Abstract
sACE (somatic angiotensin-converting enzyme) consists of two homologous, N and C domains, whereas the testis isoenzyme [tACE (testis ACE)] consists of a single C domain. Both isoenzymes are shed from the cell surface by a sheddase activity, although sACE is shed much less efficiently than tACE. We hypothesize that the N domain of sACE plays a regulatory role, by occluding a recognition motif on the C domain required for ectodomain shedding and by influencing the catalytic efficiency. To test this, we constructed two mutants: CNdom-ACE and CCdom-ACE. CNdom-ACE was shed less efficiently than sACE, whereas CCdom-ACE was shed as efficiently as tACE. Notably, cleavage occurred both within the stalk and the interdomain bridge in both mutants, suggesting that a sheddase recognition motif resides within the C domain and is capable of directly cleaving at both positions. Analysis of the catalytic properties of the mutants and comparison with sACE and tACE revealed that the kcat for sACE and CNdom-ACE was less than or equal to the sum of the kcat values for tACE and the N-domain, suggesting negative co-operativity, whereas the kcat value for the CCdom-ACE suggested positive co-operativity between the two domains. Taken together, the results provide support for (i) the existence of a sheddase recognition motif in the C domain and (ii) molecular flexibility of the N and C domains in sACE, resulting in occlusion of the C-domain recognition motif by the N domain as well as close contact of the two domains during hydrolysis of peptide substrates.
Keywords: angiotensin-converting enzyme, catalytic activity, domain selectivity, ectodomain shedding, proteolytic cleavage, sheddase
Abbreviations: ACE, angiotensin-converting enzyme; CHO cells, Chinese-hamster ovary cells; HHL, hippuryl-L-histidyl-L-leucine; PDBu, phorbol 12,13-dibutyrate; sACE, somatic ACE; tACE, testis ACE; TM, transmembrane; TNF, tumour necrosis factor; TAPI, TNF-α protease inhibitor; Z, benzyloxycarbonyl; Z-FHL, Z-phenylalanyl-L-histidyl-L-leucine
INTRODUCTION
ACE (angiotensin-converting enzyme), a zinc dipeptidyl carboxypeptidase, is responsible for the hydrolysis of angiotensin I and bradykinin, which are important modulators of blood pressure regulation. There are two ACE isoenzymes: sACE (somatic ACE) and tACE (testis ACE). sACE comprises two domains, the N and C domains, which have 60% sequence identity. tACE is identical with the C domain except for 36 amino acid residues at its N-terminus [1], and hence both isoenzymes share the same C-terminal stalk, TM (transmembrane) and cytoplasmic domains. Both sACE and tACE are type I ectoproteins that are shed by proteolytic cleavage at identical stalk cleavage sites by an unidentified sheddase. However, shedding of sACE is inefficient compared with that of tACE, but is increased 10-fold after deletion of the N domain [2,3].
Evidence for the existence of a C-domain recognition motif is based on the data derived from fusion of the tACE ectodomain to the stalk, TM and cytoplasmic domains of CD4, which resulted in shedding of the tACE-CD4 chimaera (wild-type CD4 is not shed), whereas fusion of the CD4 ectodomain to the stalk, TM and cytoplasmic domains of tACE produced a chimaera that was not shed [4]. Moreover, fusion of the sACE N domain to the stalk, TM and cytoplasmic domains of tACE also abrogated shedding [5], strongly suggesting that the C domain, but not the N domain, contains a putative sheddase recognition motif.
An alternative explanation for the different rates of shedding observed for sACE and tACE [3] is that the N domain partially obstructs the stalk cleavage site. This concept finds some support from evidence for domain co-operativity in sACE.
Although both domains are catalytically active in isolation [6], several lines of evidence indicate that the N and C domains do not function independently in full-length sACE [7–9]. By active-site titration with tight-binding inhibitors and by isothermal titration calorimetry, evidence has been presented for negative co-operativity between the N and C domains [8,9]. However, the structural basis for such interdomain co-operativity is unknown and a crystal structure of full-length sACE is not available. Of note, however, is that the N and C domains are linked by a 14-residue hinge or bridge region, which is susceptible to proteolysis [10] and which is presumed to enable considerable freedom of movement of the two domains relative to each other.
In the present study, we investigated the role of the N domain in regulating the ectodomain shedding and catalytic activity of sACE, by constructing domain swap mutants. Our results reveal that the presence of the N domain strongly reduces shedding and exerts negative co-operativity in terms of catalytic activity. In contrast, a mutant consisting of two C domains in tandem is rapidly shed and reveals positive co-operativity. Taken together, the present results suggest that the N domain plays a regulatory role in the function of sACE.
