Abstract
The secreted, multifunctional enzyme PLB1 (phospholipase B1 protein encoded by the PLB1 gene) is a virulence determinant of the pathogenic fungus Cryptococcus neoformans, but the mechanism of its secretion is unknown. The cryptococcal PLB1 gene encodes putative, N-terminal LP (leader peptide) and C-terminal GPI (glycosylphosphatidylinositol) anchor attachment motifs, suggesting that PLB1 is GPI-anchored before secretion. To investigate the role of these motifs in PLB1 secretion, four cDNA constructs were created encoding the full-length construct (PLB1) and three truncated versions without the LP and/or the GPI anchor attachment motifs [LP−PLB1 (PLB1 expressed without the LP consensus motif), LP−PLB1GPI− (PLB1 expressed without the LP and GPI consensus motifs) and PLB1GPI− (PLB1 expressed without the GPI anchor attachment motif) respectively]. The constructs were ligated into pYES2, and galactose-induced expression was achieved in Saccharomyces cerevisiae. The LP was essential for secretion of the PLB1 protein and its three activities (PLB, lysophospholipase and lysophospholipase transacylase). Deletion of the GPI motif to create PLB1GPI− resulted in a redistribution of activity from the cell wall and membranes to the secreted and cytosolic fractions, with 36–54% of the total activity being secreted as compared with <5% for PLB1. PLB1 produced the maximum cell-associated activity (>2-fold more than that for PLB1GPI−), with 75–86% of this in the cell-wall fraction, 6–19% in the membrane fraction and 3–7% in the cytosolic fraction. Cell-wall localization was confirmed by release of activity with β-glucanase in both S. cerevisiae recombinants and wild-type C. neoformans. The dominant location of PLB1 in the cell wall via GPI anchoring may permit immediate release of the enzyme in response to changing environmental conditions and may represent part of a novel mechanism for regulating the secretion of a fungal virulence determinant.
Keywords: β-glucanase, cell wall, Cryptococcus neoformans, glycosylphosphatidylinositol (GPI) anchor, phospholipase B1, secretion
Abbreviations: App1, antiphagocytic protein 1; DPPC, dipalmitoyl phosphatidylcholine; ECL, enhanced chemiluminescence; ER, endoplasmic reticulum; GPI, glycosylphosphatidylinositol; HRP, horseradish peroxidase; IAB, imidazole assay buffer; LP, leader peptide; LPL, lysophospholipase; LPTA, lysophospholipase transacylase; lysoPC, lysophosphatidylcholine; PLB, phospholipase B; PLB1, phospholipase B1 protein encoded by the PLB1 gene; LP−PLB1, PLB1 expressed without the LP consensus motif; LP−PLB1GPI−, PLB1 expressed without the LP and GPI consensus motifs; PLB1GPI−, PLB1 expressed without the GPI anchor attachment motif
INTRODUCTION
The pathogenic basidiomycetous fungus Cryptococcus neoformans causes life-threatening neurological infection, especially in immunocompromised hosts. Dissemination of primary infection from the lungs to brain requires a functional phospholipase gene (CnPLB1) that encodes a secreted enzyme protein (CnPLB1) with PLB (phospholipase B), lysophospholipase and transacylase activities [1–3]. Secreted phospholipases with similar activities facilitate tissue invasion by the pathogenic yeast, Candida albicans [4], and are produced by other fungi of medical importance such as Aspergillus species [5]. To date, only three PLB genes have been cloned from pathogenic fungi: CnPLB1 from C. neoformans [2], and CaPLB1 [4] and CaPLB2 from C. albicans [6]. By performing gene disruption studies, both candidal and cryptococcal PLB1 have been proven to be virulence determinants. Although CnPLB1 shares 36% amino acid sequence homology with CaPLB1 and CaPLB2, only CnPLB1 has a C-terminal consensus motif specifying attachment to a GPI (glycosylphosphatidylinositol) membrane anchor.
GPI anchors are commonly used to attach proteins to cell membranes and/or cell walls. The GPI anchor is also a signal for directing the transport of GPI-anchored proteins to the cell surface. A reduced ER (endoplasmic reticulum) to Golgi transport of GPI-anchored proteins has been demonstrated by blocking GPI anchoring with specific inhibitors [7] or by employing Saccharomyces cerevisiae cell lines defective in GPI anchor biosynthesis [8]. Secretion of GPI-anchored proteins is dependent on cleavage of the functional protein from the anchor by proteases, (G)PI-specific PLC/PLDs or glucanases if the protein is localized in the cell wall [9–12].
Two features of CnPLB1 suggest that it is secreted. First, it contains an N-terminal stretch of amino acids with high hydrophobicity, indicative of a secretory LP (leader peptide). Secondly, it acquires post-translational N-linked glycosylation, which accounts for 30% of the PLB1 (PLB1 protein encoded by PLB1 gene) molecular mass as observed by SDS/PAGE and which is essential for activity of the secreted enzyme [1]. In addition, there is a stretch of 22 amino acids with high hydrophobicity at the C-terminus. This motif is a signal for the attachment of a GPI membrane anchor in all eukaryotes. It serves as a temporary ER membrane anchor that is cleaved off, allowing the protein to re-attach covalently, at the omega sequence, to a preformed GPI anchor (for a review, see [13]). The modified protein is then transported by secretory vesicles to the cell membrane and/or cell wall via the Golgi apparatus [7,8,14,15].
Little is known about the regulation of PLB1 production and secretion from pathogenic fungi, despite its proven role in virulence. In C. albicans, PLB1 mRNA expression was modulated by changes in temperature and other environmental conditions [16]. In C. neoformans, secreted PLB1 activity varied with temperature, growth medium and pH [17]. However, the temperature-dependent increase in secreted CnPLB1 activity (more at 37 °C than at 30 °C) was not accompanied by an increase in CnPLB1 mRNA expression [2,18], indicating that CnPLB1 expression is not regulated at the mRNA level. This observation, coupled with the presence of a GPI anchor consensus motif in CnPLB1, led us to postulate that CnPLB1 secretion is regulated by GPI anchoring to the cryptococcal cell membrane or cell wall. In the present study, we constructed four deletion mutants to determine the role of the N- and C-terminal hydrophobic motifs in the secretion of active CnPLB1. We conclude that GPI anchoring prevents PLB1 secretion by targeting the protein to the fungal cell membrane/cell wall in the heterologous expression system, S. cerevisiae. We also confirm that native CnPLB1 associates with the cell wall and is released by β-glucanases in a virulent, clinical strain of C. neoformans.
EXPERIMENTAL
Reagents and antibodies
Anti-Express-tagged antibody was obtained from Invitrogen (Mulgrave, Vic., Australia), anti-mouse antibody conjugated with HRP (horseradish peroxidase) from Amersham Biosciences (Castle Hill, NSW, Australia) and donkey anti-goat HRP from Santa Cruz Biotechnology (Santa Cruz, CA, U.S.A.). PLB1 antibodies were prepared by immunizing goats (Institute of Medical and Veterinary Science, Gillies Plains, South Australia, Australia) with a PLB1 peptide (synthesized by Mimotopes, Clayton, Vic., Australia). Rabbit anti-α-agglutinin polyclonal antibodies were gifts from Professor P. Lipke (Hunter College of the City University of New York, U.S.A.). Protein G–Sepharose, used to affinity-purify IgG from goat serum, was obtained from Amersham Biosciences. PLB1 peptide-affinity columns (prepared by Mimotopes) were used to purify PLB1 peptide-specific IgG. BCA (bicinchoninic acid) protein assay reagents and SuperSignal [ECL (enhanced chemiluminescence) reagent] were obtained from Pierce (Rockford, IL, U.S.A.). Standard ECL reagents were obtained from Amersham Biosciences. DPPC (dipalmitoyl phosphatidylcholine; C16:0) and lysoPC (lyso-phosphatidylcholine; C16:0) were obtained from Sigma (Castle Hill, NSW, Australia) and 1,2-di[1-14C]palmitoyl phosphatidylcholine and 1-[14C]palmitoyl lysoPC from Amersham Biosciences. β-Glucanase preparation from Trichoderma harzianum (L1412) was obtained from Sigma. The plasmid pYES2 was obtained from Invitrogen, DNA purification kits were from Qiagen (Clifton Hill, Vic., Australia) and restriction endonucleases from Promega (Madison, WI, U.S.A.).
