Abstract
Although investigations of the transcriptional regulation of the rat cytochrome P450C24 [CYP24 (25-hydroxyvitamin D3 24-hydroxylase)] gene by 1,25D (1,25-dihydroxyvitamin D3) at either the genomic, or more recently at the non-genomic, level have provided insight into the mechanism of control of 1,25D levels, this regulation is still poorly characterized. Using HEK-293T cells (human embryonic kidney 293T cells), we reported that 1,25D induction of CYP24 requires JNK (c-Jun N-terminal kinase) but not the ERK1/2 (extracellular-signal-regulated kinase 1/2). The phenomenon of synergistic up-regulation of CYP24 expression by PMA and 1,25D is well known and was found to be protein kinase C-dependent. Whereas ERK1/2 was not activated by 1,25D alone, its activation by PMA was potentiated by 1,25D also. The importance of ERK1/2 for transcriptional synergy was demonstrated by transfection of a dominant-negative ERK1(K71R) mutant (where K71R stands for Lys71→Arg), which resulted in a reduced level of synergy on a CYP24 promoter-luciferase construct. JNK was also shown to be required for synergy. We report, in the present study, the identification of a site located at −171/−163, about 30 bp upstream of the vitamin D response element-1 in the CYP24 proximal promoter. This sequence, 5′-TGTCGGTCA-3′, is critical for 1,25D induction of CYP24 and is therefore termed the vitamin D stimulatory element. The vitamin D stimulatory element, a target for the JNK module, and an Ets-1 binding site were shown to be vital for synergy between PMA and 1,25D. This is the first report to identify the DNA binding sequences required for the synergy between PMA and 1,25D and a role for JNK on the CYP24 gene promoter.
Keywords: cytochrome P450C24; 1,25-dihydroxyvitamin D3; extracellular-signal-regulated kinase 1/2 (ERK1/2); c-Jun N-terminal kinase (JNK); phorbol ester; transcriptional regulation
Abbreviations: AP-1, activator protein-1; CYP24, 25-hydroxyvitamin D3 24-hydroxylase; 1,25D, 1,25-dihydroxyvitamin D3; EBS, Ets-1 binding site; EMSA, electrophoretic mobility-shift assay; ERK, extracellular-signal-regulated kinase; GST, glutathione S-transferase; HEK-293T cell, human embryonic kidney 293T cell; JNK, c-Jun N-terminal kinase; MAP, mitogen-activated protein; MKK, MAP kinase kinase; oFSHR, ovine follicle-stimulating hormone receptor; PKC, protein kinase C; RasGRP, Ras guanine-nucleotide-releasing protein; RXR, retinoid-X receptor; VDR, vitamin D receptor; VDRE, vitamin D response element; VSE, vitamin D stimulatory element; WT, wild-type
INTRODUCTION
The biologically active form of 1,25D [1,25-dihydroxyvitamin D3 or 1,25(OH)2D3], also known as calcitriol, is a secosteroid hormone that plays important roles in a range of biological activities including calcium and phosphate homoeostasis and bone remodelling [1–4], cellular proliferation and differentiation [5] and immunoregulation [6]. Transcriptional regulation of 1,25D-target genes is mediated by VDR (vitamin D receptor), a member of the ligand-inducible nuclear receptor superfamily [4,7,8]. Liganded VDR forms a heterodimeric complex with RXR (retinoid-X receptor) and stimulates the expression of target genes by binding to promoter sequences consisting of an imperfect direct repeat of the hexamer 5′-AGGTCA-3′ separated by 3 bp, termed as VDRE (vitamin D response element). In the absence of a ligand, VDR/RXR has been shown to form a complex with DNA and interact with co-repressors such as N-CoR, RIP13Δ1 and Alien [9,10], thereby resulting in the repression of basal transcription from 1,25D-responsive genes.
A balance between the bioactivation and degradation of 1,25D is responsible for maintaining homoeostatic levels of 1,25D at the correct set-point. The predominant site of synthesis for circulating 1,25D is the kidney, where the cytochrome P450 enzyme CYP27B1 (25-hydroxyvitamin D3 1α-hydroxylase) catalyses the hydroxylation of the precursor 25D to generate 1,25D [11–13]. Catabolic inactivation of 1,25D and conversion into water-soluble products such as calcitroic acid is achieved through side chain oxidation and cleavage reactions catalysed by the mitochondrial cytochrome P450 enzyme CYP24 (25-hydroxyvitamin D3 24-hydroxylase, also referred to as CYP24A1), which is highly expressed in bone and kidney cells [12–14]. 1,25D is the most potent known inducer of CYP24 expression and the level of induction is the highest for any known 1,25D target gene [13]. This potent up-regulatory system constitutes a negative feedback mechanism pivotal for controlling ambient and cellular concentrations of 1,25D [13]. In vivo, CYP24 null mice exhibit abnormal intramembranous bone development, hypercalcaemia and elevated serum 1,25D levels [15,16]. Therefore co-ordinated expression of CYP24 activity and hence 1,25D levels provide a protective barrier against 1,25D-induced toxicity.
We investigated the molecular mechanism governing the transcriptional control of the rat CYP24 gene by 1,25D. We and others have identified in the rat CYP24 promoter two functional VDREs, termed VDRE-1 and VDRE-2, localized to the antisense strand at positions −150/−136 and −258/−244 respectively [17–21]. Recent studies with 1,25D have shown that, in addition to the activation of transcription through VDR/RXR binding to VDREs, 1,25D triggers more immediate and rapid non-genomic effects, including increases in intracellular calcium and activation of several kinases including PKC (protein kinase C) [22]. In particular, we reported a functional EBS (Ets-1 binding site) downstream of the proximal VDRE at −128/−119 and demonstrated this to be important for maximal 1,25D activity of the rat CYP24 promoter [17,18]. Using COS-1 cells, activation of Ets-1 in response to 1,25D stimulation was demonstrated to involve the phosphorylation of Ets-1 by a MAP (mitogen-activated protein) kinase, ERK5 (extracellular-signal-regulated kinase 5), thereby establishing a link between the non-genomic and the genomic activation of the CYP24 promoter in response to 1,25D stimulation [18].
Previous studies in rat renal and rat intestinal epithelial cells have shown that the tumour-promoting phorbol ester PMA potentiates the effect of 1,25D in the up-regulation of the CYP24 promoter and that this synergistic effect corresponds to an increase in CYP24 mRNA levels [23–27]. PMA has traditionally been used as a tool to investigate PKC activation and it was shown that the synergy between PMA and 1,25D in intestinal epithelial cells was PKC-dependent [24]. A study by Pike et al. [28] determined that the proximal 250 bp of the human CYP24 promoter were sufficient for synergy to occur but the binding sites responsible could not be identified. Likewise, the mechanism of synergy at the non-genomic level has not been elucidated.
In the present study, we report the identification of the sequence 5′-TGTCGGTCA-3′ located approx. 30 bp upstream of VDRE-1 at −171/−163 in the CYP24 proximal promoter. The site is critical for the induction of the CYP24 promoter by 1,25D and is hence termed the VSE (vitamin D stimulatory element). In contrast with COS-1 cells, we show that in HEK-293T cells (human embryonic kidney 293T cells) 1,25D induction of the CYP24 promoter requires activation of the MAP kinase JNK (c-Jun N-terminal kinase) but not ERK1/2. The synergy between PMA and 1,25D requires the VSE and the EBS and is largely dependent on ERK1/2 and JNK activities.
EXPERIMENTAL
Reagents
1,25D was obtained from Tetrionics (Madison, WI, U.S.A.). PMA and 4α-PMA were purchased from Sigma–Aldrich (Sydney, Australia). The inhibitor calphostin C was obtained from Biomol (Plymouth Meeting, PA, U.S.A.). DOTAP (dioleoyl trimethylammonium propane) was purchased from Roche Diagnostics (Castle Hill, NSW, Australia). The DLR (dual-luciferase reporter) assay system was obtained from Promega (Madison, WI, U.S.A.). Phospho-p44/42 MAP kinase (Thr202/Tyr204) antibody and p44/42 MAP kinase antibody were obtained from New England Biolabs (Beverly, MA, U.S.A.). Anti-ERK2 (sc-154) antibody was obtained from Santa Cruz Biotechnology (Santa Cruz, CA, U.S.A.). [γ-32P]dATP and [α-32P]dCTP were supplied by Geneworks (Adelaide, Australia).
Promoter luciferase constructs and expression vectors
The rat CYP24 promoter luciferase constructs in the present study contain the promoter sequence from either −298 or −186 bp to the transcription start site as well as 74 bp of 5′-untranslated region cloned upstream of the firefly luciferase reporter gene in the pGL3 basic vector, as described previously [17]. The pCYP24(mEBS)-Luc (where Luc stands for luciferase) reporter construct containing a mutation in the EBS has been described previously [17]. The mutation in the VSE sequence was achieved using the Quik Change site-directed mutagenesis kit (Stratagene, La Jolla, CA, U.S.A.). The sequence of the generated pCYP24(M2-VSE)-Luc plasmid was verified by automated DNA sequencing. The promoterless pRL-null Renilla luciferase vector was purchased from Promega. The dominant-negative ERK1 expression vector [ERK1(K71R), where K71R stands for Lys71→Arg] has been described in [29]. The dominant-negative MKK4(K116R) (where MKK stands for MAP kinase kinase) construct was a gift from Dr D. Riches (National Jewish Medical and Research Center, Denver, CO, U.S.A.) [30]. The dominant-negative PKC constructs were kindly provided by Professor I. B. Weinstein [31].