METHODS
Construction of the C and N domain swap mutants
CCdom-ACE
The interdomain bridge region of sACE (Pro602–Asp616) was amplified by PCR and ligated to the previously described ACE construct pBS–ΔR627 [11], allowing fusion of the PCR product to the C-terminus of tACE. This construct was then ligated to the previously described tACE construct pBS–Δ36N [12]. PBS–Δ36N was used as it lacked the O-glycosylated 36-amino-acid residue N-terminal region of tACE and thus was identical with the C domain of sACE. The final CCdom-ACE construct was subcloned into the mammalian expression vector pLEN-ACEVII [13] (Figure 1).
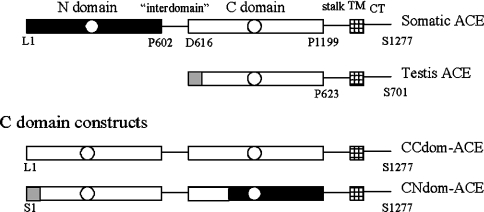
Figure 1. Schematic representation of sACE, CCdom-ACE and CNdom-ACE.
sACE comprises an N domain (black bar) and a C domain (open bar); tACE is identical with the C domain of sACE except for 36 amino acid residues at its N-terminus (grey box). All constructs contain the tACE signal peptide. The C domain of sACE was fused to an N-terminal C domain to form CCdom-ACE using the interdomain region of sACE. The positions of the N and C domains were exchanged to generate the construct CNdom-ACE: this construct comprised tACE residues Ser1–Pro623 and sACE residues Pro604–Pro739, Pro141–Pro601 and Asn1200–Ser1277. The N-terminal C domain in this construct is identical with tACE. The stalk, TM and cytoplasmic tail (CT) were unaltered.
CNdom-ACE
The previously described vector pLEN-SomNdom is a construct in which the N domain (Leu1–Pro601) was fused directly to the stalk, TM and cytoplasmic tail of tACE (Asn624–Ser701) [5]. On the basis of the available restriction sites, the region Asp142–Ser701 of this construct was ligated to the above-mentioned PCR product of CCdom-ACE in pBS, generating a construct comprising Pro604–Pro739 of sACE fused to Asp142–S701 of the SomNdom construct. The product, a membrane-anchored N domain with an N-terminal-linked interdomain bridge and a short stretch of chimaeric C domain, was subcloned into pLEN–ΔR627 using the EcoRI and ClaI restriction enzyme sites, resulting in the CNdom-ACE mutant (Figure 1).
Expression and ectodomain shedding of mutant proteins by CHO (Chinese-hamster ovary) cells
CCdom-ACE and CNdom-ACE were stably expressed in CHO cells, by methods outlined previously [13]. Positive clones were grown to confluence and ACE expression was induced overnight in 2% (v/v) foetal calf serum in complete medium supplemented with 40 μM ZnCl2. At the start of each experiment, fresh complete medium was added and the cells were grown for 4 h in the absence or presence of either 1 μM PDBu (phorbol 12,13-dibutyrate) or 100 μM TAPI [TNF-α (tumour necrosis factor α) protease inhibitor]. The culture medium was then removed and the cells were lysed in 1% Triton X-100 lysis buffer containing 50 mM Hepes (pH 7.5), 0.5 M NaCl and 1.0 mM PMSF.
Aliquots of culture media and cell lysates were resolved on a 7% (w/v) SDS/polyacrylamide gel and analysed by Western blotting using a tACE polyclonal antibody. CHO cells expressing wild-type sACE cDNA were treated in the same manner. To determine the kinetics of shedding in the presence and absence of PDBu and/or TAPI, 50 μl aliquots were removed from the media and cell lysate samples and tested for ACE activity using HHL (hippuryl-L-histidyl-L-leucine or Hip-His-Leu), as described previously [11].
Purification and cleavage-site analysis
Soluble CCdom-ACE was purified from the culture medium of CHO cells using lisinopril–Sepharose affinity chromatography, as described previously [13]. Purified protein was reduced, protected with vinyl pyridine and digested with endoproteinase Lys-C. The total digest was subjected to MALDI–TOF (matrix-assisted laser-desorption ionization–time-of-flight) MS [14,15]. In addition, soluble CCdom-ACE was resolved by SDS/PAGE and bands were eluted and subjected to N-terminal peptide sequencing by automated Edman degradation (six cycles) [15].
Catalytic properties of mutant proteins
Kinetic constants for the hydrolysis of Z-FHL (Z-phenylalanyl-L-histidyl-L-leucine; where Z stands for benzyloxycarbonyl) were determined as described previously [16,17] under initial rate conditions at 37 °C, such that substrate hydrolysis was <10%. Assays were conducted in 100 mM potassium phosphate buffer (pH 8.3) containing 300 mM NaCl. Fluorescence was measured at excitation (λex) and emission (λem) wavelengths of 360 and 486 nm respectively. The hydrolysis of the C domain-specific fluorogenic peptide AbzLFK(Dnp)-OH (where LFK stands for Leu-Phe-Lys residues) was determined in a continuous fluorimetric assay at 37 °C under initial rate conditions in 0.1 M Tris/HCl buffer (pH 7.0) containing 50 mM NaCl and 10 μM ZnCl2 [18]. Fluorescence was measured at λex=320 nm and λem=420 nm.