PLB1 constructs
PLB1 cDNA cloned into PCR2.1 TOP0 vector (obtained from Dr G. Cox, Duke University, Durham, NC, U.S.A.) was used as a template to amplify PLB1 cDNA by PCR using the following pair of forward and reverse primers: 5′-CATAGTCGACGTCAATCGCCACGGGTACTTTTGC-3′ and 5′-CATATCTAGATTAAAGCATCAAGCCCAAGCCAGC-3′ respectively. SalI and XbaI restriction sites (in boldface) were introduced at the 5′- and 3′-ends respectively and the position of the stop codon in the reverse primer is underlined. In addition to the parent construct, three truncated versions of PLB1 were amplified, without the putative C-terminal, 22-amino-acid GPI consensus sequence (GPI) (GAANADVSMGMVALAAGLGLML), without the 19-aminoacid, N-terminal LP (MSIATGTFAFSLFATIAFA) or without both. The pairs of forward and reverse PCR primers used to construct each truncated PLB1 were 5′-CATAGTCGACGTCAATCGCCACGGGTACTTTTGC-3′ and 5′-CATATCTAGATTAACTGGACGCTGTACCAGCAG-3′, and 5′-CATGTCGACTGTTCCTCCCGAGACTCCGCGGATTG-3′ and 5′-CATATCTAGATTAAAGCATCAAGCCCAAGCCAGC-3′, and 5′-CATGTCGACTGTTCCTCCCGAGACTCCGCGGATTG-3′ and 5′-CATATCTAGATTAACTGGACGCTGTACCAGCAG-3′ respectively. In each primer pair, the restriction sites are in boldface and the stop codon is underlined. PLB1 PCR products (1.9 kb) were digested with SalI and XbaI and gel-purified using a Qiagen gel extraction kit. The expression plasmid pYES2-App1 (where App1 is antiphagocytic protein 1), which is pYES2 (Invitrogen) containing an in-frame, His6/Express-cryptococcal App1 cDNA fusion (prepared as described in [19]), was digested with XhoI and XbaI to release the App1 cDNA. The linearized vector was gel-purified using a Qiagen gel extraction kit and ligated with each of the SalI/XbaI-digested PLB1 PCR products to create in-frame, His6/Express-PLB1 cDNA fusions (pYES-PLB1). The ligations were used to transform chemically competent Escherichia coli DH5α-T1 (Invitrogen). Qiagen miniprep spin columns were used to purify plasmids from colonies growing on Luria–Bertani plates containing ampicillin (100 μg/ml). Plasmids were digested with XbaI and XhoI and screened for the release of a 930 bp insert (made shorter due to the presence of an XhoI site at nucleotide position 1018 within the PLB1 cDNA) by 0.8% agarose gel electrophoresis. The cloned PLB1 sequences were verified by DNA sequence analysis (courtesy of the DNA Sequencing Laboratory, Department of Biochemistry and Molecular Biology, Medical University of South Carolina). pYES-PLB1 recombinants were then used to transform S. cerevisiae strains JK93Dα and MF17 (Δplb1,2,3) [20] using the lithium acetate/single-stranded carrier DNA/PEG method [where PEG stands for poly(ethylene glycol) 4000]. Recombinants were selected by growth on agar medium containing 7.4 g/l yeast nitrogen base without amino acids (YNB; Difco, Detroit, MI, U.S.A.), 0.77 g/l uracil drop-out supplement (BD Biosciences, North Ryde, NSW, Australia), 100 μg/ml threonine and 40 μg/ml tryptophan containing either 2% (w/v) glucose (URA−glu) or 2% (w/v) galactose (URA−gal).
Expression of cryptococcal PLB1 in S. cerevisiae
PLB1 expression was driven by the galactose-inducible promoter (Gal 1) present in pYES2. Starter cultures were prepared by inoculating recombinants into 5 or 20 ml of URA−glu medium followed by incubation of the cultures for 16–22 h at 30 °C (250 rev./min). Cells were harvested, washed with 20 ml of 0.9% (w/v) saline and resuspended in 10 or 60 ml of URA−gal medium, to induce PLB1 expression. When testing for intracellular PLB1 expression by Western blotting, 5 ml starter cultures were prepared and expression was performed in 10 ml of URA−gal for 16 h. When testing for PLB1 secretion, 20 ml starter cultures were prepared in URA−glu medium and induction of PLB1 expression was performed in 60 ml of URA−gal. Cells were then harvested by centrifugation, resuspended in 450 μl of ISB (imidazole secretion buffer; 10 mM imidazole, 2 mM CaCl2 and 2 mM MgCl2, pH 5.5) containing 2% galactose (ISBgal) and incubated for 22 h at 30 °C (250 rev./min). Cells were pelleted by centrifugation and supernatants (secretions) were assayed for protein. PLB1 activity assays were performed, as described below, or protein was precipitated with 10% (w/v) trichloroacetic acid and subjected to SDS/PAGE and Western blotting, also as described below.
Preparing cell lysates for detection of intracellular PLB1 expression
Cells, grown and induced as described above, were harvested by centrifugation and the pellet was resuspended in 300 μl of lysis buffer (50 mM Tris/HCl, pH 6.8, 1 mM EDTA, 300 mM sucrose and 0.1% Triton X-100), containing 2-mercaptoethanol (1 μl), protease inhibitor cocktail {P 8215 for fungal and yeast cells; 4-(2-aminoethyl)benzenesulphonyl fluoride hydrochloride (100 mM), 1,10-phenanthroline (500 mM), pepstatin A (2.2 mM) and E-64 [trans-epoxysuccinyl-L-leucylamido-(4-guanidino)butane] (1.4 mM); Sigma}, PMSF (2 μl of a 1 mM solution) and antipain (0.15 mg/ml). To achieve cell lysis, three-quarters of a volume of acid-washed glass beads (425–600 μm) were added and homogenization was performed in a MiniBeadbeater-8 cell disrupter (MBB-8; Daintree Scientific, Tasmania, Australia) for 3 cycles of 1 min, alternating with a 1 min cooling period on ice. Particulates were removed by two rounds of centrifugation at 2500 g for 10 min. Supernatants containing 30 μg of protein were subjected to SDS/PAGE and Western blotting as described below.
Preparing subcellular fractions for activity assays
The cell pellet from the preparation of the supernatant mentioned above was washed twice with 0.9% saline and frozen at −70 °C. The cell pellets were thawed and then resuspended in 700 μl of IAB (imidazole assay buffer; 125 mM imidazole, pH 4) containing protease inhibitors (see above), and disrupted with glass beads using a MiniBeadbeater as described above. The homogenate was centrifuged at 14000 g for 10 min at 4 °C to obtain a pellet (mainly cell wall) and a supernatant (membranes and cytosol). The supernatant was subjected to further centrifugation at 100000 g for 1 h at 4 °C to obtain membrane (pellet) and cytosolic (supernatant) fractions. Fractions were assayed for protein and then for enzyme activity as described below.
β-Glucanase digestion of cell walls
Cell walls were pelleted by centrifugation as described above. Pellets were washed by resuspension in 1.5 ml of IAB containing protease inhibitors (see above), followed by centrifugation at 14000 g for 10 min at 4 °C. The cell pellets were incubated with 150 μl of a 12 mg/ml solution of β-glucanase (lysing enzymes from Trichoderma harzianum; Sigma L1412) made up in IAB (pH adjusted to 5.8), with or without protease inhibitors. Incubation was performed for 2 h at 37 °C and samples for assay were collected at 0, 1 and 2 h. To collect these samples, cell walls were pelleted by centrifugation as described above and aliquots of the supernatant were removed for enzyme assay as described below.