Cell maintenance and transfection
HEK-293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal bovine serum and kept at 37 °C in an atmosphere of 5% CO2 in air. Cells were transiently co-transfected with a CYP24 promoter luciferase construct and pRL-null Renilla luciferase vector as a control for transfection efficiency using DOTAP, essentially as described previously [18]. Where indicated, 200 ng of dominant-negative expression or control vector was also transfected. On the following day, cells were treated with 10−7 M PMA or vehicle (ethanol) for 2 h. Cells were then washed and the medium was replaced with RPMI 1640. For inhibitor studies, the inhibitor was diluted in RPMI 1640 and added to the cells at this point. 1,25D (10−8 M) or vehicle (ethanol) was then added and cells incubated for a further 22 h before harvesting. Each treatment was performed in triplicate. The DLR assay kit and a luminometer (model TD 20/20) were used to determine the luciferase activity of cell lysates.
EMSAs (electrophoretic mobility-shift assays)
The double-stranded oligonucleotide probe containing the VSE [VSE-WT (wild-type)] was end-filled with [α-32P]dCTP using a Klenow fragment of DNA polymerase I. The sequences of the oligonucleotides used in this study are shown below: VSE-WT, 5′-GGCGTGTCGGTCACCG-3′ and 3′-CGCACAGCCAGTGGCG-5′; M2-VSE, 5′-GGCGTGTAAGCTTCCG-3′ and 3′-CGCACATTCGAAGGCG-5′. Nuclear extracts were prepared from HEK-293T cells [32] and EMSA was performed as described previously [17].
Cell lysate preparation
HEK-293T cells grown to confluency in 10 cm dishes were treated with 10−7 M PMA, 10−7 M 1,25D or a vehicle for the times indicated. The cells were scraped and lysed at 4 °C in 300 μl of lysis buffer (25 mM Tris/HCl, pH 7.5, 2 mM EGTA, 25 mM NaCl, 1 mM Na3VO4, 38 mM p-nitrophenyl phosphate, 10 μg/ml each of pepstatin A, benzamidine, aprotinin and leupeptin, 2 mM PMSF and 1 mM dithiothreitol) as described previously [33]. After 2 h, the samples were centrifuged (16000 g, 3 min, 4 °C) and the protein concentration of the soluble fractions was determined using Lowry's method [54]. Samples were then stored at −20 °C until assayed.
Western-blot analyses
To assay for ERK1/ERK2 activation, equal amounts of denatured proteins were subjected to Western-blot analysis using the anti-ACTIVE ERK antibody and the immunocomplexes were visualized by enhanced chemiluminescence as described previously [33–35]. A Western-blot recycling kit (Alpha Diagnostic International, San Antonio, TX, U.S.A.) was used to strip the blots that were reprobed with an anti-ERK2 or anti-ERK1/2 antibody.
JNK activity assays
A solid-phase assay using GST (glutathione S-transferase)-conjugated jun (1–79) was employed to assay JNK activity in the presence of [γ-32P]dATP (8–10 μCi/sample) [34]. Phosphorylated GST–jun (1–79) was fractionated on SDS/12% (w/v) polyacrylamide gels, and the bands were detected and radioactivity determined using an Instant Imager (Packard Instruments, Canberra, Australia).
Statistical analysis
Statistical analyses were performed by either Student's two-tailed unpaired t test or ANOVA, followed by the Tukey–Kramer multiple comparisons test using GraphPAD Software (Cricket Software, 1985; Philadelphia, PA, U.S.A.).
RESULTS
1,25D and PMA action on the CYP24 promoter in HEK-293T cells
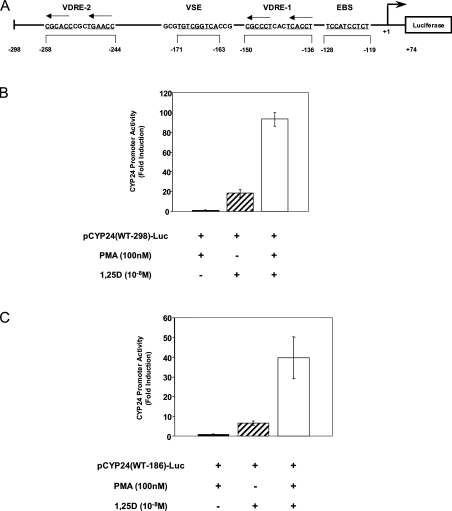
We first investigated the effect of added 1,25D on the expression of rat CYP24 promoter in HEK-293T cells. Cells were transiently transfected with luciferase reporter gene constructs driven by either the proximal 298 or 186 bp of the CYP24 promoter [pCYP24(WT-298)-Luc or pCYP24(WT-186)-Luc respectively]. The −298 CYP24 promoter region contains the two previously identified functional VDREs, a proximal VDRE denoted VDRE-1 at −150/−136 bp and a distal VDRE-2 at −258/−244 bp [19,21], whereas the –186 CYP24 promoter length contains only VDRE-1 (Figure 1A). With the pCYP24(WT-298)-Luc construct, treatment of cells with 1,25D resulted in an 18.4-fold increase in reporter gene activity relative to treatment with the vehicle ethanol (Figure 1B). 1,25D induction of the pCYP24(WT-186)-Luc promoter was lowered to 6.6-fold as expected since this promoter sequence contains only one VDRE (Figure 1C).
Figure 1. PMA and 1,25D synergy requires the first 186 bp of the CYP24 promoter.
(A) Schematic representation of the WT pCYP24(WT-298)-luciferase reporter construct. The sequence and relative locations of the VDRE-1 and VDRE-2, EBS and VSE on the antisense strand of the rat CYP24 promoter are shown. The construct contains 74 bp of 5′-untranslated region and −298 bp of promoter sequence. (B) pCYP24(WT-298)-Luc or (C) pCYP24(WT-186)-Luc luciferase constructs were transiently transfected into HEK-293T cells. pRL-Null-Luc was co-transfected as an internal control for transfection efficiency. Each construct was tested for transactivation by 100 nM PMA, 10−8 M 1,25D or by both 100 nM PMA and 10−8 M 1,25D together (see the Experimental section). The level of induction shown (presented as fold induction) is the ratio of luciferase activity from treated to untreated (vehicle/ethanol-treated) cells. Data presented here represent the average of six replicates for two independent experiments, ±S.D.
It was reported that the proximal 250 bp of the CYP24 promoter are sufficient for a synergistic effect of PMA on 1,25D induction, although attempts to further localize the PMA responsive element were unsuccessful [28]. We have revisited these studies in an attempt to further elucidate the mechanism of this synergy. HEK-293T cells were therefore treated with PMA or the vehicle ethanol for 2 h, washed and then 1,25D was added for a further 22 h before luciferase assay determinations. Treatment of cells with PMA did not alter basal promoter activity of either the pCYP24(WT-298)-Luc (Figure 1B) or pCYP24(WT-186)-Luc construct (Figure 1C). Occasionally, some induction by PMA alone was observed (up to 2-fold) but this was inconsistent. It can be seen that treatment with both PMA and 1,25D resulted in strong induction of the pCYP24(WT-298)-Luc construct (93.3-fold) representing a transcriptional synergistic action of 4.8-fold (Figure 1B). PMA and 1,25D treatment produced a 39.7-fold induction on the pCYP24(WT-186)-Luc construct, which, despite the expected decrease in the magnitude of response compared with pCYP24(WT-298)-Luc, corresponds to a 5.3-fold level of synergy (Figure 1C). These results demonstrate that in HEK-293T cells, the CYP24 promoter can be induced by 1,25D and there is synergistic activation of the promoter by PMA and 1,25D. The −186 bp promoter is sufficient for the synergistic effect, suggesting that the elements responsible for this PMA-directed synergy lie within this region. For the remainder of the studies, we have therefore employed the −186 bp promoter construct.
PKC is involved in PMA and 1,25D synergistic activation
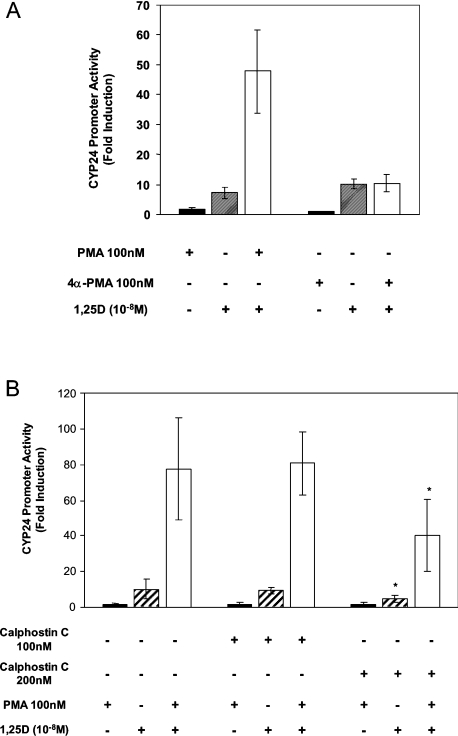
We next investigated whether the synergistic response involves PKC activity. It is well established that acute treatment of cells with PMA leads to PKC activation, whereas chronic PMA treatment results in PKC down-regulation [35]. In keeping with this, we found that 2 h PMA treatment followed by 1,25D treatment for 24 h produced a 48.4±4.0-fold induction of pCYP24(WT-186)-Luc, but 24 h PMA treatment followed by 24 h 1,25D treatment resulted in a much reduced 11.7±2.0-fold level of induction (P=0.0001 by Student's unpaired t test). This corresponded to a reduction in fold synergy from 6- to 1.9-fold, consistent with PKC participation in the synergistic activation. The remainder of the present study employed 2 h pretreatment with PMA followed by 1,25D treatment to examine the synergistic response.
