. However, future studies will be needed for mapping the putative cofactor-binding sites and identifying the interacting Sox partner factors on the Pe1 element.
Acknowledgments
We are grateful to V. Lefebvre for critically reading this paper, to B. de Crombrugghe and V. Lefebvre for kindly providing antisera specifically recognizing SOX9, L-Sox5 and Sox6, and to P. Berta for the GST–SOX9 plasmid. We thank P. Szabó for introducing O. R. to genomic footprinting, A. Simon, K. Kovács and I. Kravjár for excellent technical assistance and M. Tóth for the artwork. We are also thankful to our colleagues at the Institute of Genetics for the opportunity to photograph the whole mount stained embryos with the Leica DC 300 F camera. This work was supported by grants OTKA T029142, T034399 and T049608 from the Hungarian National Scientific Research Foundation, a grant ETT 256/2003 from the Medical Research Council of Hungary and grant GVOP-3.1.1.-2004-05-0290/3.0 from the Economic Competitiveness Operative Programme of the National Development Plan to I. K.
References
- 1.Karsenty G., Wagner E. F. Reaching a genetic and molecular understanding of skeletal development. Dev. Cell. 2002;2:389–406. doi: 10.1016/s1534-5807(02)00157-0. [DOI] [PubMed] [Google Scholar]
- 2.Cancedda R., Descalzi F., Castagnola P. Chondrocyte differentiation. Int. Rev. Cytol. 1995;159:265–358. doi: 10.1016/s0074-7696(08)62109-9. [DOI] [PubMed] [Google Scholar]
- 3.Ducy P., Karsenty G. Genetic control of cell differentiation in the skeleton. Curr. Opin. Cell Biol. 1998;10:614–619. doi: 10.1016/s0955-0674(98)80037-9. [DOI] [PubMed] [Google Scholar]
- 4.Franzén A., Heinegard D., Solursh M. Evidence for sequential appearance of cartilage matrix proteins in developing mouse limbs and in cultures of mouse mesenchymal cells. Differentiation. 1987;36:199–210. doi: 10.1111/j.1432-0436.1987.tb00194.x. [DOI] [PubMed] [Google Scholar]
- 5.Stirpe N. S., Goetinck P. F. Gene regulation during cartilage differentiation: temporal and spatial expression of link protein and cartilage matrix protein in the developing limb. Development. 1989;107:23–33. doi: 10.1242/dev.107.1.23. [DOI] [PubMed] [Google Scholar]
- 6.Muratoglu S., Bachrati C., Malpeli M., Szabó P., Neri M., Dozin B., Deák F., Cancedda R., Kiss I. Expression of the cartilage matrix protein gene at different chondrocyte developmental stages. Eur. J. Cell Biol. 1995;68:411–419. [PubMed] [Google Scholar]
- 7.Szüts V., Möllers U., Bittner K., Schürmann G., Muratoglu S., Deák F., Kiss I., Bruckner P. Terminal differentiation of chondrocytes is arrested at distinct stages identified by their expression repertoire of marker genes. Matrix Biol. 1998;17:435–448. doi: 10.1016/s0945-053x(98)90103-2. [DOI] [PubMed] [Google Scholar]
- 8.de Crombrugghe B., Lefebvre V., Behringer R. R., Bi W., Murakami S., Huang W. Transcriptional mechanisms of chondrocyte differentiation. Matrix Biol. 2000;19:389–394. doi: 10.1016/s0945-053x(00)00094-9. [DOI] [PubMed] [Google Scholar]
- 9.Lefebvre V. Toward understanding the functions of the two highly related Sox5 and Sox6 genes. J. Bone Miner. Metab. 2002;20:121–130. doi: 10.1007/s007740200017. [DOI] [PubMed] [Google Scholar]
- 10.Wegner M. From head to toes: the multiple facets of Sox proteins. Nucleic Acids Res. 1999;27:1409–1420. doi: 10.1093/nar/27.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamachi Y., Uchikawa M., Kondoh H. Pairing Sox off with partners in the regulation of embryonic development. Trends Genet. 2000;16:182–187. doi: 10.1016/s0168-9525(99)01955-1. [DOI] [PubMed] [Google Scholar]