Radiometric phospholipase assays
Enzyme activities were measured as described previously [1] in a final volume of 125 μl at 37 °C. For the determination of secreted, insoluble, membrane and cytosolic PLB activities, carrier DPPC and 1,2-di[1-14C]palmitoyl phosphatidylcholine (20000 d.p.m.) were dried under nitrogen and suspended in IAB by sonication using a Branson 450 sonifier. Linearity of activity with protein concentration, assay time and substrate concentration was determined. The optimal assay conditions for secreted PLB activity were determined to be 22 min assay time, 13 μg of protein and a substrate (DPPC) concentration of 800 μM. PLB activity was determined by the rate of decrease in the radiolabelled phosphatidylcholine substrate with the appearance of the label in non-esterified fatty acid. Variations in the assay conditions, established during the optimization procedure, for the cell-wall, membrane and cytosolic fractions are indicated in the Figure legends. Secreted LPL (lysophospholipase) and LPTA (lysophospholipase transacylase) activities were measured simultaneously in a reaction mixture containing 1-[14C]palmitoyl lysoPC (25000 d.p.m.) and carrier lysoPC in IAB. The optimal reaction time chosen was 15 s with 2.5 μg of total protein and a substrate (lysoPC) concentration of 200 μM. LPL activity was measured by the rate of loss of 1-[14C]palmitoyl lysoPC with the release of radiolabelled fatty acids. LPTA was estimated from the rate of formation of radiolabelled phosphatidylcholine. Variations in these conditions, established during the optimization procedure, for cell-wall, membrane and cytosolic fractions are indicated in the Figure legends. All reactions were terminated by adding 0.5 ml of chloroform/methanol (2:1, v/v). The reaction products were extracted by the method of Bligh and Dyer [21], separated by TLC using a chloroform/methanol/water (65:25:4, by vol.) solvent mixture and then quantified as described previously [22].
Western blotting
Protein samples were dissolved in SDS/PAGE sample buffer [100 mM Tris, pH 7.0, 0.2 mM dithiothreitol, 4% (w/v) SDS, 20% (v/v) glycerol and 0.05% (w/v) Bromophenol Blue], boiled for 7 min and subjected to SDS/PAGE on a precast 4–20% Novex gel (Invitrogen, Mulgrave, Vic., Australia). Proteins were then electroblotted on to nitrocellulose. For Western blotting with anti-Express-tagged antibody, nitrocellulose blots were blocked in PBST [PBS/0.1% (v/v) Tween 20] containing 5% (w/v) skimmed milk and incubated with anti-Express-tagged antibody (1:2500 dilution) in PBST containing 3% (w/v) BSA, overnight at 4 °C. After 3×10 min washes with PBST, blots were incubated with anti-mouse HRP (1:4000 dilution) in PBST containing 5% skimmed milk. For Western blotting with anti-α-agglutinin, the procedure was the same as that for anti-Express except for the following modifications: 3% BSA was used instead of skimmed milk as the blocking reagent, 0.25% Tween 20 was used instead of 0.1% and 0.2 M α-methyl D-mannoside (Sigma M-6882) was present in the antibody incubation buffer. For Western blotting with anti-PLB1 peptide antibodies, nitrocellulose blots were blocked in TBS (Tris-buffered saline; pH 8.0) containing 0.1% Tween 20 (TBST) and 3% BSA and then incubated with anti-PLB1 peptide antibodies (1 μg/ml) in TBST containing 3% BSA. After 3×10 min washes with TBST, the blots were incubated with donkey anti-goat HRP (1:1000 dilution) in TBST containing 3% BSA. Proteins were detected by ECL after exposure to an X-ray film. When SuperSignal was used as the ECL reagent, the secondary antibody was diluted to 1:4000.
RESULTS
Detection of PLB1-expressing recombinants
Due to the non-availability of cryptococcal expression vectors, we chose to study cryptococcal PLB1 secretion in a heterologous S. cerevisiae expression system. S. cerevisiae has low endogenous PLB activity [20], performs post-translational modifications such as glycosylation (that is essential for the activity of cryptococcal PLB1) and has a well-characterized GPI-anchor biosynthetic pathway that is highly conserved in all eukaryotes.
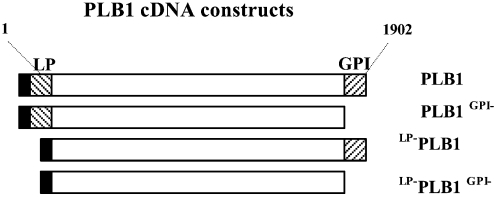
To characterize the role of the N- and C-terminal hydrophobic motifs in PLB1 secretion, a full-length, wild-type cDNA construct, referred to as PLB1, and three constructs truncated by deletion of the putative N-terminal LP and/or C-terminal GPI anchor attachment motifs were generated by PCR amplification (Figure 1). The position of the truncation was determined using the LP and GPI anchor prediction programs available at www.expasy.org. At the C-terminus, the truncation was made after the amino acid residue at position 612 and included the last residue (Gly) at position 613, which is part of the predicted Ser-Ser-Gly omega consensus sequence. All constructs were inserted into the expression vector, pYES2, downstream of the Gal 1 promoter and sequences encoding in-frame His/Express tags. The pYES2-PLB1 hybrid vectors were used to transform S. cerevisiae, and recombinants were selected on URA−glu agar plates.
Figure 1. Schematic representation of PLB1 cDNA constructs.
The N- and C-terminal hatched regions encode the putative 19-amino-acid LP motif (MSIATGTFAFSLFATIAFA) (LP) and the 22-amino-acid GPI anchor attachment motif GAANADVSMGMVALAAGLGLML (GPI) respectively. PLB1 cDNA (1902 bp) was amplified by PCR, incorporating both (PLB1), one (PLB1GPI− or LP−PLB1) or neither (LP−PLB1GPI−) motifs, and each construct was inserted into the pYES2 expression vector. The solid box at the N-terminus represents the His/Express recognition motif that is already present in the pYES2 expression vector.
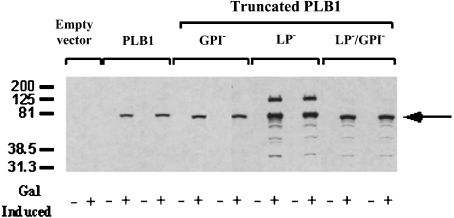
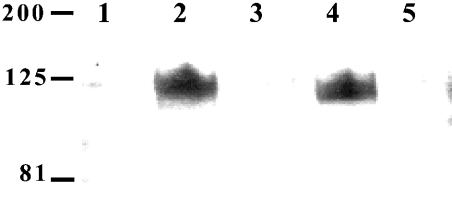
Two transformants were selected from each of the four PLB1 recombinants, and PLB1 expression was induced by growth in URA−gal medium. Cell lysates were prepared in the presence of 0.1% Triton X-100 and analysed by SDS/PAGE and Western blotting with the anti-Express antibody (Figure 2). For all constructs, the anti-Express antibody and an anti-PLB1 peptide antibody (results not shown) recognized a band with a molecular mass of approx. 75 kDa, consistent with the molecular mass predicted from the PLB1 cDNA sequence, when galactose (but not glucose) was used as the sole carbon source. No bands were detected in the lysates from cells transfected with empty vector. Quantification of the 75 kDa PLB1 bands by densitometry revealed a 4-fold increase in the amount of LP−PLB1 (PLB1 expressed without the LP consensus motif) and LP−PLB1GPI− (PLB1 expressed without the LP and GPI consensus motifs) compared with PLB1 and PLB1GPI− (PLB1 expressed without the GPI anchor attachment motif). A small amount of proteolytic breakdown was observed for all constructs and was most evident in the more highly expressed LP− constructs. This was despite the presence of a protease inhibitor cocktail covering an extensive range of protease inhibitor classes. A second, higher-molecular-mass band (∼125 kDa) was detected, especially in the LP−PLB1 construct. Although this band corresponded to the molecular mass of glycosylated, secreted PLB1, which was previously demonstrated to be 110–120 kDa [23], it quite probably represents an SDS-resistant, PLB1-containing aggregate.
Figure 2. Detection of intracellular PLB1 expression by Western blotting.
PLB1-expressing recombinants (and empty vector control) were cultured in URA−gal (+) or URA−glu (−) medium to induce and suppress PLB1 expression respectively. Except for the empty vector, two transformants of each recombinant were analysed. Cell lysates were prepared with 0.1% Triton X-100 as described in the Experimental section, and aliquots, containing 30 μg of protein, were subjected to SDS/PAGE and Western blotting. PLB1 constructs were detected with the anti-Express-tagged antibody as described in the Experimental section using ECL and exposure to an X-ray film. The molecular-mass standards (in kDa) are indicated, and the arrow indicates the position of the PLB1 constructs.