The role of PKC in the synergistic induction of the CYP24 promoter was further investigated using the PMA analogue, 4α-PMA. This compound is a stereoisomer of PMA that does not activate PKC [36,37] but can activate pathways not dependent on PKC [36]. Consistent with a reliance on PKC activation for synergy, 4α-PMA failed to synergize with 1,25D (Figure 2A). To further examine the possible roles of PKC in the synergistic response, we also investigated the effect of the PKC inhibitor calphostin C. At the concentration of 200 nM, this inhibitor reduced the 1,25D response of pCYP24(WT-186)-Luc from 10-fold induction to 4.4-fold (Figure 2B). Fold induction by PMA and 1,25D was also lowered in the presence of calphostin C (77.5- to 40-fold at 200 nM). 1,25D induction and synergy were also similarly reduced using the PKC inhibitor GF 109203X at 1 μM (results not shown). These inhibitors are known to inhibit PKC activity and PKC-dependent phosphorylation of target proteins in intact cells at the concentrations tested here, with IC50 in cell-free systems of 50 and 20 nM for calphostin C [38] and GF 109203X [39] respectively.
Figure 2. PKC is required for PMA and 1,25D synergy.
(A) pCYP24(WT-186)-Luc was transfected into HEK-293T cells. Cells were treated for 2 h with 100 nM PMA, 100 nM 4α-PMA or their respective solvents (ethanol or DMSO). Treatment for 24 h with 10−8 M 1,25D or ethanol was then carried out. Data are shown as fold induction of relative luciferase activity compared with vehicle-treated cells and represent the means±S.D. for three independent experiments. (B) HEK-293T cells transfected with pCYP24(WT-186)-Luc were treated for 2 h with 100 nM PMA or vehicle. After washing the cells, 100 or 200 nM calphostin C or the control solvent DMSO was added to the cells. Cells were then treated with 10−8 M 1,25D or vehicle for 24 h. The experiment was repeated three times. Data represent the means±S.D. of six replicates for three experiments and are presented as fold induction of relative luciferase activity compared with vehicle-treated cells. Significance of difference between treatment with DMSO and calphostin C (200 nM) in the presence of 1,25D alone or PMA+1,25D: *P<0.05 by Tukey–Kramer multiple comparisons test.
1,25D has generally been reported to act through PKCα in a number of cell lines, including intestinal epithelial Caco-2 cells [40], chick myoblasts [41] and rat osteosarcoma cells [42], although reports in other cell types such as rat adipocytes have also shown an involvement of PKCβ [43]. On the other hand, any of the classical or novel PKC isoenzymes could mediate the actions of PMA [44]. To investigate which PKC isoenzymes were involved in mediating the action of 1,25D and PMA in synergy, constructs encoding the dominant-negative forms of PKCα, PKCβ1, PKCϵ and PKCδ were each co-transfected with pCYP24(WT-186)-Luc. These dominant-negative PKC mutants have previously been demonstrated to lack kinase activity and block the action of the WT isoenzyme [31]. In these experiments, the pcDNA3 empty vector was co-transfected as a control for the plasmid carrying the PKC mutants (Table 1). This repeatedly led to an increased response to PMA alone, which was capable of producing an 8.8-fold induction of luciferase activity (compare with the 2-fold induction seen in non-pcDNA3 co-transfected cells in Figures 1, 2A, 2B and later in Figure 7). This phenomenon may be due to the presence of viral promoters in these vectors (see the Discussion section). The results presented in Table 1 show that the kinase-dead mutants of PKCβ1, PKCδ and PKCϵ each reduced the response to PMA alone, confirming that this effect of PMA on the CYP24 promoter requires PKC activation. In contrast, PKCβ1 was the only mutant to reduce significantly the response to 1,25D alone. This demonstrates the specificity of the PKC mutants. The failure of the PKCα mutant to affect 1,25D induction implies that in this cell line, PKCβ1 is required to mediate the response to 1,25D. This is consistent with the involvement of PKCβ in the 1,25D response in rat adipocytes [43]. While each of the dominant-negative PKCs tested reduced the level of synergy between PMA and 1,25D, the most striking result is that of PKCα. This mutant failed to reduce the response to PMA or 1,25D alone yet it significantly reduced the synergistic response to PMA and 1,25D from 69.1- to 23.5-fold induction. This suggests that although PKCα is not required for the response to PMA or 1,25D alone, it is important in mediating synergism. Taken together, these results clearly establish a role for PKC in the regulation of CYP24 in HEK-293T cells. Whereas PKCβ1 is crucial for the action of 1,25D alone, PKCα is likely to be particularly important for the synergism between PMA and 1,25D.
Table 1. Effect of dominant-negative PKC mutants on the response of pCYP24(WT-186)-Luc to PMA and 1,25D.
293T cells were co-transfected with pCYP24(WT−186)-Luc together with either a dominant-negative PKC expression vector or the control vector pcDNA3 as described in the Experimental section. Cells were treated with 100 nM PMA or 10−8 M 1,25D alone or with 100 nM PMA for 2 h followed by 10−8 M 1,25D for 24 h as described in the Experimental section. Data are shown as fold induction. Fold synergy corresponds to PMA+1,25D induction divided by the sum of PMA induction and 1,25D induction. Values represent the means±S.D. for three experiments performed in triplicate. Significance of difference between pcDNA3 empty vector and dominant-negative vector-transfected cells was determined for each treatment group (PMA induction, 1,25D induction, PMA+1,25D induction and fold synergy). *P<0.05; **P<0.01; ***P<0.001 (Tukey–Kramer multiple comparisons test).
| Fold induction | ||||
|---|---|---|---|---|
| Vector | PMA | 1,25D | PMA+1,25D | Fold synergy |
| pcDNA3 | 8.82±3.29 | 5.07±0.72 | 69.09±17.79 | 4.97±1.28 |
| PKCα(K→R) | 6.88±2.93 | 5.54±1.21 | 23.53±7.90*** | 1.90±0.64*** |
| PKCβ1(K→R) | 3.06±0.54*** | 3.63±0.99* | 15.12±7.02*** | 2.26±1.05*** |
| PKCδ(K→R) | 4.87±1.86** | 5.18±0.83 | 18.64±5.18*** | 1.86±0.52*** |
| PKCϵ(K→R) | 3.17±1.16*** | 4.51±1.25 | 15.73±4.56*** | 2.05±0.59*** |
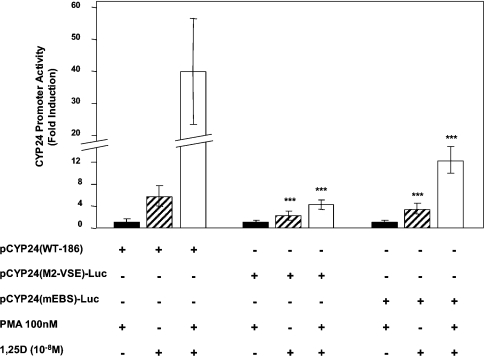
Figure 7. A VSE binding site is required for PMA and 1,25D synergy.
HEK-293T cells were transfected with the luciferase construct pCYP24(WT-186)-Luc, pCYP24(mEBS)-Luc containing a mutation in the EBS or pCYP24(M2-VSE)-Luc containing a mutation in the VSE. The constructs were tested for their response to PMA (100 nM for 2 h) or 1,25D (10−8 M for 24 h) treatments or PMA (2 h) followed by 1,25D (24 h). Results represent the means±S.D. for five independent experiments performed in triplicate. Significance of difference between 1,25D induction of pCYP24(WT-186)-Luc and 1,25D induction of pCYP24(mEBS)-Luc or pCYP24(M2-VSE)-Luc: ***P<0.001 (Tukey–Kramer multiple comparisons test). Significance of difference between PMA+1,25D induction of pCYP24(WT-186)-Luc and PMA+1,25D induction of pCYP24(mEBS)-Luc or pCYP24(M2-VSE)-Luc: ***P<0.001 (Tukey–Kramer multiple comparisons test).