- 12.Pöhler J. R., Norman D. G., Bramhan J., Bianchi M. E., Lilley D. M. HMG box proteins bind to four-way DNA junctions in their open conformation. EMBO J. 1998;17:817–826. doi: 10.1093/emboj/17.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mertin S., McDowall G. S., Harley V. R. The DNA binding specificity of SOX9 and other SOX proteins. Nucleic Acids Res. 1999;27:1359–1364. doi: 10.1093/nar/27.5.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foster J. W., Dominguez-Steglich M. A., Guioli S., Kowk G., Weller P. A., Stevanovic M., Weissenbach J., Mansour S., Young I. D., Goodfellow P. N., et al. Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature (London) 1994;372:525–530. doi: 10.1038/372525a0. [DOI] [PubMed] [Google Scholar]
- 15.Wagner T., Wirth J., Meyer J., Zabel B., Held M., Zimmer J., Pasantes J., Dagna Bricarelli F., Keutel J., Hustert E., et al. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX-9. Cell (Cambridge, Mass.) 1994;79:1111–1120. doi: 10.1016/0092-8674(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 16.Bi W., Deng J. M., Zhang Z., Behringer R. R., de Crombrugghe B. Sox9 is required for cartilage formation. Nat. Genet. 1999;22:85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 17.Smits P., Li P., Mandel J., Zhang Z., Deng J. M., Behringer R. R., de Crombrugghe B. The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev. Cell. 2001;1:277–290. doi: 10.1016/s1534-5807(01)00003-x. [DOI] [PubMed] [Google Scholar]
- 18.Lefebvre V., Li P., de Crombrugghe B. A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J. 1998;17:5718–5733. doi: 10.1093/emboj/17.19.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiss I., Deák F., Holloway R. G., Delius H., Mebust K. A., Frimberger E., Argraves W. S., Tsonis P. A., Winterbottom N., Goetinck P. F. Structure of the gene for cartilage matrix protein, a modular protein of the extracellular matrix. J. Biol. Chem. 1989;264:8126–8134. [PubMed] [Google Scholar]
- 20.Deák F., Wagener R., Kiss I., Paulsson M. The matrilins: a novel family of oligomeric extracellular matrix proteins. Matrix Biol. 1999;18:55–64. doi: 10.1016/s0945-053x(98)00006-7. [DOI] [PubMed] [Google Scholar]
- 21.Hauser N., Paulsson M., Heinegard D., Mörgelin M. Interaction of cartilage matrix protein (CMP) with aggrecan. Increased covalent crosslinking with maturation. J. Biol. Chem. 1996;271:32247–32252. doi: 10.1074/jbc.271.50.32247. [DOI] [PubMed] [Google Scholar]
- 22.Chen Q., Zhang Y., Johnson D. M., Goetinck P. F. Assembly of a novel cartilage matrix protein filamentous network: molecular basis of differential requirement of von Willebrand factor A domains. Mol. Biol. Cell. 1999;10:2149–2162. doi: 10.1091/mbc.10.7.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiss I., Bösze Z., Szabó P., Rencendorj A., Barta E., Deák F. Identification of positive and negative regulatory regions controlling expression of the cartilage matrix protein gene. Mol. Cell. Biol. 1990;10:2432–2436. doi: 10.1128/mcb.10.5.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szabó P., Moitra J., Rencendorj A., Rákhely G., Rauch T., Kiss I. Identification of a nuclear factor-I family protein-binding site in the silencer region of the cartilage matrix protein gene. J. Biol. Chem. 1995;270:10212–10221. doi: 10.1074/jbc.270.17.10212. [DOI] [PubMed] [Google Scholar]