Secretion of PLB1 protein and activity
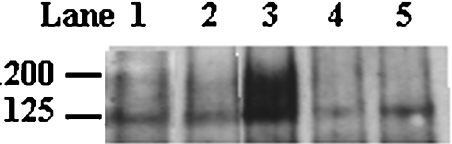
The PLB1-expressing recombinants were grown to stationary phase in URA−gal induction medium, harvested and allowed to secrete as described in the Experimental section. A lack of uptake of propidium iodide by the cells, as detected by flow cytometry, indicated that the cells were more than 90% viable after the secretion period (results not shown). Proteins in the secretions were subjected to SDS/PAGE and Western blotting with anti-PLB1 (Figure 3). A broad band with a molecular mass range of 130–200 kDa was prominent only in supernatants from PLB1GPI− cells (Figure 3, lane 3), and the 75 kDa band recognized in all of the constructs by the His/Express antibody (Figure 1) was absent. Reprobing of the blot with the anti-Express-tagged antibody revealed no PLB1 bands at 75 or 130–200 kDa in any of the constructs (results not shown). This is most likely due to the removal of the Express/His tag along with the adjacent LP (see Figure 1) in the ER by signal peptidase cleavage, before secretion, and suggests that the 75 kDa band recognized in Figure 1 is cytoplasmically expressed due to saturation of the secretion machinery. The increase in the secreted molecular mass of PLB1GPI−, compared with cell-associated PLB1 as detected with anti-Express, suggests that the protein is glycosylated before secretion. The detection of PLB1GPI−, but not PLB1, demonstrates that the presence of the GPI anchor attachment sequence during expression retards PLB1 secretion. The PLB1 band was absent from LP−PLB1 and LP−PLB1GPI− cells (lanes 4 and 5), demonstrating that the LP is essential for PLB1 secretion. A lower band at approx. 125 kDa is present in all samples, including the empty vector control (lane 1), and most probably represents an endogenous protein secreted by S. cerevisiae, which is recognized non-specifically in the Western blot.
Figure 3. Detection of secreted PLB1 by Western blotting.
PLB1 recombinants were cultured in URA−gal medium to induce PLB1 expression and then allowed to secrete for 22 h. The supernatants (secretions) were collected, and protein (40 μg) was subjected to SDS/PAGE and Western blotting with anti-PLB1 peptide antibody as described in the Experimental section. PLB1 was detected by ECL after exposure to an X-ray film. The molecular mass standards (in kDa) are indicated. Lane 1 contains empty vector control and lanes 2–5 contain PLB1, PLB1GPI−, LP−PLB1 and LP−PLB1GPI− recombinants respectively.
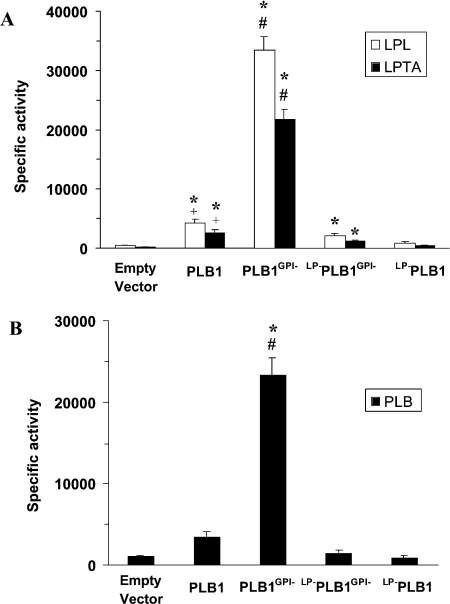
Secreted PLB, LPL and LPTA activities were measured and correlated directly with levels of secreted protein (Figure 3) when expressed as either specific (Figure 4) or total activities. All three activities secreted by PLB1GPI− recombinants were 9-fold higher than recombinants expressing PLB1, after correcting for the low level of endogenous activity secreted by the empty vector control. Almost no activity was produced by the LP-deficient constructs LP−PLB1 and LP−PLB1GPI−, again indicating the essential role of the LP for secretion of active PLB1. Identical secretion profiles were obtained when the results were expressed as total secreted activity (results not shown) and when each construct was expressed in an S. cerevisiae (plb1, 2 and 3) triple-knockout strain [20] (results not shown). However, due to overall lower levels of secreted enzyme activity in this strain (>3-fold), we continued to use the wild-type strain. The ratios of PLB/LPL/LPTA activities secreted by PLB1 and PLB1GPI− recombinants remained the same (0.001:1.6:1), demonstrating that the absence of the GPI anchor attachment motif does not affect the proportions of secreted enzyme activities. Similar ratios (results not shown) were obtained for native PLB1 secreted from C. neoformans cultured under similar conditions, demonstrating that the properties of PLB1 are not altered in the S. cerevisiae expression system.
Figure 4. Comparison of secreted PLB1 specific activities by radiometric assay.
PLB1 recombinants were cultured in URA−gal medium to induce PLB1 expression and then allowed to secrete for 22 h. The supernatants (secretions) were collected and radiometric assays were performed as described in the Experimental section. For LPL/LPTA (A) and PLB (B) assays, 2.5 and 13 μg of protein were used respectively. Results are expressed as mean specific activity±S.E.M. with n=3 [nmol·min−1·(mg of protein)−1 for LPL and LPTA activities and pmol·min−1·mg−1 for PLB activity]. The * indicates that the higher activity relative to the empty vector control is statistically significant (P<0.05). The # indicates that the higher activity for PLB1GPI− relative to PLB1, LP−PLB1 and LP−PLB1GPI− is statistically significant (P<0.05). The + indicates that the higher activity for PLB1 relative to LP−PLB1GPI− and LP−PLB1 is statistically significant (P<0.05). Statistical significance was determined using a paired, two-tailed parametric t test.
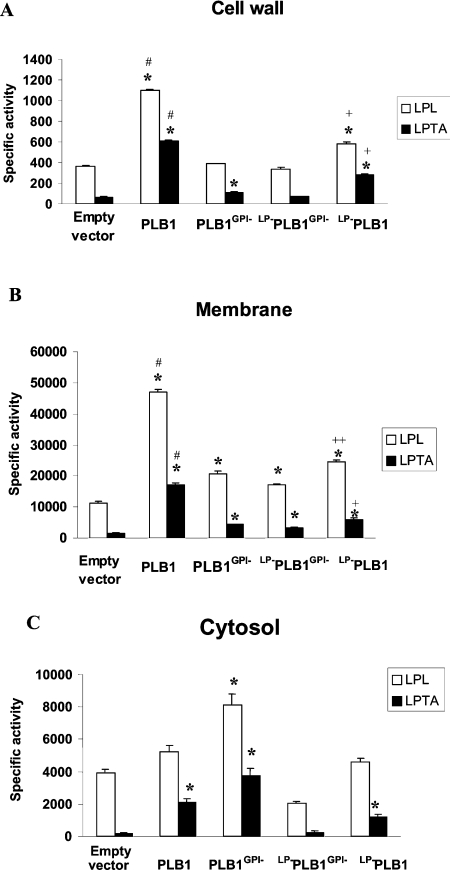
Subcellular localization of PLB1 and truncated forms
Subcellular fractions enriched in cell-wall (including unbroken cells, organelles and some membranes), cell-membrane (including transport vesicles, ER and mitochondrial and plasma membranes) and cytosolic components were prepared and assayed for PLB1 activity as described in the Experimental section. Both LPL and LPTA specific activities (Figures 5A and 5B) and PLB specific activities (results not shown) were significantly higher in the cell-wall and cell-membrane fractions prepared from PLB1- and LP−PLB1-expressing recombinants when compared with the corresponding fractions prepared from recombinants expressing PLB1GPI−. The cell-wall-associated specific activity was substantially lower than the membrane-associated specific activity; however, the cell wall contained the majority of the total enzyme activity (see Figure 7), due to the large amount of protein present in this fraction (Table 1). Little or no activity (above empty vector levels) was observed in the membrane- and cell- wall-enriched fractions of recombinants expressing PLB1GPI− or LP−PLB1GPI−. PLB1 protein was difficult to detect in anti-PLB1 Western blots of the cell-wall fractions, due to high non-specific binding. However, PLB1 protein, but not PLB1GPI−, was detected by anti-PLB1 in membrane fractions (Figure 6). The PLB1 band had a molecular mass of approx. 120 kDa (lane 2), similar to that of native PLB1 secreted by C. neoformans [23], suggesting that it was glycosylated. Interestingly, LP−PLB1 also had a molecular mass consistent with the glycosylated form (Figure 6, lane 4).