Role of ERK1/2 and JNK in 1,25D induction
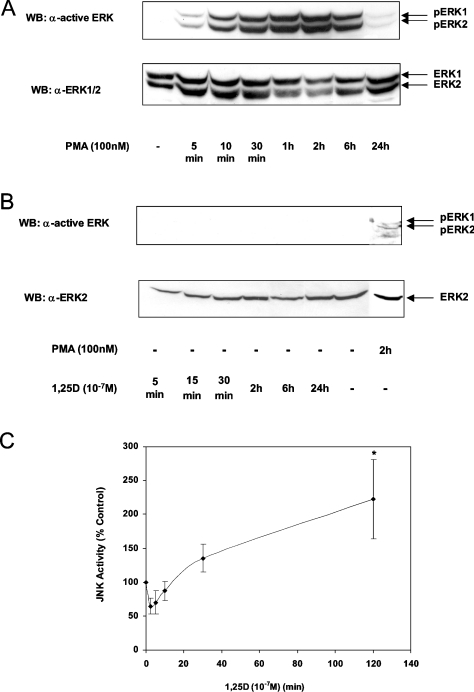
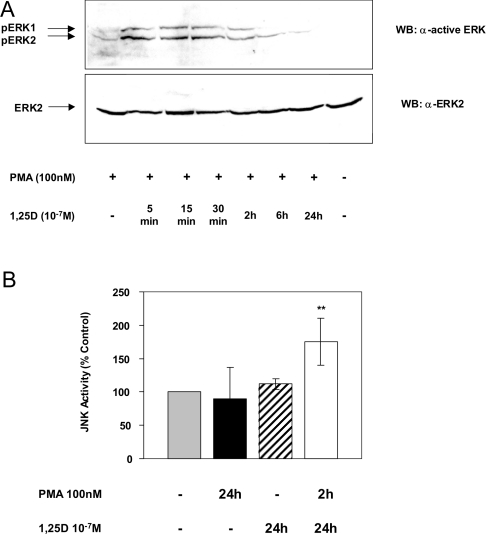
In our earlier studies, we established that activation of the MAP kinases ERK1/2 by 1,25D was critical for induction of the transfected CYP24 promoter in COS-1 cells [18]. Neither JNK nor p38 pathways were involved in 1,25D induction in this cell line. We investigated whether ERK1/2 activity was also increased by 1,25D in HEK-293T cells. The activation of ERK1/2 was determined by immunoblotting using an antibody raised against activated dual- phosphorylated ERK1/2. As expected, PMA treatment stimulated the activation of ERK1/2 after 10 min. Activation was at its strongest after 2 h before returning almost to basal levels after 24 h (Figure 3A). Surprisingly, 1,25D treatment (5 min to 24 h) did not increase ERK1/2 activation (Figure 3B). This result contrasts with the previous study in COS-1 cells where 5–15 min of 1,25D treatment was shown to induce ERK1/2 activity significantly [18].
Figure 3. Activation of ERK1/2 and JNK in response to 1,25D.
(A, B) HEK-293T cells were treated with DMSO control, 100 nM PMA or 10−7 M 1,25D for the indicated times. Proteins from cell lysates were fractionated by SDS/PAGE and transferred on to a nitrocellulose membrane. To detect activated ERK1/2, the blots were probed with anti-ACTIVE ERK (Thr202/Tyr204) antibody (upper panel). The membranes were stripped and reprobed with anti-ERK1/2 antibody (lower panel). Results shown are representative of two independent experiments. (C) Lysates were prepared from HEK-293T cells treated with 10−7 M 1,25D for the indicated time periods. JNK activity was determined by a solid-phase assay in the presence of [γ-32P]dATP (see the Experimental section). After separating by SDS/PAGE, bands were quantified as net c.p.m. Data (means±S.D. for three independent experiments) are shown as percentage c.p.m. relative to ethanol-treated control cells. Significance of difference between control and 1,25D treatment for 120 min: *P<0.01 (Tukey–Kramer multiple comparisons test). WB, Western blot.
Since ERK1/2 activity was not increased by 1,25D alone, we investigated whether JNK activity was stimulated in HEK-293T cells. To investigate the profile of JNK activation, HEK-293T cells were treated with either ethanol or 1,25D, lysed and JNK was adsorbed on to GST–jun (1–79) conjugated with glutathione–Sepharose. The GST–jun (1–79) also served as a substrate for JNK in these solid-phase assays. As shown in Figure 3(C), an initial suppression of JNK activity by 1,25D was observed between 2.5 and 10 min of treatment before stimulation of JNK activity occurred after 30 min. This activation was further increased after 2 h. However, by 24 h, kinase activity had returned to basal levels (see Figure 5B). The initial inhibition of JNK activity by 1,25D is surprising because we are not aware of such inhibition with other JNK activators. Although the reason for this is unclear, these results are consistent with reports in other cell lines showing JNK activation in response to 1,25D [40,45] and suggest that JNK may be involved with the 1,25D induction of CYP24 in HEK-293T cells.
Figure 5. ERK1/2 activation is potentiated and JNK activation is prolonged in response to PMA and 1,25D.
(A) HEK-293T cells were treated with DMSO vehicle, 100 nM PMA alone, 1,25D (see Figure 3A) or 100 nM PMA for 2 h followed by 10−7 M 1,25D for the indicated times. Proteins from cell lysates were fractionated by SDS/PAGE and transferred on to a nitrocellulose membrane. The blot was probed with anti-ACTIVE ERK antibody (upper panel), stripped and reprobed with anti-ERK2 antibody (lower panel). Results shown are representative of three independent experiments. (B) Lysates were prepared from HEK-293T cells treated with ethanol, 100 nM PMA for 24 h, 10−7 M 1,25D for 24 h or 100 nM PMA for 2 h followed by 10−7 M 1,25D for 24 h. JNK activity was determined using a solid-phase assay. Results, presented as percentage of control (ethanol-treated) cells, are the means±S.D. for five independent experiments. Significance of difference between control and PMA+1,25D treatments: **P<0.01 (Tukey–Kramer multiple comparisons test). WB, Western blot.
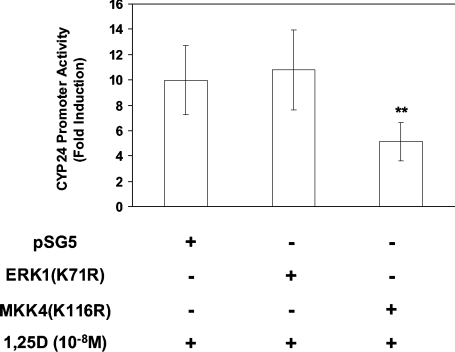
To investigate further whether ERK1/2 and JNK participated in 1,25D induction, cells were transfected with pCYP24(WT-186)-Luc and either the empty control vector (pSG5), or a construct encoding a dominant-negative ERK1 mutant [ERK1(K71R)] to block ERK1/2 [29] or a construct encoding a dominant-negative MKK4 [MKK4(K116R)] [30], one of the two immediate upstream regulators of JNK [46], to block the JNK pathway. The 10-fold level of 1,25D induction seen in control vector-transfected cells was unaffected by the ERK1(K71R) dominant-negative mutant employed at 200 ng (Figure 4). Similar results were obtained using 500 ng of this dominant-negative construct (results not shown), a concentration at which there is marked inhibition of 1,25D-induced promoter activity in COS-1 cells [18]. However, 1,25D induction was reduced to 5.1-fold by 200 ng of MKK4(K116R) (Figure 4). These results demonstrate that ERK1/2 activity is not required for induction by 1,25D on the −186 promoter in HEK-293T cells and suggests that JNK activation is predominantly required for 1,25D induction in these cells, a result that contrasts distinctly with our previous findings in COS-1 cells, where JNK was not required for 1,25D induction [18].
Figure 4. 1,25D-induced activation of the CYP24 promoter requires the JNK module.
pCYP24(WT-186)-Luc luciferase constructs were co-transfected into HEK-293T cells along with 200 ng of expression vector for dominant-negative MKK4 [an upstream regulator of JNK, MKK4(K116R)] or for dominant-negative ERK1 [ERK1(K71R)], as shown. Cells were treated with 10−8 M 1,25D for 24 h and luciferase activity was determined. Data (means±S.D. for three experiments performed in triplicate) are presented as fold induction. Significance of difference between pSG5 and MKK4(K116R): **P<0.01 (Tukey–Kramer multiple comparisons test).
Role of ERK1/2 and JNK activities in PMA and 1,25D synergy
Although ERK1/2 activity is not necessary for induction by 1,25D, it was still possible that this activity contributed to synergy by PMA and 1,25D. As seen above in Figure 3(A), PMA treatment alone for 2 h elevated ERK1/2 activity. This result is also seen in Figure 5(A) (compare lanes 1 and 8). Significantly, when cells were pretreated with PMA for 2 h and then 1,25D for 5 min, there was a further increase in ERK1/2 activation compared with PMA alone (lane 2, Figure 5A). This increased activation gradually declined over 24 h after 1,25D treatment (lanes 3–7, Figure 5A). Hence the potentiated activation of ERK1/2 could be important for synergy even though this ERK1/2 activity does not contribute to induction by the hormone alone.
The requirement for JNK activity in 1,25D induction suggested that JNK also plays a role in the synergy between PMA and 1,25D. PMA treatment between 5 and 10 min was able to activate JNK activity (163±28% of control, P<0.05: Student's unpaired t test); however, as seen in Figure 5(B), 24 h of treatment with PMA alone resulted in JNK activation at or below vehicle-treated cells. The 1,25D-induced activation of JNK observed between 30 min and 2 h (Figure 3B) had returned to basal levels after 24 h (Figure 5B). Interestingly, after treatment with PMA for 2 h followed by 1,25D for 24 h, JNK activity remained elevated at 24 h (Figure 5B). This prolonged activation of JNK indicated a potential role for JNK in synergy.