Kinetic constants were calculated by non-linear regression analysis of Michaelis–Menten plots. A standard calibration curve was generated by total peptide hydrolysis. Catalytic-centre activity (kcat) and specificity constants [kcat/Km (Michaelis constant)] were determined for both substrates using a calculated molecular mass of 170 kDa.
RESULTS
Expression and ectodomain shedding of mutant proteins by CHO cells
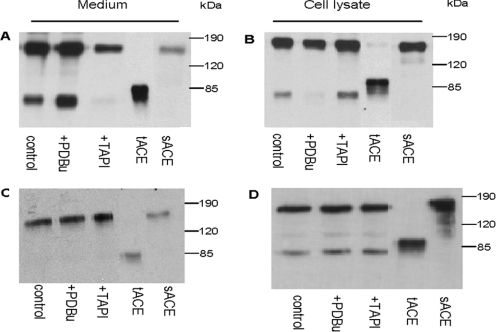
The expression and ectodomain shedding of CCdom-ACE and CNdom-ACE was investigated by Western-blot analysis (Figure 2). Transfected CHO cells grown to confluence, were washed and refed at the start of each experiment. After 4 h, cell lysates and media of CCdom-ACE (Figures 2A and 2B) revealed the presence of an approx. 170 kDa protein band with a similar electrophoretic mobility compared with that of sACE, confirming the expression of the mutant proteins by CHO cells. Interestingly, both lysates and media revealed the presence of an additional smaller protein band of approx. 80 kDa (Figures 2A and 2B). The appearance of the lower band, which corresponded to the molecular mass of tACE suggested that CCdom-ACE was cleaved to generate two products: full-length soluble CCdom-ACE and soluble C domain. The lower band was smaller than the tACE protein, which has an intact O-glycosylated N-terminal region. Thus cleavage seemed to be occurring not only within the stalk region, but also within, or close to, the interdomain bridge of CCdom-ACE. Densitometric analysis showed that these two cleavage events of CCdom-ACE were up-regulated in the presence of PDBu (Figure 2A, lane 2), by 10% for full-length soluble CCdom ACE and 92% for soluble C domain. There was a concomitant disappearance of the cell-associated CCdom-ACE and C domain by 14 and 88% respectively (Figure 2B, lane 2). This suggested that both C domains of CCdom-ACE, distal and proximal to the TM, were shed into the medium. Furthermore, proteolysis of CCdom-ACE at both sites was inhibited by TAPI, reducing the levels of soluble CCdom-ACE by 51% and soluble C domain by 93% (Figure 2A, lane 3). The levels of the corresponding cell-associated proteins were increased by 33 and 85% respectively (Figure 2B, lane 3).
Figure 2. ACE mutant expression and shedding in CHO cells.
Confluent CHO cells expressing CCdom-ACE (A, B) and CNdom-ACE (C, D) were refed with fresh media and grown for 4 h (A, B) and 24 h (C, D) in the absence and presence of either 1 μM PDBu or 100 μM TAPI, before the culture medium and the cell lysate aliquots were resolved by SDS/PAGE and analysed by Western blotting with a tACE polyclonal antibody. CHO cells expressing sACE were treated in a similar manner [3]. Purified soluble tACE was included to control for the molecular mass of the proteins. Quantification of the CC dom-ACE data (A, B) can be seen as supplementary online data at http://www.BiochemJ.org/bj/389/bj3890739add.htm.
In contrast, CNdom-ACE was not identified in the culture medium of cells grown for 4 h after refeeding, in the absence or presence of PDBu and TAPI (results not shown). However, after growth for 24 h, two proteolytic products were produced, similar to those for CCdom-ACE (Figures 2C and 2D). A lower band (∼85 kDa), which was only observed after prolonged exposure (results not shown), probably corresponded to the soluble form of the N-terminal C domain. Thus cleavage at the bridge site of CNdom-ACE was far less efficient than that in CCdom-ACE. The cell lysate samples revealed the residual membrane-bound N domain (∼80 kDa) as well as the full-length construct (Figure 2D). The results for the CCdom- and CNdom-ACE swap mutants suggest that when the C domain is N-terminal to the inter-domain region, cleavage occurs within this protein sequence. In sACE, by contrast, no proteolysis of the interdomain bridge was observed after 4 h (Figures 2A–2D, lane 5). Metabolic labelling of sACE and pulse chase for 24 h indicated only full-length soluble sACE in the medium [3], supporting the notion that sACE is not cleaved within the interdomain bridge. The presence of full-length soluble CNdom-ACE in the medium samples after 24 h (Figure 2C) indicated that cleavage also occurred within the stalk region, albeit inefficiently. Unlike the CCdom-ACE construct, however, phorbol ester and TAPI appeared to have a negligible effect on the shedding of CNdom-ACE (Figures 2C and 2D, lanes 2 and 3).