- 25.Karcagi I., Rauch T., Hiripi L., Rentsendorj O., Nagy A., Bösze Zs., Kiss I. Functional analysis of the regulatory regions of the matrilin-1 gene in transgenic mice reveals modular arrangement of tissue-specific control elements. Matrix Biol. 2004;22:605–618. doi: 10.1016/j.matbio.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 26.Hammond M. P., Birney E. Genome information resources – developments at ENSEMBL. Trends Genet. 2004;20:268–272. doi: 10.1016/j.tig.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 27.Kulikova T., Aldebert P., Althorpe N., Baker W., Bates K., Browne P., van den Broek A., Cochrane G., Duggan K., Eberhardt R., et al. The EMBL Nucleotide Sequence Database. Nucleic Acids Res. 2004;32:D27–D30. doi: 10.1093/nar/gkh120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rice P., Longden I., Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 29.Morgenstem B. DIALIGN 2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics. 1999;15:211–218. doi: 10.1093/bioinformatics/15.3.211. [DOI] [PubMed] [Google Scholar]
- 30.Bailey T. L., Elkan C. The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Instell. Syst. Mol. Biol. 1995;3:21–29. [PubMed] [Google Scholar]
- 31.Pfeifer G. P., Chen H. H., Komura J., Riggs A. D. Chromatin structure analysis by ligation-mediated and terminal transferase-mediated polymerase chain reaction. Methods Enzymol. 1999;304:548–571. doi: 10.1016/s0076-6879(99)04032-x. [DOI] [PubMed] [Google Scholar]
- 32.De Santa Barbara P., Bonneaud N., Boizet B., Desclozeaux M., Moniot B., Sudbeck P., Scherer G., Poulat F., Berta P. Direct interaction of SRY-related protein SOX9 and steroidogenic factor 1 regulates transcription of the human anti-Mullerian hormone gene. Mol. Cell. Biol. 1998;11:6653–6655. doi: 10.1128/mcb.18.11.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hames B. D., Higgins S. J. Oxford University Press, Oxford: The Practical Approach Series; 1993. Gene Transcription: A Practical Approach. [Google Scholar]
- 34.Smits P., Dy P., Mitra S., Lefebvre V. Sox5 and Sox6 are needed to develop and maintain source, columnar, and hypertrophic chondrocytes in the cartilage growth plate. J. Cell Biol. 2004;164:747–758. doi: 10.1083/jcb.200312045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bridgewater L. C., Walker M. D., Miller G. C., Ellison T. A., Holsinger L. D., Potter J. L., Jackson T. L., Chen R. K., Winkel V. L., Zhang Z., et al. Adjacent DNA sequences modulate Sox9 transcriptional activation at paired Sox sites in three chondrocyte-specific enhancer elements. Nucleic Acids Res. 2003;31:1541–1553. doi: 10.1093/nar/gkg230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie W. F., Zhang X., Sakano S., Lefebvre V., Sandell L. J. Trans-activation of the mouse cartilage-derived retinoic acid-sensitive protein gene by Sox9. J. Bone Miner. Res. 1999;14:757–763. doi: 10.1359/jbmr.1999.14.5.757. [DOI] [PubMed] [Google Scholar]
- 37.Sekiya I., Tsuji K., Koopman P., Watanabe H., Yamada Y., Sinomiya K., Nifuji A., Noda M. Sox9 enhances aggrecan gene promoter/enhancer activity and is up-regulated by retinoic acid in cartilage-derived cell line, TC6. J. Biol. Chem. 2000;275:10738–10744. doi: 10.1074/jbc.275.15.10738. [DOI] [PubMed] [Google Scholar]
- 38.Uchikawa M., Ishida Y., Takemoto T., Kamachi Y., Kondoh H. Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Dev. Cell. 2003;4:509–519. doi: 10.1016/s1534-5807(03)00088-1. [DOI] [PubMed] [Google Scholar]