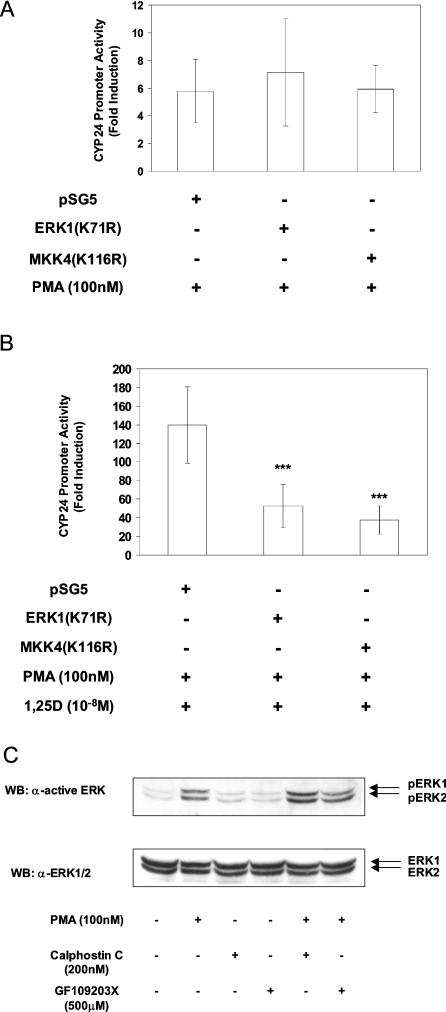
These results prompted us to investigate directly whether ERK1/2 and JNK activities are important for the synergistic response. Cells were co-transfected with the pCYP24(WT-186)-Luc promoter construct and either ERK1(K71R), MKK4(K116R) or pSG5. In these experiments, the presence of co-transfected empty pSG5 vector again led to PMA activation of the pCYP24(WT-186)-Luc reporter construct that amounted to 5.8-fold (Figure 6A). Likewise, the ERK1(K71R) and MKK4(K116R) vectors respectively produced 7.2- and 5.9-fold responses to PMA (Figure 6A), which were not significantly different from that seen with the control vector. The elevated response to PMA alone hence produced a high level of induction by PMA and 1,25D (139.8-fold) in pSG5-transfected cells, equivalent to a 9.3-fold level of synergy (Figure 6B). Interestingly, MKK4(K116R) greatly reduced CYP24 promoter induction to 37.6-fold in the presence of PMA and 1,25D, resulting in only a 3.7-fold synergy. The ERK1(K71R) mutant also strongly reduced the fold induction by PMA and 1,25D from 139.8- to 52.7-fold and hence synergy to 3.2-fold (Figure 6B). Co-transfecting a combination of both the ERK1(K71R) and MKK4(K116R) mutants did not produce a significant effect either on 1,25D induction or on synergy (results not shown). Therefore it can be concluded that JNK activity is important for 1,25D induction of the CYP24 promoter in HEK-293T cells and that it also plays a role in PMA and 1,25D synergy. Conversely, although ERK1/2 activation is not required for 1,25D induction, it is important for the synergistic activation of the promoter by PMA and 1,25D, consistent with the potentiation of PMA-stimulated activation of ERK1/2 by 1,25D.
Figure 6. Role of MAP kinase in PMA and 1,25D synergy on the CYP24 promoter.
pCYP24(WT-186)-Luc luciferase constructs were co-transfected into HEK-293T cells together with dominant-negative ERK1 or MKK4 expression vectors (200 ng) as indicated. (A) Cells were treated with 100 nM PMA or (B) with both 100 nM PMA and 10−8 M 1,25D as described in the Experimental section. Results are presented as fold induction and represent the means±S.D. for three experiments performed in triplicate. Significance of difference between pSG5 control vector and dominant-negative vector-transfected cells: ***P<0.001 (Tukey–Kramer multiple comparisons test). (C) HEK-293T cells were pretreated with DMSO, 200 nM calphostin C or 500 nM GF 109203X for 10 min followed by treatment with ethanol or 100 nM PMA for 10 min as indicated. Proteins from cell lysates were separated by SDS/PAGE and transferred on to nitrocellulose membrane. The membrane was probed with anti-ACTIVE ERK1/2 (Thr202/Tyr204) (upper panel), stripped and then reprobed with anti-ERK1/2 antibody (lower panel). Results are representative for two independent experiments. WB, Western blot.
An investigation was also conducted to determine whether the activation of ERK1/2 and JNK by PMA alone was dependent on PKC. Although PMA is a classical activator of PKC [44], it is now known that there are other intracellular targets for PMA. These include the RasGRPs (Ras guanine-nucleotide-releasing proteins), which activate Ras [47]. We therefore incubated the cells with 200 nM calphostin C or 500 nM GF 109203X for 10 min followed by the addition of 100 nM PMA for 10 min. The results demonstrated that neither calphostin C nor GF 109203X inhibited the ability of PMA to stimulate ERK1/2 activation (Figure 6C). The inhibitors also did not prevent PMA from activating JNK activity (results not shown). These results imply that PKC is not required for the activation of the MAP kinases by PMA in HEK-293T cells, making this one of the rare cell types in which PMA-mediated stimulation of the ERK1/2 module is through a PKC-independent route.
Identification of a sequence that mediates PMA and 1,25D synergy
An important question relates to the target sites within the −186 bp promoter that underlie synergy between PMA and 1,25D. The EBS located at −128/−119 (see Figure 1A) has been shown to contribute to hormone induction [17,18]. In order to evaluate any potential contribution of the EBS to synergy, the pCYP24(mEBS)-Luc construct containing a mutation within the EBS was utilized. As expected, mutagenesis of the EBS reduced 1,25D induction from 5.8- to 3.5-fold (Figure 7). The response to PMA and 1,25D treatment was reduced from 40-fold on pCYP24(WT-186) to 12.3-fold by mEBS (Figure 7). This equated to the loss of approximately half the fold synergy (from 5.8- to 2.7-fold), suggesting that EBS is partially responsible for mediating the synergy between PMA and 1,25D.
It is well known that PMA can function through AP-1 (activator protein-1) sites, but no consensus AP-1 site exists within the −186 promoter sequence. Interestingly, however, we have noted the sequence 5′-TGTCGGTCA-3′ at position –171/–163 (see Figure 1A) that strongly resembles the complement of the identified RE-3 sequence (5′-TGACCCACA-3′) present in the oFSHR (ovine follicle-stimulating hormone receptor) gene and which can respond to PMA [48]. When the 5′-TGTCGGTCA-3′ sequence was mutated to 5′-TGTAAGCTT-3′ within the CYP24–186 promoter construct designated pCYP24(M2-VSE)-Luc, basal expression was not altered but 1,25D induction was substantially reduced from 5.8- to 2.3-fold (Figure 7). This site is referred to as a 1,25D stimulatory element (VSE). Importantly, inactivation of the VSE also limited the response to treatment with both PMA and 1,25D to 4.8-fold induction (Figure 7). This corresponds to a complete loss of synergy. These results establish that the newly identified VSE site plays a significant role in the induction of the promoter by 1,25D and also in the synergistic response to PMA and 1,25D. When the VDRE-1 alone was mutated within the −186 WT construct, the intact VSE could not itself respond to 1,25D (results not shown).
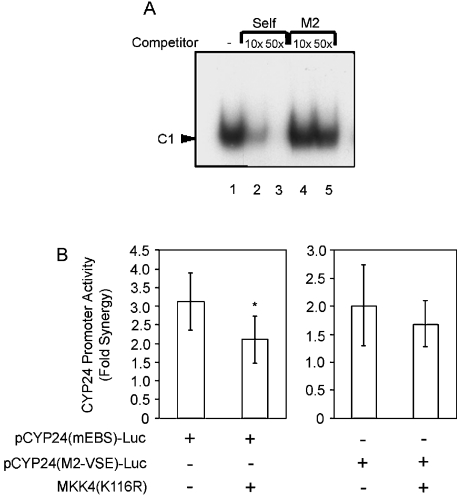
The VSE was shown to bind a single major protein complex as determined by EMSA using nuclear extracts derived from HEK-293T cells (Figure 8A). Self-competition with an unlabelled VSE oligonucleotide prevented the formation of C1 (lanes 2 and 3), whereas an unlabelled oligonucleotide containing a mutation in the VSE (M2) failed to compete for binding (lanes 4–5). This indicates specific binding of the protein complex C1 to the VSE site. Binding to the VSE was also examined using nuclear extracts prepared from cells that had been treated with PMA, 1,25D or both PMA and 1,25D. These treatments had no effect on either the pattern or intensity of binding to the VSE (results not shown). This suggests that the identity of the complex acting through the VSE to mediate promoter induction is not altered by these treatments. However, this does not exclude post-translational modification of the bound proteins in response to either PMA or 1,25D being necessary for the functionality of the VSE.
Figure 8. VSE binds a specific protein complex and mediates the action of JNK in synergy.
(A) A double-stranded oligonucleotide probe spanning the region of CYP24 promoter containing the VSE was labelled by end-filling with [α-32P]dCTP. The radiolabelled probe was incubated with nuclear extracts from HEK-293T cells. Self-competition was performed using a 10- and 50-fold molar excess of unlabelled VSE oligonucleotide (lanes 2 and 3). Cross-competition with 10- and 50-fold molar excess of the unlabelled oligonucleotide M2-VSE (lanes 4 and 5) was also performed. The retarded complex on the VSE probe is indicated by an arrowhead and labelled C1. The experiment was performed three times with similar results. (B) HEK-293T cells were co-transfected with the pCYP24(mEBS)-Luc or the pCYP24(M2-VSE)-Luc reporter construct along with either the pSG5 empty vector or the dominant-negative MKK4 mutant, MKK4(K116R). Fold synergy was calculated by dividing the PMA+1,25D fold induction by the sum of the fold inductions of PMA alone and 1,25D alone. Data are presented as the average level of fold synergy ±S.D. of six replicates for two independent experiments. Significance of difference between pSG5 control vector and MKK4(K116R) on pCYP24(mEBS)-Luc fold synergy: *P<0.05 (Student's unpaired two-tailed t test).