Figure 5. Comparison of cell-associated PLB1 specific activities by radiometric assay.
PLB1 expression was induced as described for Figure 4. After the 22 h secretion period, the cells were harvested, homogenized and centrifuged at 14000 g. The supernatant was recentrifuged at 100000 g. The 14000 g pellet (cell walls) (A) and the 100000 g pellet (cell membranes) (B) were resuspended in IAB and, together with the 100000 g supernatant (cytosol) (C), were assayed for LPL/LPTA activity. For each fraction, 40, 1 and 2 μg of protein was used respectively. Results are expressed as mean specific activity [nmol·min−1·(mg of protein)−1]±S.E.M with n=3. The * indicates that the higher specific activity relative to the empty vector control is statistically significant (P<0.05). The # indicates that the higher specific activity relative to PLB1GPI−, LP−PLB1 and LP−PLB1GPI− is statistically significant. The + indicates that the higher specific activity relative to PLB1GPI− and LP−PLB1GPI− is statistically significant. The ++ indicates that the higher specific activity relative to LP−PLB1GPI− is statistically significant. For the cytosol, the increased mean PLB1GPI− specific activities compared with the mean PLB1 specific activities are not statistically significant. Statistical significance was determined using a paired, two-tailed parametric t test.
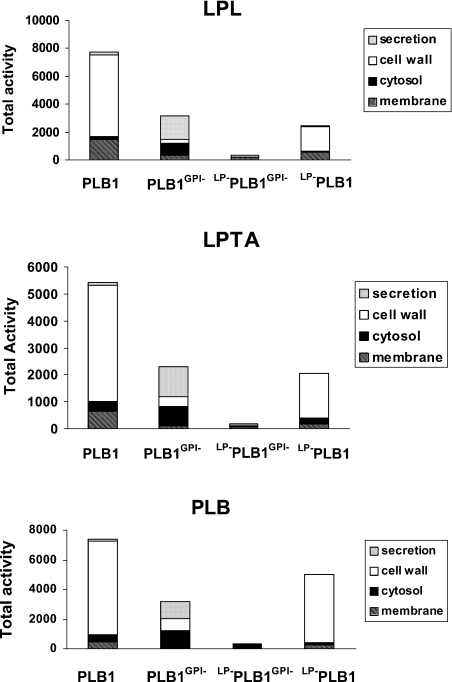
Figure 7. Comparison of the cellular distribution of total PLB1 activity.
PLB1 expression was induced as described for Figure 4. After a 22 h secretion period, the cells were pelleted by centrifugation and the supernatants (secretions) were undisturbed. The cell pellets were fractionated into cell wall, membranes and cytosol as described for Figure 5. Each fraction was assayed for LPL, LPTA and PLB activities as described in the Experimental section. The amount of protein used for the LPL/LPTA assays is indicated in Figures 4 and 5. To assay PLB activity, 13, 60, 20 and 20 μg of protein was used for the secreted, cell-wall, membrane and cytosolic fractions respectively. The results were corrected for background from the empty vector control and expressed as the total activity (nmol·min−1·fraction−1 for LPL/LPTA activity and pmol·min−1·fraction−1 for PLB activity).
Table 1. Subcellular protein distribution in S. cerevisiae PLB1 recombinants.
After a 22 h secretion period in URA−gal medium, cells were fractionated into membrane, cell wall and cytosol. Each fraction and the secretion were assayed as described in the Experimental section. Results are expressed as μg of protein per fraction and as a percentage of the total. Each fraction was assayed in duplicate or triplicate.
| Fraction | Total protein (μg) | Protein (% of total) |
|---|---|---|
| Secreted | 103 | 1 |
| Membrane | 82 | 1 |
| Cell wall | 7844 | 93 |
| Cytosol | 408 | 5 |
Figure 6. Detection of membrane-associated PLB1 by Western blotting.
Total membranes from recombinants were prepared as described for Figure 5 and subjected to SDS/PAGE and Western blotting with anti-PLB1 peptide antibody as described in the Experimental section. PLB1 was detected by ECL after exposure to an X-ray film. The masses of the molecular-mass standards (in kDa) are indicated. Lane 1 contains empty vector control and lanes 2–5 contain PLB1, PLB1GPI−, LP−PLB1 and LP−PLB1GPI− recombinants respectively.
In contrast, the highest LPL and LPTA activities in the cytosolic fraction were present in recombinants expressing PLB1GPI− followed by those expressing PLB1 and LP−PLB1 (Figure 5C). No activity (relative to empty vector control) was expressed by LP−PLB1GPI−. Similar results were obtained for cytosolic PLB activity (results not shown). The appearance of some PLB1 activity in the cytosol was probably due to the activity of endogenous (G)PI-specific phospholipases and/or glucanases, and/or due to mechanical shearing of PLB1 from the cell membranes/cell walls during preparation of the subcellular fractions. High endogenous LPL activity, but not LPTA or PLB activity, was observed, particularly in the cytosolic fraction, and it may be due to the three S. cerevisiae PLB enzymes (encoded by the plb 1, 2 and 3 genes) that are located in membranes and the periplasmic space [20]. In summary, these results demonstrate that the C-terminal 22 amino acids are required to localize PLB1 to the cell membrane and cell wall.
Comparison of total LPL, LPTA and PLB activities
The cellular distribution of total activities (comprising the cell wall, membrane, cytosol and secreted activities) for each clone is represented in Figure 7. Total LPL, LPTA and PLB activities produced by the full-length PLB1-expressing recombinants were >2-fold higher than for recombinants expressing PLB1GPI−. LP−PLB1GPI− produced virtually no active enzyme, despite the detection of cell-associated PLB1 protein by Western blotting (Figure 2), indicating that both the LP and GPI motifs are important for the expression of a functional protein. Most of the LPL, LPTA and PLB activities in the PLB1-expressing recombinants (containing the GPI anchor motif) were cell-wall-associated (75, 79 and 86% respectively) with <5% secreted after 16 h (results not shown) and 22 h of galactose-induced growth; 7% or less of all activities was cytosolic. In contrast, most of the LPL, LPTA and PLB activities in recombinants expressing PLB1 without the GPI motif (PLB1GPI−) were secreted (54, 48 and 36% respectively) or cytosolic (26, 31 and 38% respectively). Only 8–25% of the activities were in the cell-wall fraction, despite this fraction containing 90% of the total protein (results not shown). The low levels of activity secreted by the PLB1-expressing recombinants (<5%) prompted us to compare the secreted and cell-associated activity in C. neoformans strain H99, cultured under similar conditions. Secreted LPL, LPTA and PLB activities were all found to be higher, representing 15, 13 and 36% of the total activity respectively (Table 2). It has been shown previously that PLB1 is responsible for nearly all of the secreted activity in C. neoformans [2].
Table 2. Comparison of the cell-associated and secreted LPL, LPTA and PLB activities in C. neoformans broth cultures.
After a 22 h secretion period in YNB broth, the cells were fractionated into membrane, cell wall and cytosol, and each fraction was assayed in triplicate for LPL, LPTA and PLB activities as described in the Experimental section. The secretions were dialysed, freeze-dried and resuspended in IAB, before assaying. The activities in the cell-wall, membrane and cytosolic fractions were added together (cell-associated). The cell-associated and secreted activities are expressed as a percentage of the total (secreted + cell-associated) activities. Values in paratheses indicate the S.E.M. with n=3.