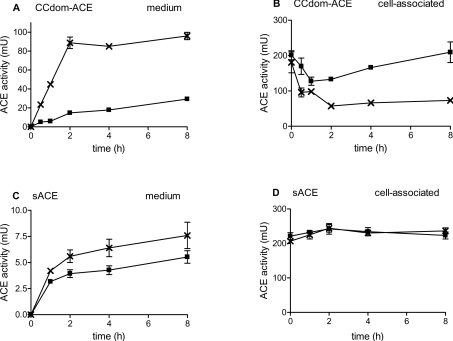
In light of the efficient ectodomain cleavage of CCdom-ACE from CHO cells, the rate at which shedding occurred relative to sACE was determined. Transfected cells grown to confluence were refed and grown for 8 h, with determination of ACE activity performed at various time points for both the medium and cell lysate samples, using HHL as the substrate (Figure 3). There was a progressive increase in the ACE activity in the culture medium over an 8 h period, indicating that CCdom-ACE was shed into the medium.
Figure 3. Rate of CCdom-ACE and sACE ectodomain shedding by CHO cells.
CHO cells stably expressing CCdom-ACE and sACE were grown to confluence and then refed with fresh medium supplemented with (×) or without (■) PDBu and grown for a further 8 h. At the indicated times, culture media (A, C) and cell lysates (B, D) were collected and assayed for ACE activity using the ACE substrate HHL (n=3±S.D.).
In the presence of PDBu, there was a 6-fold increase in shedding, with a concomitant decrease in cell-associated ACE (Figures 3A and 3B). In contrast, levels of soluble sACE were approx. 10-fold lower at all time points, a difference that became even more pronounced in the presence of PDBu (Figures 3C and 3D). CNdom-ACE activity was only detected in the medium after 24 h (results not shown), indicating that this mutant was shed even less efficiently than sACE.
Expression of the culture-medium ACE activity as a percentage of the total cell-associated plus soluble activity indicated that CCdom-ACE was shed as efficiently as wild-type tACE (13.2 and 8.9% respectively). This suggested that the presence of an additional C domain did not hinder the proteolysis of the C domain. In contrast, CNdom-ACE was not shed after 4 h and <1% shedding was observed after 24 h. Therefore the presence of the N domain decreased the efficiency at which the C domain was shed from CHO cells.
Cleavage-site analysis
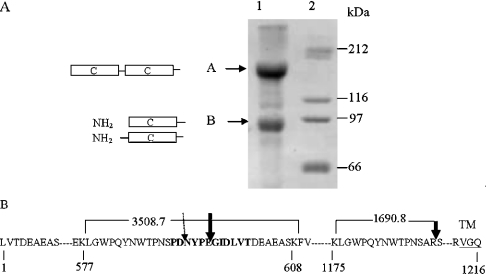
To determine the peptide bond at which CCdom-ACE was cleaved and released from the membrane, soluble protein was purified by lisinopril–Sepharose affinity chromatography from the conditioned medium of CCdom-ACE-CHO cells and analysed by SDS/PAGE. Two populations of soluble CCdom-ACE were obtained: full-length CCdom-ACE, comprising both the C domains (∼170 kDa) and an approx. 80 kDa band corresponding to the single C domain (Figure 4A). The two protein fractions were eluted from the gel, digested with endoproteinase Lys-C and the [M+H]+ ions were determined by MALDI–TOF MS (Table 1).
Figure 4. Purification and cleavage-site determination of CCdom-ACE.
(A) Purified CCdom-ACE was resolved on a 10% (w/v) polyacrylamide gel and stained with Coomassie Blue (lane 1). Two protein bands were observed: ‘A’ representing full-length soluble CCdom-ACE (170 kDa) and ‘B’ comprising the cleaved soluble C domains (80 kDa). Both C domains are represented as open boxes. Molecular-mass standards are included in lane 2. (B) Cleavage-site determination of CCdom-ACE. The sequence depicts CCdom-ACE from Leu1 to Gln1216 with the start of the TM underlined. Two peptides generated by Lys-C digestion are shown with the expected m/z: the intact bridge peptide with an m/z of 3508.7 and the C-terminal peptide generated by cleavage within the stalk with an m/z of 1690.8. The major cleavage sites are indicated with boldface arrows and the minor site is shown by a broken-line with an arrowhead. The sequence of the bridge region is shown in boldface.
Table 1. Observed [M+H]+ ions of the peptides generated by endoproteinase Lys-C digestion of purified soluble CCdom-ACE.
* The intact bridge peptide.
† The C-terminal stalk peptide.