Given the requirement for the EBS, the VSE and the JNK pathway in synergy, it was of interest to investigate the potential site of JNK action on the CYP24 promoter. Cells were co-transfected with either pCYP24(mEBS)-Luc or pCYP24(M2-VSE)-Luc. The overall level of fold synergy on these constructs was determined in the presence of the dominant-negative MKK4(K116R) or an empty vector control (Figure 8B). The 3.1-fold synergy on the pCYP24(mEBS)-Luc construct was further reduced to 2.1-fold synergy by the dominant-negative MKK4, suggesting that JNK acts on a site other than the EBS. Interestingly, the 2-fold synergy observed on the pCYP24(M2-VSE)-Luc construct was not further decreased by dominant-negative MKK4. This result implicates the VSE as the potential site of JNK action in the mechanism of synergy.
DISCUSSION
We have demonstrated that, as observed previously in primary rat renal cells and rat intestinal epithelial cell lines [23–27], there is synergy between PMA and 1,25D in the activation of the CYP24 promoter in HEK-293T cells and determined that the phorbol ester sensitive site lies within the first −186 bp. This is consistent with previous reports that the target sequence for the phorbol ester lies within the first 250 bp of the promoter [28]. The previously characterized EBS [17,18], located at –128/–119, was found to play a role in synergy since inactivation of the EBS reduced PMA and 1,25D synergy by half. The 50% of synergy remaining on mutagenesis of the EBS suggested the involvement of other factors in the mechanism of synergy. This has led to the identification of a new transcription-factor-binding site termed VSE located at −171/−163 in the CYP24 promoter, which encompasses the sequence 5′-TGTCGGTCA-3′. A similar requirement for this site for maximal induction has been noted in COS-1 cells and the osteoblast cell line ROS17/2.8 (B. K. Nutchey, P. P. Dwivedi and B. K. May, unpublished work). Basal expression of the CYP24 promoter was not altered by mutagenesis of the VSE.
Sequence analysis of the VSE showed it to be similar to a site previously identified in the promoter of the oFSHR gene, termed RE-3 [48]. The RE-3 sequence (5′-TGACCCACA-3′) has only a single nucleotide difference from the VSE sequence on the non-coding strand (5′-TGACCGACA-3′). It was demonstrated that the RE-3 site was able to mediate the PMA response of the oFSHR gene and that an oligonucleotide containing the RE-3 site (5′-AATAGTGACCCACAGGGACAGTCTTA-3′) was able to bind the AP-1 proteins c-Jun/c-Fos contained in JC-410 cell nuclear extracts [48]. We found that overexpression of c-Jun in HEK-293T cells increases 1,25D induction of pCYP24(WT-186)-Luc by 2-fold (results not shown). This suggests that, like the RE-3 site, the VSE may be an AP-1 binding site. However, repeated EMSA experiments using supershifting antibodies to AP-1 family members (c-Jun, Jun B, Jun D, c-Fos, Fos B, Fra-1 and Fra-2) and to the cAMP-responsive-element-binding ATF2 and CREB-1 failed to supershift the single major complex (C1) shown in the present study to bind to the VSE (result not shown). A role for other targets of JNK such as Elk-1 and Sap1a [49] remains to be investigated.
There is now substantial evidence that 1,25D and other steroid hormones can activate MAP kinases and other signalling molecules, including PKC, through a non-genomic action directed by a receptor activity associated with the plasma membrane [22,50]. Unlike its effect in COS-1 cells, 1,25D surprisingly did not affect the activity of ERK1/2 in HEK-293T cells. Consistent with this, our results demonstrate that ERK1/2 plays no role in the activation of the CYP24 promoter by 1,25D alone in HEK-293T cells. However, we have demonstrated that JNK activity is important for the induction of the CYP24 promoter by 1,25D in HEK-293T cells. Thus 1,25D stimulated the activity of JNK and in the presence of the dominant-negative MKK4 mutant, promoter induction by 1,25D was inhibited by 54%. These results imply that JNK but not ERK1/2 is the predominant regulator of the −186 promoter in HEK-293T cells. This is in direct contrast with our previous observation in COS-1 cells that JNK played no role in the action of 1,25D [18].
It is well established that the activation and extent of activation of the different MAP kinase pathways occur in a cell type-specific fashion. For example, in contrast with our findings in COS-1 [18] and HEK-293T cells, it has been reported that 1,25D increased the activities of both JNK and ERK1/2 in Caco-2 cells and keratinocytes [40,45]. In both cases, JNK was then able to activate c-Jun to effect the up-regulation of AP-1-regulated promoters. It is likely that differences in the levels of signalling molecule expression between cell lines play a role in the variety of responses observed. In the case of 1,25D response, variation in the expression of the VDR involved in non-genomic signalling may also contribute to such differences. Indeed, it has been proposed that an increase in VDR expression following PMA treatment may be involved in the mechanism of synergy in LLCPK-1 cells [25]. However, we have been unable to show such an increase in VDR in HEK-293T cells (P. P. Dwivedi and B. K. May, unpublished work). Regardless, the different profiles of signalling molecule expression between cell lines highlight the importance of investigating intracellular signalling pathways in different cell types. It seems reasonable to assume that such differences between HEK-293T cells and COS-1 cells will underlie the fact that the synergy reported here between PMA and 1,25D in HEK-293T cells is not observed in COS-1 cells (P. P. Dwivedi and B. K. May, unpublished work).
Our results also demonstrate a role for PKC in the regulation of the CYP24 promoter. Thus we have not only found that PKCβ1 was necessary for the action of 1,25D alone but we also present several lines of evidence implicating the involvement of PKC in the synergistic induction of the −186 CYP24 promoter by PMA and 1,25D. Very importantly, 4α-PMA, which does not activate PKC [36,37], was not able to amplify the response to 1,25D. When the effects of a number of dominant-negative PKC isoenzyme mutants were examined, the result with dominant-negative PKCα is the most striking. While this mutant did not affect the response to PMA or 1,25D alone, it significantly reduced the level of synergy observed. This suggests that the activation of PKCα is critical for the synergistic up-regulation of the CYP24 promoter.
Significantly, we have also demonstrated that both ERK1/2 and JNK MAP kinases are critical for synergy. Although ERK1/2 activity was not required for the action of 1,25D or PMA alone, this kinase was required for synergy. Not only did PMA increase ERK1/2 phosphorylation and activation but in the presence of 1,25D, a greater degree of ERK1/2 activation was also observed. Consistent with ERK1/2 being involved in the synergistic activation of the −186 promoter, the dominant-negative ERK1 mutant inhibited the combined effect of PMA and 1,25D on promoter activity. Interestingly, PKC inhibitors did not prevent ERK1/2 or JNK activation by PMA alone. This suggests that PMA acts independent of PKC but perhaps through RasGRP to activate the Ras/Raf/MEK (MAP kinase/ERK kinase)/ERK signalling pathway [47]. An important issue concerns the target of the activated ERK1/2 within the −186 promoter. ERK1/2 could act on the unknown protein (C1) bound to the VSE. Ets-1, which binds to a site nearby at −128/−119 and is important for 1,25D induction [17], is also required for synergy and hence provides another possible site of ERK1/2 activity [51].
Our results demonstrate that JNK is also involved in the synergistic induction of the −186 promoter by PMA and 1,25D. Thus, although 1,25D and PMA could each stimulate the activity of JNK, this effect had disappeared by 24 h of treatment. In contrast, when the cells were co-incubated with PMA and 1,25D, enhanced JNK activity was still evident at 24 h. When the cells were transfected with MKK4(K116R), a reduction in 1,25D- and PMA-induced promoter activation was observed. The MKK4 dominant-negative mutant did not inhibit the effect of PMA alone. In the synergy state, the action of JNK is at least partially mediated by the VSE. Thus, whereas co-transfection of MKK4(K116R) resulted in a further decrease in the level of fold synergy seen on the CYP24 reporter construct containing a mutated EBS, MKK4(K116R) did not further decrease the fold synergy observed with the mutated VSE reporter construct.
An unexpected result was the increased response of the pCYP24(WT-186)-Luc construct to PMA when other vectors were co-transfected. A recent paper by Gardiner et al. [52] reported that vectors containing viral promoters affected the transactivating ability of the two VDR isoforms. Two reasons were proposed. First, the functional retinoic acid response elements in the cytomegalovirus promoter were proposed to compete for RXR or other factors [53], and secondly, VDR protein levels were markedly lowered by co-transfection of vectors such as pSG5 [52]. Similar mechanisms may operate in HEK-293T cells, resulting in suboptimal recruitment of the repressor complex to the CYP24 promoter in the absence of 1,25D [9,10] such that PMA-activated kinases are then able to promote CYP24 expression.
Overall, we have identified a new transcription-factor-binding site in the −186 promoter, which resembles an AP-1 site. This site contributes substantially to induction of the promoter by 1,25D and to the synergistic activity in the presence of both phorbol ester and 1,25D. This binding site may well be the target of activated JNK and ERK1/2. This work provides further evidence as to the importance of non-genomic actions of 1,25D in modulating CYP24 expression.