| Activity (% of total) | |||
|---|---|---|---|
| Cell fraction | LPL | LPTA | PLB |
| Secreted | 15 (1) | 13 (1) | 36 (2) |
| Cell-associated | 85 (6) | 87 (4) | 64 (4) |
β-Glucanase digestion of S. cerevisiae recombinant and cryptococcal cell walls
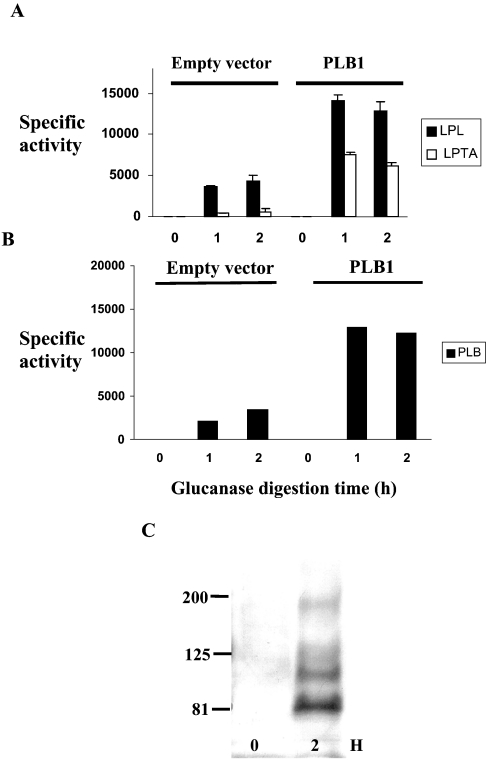
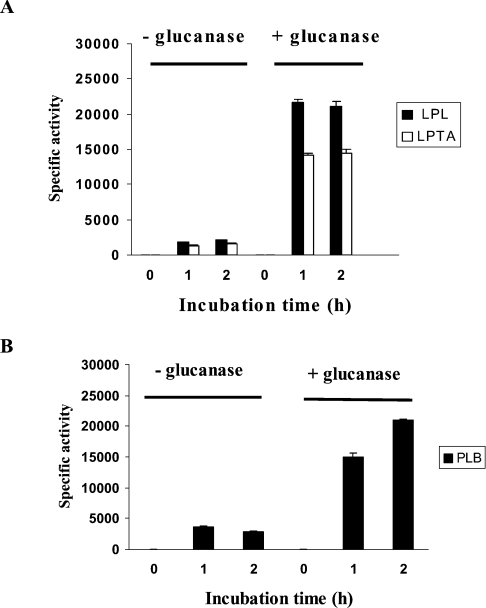
After exit from the Golgi apparatus, GPI-anchored proteins produced by S. cerevisiae are transported to the plasma membrane where they remain or are rerouted to the cell wall [12], becoming covalently attached to the intricate β-glucan network [24]. To determine whether PLB1 takes this route, cell-wall preparations from the PLB1-expressing S. cerevisiae recombinant and C. neoformans (prepared as described in Figure 5) were incubated with a β-glucanase preparation containing protease inhibitors. The cell walls were pelleted by centrifugation and the supernatants were assayed for the released PLB1 enzyme activity. Results for the PLB1-expressing S. cerevisiae recombinant (containing the GPI anchor motif) and the empty vector control are shown in Figures 8(A) and 8(B). For PLB1-expressing recombinants, the release of activity was complete after 1 h. As expected, more activity was released from PLB1-containing cell walls compared with those of the empty vector. The released activity represented approx. 12% of the total cell-wall-associated activity determined in Figures 5 and 7 and the ratio of the released activities was similar to the ratio of the secreted activities (Figure 4), confirming that the secreted activity is derived from the cell wall. The activity released from empty vector cell walls is most likely due to the endogenous GPI-anchored PLB1 and PLB3 in S. cerevisiae [20]. β-Glucanase-released proteins were also subjected to Western blotting with an α-agglutinin antibody. α-Agglutinin is a cell- wall-associated GPI-anchored protein involved in mating. After 2 h of β-glucanase treatment, the antibody recognized an α-agglutinin banding profile similar to that obtained previously with the lower bands suspected to be breakdown products [25], confirming that the fraction we are treating with β-glucanase contains cell walls.
Figure 8. Analysis of β-glucanase-released proteins from recombinant cell walls.
Cell-wall fractions from empty vector control and PLB1-expressing recombinants, prepared as described for Figure 5, were incubated with β-glucanase for 2 h at 37 °C. To assay PLB1 activity released at 0, 1 and 2 h, cell walls were pelleted by centrifugation and aliquots of the supernatant were removed for enzyme assay as described in the Experimental section. For the LPL and LPTA activity assays (A), 1 μl was used in a triplicate assay; for the PLB activity assays (B), 5 μl was used in a duplicate assay. Results are expressed as the mean activity released±S.E.M. with n=3 (nmol·min−1·ml−1 for LPL and LPTA activities and pmol·min−1·ml−1 for PLB activity). Protein was not estimated, due to interference from the added glucanase. The β-glucanase-released proteins were also subjected to SDS/PAGE and Western blotting with an anti-α-agglutinin antibody as described in the Experimental section (C). The GPI-anchored cell-wall marker protein, α-agglutinin, was detected by ECL after exposure to an X-ray film. The masses of the molecular-mass standards (in kDa) are indicated. Lanes 1 and 2 represent 0 and 2 h of β-glucanase digestion respectively.
β-Glucanase also released activity from C. neoformans cell-wall preparations (Figure 9). After correcting for the activity released in the presence of digestion buffer without β-glucanase, the enzyme was calculated to have released 90% of the activity detected in intact cell walls (results not shown). No difference was obtained when cell walls were incubated with β-glucanase in the absence of protease inhibitors (results not shown).
Figure 9. Release of CnPLB1 activity from cryptococcal cell walls by β-glucanase.
Cryptococcal cell walls, prepared as described for Figure 5, were incubated with β-glucanase as described in Figure 8. For the LPL and LPTA activity assays (A), 1 μl was used; for the PLB activity assays (B), 5 μl was used. Results are expressed as the mean activity released±S.E.M. with n=3 (nmol·min−1·ml−1 for LPL and LPTA activities and pmol·min−1·ml−1 for PLB activity). Protein was not estimated due to interference from the added glucanase.
DISCUSSION
In the present study, we expressed the cryptococcal virulence determinant, PLB1, in S. cerevisiae and determined that GPI anchoring prevents PLB1 secretion by targeting the protein to the fungal cell membrane/cell wall. Deletion of the GPI anchor motif from PLB1 resulted in redistribution of active LPL, LPTA and PLB from the cell wall and membranes to the secretions. A 9-fold increase in the secretion of all three enzyme activities was observed. This mechanism for secretion of a functional enzymic virulence determinant has not been described previously. It is consistent with previous observations in fungi such as S. cerevisiae and Fusarium oxysporum whereby deletion of the hydrophobic GPI anchor attachment motif from cell wall-associated, GPI-anchored proteins led to secretion of a functional protein [25,26]. We confirmed that the typical cryptococcal LPL/LPTA/PLB activity ratio, and the high specific activity of secreted cryptococcal PLB1 [1] relative to those of endogenous PLBs in S. cerevisiae [20], were retained in this model system.
PLB1 N-terminal hydrophobic motif is a bona fide secretory LP
Secretion of PLB1 activity and protein was dependent on the 19 N-terminal amino acids, which therefore represent a bona fide LP, directing PLB1 into the secretory pathway. Secreted PLB1 protein was most abundant in the recombinant strain PLB1GPI−. It had a molecular mass in the range 130–200 kDa compared with 75 kDa demonstrated for the cytosolic form, suggesting that it was glycosylated. The broad and high molecular mass of secreted recombinant PLB1 relative to that of native CnPLB1 (110–120 kDa) is indicative of the heterogeneous hyperglycosylation that is characteristic of protein expression in S. cerevisiae [27]. Secreted, endogenous, S. cerevisiae PLB1 is also hyperglycosylated. It has a molecular mass of 220 kDa, despite having a similar number of amino acids as CnPLB1 [20]. Interestingly, membrane-associated recombinant PLB1 had a molecular mass of approx. 120 kDa, greater than that of the cytosolic form but less than that of the secreted form. The membrane form of a phospholipase that is produced by S. cerevisiae, and is quite probably PLB1, is also smaller than the secreted form [28]. Incorporation of hexose moieties has been reported to occur in the yeast plasma membrane [29,30]; thus it is possible that the size discrepancy between the membrane and secreted forms of recombinant PLB1 is due to additional glycosylation in the periplasmic space and/or cell wall before secretion. Similar conclusions have been drawn with other secreted yeast glycoproteins [31,32].
Reduced secretion from PLB1, LP−PLB1 and LP−PLB1GPI− relative to PLB1GPI− was not due to reduced synthesis. On the contrary, PLB1 protein detected by Western blotting was increased 4-fold in LP-deficient constructs, as determined using the anti-Express antibody, compared with PLB1 and PLB1GPI−.