The intact interdomain peptide Leu578–Lys608 with an observed m/z of 3508.4 was identified in the peptides obtained from the 170 kDa protein (Figure 4A and Table 1). This suggested that the higher molecular mass band corresponded to full-length CCdom-ACE with an intact interdomain bridge. In contrast, the interdomain peptide was not found in the peptides derived from the 80 kDa protein, indicating that it had been cleaved into two peptides. The proteins eluted from these two bands were sequenced to determine where cleavage occurred within the interdomain bridge. Two sequences were obtained from the protein from the lower band: (i) a major sequence, GIDVLT, and (ii) a minor sequence, NYPEG. This suggested that cleavage occurred predominantly at the Glu595–Gly596 bond within the bridge region and to a lesser extent after Asp591 (Figure 4B). Peptides with m/z of 1690.8 were identified in the Lys-C digest of both fractions (Figure 4B and Table 1). These correspond to the C-terminal peptide, Leu614–Arg627 of wild-type tACE. Thus juxtamembrane cleavage of CCdom-ACE occurred at the Arg1189–Ser1190 bond, the same site as in tACE (Figure 4B).
Kinetic characterization of the mutant proteins
To determine the effects of domain swaps on the catalytic properties of sACE, CCdom-ACE and CNdom-ACE, these enzymes were characterized with respect to the hydrolysis of the substrate Z-FHL and the C-domain-specific fluorogenic peptide AbzLFK(Dnp)-OH. The kcat of CCdom-ACE was approximately twice that for tACE using Z-FHL as the substrate (Table 2). For the hydrolysis of AbzLFK(Dnp)-OH, the kcat of CCdom-ACE was more than twice the kcat for tACE (Table 2). This suggested that the two domains of CCdom-ACE function independently or exhibit positive co-operativity.
Table 2. Kinetic parameters for the hydrolysis of Z-FHL and AbzLFK(Dnp)-OH (LFK).
The kinetic constants of the various ACE forms were determined as described in the Methods section. Data shown are the means for three determinations±S.E.M.
| Km (μM) | kcat (s−1) | kcat/Km (μM−1·s−1) | ||||
|---|---|---|---|---|---|---|
| Z-FHL | LFK | Z-FHL | LFK | Z-FHL | LFK | |
| N domain | 1180.7±118.6 | 5.0±0.5 | 73.2±5.8 | 1.0±0.1 | 0.06±0.01 | 0.2±0.0 |
| tACE | 318.35±11.8 | 3.3±0.5 | 107.0±3.1 | 20.6±0.5 | 0.34±0.02 | 6.3±1.0 |
| sACE | 406.8±75.2 | 3.6±0.3 | 116.9±3.4 | 13.3±0.49 | 0.29±0.06 | 3.7±1.7 |
| CNdom-ACE | 158.4±16.4 | 4.7±1.4 | 106.6±2.6 | 19.2±2.8 | 0.68±0.05 | 4.1±2.0 |
| CCdom-ACE | 357.6±89.7 | 5.0±0.8 | 315.8±27.4 | 69.4±5.7 | 0.90±0.15 | 13.9±7.1 |
In contrast, for both sACE and CNdom-ACE, the kcat values for both Z-FHL and AbzLFK(Dnp)-OH were less than the sum of the kcat values for the individual N and C (tACE) domains (Table 2). These results are suggestive of negative co-operativity between the C and N domains irrespective of their positions relative to each other (NC or CN).
DISCUSSION
The physiological significance of the two-domain structure of sACE remains uncertain. In some respects, the N domain appears to be redundant because the C domain is sufficient for the hydrolysis of ‘conventional’ substrates such as angiotensin I and bradykinin (reviewed in [19]). However, the N domain may hydrolyse unconventional substrates, such as luteinizing-hormone-releasing hormone and the haemoregulatory peptide AcSDKP [7,20]. Another possibility is that the N domain has a regulatory role, affecting the rate of ectodomain shedding and the catalytic properties of the C domain. To examine this possibility, we constructed two domain swap mutants, CCdom- and CNdom-ACE, and compared their shedding rates and catalytic activity with sACE, tACE and isolated N domain.
The poor ectodomain shedding of sACE compared with tACE is probably due to either steric interference of the sheddase cleavage site by the N domain or occlusion of a C domain sheddase recognition motif [2,3]. To determine whether the presence of an additional domain by itself causes steric hindrance to the shedding of the C domain, two C domains were fused together using the bridge region of sACE. This ACE construct, CCdom-ACE, was shed as efficiently as tACE from the membrane, which makes it less likely that the mere presence of an additional domain (N domain) in sACE sterically interferes with the ACE sheddase. Although it is possible that the native conformation of sACE is not mimicked in the CCdom-ACE construct, there is evidence for independent folding of the two domains in sACE [21]. Thus steric hindrance of the stalk cleavage site by the N domain does not appear to play a major role in the poor shedding efficiency of sACE.
An alternative hypothesis is that the N domain occludes the sheddase recognition motif present in the C domain of sACE (as opposed to non-specific hindrance of access to the stalk cleavage site). Evidence for the presence of a recognition motif was obtained by Western-blot analysis, which indicated that CCdom-ACE was cleaved not only within the stalk region to release full-length soluble CCdom-ACE, but also within the interdomain bridge region. In contrast, the ACE sheddase did not cleave sACE within this interdomain bridge region; this we interpret as implying that the N domain lacks the recognition motif needed for the orientation of the sheddase.