Acknowledgments
This work was supported by grants from the Australian Research Council, the National Health and Medical Research Council, and the Women's and Children's Hospital Research Foundation. We thank Professor I. B. Weinstein for the gift of dominant-negative PKC constructs.
References
- 1.Brown E. M., MacLeod R. J. Extracellular calcium sensing and extracellular calcium signaling. Physiol. Rev. 2001;81:239–297. doi: 10.1152/physrev.2001.81.1.239. [DOI] [PubMed] [Google Scholar]
- 2.Holick M. F. Vitamin D: a millennium perspective. J. Cell. Biochem. 2003;88:296–307. doi: 10.1002/jcb.10338. [DOI] [PubMed] [Google Scholar]
- 3.Jones G., Strugnell S. A., DeLuca H. F. Current understanding of the molecular actions of vitamin D. Physiol. Rev. 1998;78:1193–1231. doi: 10.1152/physrev.1998.78.4.1193. [DOI] [PubMed] [Google Scholar]
- 4.Sutton A. L. M., MacDonald P. N. Vitamin D: more than a ‘Bone-a fide’ hormone. Mol. Endocrinol. 2003;17:777–791. doi: 10.1210/me.2002-0363. [DOI] [PubMed] [Google Scholar]
- 5.Ylikomi T., Laaksi I., Lou Y. R., Martikainen P., Miettinen S., Pennanen P., Purmonen S., Syvala H., Vienonen A., Tuohimaa P. Antiproliferative action of vitamin D. Vitam. Horm. 2002;64:357–406. doi: 10.1016/s0083-6729(02)64010-5. [DOI] [PubMed] [Google Scholar]
- 6.Griffin M. D., Xing N., Kumar R. Vitamin D and its analogs as regulators of immune activation and antigen presentation. Annu. Rev. Nutr. 2003;23:117–145. doi: 10.1146/annurev.nutr.23.011702.073114. [DOI] [PubMed] [Google Scholar]
- 7.Barry J. B., Leong G. M., Church W. B., Issa L. L., Eisman J. A., Gardiner E. M. Interactions of SKIP/NCoA-62, TFIIB, and retinoid X receptor with vitamin D receptor helix H10 residues. J. Biol. Chem. 2003;278:8224–8228. doi: 10.1074/jbc.C200712200. [DOI] [PubMed] [Google Scholar]
- 8.Rachez C., Freedman L. P. Mechanisms of gene regulation by vitamin D3 receptor: a network of coactivator interactions. Gene. 2000;246:9–21. doi: 10.1016/s0378-1119(00)00052-4. [DOI] [PubMed] [Google Scholar]
- 9.Dwivedi P. P., Muscat G. E. O., Bailey P. J., Omdahl J. L., May B. K. Repression of basal transcription by vitamin D receptor: evidence for interaction of unliganded vitamin D receptor with two receptor interaction domains in RIP13delta1. J. Mol. Endocrinol. 1998;20:327–335. doi: 10.1677/jme.0.0200327. [DOI] [PubMed] [Google Scholar]
- 10.Polly P., Herdick M., Moehren U., Baniahmad A., Heinzel T., Carlberg C. VDR-Alien: a novel, DNA-selective vitamin D(3) receptor-corepressor partnership. FASEB J. 2000;14:1455–1463. doi: 10.1096/fj.14.10.1455. [DOI] [PubMed] [Google Scholar]
- 11.Hewison M., Zehnder D., Bland R., Stewart P. M. 1α-Hydroxylase and the action of vitamin D. J. Mol. Endocrinol. 2000;25:141–148. doi: 10.1677/jme.0.0250141. [DOI] [PubMed] [Google Scholar]
- 12.Omdahl J. L., Bobrovnikova E. A., Choe S., Dwivedi P. P., May B. K. Overview of regulatory cytochrome P450 enzymes of the vitamin D pathway. Steroids. 2001;66:381–389. doi: 10.1016/s0039-128x(00)00157-4. [DOI] [PubMed] [Google Scholar]
- 13.Omdahl J. L., Morris H. A., May B. K. Hydroxylase enzymes of the vitamin D pathway: expression, function, and regulation. Annu. Rev. Nutr. 2002;22:139–166. doi: 10.1146/annurev.nutr.22.120501.150216. [DOI] [PubMed] [Google Scholar]
- 14.Omdahl J. L., May B. K. The 25-hydroxyvitamin D 24-hydroxylase. In: Feldman D., Glorieux F. H., Pike J. W., editors. Vitamin D. London: Academic Press; 1997. pp. 69–85. [Google Scholar]
- 15.St-Arnaud R., Arabian A., Travers R., Barletta F., Raval-Pandya M., Chapin K., Depovere J., Mathieu C., Christakos S., Demay M. B., et al. Deficient mineralization of intramembranous bone in vitamin D-24-hydroxylase-ablated mice is due to elevated 1,25-dihydroxyvitamin D and not to the absence of 24,25-dihydroxyvitamin D. Endocrinology. 2000;141:2658–2666. doi: 10.1210/endo.141.7.7579. [DOI] [PubMed] [Google Scholar]
- 16.St-Arnaud R. Targeted inactivation of vitamin D hydroxylases in mice. Bone. 1999;25:127–129. doi: 10.1016/s8756-3282(99)00118-0. [DOI] [PubMed] [Google Scholar]
- 17.Dwivedi P. P., Omdahl J. L., Kola I., Hume D. A., May B. K. Regulation of rat cytochrome P450C24 (CYP24) gene expression: evidence for functional cooperation of Ras-activated Ets transcription factors with the vitamin D receptor in 1,25-dihydroxyvitamin D3-mediated induction. J. Biol. Chem. 2000;275:47–55. doi: 10.1074/jbc.275.1.47. [DOI] [PubMed] [Google Scholar]
- 18.Dwivedi P. P., Hii C. S. T., Ferrante A., Tan J., Der C. J., Omdahl J. L., Morris H. A., May B. K. Role of MAP kinases in the 1,25-dihydroxyvitamin D3-induced transactivation of the rat cytochrome P450C24 (CYP24) promoter: specific functions for ERK1/ERK2 and ERK5. J. Biol. Chem. 2002;277:29643–29653. doi: 10.1074/jbc.M204561200. [DOI] [PubMed] [Google Scholar]
- 19.Kerry D. M., Dwivedi P. P., Hahn C. N., Morris H. A., Omdahl J. L., May B. K. Transcriptional synergism between vitamin D-responsive elements in the rat 25-hydroxyvitamin D3 24-hydroxylase (CYP24) promoter. J. Biol. Chem. 1996;271:29715–29721. doi: 10.1074/jbc.271.47.29715. [DOI] [PubMed] [Google Scholar]
- 20.Ohyama Y., Ozono K., Uchida M., Yoshimura M., Shinki T., Suda T., Yamamoto O. Functional assessment of two vitamin D-responsive elements in the rat 25-hydroxyvitamin D3 24-hydroxylase gene. J. Biol. Chem. 1996;271:30381–30385. doi: 10.1074/jbc.271.48.30381. [DOI] [PubMed] [Google Scholar]
- 21.Zierold C., Darwish H. M., DeLuca H. F. Two vitamin D response elements function in the rat 1,25-dihydroxyvitamin D 24-hydroxylase promoter. J. Biol. Chem. 1995;270:1675–1678. doi: 10.1074/jbc.270.4.1675. [DOI] [PubMed] [Google Scholar]
- 22.Falkenstein E., Tillmann H.-C., Christ M., Feuring M., Wehling M. Multiple actions of steroid hormones – a focus on rapid, nongenomic effects. Pharmacol. Rev. 2000;52:513–555. [PubMed] [Google Scholar]
- 23.Armbrecht H. J., Hodam T. L., Boltz M. A., Chen M. L. Phorbol ester markedly increases the sensitivity of intestinal epithelial cells to 1,25-dihydroxyvitamin D3. FEBS Lett. 1993;327:13–16. doi: 10.1016/0014-5793(93)81028-x. [DOI] [PubMed] [Google Scholar]
- 24.Armbrecht H. J., Boltz M. A., Hodam T. L., Kumar V. B. Differential responsiveness of intestinal epithelial cells to 1,25-dihydroxyvitamin D3 – role of protein kinase C. J. Endocrinol. 2001;169:145–151. doi: 10.1677/joe.0.1690145. [DOI] [PubMed] [Google Scholar]
- 25.Barletta F., Dhawan P., Christakos S. Integration of hormone signaling in the regulation of human 25(OH)D3 24-hydroxylase transcription. Am. J. Physiol. Endocrinol. Metab. 2004;286:E598–E608. doi: 10.1152/ajpendo.00214.2003. [DOI] [PubMed] [Google Scholar]
- 26.Chen M. L., Boltz M. A., Armbrecht H. J. Effects of 1,25-dihydroxyvitamin D3 and phorbol ester on 25-hydroxyvitamin D3 24-hydroxylase cytochrome P450 messenger ribonucleic acid levels in primary cultures of rat renal cells. Endocrinology. 1993;132:1782–1788. doi: 10.1210/endo.132.4.7681765. [DOI] [PubMed] [Google Scholar]
- 27.Koyama H., Inaba M., Nishizawa Y., Ohno S., Morii H. Protein kinase C is involved in 24-hydroxylase gene expression induced by 1,25(OH)2D3 in rat intestinal epithelial cells. J. Cell. Biochem. 1994;55:230–240. doi: 10.1002/jcb.240550210. [DOI] [PubMed] [Google Scholar]