- 39.Kamachi Y., Uchikawa M., Collignon J., Lovell-Badge R., Kondoh H. Involvement of Sox1, 2 and 3 in the early and subsequent molecular events of lens induction. Development. 1998;125:2521–2532. doi: 10.1242/dev.125.13.2521. [DOI] [PubMed] [Google Scholar]
- 40.Nishiguchi S., Wood H., Kondoh H., Lovell-Badge R., Episkopou V. Sox1 directly regulates the γ-crystallin genes and is essential for lens development in mice. Genes Dev. 1998;12:776–781. doi: 10.1101/gad.12.6.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Furlong E. E, Rein T., Martin F. YY1 and NF1 both activate the human p53 promoter by alternatively binding to a composite element, and YY1 and E1A cooperate to amplify p53 promoter activity. Mol. Cell. Biol. 1996;16:5933–5945. doi: 10.1128/mcb.16.10.5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niehof M., Kubicka S., Zender L., Manns M. P., Trautwein C. Autoregulation enables different pathways to control CCAAT/enhancer binding protein beta (C/EBP beta) transcription. J. Mol. Biol. 2001;309:855–868. doi: 10.1006/jmbi.2001.4708. [DOI] [PubMed] [Google Scholar]
- 43.Ducy P., Karsenty G. Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol. Cell. Biol. 1995;15:1858–1869. doi: 10.1128/mcb.15.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou G., Garofalo S., Mukhopadhyay K., Lefebvre V., Smith C. N., Eberspaecher H., de Crombrugghe B. A 182 bp fragment of the mouse proα1(II) collagen gene is sufficient to direct chondrocyte expression in transgenic mice. J. Cell Sci. 1995;108:3677–3684. doi: 10.1242/jcs.108.12.3677. [DOI] [PubMed] [Google Scholar]
- 45.Tsumaki N., Kimura T., Matsui Y., Nakata K., Ochi T. Separable cis-regulatory elements that contribute to tissue- and site-specific α2(XI) collagen gene expression in the embryonic mouse cartilage. J. Cell Biol. 1996;134:1573–1582. doi: 10.1083/jcb.134.6.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xie W. F., Zhang X., Sandell L. J. The 2.2-kb promoter of cartilage-derived retinoic acid-sensitive protein controls gene expression in cartilage and embryonic mammary buds of transgenic mice. Matrix Biol. 2000;19:501–509. doi: 10.1016/s0945-053x(00)00087-1. [DOI] [PubMed] [Google Scholar]
- 47.Zhou G., Lefebvre V., Zhang Z., Eberspaecher H., de Crombrugghe B. Three high mobility group-like sequences within a 48-base pair enhancer of the Col2α1 gene are required for cartilage-specific expression in vivo. J. Biol. Chem. 1998;273:14989–14997. doi: 10.1074/jbc.273.24.14989. [DOI] [PubMed] [Google Scholar]
- 48.Sock E., Pagon R. A., Keymolen K., Lissens W., Wegner M., Scherer G. Loss of DNA-dependent dimerization of the transcription factor SOX9 as a cause for campomelic dysplasia. Hum. Mol. Genet. 2003;12:1439–1447. doi: 10.1093/hmg/ddg158. [DOI] [PubMed] [Google Scholar]
- 49.Peirano R. I., Wegner M. The glial transcription factor Sox10 binds to DNA both as monomer and dimer with different functional consequences. Nucleic Acids Res. 2000;28:3047–3055. doi: 10.1093/nar/28.16.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bernard P., Tang P., Liu S., Dewing P., Harley V. R., Vilain E. Dimerization of SOX9 is required for chondrogenesis, but not for sex determination. Hum. Mol. Genet. 2003;12:1755–1765. doi: 10.1093/hmg/ddg182. [DOI] [PubMed] [Google Scholar]
- 51.Huang W., Lu N., Eberspaecher H., de Crombrugghe B. A new long form of c-Maf cooperates with Sox9 to activate the type II collagen gene. J. Biol. Chem. 2002;277:50668–50675. doi: 10.1074/jbc.M206544200. [DOI] [PubMed] [Google Scholar]