Earlier work suggested that the ACE sheddase positions itself with respect to the extracellular domain, proximal to the stalk region [11]. Western-blot analysis of CCdom-ACE indicated that the remaining cell-associated protein fragment corresponded to the membrane-proximal C domain. The presence of a second C domain was capable of directing cleavage by the sheddase to the interdomain bridge, which acted as a surrogate stalk. The bridge cleavage probably involved the ACE sheddase or related protease because it was stimulated by phorbol ester and inhibited by TAPI, typical characteristics of regulated ectodomain shedding.
To investigate this model further, the two domains of sACE were interchanged to generate a construct with an N-terminal C domain and a membrane-proximal N domain. This construct, CNdom-ACE, was shed even less efficiently than sACE, indicating that the presence of the N domain adjacent to the stalk down-regulated shedding. Interestingly, cleavage also occurred in the interdomain bridge region, similar to CCdom-ACE, albeit to a much lesser extent. These results suggest that the C domain provided the recognition motif for cleavage both within the interdomain bridge of CNdom-ACE and probably for the stalk region as well.
We have shown previously that a construct consisting of the N domain only, fused to the stalk, TM and cytoplasmic domains, is shed extremely inefficiently [5,22]. Therefore the C domain appears to be required for enabling cleavage by the ACE sheddase, and the C domain cannot be substituted with the N domain. The most likely explanation for these results is the presence of a recognition motif in the C domain. To study this further, discrete regions of the C domain were substituted for by corresponding regions of the N domain and the ectodomain shedding of these chimaeras was investigated, allowing delineation of the motif to the region Asp164–Val416 of the C domain (Z. L. Woodman and E. D. Sturrock, unpublished work).
The presence of a recognition motif in the ectodomain distal to the stalk of tACE has been previously postulated to direct the shedding of the protein, based on the shedding of ACE-CD4 chimaeras [4]. This ‘distal interaction’ hypothesis has also been used to explain the up-regulated shedding of murine L-selectin, which requires the presence of an epidermal growth factor domain remote from the cleavage site [23]. Similar studies have highlighted the importance of such distal interactions in the shedding of TNFα [24,25] and CD30 [26] by ADAM 17 (a disintegrin and metalloproteinase 17; TNFα-converting enzyme).
The interdomain co-operativity, both negative and positive, observed for sACE, CCdom-ACE and CNdom-ACE suggests that the two domains are in close contact during the binding and hydrolysis of peptide substrates. This is consistent with our hypothesis that the N domain of sACE occludes the sheddase recognition motif present in the C domain. For this to occur, flexibility within the interdomain bridge must permit folding of the N domain over the C domain, while the ectodomain remains membrane-bound. This flexibility persists after the protein is solubilized, placing the two domains in sufficiently close contact to co-operate in a positive or negative manner.
A number of studies have indicated that, in the context of full-length sACE, the N and C domains do not function as independent catalytic domains. With respect to several conventional substrates, the kcat determined for sACE is not significantly different from that determined for tACE, suggesting that the C domain (in both sACE and tACE) is the principal angiotensin-converting domain [7]. Other studies have reported strong negative co-operativity for sACE, such that the kcat for Z-FHL, angiotensin I and angiotensin 1–7 was approximately the mean of the kcat values determined for the individual domains [8,27]. A recent in vivo study in the mouse indicated that angiotensin I hydrolysis was completely abolished by selective inhibition of either the N or C domain [28], suggesting some kind of domain interaction. Moreover, by active-site titration with the tight-binding inhibitor lisinopril, only 1 mol/mol was sufficient to abolish catalytic activity in both sACE and tACE [7,9], and by isothermal titration calorimetry, only 1 mol/mol of inhibitor bound to sACE [9]. Our results reinforce this notion, in that we found negative co-operativity between the N and C domains in both sACE and CNdom-ACE. However, in contrast with sACE and CNdom-ACE, we found that two C domains in tandem (CCdom-ACE) resulted in either additive catalytic activity or positive co-operativity. Therefore the N domain exerts a negative regulatory effect on both shedding and catalytic activity of two-domain enzymes.
In conclusion, ectodomain shedding of human sACE is inhibited by the presence of the N domain. This effect may be the result of occlusion of a putative ACE sheddase recognition motif present in the C domain, which directs cleavage within the juxtamembrane stalk as well as the interdomain bridge, depending on the relative positioning of the C domain with respect to the N domain. For this to occur, a flexible interdomain bridge is required that permits close contact between the two domains, at the same time also facilitating interdomain co-operativity in the hydrolysis of peptide substrates.
Online data
Acknowledgments
We are grateful for the financial support of the Wellcome Trust (London, U.K.) and the N. R. F. (Pretoria, South Africa).