- 28.Pike J. W., Kerner S. A., Jin C. H., Allegretto E. A., Elgort M. Direct activation of the human 25-hydroxyvitamin D3-24-hydroxylase promoter by 1,25(OH)2D3 and PMA: identification of cis elements and transactivators. J. Bone Miner. Res. 1994;9(suppl. 1) Abstract S144. [Google Scholar]
- 29.Westwick J. K., Cox A. D., Der C. J., Cobb M. H., Hibi M., Karin M., Brenner D. A. Oncogenic Ras activates c-Jun via a separate pathway from the activation of extracellular signal-regulated kinases. Proc. Natl. Acad. Sci. U.S.A. 1994;91:6030–6034. doi: 10.1073/pnas.91.13.6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan E. D., Winston B. W., Uh S.-T., Wynes M. W., Rose D. M., Riches D. W. H. Evaluation of the role of mitogen-activated protein kinases in the expression of inducible nitric oxide synthase by IFN-γ and TNF-α in mouse macrophages. J. Immunol. 1999;162:415–422. [PubMed] [Google Scholar]
- 31.Soh J.-W., Weinstein I. B. Roles of specific isoforms of protein kinase C in the transcriptional control of cyclin D1 and related genes. J. Biol. Chem. 2003;278:34709–34716. doi: 10.1074/jbc.M302016200. [DOI] [PubMed] [Google Scholar]
- 32.Andrews N. C., Faller D. V. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hii C. S. T., Moghadammi N., Dunbar A., Ferrante A. Activation of the phosphatidylinositol 3-kinase-Akt/protein kinase B signaling pathway in arachidonic acid-stimulated human myeloid and endothelial cells: involvement of the ErbB receptor family. J. Biol. Chem. 2001;276:27246–27255. doi: 10.1074/jbc.M103250200. [DOI] [PubMed] [Google Scholar]
- 34.Hii C. S. T., Huang Z. H., Bilney A., Costabile M., Murray A. W., Rathjen D. A., Der C. J., Ferrante A. Stimulation of p38 phosphorylation and activity by arachidonic acid in HeLa cells, HL60 promyelocytic leukemic cells, and human neutrophils: evidence for cell type-specific activation of mitogen-activated protein kinases. J. Biol. Chem. 1998;273:19277–19282. doi: 10.1074/jbc.273.30.19277. [DOI] [PubMed] [Google Scholar]
- 35.Hii C. S., Ferrante A., Edwards Y. S., Huang Z. H., Hartfield P. J., Rathjen D. A., Poulos A., Murray A. W. Activation of mitogen-activated protein kinase by arachidonic acid in rat liver epithelial WB cells by a protein kinase C-dependent mechanism. J. Biol. Chem. 1995;270:4201–4204. doi: 10.1074/jbc.270.9.4201. [DOI] [PubMed] [Google Scholar]
- 36.Fischer S. M., Patrick K. E., Lee M. L., Cameron G. S. 4β- and 4α-12-O-tetradecanoylphorbol-13-acetate elicit arachidonate release from epidermal cells through different mechanisms. Cancer Res. 1991;51:850–856. [PubMed] [Google Scholar]
- 37.Jeffrey A. M., Liskamp R. M. J. Computer-assisted molecular modeling of tumor promoters: rationale for the activity of phorbol esters, teleocidin B, and aplysiatoxin. Proc. Natl. Acad. Sci. U.S.A. 1986;83:241–245. doi: 10.1073/pnas.83.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kobayashi E., Nakano H., Morimoto M., Tamaoki T. Calphostin C (UCN-1028C), a novel microbial compound, is a highly potent and specific inhibitor of protein kinase C. Biochem. Biophys. Res. Commun. 1989;159:548–553. doi: 10.1016/0006-291x(89)90028-4. [DOI] [PubMed] [Google Scholar]
- 39.Toullec D., Pianetti P., Coste H., Bellevergue P., Grand-Perret T., Ajakane M., Baudet V., Boissin P., Boursier E., Loriolle F., et al. The bisindolylmaleimide GF 109203X is a potent and selective inhibitor of protein kinase C. J. Biol. Chem. 1991;266:15771–15781. [PubMed] [Google Scholar]
- 40.Chen A., Davis B. H., Bissonnette M., Scaglione-Sewell B., Brasitus T. A. 1,25-dihydroxyvitamin D3 stimulates activator protein-1-dependent Caco-2 cell differentiation. J. Biol. Chem. 1999;274:35505–35513. doi: 10.1074/jbc.274.50.35505. [DOI] [PubMed] [Google Scholar]
- 41.Buitrago C. G., Pardo V. G., de Boland A. R., Boland R. Activation of RAF-1 through Ras and protein kinase Cα mediates 1α,25(OH)2-vitamin D3 regulation of the mitogen-activated protein kinase pathway in muscle cells. J. Biol. Chem. 2003;278:2199–2205. doi: 10.1074/jbc.M205732200. [DOI] [PubMed] [Google Scholar]
- 42.Rivera-Bermúdez M. A., Bertics P. J., Albrecht R. M., Mosavin R., Mellon W. S. 1,25-Dihydroxyvitamin D3 selectively translocates PKCα to nuclei in ROS 17/2.8 cells. Mol. Cell. Endocrinol. 2002;188:227–239. doi: 10.1016/s0303-7207(01)00670-0. [DOI] [PubMed] [Google Scholar]
- 43.Natsume Y., Ishizuka T., Yamamoto Y., Miura A., Kajita K., Ishizawa M., Kawai Y., Huang Y., Morita H., Uno Y., et al. Dominant negative protein kinase Cβ improves 1α, 25-dihydroxy vitamin D3-induced insulin resistance. Endocr. Res. 2003;29:457–464. doi: 10.1081/erc-120026951. [DOI] [PubMed] [Google Scholar]
- 44.Tanaka C., Nishizuka Y. The protein kinase C family for neuronal signaling. Annu. Rev. Neurosci. 1994;17:551–567. doi: 10.1146/annurev.ne.17.030194.003003. [DOI] [PubMed] [Google Scholar]
- 45.Johansen C., Kragballe K., Henningsen J., Westergaard M., Kristiansen K., Iversen L. 1α,25-dihydroxyvitamin D3 stimulates activator protein 1 DNA-binding activity by a phosphatidylinositol 3-kinase/Ras/MEK/extracellular signal regulated kinase 1/2 and c-Jun N-terminal kinase 1-dependent increase in c-Fos, Fra1, and c-Jun expression in human keratinocytes. J. Invest. Dermatol. 2003;120:561–570. doi: 10.1046/j.1523-1747.2003.12095.x. [DOI] [PubMed] [Google Scholar]
- 46.Cuenda A. Mitogen-activated protein kinase kinase 4 (MKK4) Int. J. Biochem. Cell Biol. 2000;32:581–587. doi: 10.1016/s1357-2725(00)00003-0. [DOI] [PubMed] [Google Scholar]
- 47.Brose N., Betz A., Wegmeyer H. Divergent and convergent signaling by the diacylglycerol second messenger pathway in mammals. Curr. Opin. Neurobiol. 2004;14:328–340. doi: 10.1016/j.conb.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 48.Xing W., Danilovich N., Sairam M. R. Orphan receptor chicken ovalbumin upstream promoter transcription factors inhibit steroid factor-1, upstream stimulatory factor, and activator protein-1 activation of ovine follicle-stimulating hormone receptor expression via composite cis-elements. Biol. Reprod. 2002;66:1656–1666. doi: 10.1095/biolreprod66.6.1656. [DOI] [PubMed] [Google Scholar]
- 49.Janknecht R., Hunter T. Activation of the Sap-1a transcription factor by the c-Jun N-terminal kinase (JNK) mitogen-activated protein kinase. J. Biol. Chem. 1997;272:4219–4224. doi: 10.1074/jbc.272.7.4219. [DOI] [PubMed] [Google Scholar]
- 50.Simoncini T., Genazzani A. R. Non-genomic actions of sex steroid hormones. Eur. J. Endocrinol. 2003;148:281–292. doi: 10.1530/eje.0.1480281. [DOI] [PubMed] [Google Scholar]
- 51.Waas W. F., Dalby K. N. Transient protein-protein interactions and a random-ordered kinetic mechanism for the phosphorylation of a transcription factor by extracellular-regulated protein kinase 2. J. Biol. Chem. 2002;277:12532–12540. doi: 10.1074/jbc.M110523200. [DOI] [PubMed] [Google Scholar]
- 52.Gardiner E. M., Esteban L. M., Fong C., Allison S. J., Flanagan J. L., Kouzmenko A. P., Eisman J. A. Vitamin D receptor B1 and exon 1d: functional and evolutionary analysis. J. Steroid Biochem. Mol. Biol. 2004;89–90:233–238. doi: 10.1016/j.jsbmb.2004.03.078. [DOI] [PubMed] [Google Scholar]
- 53.Angulo A., Suto C., Heyman R. A., Ghazal P. Characterization of the sequences of the human cytomegalovirus enhancer that mediate differential regulation by natural and synthetic retinoids. Mol. Endocrinol. 1996;10:781–793. doi: 10.1210/mend.10.7.8813719. [DOI] [PubMed] [Google Scholar]
- 54.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]