LP not essential for GPI anchoring
Although essential for secretion, the LP is not essential for cell-associated PLB1 activity provided that the GPI anchor motif is present. This was indicated by LP−PLB1, but not by LP−PLB1GPI−, exhibiting a high level of cell-associated activity (especially PLB activity) relative to constructs containing the LP. The results suggest that there is a hierarchy of importance of the hydrophobic motifs with respect to intracellular activity, i.e. activity with both > activity with one > activity with none. The high level of intracellular activity exhibited by LP−PLB1 was surprising. In the absence of an LP, PLB1 should not enter the secretory pathway and therefore neither acquire N-linked glycosylation, which is essential for activity of the secreted enzyme [1], nor become GPI-anchored. We found, however, that membrane-associated LP−PLB1 and a portion of LP−PLB1 in cell lysates had a molecular mass consistent with the glycosylated form, and that LP−PLB1 activity was mainly membrane- and cell-wall-associated rather than cytosolic, suggesting that the GPI anchor attachment motif was active despite the absence of an LP. Thus our findings provide new evidence that GPI anchoring may proceed in the absence of an LP.
Cell-associated PLB1 was mostly in the cell wall
After exit from the Golgi apparatus, GPI-anchored proteins are typically transported to the plasma membrane where they remain or are rerouted to the cell wall, becoming covalently attached to glucan moieties [12]. Approximately 12% of the activity in the PLB1 recombinant was in the cell membrane-enriched fraction, consistent with a role in membrane remodelling or homoeostasis and/or as an intermediate location before transport to the cell wall. Most of the enzyme activity in the PLB1 recombinant (80%) was associated with the β-glucan-rich cell wall. Incubation of this fraction with β-glucanase, however, released only 12% of the activity, compared with 90% for C. neoformans cell-wall preparations. Failure to release the remaining activity may be due to a difference in cell-wall composition between S. cerevisiae and C. neoformans [33]. However, the similar ratio of released LPL/LPTA/PLB activities from both fungi confirmed that the same multifunctional enzyme was produced in each organism. Little is known about the molecular basis of cell-membrane and cell-wall targeting except that GPI-anchored proteins destined to remain in the plasma membrane contain a dibasic motif immediately upstream of the GPI anchor attachment site [8]. Since PLB1 does not contain this motif, we predicted that it would be in the cell-wall fraction. Positioning PLB1 in the cell wall may be advantageous to the pathogen, allowing rapid release of the enzyme in response to changing environmental conditions. Altered environmental conditions, which triggered the rerouting of GPI-linked proteins to the cell wall in S. cerevisiae [8,34], also accelerated PLB1 enzyme release from C. neoformans. In C. neoformans, we showed previously that PLB1 secretion from strain H99 was enhanced by incubating at 37 °C instead of 22 and 30 °C and in tissue culture medium instead of standard assay buffer [17].
Release of secreted PLB1 activity
Most of the total LPL, LPTA and PLB activities in the PLB1-expressing recombinants and in C. neoformans (contributed by PLB1 CnLYSO1, LPL1 and other unidentified gene products) were cell-associated. However, C. neoformans still produced more secreted activity (15–34%) compared with 5% for the PLB1 recombinant, which is mainly attributed to the PLB1 gene [2]. This may be due to one or a combination of factors. First, the enzymes initiating cleavage of GPI-anchored proteins in S. cerevisiae may not efficiently recognize cryptococcal GPI-anchored proteins. Secondly, due to overexpression of recombinant PLB1, sufficient amounts of cleaving enzyme may not be available. Thirdly, it has been reported that the activity of endogenous enzymes responsible for cleaving yeast cell-surface proteins, such as (G)PI-PLC, is stimulated by growth in the presence of glucose [35]. These enzymes may therefore not be active in the S. cerevisiae expression system where protein expression is switched on by using galactose as the sole carbon source. Consequently, a backlog of protein would be expected in the membranes and the cell wall. Finally, GPI-anchor-cleaving enzymes may not be abundant in S. cerevisae. This is suggested by the finding that very little endogenous PLB1 and PLB3, produced from genes that encode GPI anchor consensus motifs, is secreted from S. cerevisae [20,28].
Enzymes responsible for the release of CnPLB1 from the cell surface of C. neoformans have not been determined, but could include glucanases, proteases, (G)PI-PLC/PLD or a combination thereof, as in the case of the plasma-membrane-localized GPI-anchored cellular prion protein (PrPc), which is released by the action of both proteases and phospholipases [9]. Secretion of CnPLB1 is not protease-driven, since the presence of protease inhibitors failed to reduce the level of secreted activity [22]. Genes encoding (G)PI-PLC are present in S. cerevisiae and C albicans. A BLAST search of the cryptococcal (serotype A) genomic database using these sequences revealed the existence of a putative PI-PLC. We are currently investigating whether this enzyme regulates PLB1 secretion. Extracellular environments may also directly affect the enzymes responsible for cleaving cell-surface proteins.
In summary, we report a novel finding that GPI anchoring of PLB1 promotes cell-membrane and cell-wall attachment and regulates secretion of this cryptococcal virulence factor. We also show that the LP is not required for cell-associated PLB1 activity and that some glycosylation of PLB1 occurs in the membrane and/or the cell wall before secretion. The dominant location of PLB1 in the cell wall may permit the immediate release of this virulence factor in response to changing environmental conditions.
Acknowledgments
We thank M. Larsen and C. Wilson for technical assistance and Dr G. Cox for providing PLB1 cDNA. This work was supported by a National Health and Medical Research Council grant (211040) and in part by a Burroughs Wellcome Fund, by grants AI56168 from the National Institutes of Health and RR17677 Project 2 from the Center for Research Excellence Program of the National Center for Research Resources to M.D.P. M.D.P. is a Burroughs Wellcome New Investigator in Pathogenesis of Infectious Diseases.
References
- 1.Chen S. C. A., Wright L. C., Golding J. C., Sorrell T. C. Purification and characterisation of secretory phospholipase B, lysophospholipase-transacylase from a virulent strain of the pathogenic fungus Cryptocococcus neoformans. Biochem. J. 2000;347:431–439. doi: 10.1042/0264-6021:3470431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cox G. M., McDade H. C., Chen S. C. A., Tucker S. C., Gottfredsson M., Wright L. C., Sorrell T. C., Leidich S. D., Casadevall A., Ghannoum M. A., et al. Extracellular phospholipase activity is a virulence factor for Cryptococcus neoformans. Mol. Microbiol. 2001;39:166–175. doi: 10.1046/j.1365-2958.2001.02236.x. [DOI] [PubMed] [Google Scholar]
- 3.Santangelo R., Zoellner H., Sorrell T., Wilson C., Donald C., Djordjevic J. T., Shounan Y., Wright L. C. The role of extracellular phospholipases and mononuclear phagocytes in the dissemination of cryptococcosis in a murine model. Infect. Immun. 2004;72:2229–2239. doi: 10.1128/IAI.72.4.2229-2239.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leidich S. D., Ibrahim A. S., Fu Y., Koul A., Jessup C., Vitullo J., Fonzi W., Mirbod F., Nakashima S., Nozawa Y., et al. Cloning and disruption of caPLB1, a phospholipase B gene involved in the pathogenicity of Candida albicans. J. Biol. Chem. 1998;273:26078–26086. doi: 10.1074/jbc.273.40.26078. [DOI] [PubMed] [Google Scholar]