References
- 1.Ehlers M. R., Fox E. A., Strydom D. J., Riordan J. F. Molecular cloning of human testicular angiotensin-converting enzyme: the testis isozyme is identical to the C-terminal half of endothelial angiotensin-converting enzyme. Proc. Natl. Acad. Sci. U.S.A. 1989;86:7741–7745. doi: 10.1073/pnas.86.20.7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beldent V., Michaud A., Bonnefoy C., Chauvet M. T., Corvol P. Cell surface localization of proteolysis of human endothelial angiotensin I-converting enzyme. Effect of the amino-terminal domain in the solubilization process. J. Biol. Chem. 1995;270:28962–28969. doi: 10.1074/jbc.270.48.28962. [DOI] [PubMed] [Google Scholar]
- 3.Woodman Z. L., Oppong S. Y., Cook S., Hooper N. M., Schwager S. L., Brandt W. F., Ehlers M. R., Sturrock E. D. Shedding of somatic angiotensin-converting enzyme (ACE) is inefficient compared with testis ACE despite cleavage at identical stalk sites. Biochem. J. 2000;347:711–718. [PMC free article] [PubMed] [Google Scholar]
- 4.Sadhukhan R., Sen G. C., Ramchandran R., Sen I. The distal ectodomain of angiotensin-converting enzyme regulates its cleavage-secretion from the cell surface. Proc. Natl. Acad. Sci. U.S.A. 1998;95:138–143. doi: 10.1073/pnas.95.1.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pang S., Chubb A. J., Schwager S. L., Ehlers M. R., Sturrock E. D., Hooper N. M. Roles of the juxtamembrane and extracellular domains of angiotensin-converting enzyme in ectodomain shedding. Biochem. J. 2001;358:185–192. doi: 10.1042/0264-6021:3580185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei L., Alhenc-Gelas F., Corvol P., Clauser E. The two homologous domains of human angiotensin I-converting enzyme are both catalytically active. J. Biol. Chem. 1991;266:9002–9008. [PubMed] [Google Scholar]
- 7.Ehlers M. R., Riordan J. F. Membrane proteins with soluble counterparts: role of proteolysis in the release of transmembrane proteins. Biochemistry. 1991;30:10065–10074. doi: 10.1021/bi00106a001. [DOI] [PubMed] [Google Scholar]
- 8.Binevski P. V., Sizova E. A., Pozdnev V. F., Kost O. A. Evidence for the negative cooperativity of the two active sites within bovine somatic angiotensin-converting enzyme. FEBS Lett. 2003;550:84–88. doi: 10.1016/s0014-5793(03)00825-1. [DOI] [PubMed] [Google Scholar]
- 9.Andujar-Sanchez M., Camara-Artigas A., Jara-Perez V. A calorimetric study of the binding of lisinopril, enalaprilat and captopril to angiotensin-converting enzyme. Biophys. Chem. 2004;111:183–189. doi: 10.1016/j.bpc.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 10.Sturrock E. D., Danilov S. M., Riordan J. F. Limited proteolysis of human kidney angiotensin-converting enzyme and generation of catalytically active N- and C-terminal domains. Biochem. Biophys. Res. Commun. 1997;236:16–19. doi: 10.1006/bbrc.1997.6841. [DOI] [PubMed] [Google Scholar]
- 11.Ehlers M. R., Schwager S. L., Scholle R. R., Manji G. A., Brandt W. F., Riordan J. F. Proteolytic release of membrane-bound angiotensin-converting enzyme: role of the juxtamembrane stalk sequence. Biochemistry. 1996;35:9549–9559. doi: 10.1021/bi9602425. [DOI] [PubMed] [Google Scholar]
- 12.Ehlers M. R., Chen Y. N., Riordan J. F. The unique N-terminal sequence of testis angiotensin-converting enzyme is heavily O-glycosylated and unessential for activity or stability. Biochem. Biophys. Res. Commun. 1992;183:199–205. doi: 10.1016/0006-291x(92)91628-4. [DOI] [PubMed] [Google Scholar]
- 13.Ehlers M. R., Chen Y. N., Riordan J. F. Spontaneous solubilization of membrane-bound human testis angiotensin-converting enzyme expressed in Chinese hamster ovary cells. Proc. Natl. Acad. Sci. U.S.A. 1991;88:1009–1013. doi: 10.1073/pnas.88.3.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwager S. L., Chubb A. J., Scholle R. R., Brandt W. F., Eckerskorn C., Sturrock E. D., Ehlers M. R. Phorbol ester-induced juxtamembrane cleavage of angiotensin-converting enzyme is not inhibited by a stalk containing intrachain disulfides. Biochemistry. 1998;37:15449–15456. doi: 10.1021/bi981260k. [DOI] [PubMed] [Google Scholar]