- 5.Ghannoum M. A. Potential role of phospholipases in virulence and fungal pathogenesis. Clin. Microbiol. Rev. 2000;13:122–143. doi: 10.1128/cmr.13.1.122-143.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sugiyama Y., Nakashima S., Mirbod F., Kanoh H., Kitajima Y., Ghannoum M. A., Nozawa Y. Molecular cloning of a second phospholipase B gene, caPLB2 from Candida albicans. Med. Mycol. 1999;37:61–67. [PubMed] [Google Scholar]
- 7.Sütterlin C., Horvath A., Gerold P., Schwarz R. T., Wang Y., Dreyfuss M., Riezman H. Identification of a species-specific inhibitor of glycosylphosphatidylinositol synthesis. EMBO J. 1997;16:6374–6383. doi: 10.1093/emboj/16.21.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vossen J. H., Müller W. H., Lipke P. N., Klis F. M. Restrictive glycosylphosphatidylinositol anchor synthesis in cwh6/gpi3 yeast cells causes aberrant biogenesis of cell wall proteins. J. Bacteriol. 1997;179:2202–2209. doi: 10.1128/jb.179.7.2202-2209.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parkin E. T., Watt N. T., Turner A. J., Hooper N. M. J. Dual mechanisms for shedding of the cellular prion protein. J. Biol. Chem. 2004;279:11170–11178. doi: 10.1074/jbc.M312105200. [DOI] [PubMed] [Google Scholar]
- 10.Kristiansen S., Richter E. A. GLUT4-containing vesicles are released from membranes by phospholipase D cleavage of a GPI anchor. Am. J. Physiol. Endocrinol. Metab. 2002;283:E374–E382. doi: 10.1152/ajpendo.00441.2001. [DOI] [PubMed] [Google Scholar]
- 11.Mann K. J., Hepworth M. R., Raikwar N. S., Deeg M. A., Sevlever D. Effect of glycosylphosphatidylinositol (GPI)-phospholipase D overexpression on GPI metabolism. Biochem. J. 2004;378:641–648. doi: 10.1042/BJ20031326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brul S., King A., van der Vaart J. M., Chapman J., Klis F., Verrips C. T. The incorporation of mannoproteins in the cell wall of. S. cerevisiae and filamentous Ascomycetes. Antonie van Leeuwenhoek. 1997;72:229–237. doi: 10.1023/a:1000429208049. [DOI] [PubMed] [Google Scholar]
- 13.Englund P. T. The structure and biosynthesis of glycosyl phosphatidylinositol protein anchors. Annu. Rev. Biochem. 1993;62:121–138. doi: 10.1146/annurev.bi.62.070193.001005. [DOI] [PubMed] [Google Scholar]
- 14.Horvath A., Sütterlin C., Manning-Krieg U., Movva N. R., Riezman H. Ceramide synthesis enhances transport of GPI-anchored proteins to the Golgi apparatus in yeast. EMBO J. 1994;13:3687–3695. doi: 10.1002/j.1460-2075.1994.tb06678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sütterlin C., Doering T. L., Schimmöller F., Schröder S., Riezman H. Specific requirements for the ER to Golgi transport of GPI-anchored proteins in yeast. J. Cell Sci. 1997;110:2703–2714. doi: 10.1242/jcs.110.21.2703. [DOI] [PubMed] [Google Scholar]
- 16.Mukherjee P. K., Chandra J., Kuhn D. M., Ghannoum M. A. Differential expression of Candida albicans phospholipase B (PLB1) under various environmental and physiological conditions. Microbiology. 2003;149:261–267. doi: 10.1099/mic.0.25829-0. [DOI] [PubMed] [Google Scholar]
- 17.Wright L. C., Chen S. C. A., Wilson C. F., Simpanya M. F., Blackstock R., Cox G. M., Murphy J. W., Sorrell T. C. Strain-dependent effects of environmental signals on the production of extracellular phospholipase by Cryptococcus neoformans. FEMS Microbiol. Lett. 2002;209:175–181. doi: 10.1111/j.1574-6968.2002.tb11128.x. [DOI] [PubMed] [Google Scholar]
- 18.Latouche G. N., Sorrell T. C., Meyer W. Isolation and characterisation of the phospholipase B gene of C. neoformans var. gattii. FEMS Yeast Res. 2002;2:551–561. doi: 10.1111/j.1567-1364.2002.tb00122.x. [DOI] [PubMed] [Google Scholar]
- 19.Luberto C., Martinez-Marino B., Taraskiewicz D., Bolanos B., Chitano P., Toffaletti D. L., Cox G. M., Perfect J. R., Hannun Y. A., Balish E., et al. Identification of App1 as a regulator of phagocytosis and virulence of Cryptococcus neoformans. J. Clin. Invest. 2003;112:1080–1094. doi: 10.1172/JCI18309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merkel O., Fido M., Mayr J. A., Prüger H., Raab F., Zandonella G., Kohlwein S. D., Paltauf F. Characterization and function in vivo of two novel phospholipases B/lysophospholipases from Saccharomyces cerevisiae. J. Biol. Chem. 1999;274:28121–28127. doi: 10.1074/jbc.274.40.28121. [DOI] [PubMed] [Google Scholar]
- 21.Bligh E. C., Dyer W. J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 22.Chen S. C. A., Wright L. C., Santangelo R. T., Muller M., Moran V. R., Kuchel P. W., Sorrell T. C. Identification of extracellular phospholipase B, lysophospholipase, and acyltransferase produced by Cryptococcus neoformans. Infect. Immun. 1997;65:405–411. doi: 10.1128/iai.65.2.405-411.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wright L. C., Payne J., Santangelo R. T., Simpanya M. F., Chen S. C. A., Widmer F., Sorrell T. C. Cryptococcal phospholipases: a novel lysophospholipase discovered in the pathogenic fungus Cryptococcus gattii. Biochem. J. 2004;384:1–8. doi: 10.1042/BJ20041079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aguilar-Uscanga B., Francois J. M. A study of the yeast cell wall composition and structure in response to growth conditions and mode of cultivation. Letts. Appl. Microbiol. 2003;37:268–274. doi: 10.1046/j.1472-765x.2003.01394.x. [DOI] [PubMed] [Google Scholar]
- 25.Wojciechowicz D., Lu C., Kurjan J., Lipke P. N. Cell surface anchorage and ligand-binding domains of the Saccharomyces cerevisiae cell adhesion protein α-agglutinin, a member of the immunoglobulin superfamily. Mol. Cell. Biol. 1993;13:2554–2563. doi: 10.1128/mcb.13.4.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schoffelmeer E. A. M., Vossen J. H., van Doorn A. A., Cornelissen B. J. C., Haring M. A. FEM1, a Fusarium oxysporum glycoprotein that is covalently linked to the cell wall matrix and is conserved in filamentous fungi. Mol. Genet. Genomics. 2001;265:143–152. doi: 10.1007/s004380000402. [DOI] [PubMed] [Google Scholar]
- 27.Conde R., Cueva R., Pablo G., Polaina J., Larriba G. A search for hyperglycosylation signals in yeast glycoproteins. J. Biol. Chem. 2004;279:43789–43798. doi: 10.1074/jbc.M406678200. [DOI] [PubMed] [Google Scholar]
- 28.Witt W., Mertsching A., König E. Secretion of phospholipase B from Saccharomyces cerevisiae. Biochim. Biophys. Acta. 1984;795:117–124. doi: 10.1016/0005-2760(84)90111-5. [DOI] [PubMed] [Google Scholar]
- 29.Lehle L., Bauer F., Tanner W. The formation of glycosidic bonds in yeast glycoproteins. gattii. Arch. Microbiol. 1977;114:77–81. doi: 10.1007/BF00429634. [DOI] [PubMed] [Google Scholar]
- 30.Boer P. Glycosylation of yeast and fungal cell wall components. Biochem. Soc. Trans. 1979;7:331–333. doi: 10.1042/bst0070331. [DOI] [PubMed] [Google Scholar]
- 31.Meyer J., Matile P. Subcellular distribution of yeast invertase isoenzymes. Arch. Microbiol. 1975;103:51–55. doi: 10.1007/BF00436329. [DOI] [PubMed] [Google Scholar]
- 32.Holbein B. E., Forsberg C. W., Kidby D. K. A modified procedure for studying enzyme secretion in yeast sphaeroplasts: subcellular distribution of invertase. Can. J. Microbiol. 1976;22:989–995. doi: 10.1139/m76-144. [DOI] [PubMed] [Google Scholar]
- 33.Casadevall A., Perfect J. R. Washington, DC: ASM Press; 1998. Cryptococcus neoformans, chapter 4; pp. 84–85. [Google Scholar]
- 34.Kowalski L. R., Kondo K., Inouye M. Cold-shock induction of a family of TIP1-related proteins associated with the membrane in Saccharomyces cerevisiae. Mol. Microbiol. 1995;15:341–353. doi: 10.1111/j.1365-2958.1995.tb02248.x. [DOI] [PubMed] [Google Scholar]
- 35.Müller G., Gross E., Wied S., Bandlow W. Glucose-induced sequential processing of a glycosylphosphatidylinositol-anchored ectoprotein in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:442–456. doi: 10.1128/mcb.16.1.442. [DOI] [PMC free article] [PubMed] [Google Scholar]