- 15.Schwager S. L., Chubb A. J., Scholle R. R., Brandt W. F., Mentele R., Riordan J. F., Sturrock E. D., Ehlers M. R. Modulation of juxtamembrane cleavage (‘shedding’) of angiotensin-converting enzyme by stalk glycosylation: evidence for an alternative shedding protease. Biochemistry. 1999;38:10388–10397. doi: 10.1021/bi990357j. [DOI] [PubMed] [Google Scholar]
- 16.Friedland J., Silverstein E. A sensitive fluorimetric assay for serum angiotensin-converting enzyme. Am. J. Clin. Pathol. 1976;66:416–424. doi: 10.1093/ajcp/66.2.416. [DOI] [PubMed] [Google Scholar]
- 17.Piquilloud Y., Reinharz A., Roth M. Studies on the angiotensin converting enzyme with different substrates. Biochim. Biophys. Acta. 1970;206:136–142. doi: 10.1016/0005-2744(70)90090-2. [DOI] [PubMed] [Google Scholar]
- 18.Bersanetti P. A., Andrade M. C., Casarini D. E., Juliano M. A., Nchinda A. T., Sturrock E. D., Juliano L., Carmona A. K. Positional-scanning combinatorial libraries of fluorescence resonance energy transfer peptides for defining substrate specificity of the angiotensin I-converting enzyme and development of selective C-domain substrates. Biochemistry. 2004;43:15729–15736. doi: 10.1021/bi048423r. [DOI] [PubMed] [Google Scholar]
- 19.Acharya K. R., Sturrock E. D., Riordan J. F., Ehlers M. R. ACE revisited: a new target for structure-based drug design. Nat. Rev. Drug Discov. 2003;2:891–902. doi: 10.1038/nrd1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rousseau A., Michaud A., Chauvet M. T., Lenfant M., Corvol P. The hemoregulatory peptide N-acetyl-Ser-Asp-Lys-Pro is a natural and specific substrate of the N-terminal active site of human angiotensin-converting enzyme. J. Biol. Chem. 1995;270:3656–3661. doi: 10.1074/jbc.270.8.3656. [DOI] [PubMed] [Google Scholar]
- 21.Voronov S., Zueva N., Orlov V., Arutyunyan A., Kost O. Temperature-induced selective death of the C-domain within angiotensin-converting enzyme molecule. FEBS Lett. 2002;522:77–82. doi: 10.1016/s0014-5793(02)02888-0. [DOI] [PubMed] [Google Scholar]
- 22.Balyasnikova I. V., Woodman Z. L., Albrecht R. F., II, Natesh R., Acharya K. R., Sturrock E. D., Danilov S. M. The localization of an N-domain region of angiotensin-converting enzyme involved in the regulation of ectodomain shedding using monoclonal antibodies. J. Proteome Res. 2005;4:258–267. doi: 10.1021/pr049859w. [DOI] [PubMed] [Google Scholar]
- 23.Zhao L., Shey M., Farnsworth M., Dailey M. O. Regulation of membrane metalloproteolytic cleavage of L-selectin (CD62l) by the epidermal growth factor domain. J. Biol. Chem. 2001;276:30631–30640. doi: 10.1074/jbc.M103748200. [DOI] [PubMed] [Google Scholar]
- 24.Black R. A., Doedens J. R., Mahimkar R., Johnson R., Guo L., Wallace A., Virca D., Eisenman J., Slack J., Castner B., et al. Substrate specificity and inducibility of TACE (tumour necrosis factor alpha-converting enzyme) revisited: the Ala-Val preference, and induced intrinsic activity. Biochem. Soc. Symp. 2003;70:39–52. doi: 10.1042/bss0700039. [DOI] [PubMed] [Google Scholar]
- 25.Zheng Y., Saftig P., Hartmann D., Blobel C. Evaluation of the contribution of different ADAMs to tumor necrosis factor alpha (TNFalpha) shedding and of the function of the TNFalpha ectodomain in ensuring selective stimulated shedding by the TNFalpha convertase (TACE/ADAM17) J. Biol. Chem. 2004;279:42898–42906. doi: 10.1074/jbc.M403193200. [DOI] [PubMed] [Google Scholar]
- 26.Hansen H. P., Recke A., Reineke U., von Tresckow B., Borchmann P., von Strandmann E. P., Lange H., Lemke H., Engert A. The ectodomain shedding of CD30 is specifically regulated by peptide motifs in its cysteine-rich domains 2 and 5. FASEB J. 2004;18:893–895. doi: 10.1096/fj.03-0901fje. [DOI] [PubMed] [Google Scholar]
- 27.Rice G. I., Thomas D. A., Grant P. J., Turner A. J., Hooper N. M. Evaluation of angiotensin-converting enzyme (ACE), its homologue ACE2 and neprilysin in angiotensin peptide metabolism. Biochem. J. 2004;383:45–51. doi: 10.1042/BJ20040634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Georgiadis D., Beau F., Czarny B., Cotton J., Yiotakis A., Dive V. Roles of the two active sites of somatic angiotensin-converting enzyme in the cleavage of angiotensin I and bradykinin: insights from selective inhibitors. Circ. Res. 2003;93:148–154. doi: 10.1161/01.RES.0000081593.33848.FC. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.