Abstract
The insulin promoter binds a number of tissue-specific and ubiquitous transcription factors. Of these, the homoeodomain protein PDX-1 (pancreatic duodenal homeobox factor-1), the basic leucine zipper protein MafA and the basic helix–loop–helix heterodimer E47/BETA2 (β-cell E box transactivator 2; referred to here as β2) bind to important regulatory sites. Previous studies have shown that PDX-1 can interact synergistically with E47 and β2 to activate the rat insulin 1 promoter. The aim of the present study was to determine the relative contribution of PDX-1, MafA and E47/β2 in regulating the human insulin promoter, and whether these factors could interact synergistically in the context of the human promoter. Mutagenesis of the PDX-1, MafA and E47/β2 binding sites reduced promoter activity by 60, 74 and 94% respectively, in INS-1 β-cells. In the islet glucagonoma cell line αTC1.6, overexpression of PDX-1 and MafA separately increased promoter activity approx. 2.5–3-fold, and in combination approx. 6-fold, indicating that their overall effect was additive. Overexpression of E47 and β2 had no effect. In HeLa cells, PDX-1 stimulated the basal promoter by approx. 40-fold, whereas MafA, E47 and β2 each increased activity by less than 2-fold. There was no indication of any synergistic effects on the human insulin promoter. On the other hand, the rat insulin 1 promoter and a mutated version of the human insulin promoter, in which the relevant regulatory elements were separated by the same distances as in the rat insulin 1 promoter, did exhibit synergy. PDX-1 was shown further to activate the endogenous insulin 1 gene in αTC1.6 cells, whereas MafA activated the insulin 2 gene. In combination, PDX-1 and MafA activated both insulin genes. Chromatin immunoprecipitation assays confirmed that PDX-1 increased the association of acetylated histones H3 and H4 with the insulin 1 gene and MafA increased the association of acetylated histone H3 with the insulin 2 gene.
Keywords: diabetes mellitus, gene transcription, insulin gene, islets of Langerhans, PDX-1 (pancreatic duodenal homeobox factor-1)
Abbreviations: bHLH, basic helix–loop–helix; bZIP, basic leucine zipper; ChiP, chromatin immunoprecipitation; CRE, cAMP-responsive element; DMEM, Dulbecco's modified Eagle's minimum essential medium; FBS, fetal bovine serum; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HSV-1, herpes simplex virus-1; IAPP, islet amyloid polypeptide; PDX-1, pancreatic duodenal homeobox factor-1; RT, reverse transcriptase
INTRODUCTION
Expression of the insulin gene in adults is restricted to the β-cells of the islets of Langerhans [1]. This tissue specificity is conferred by cis-acting regulatory sequences located within 300–400 bp from the transcription start site, which bind β-cell-restricted and ubiquitous transcription factors. To date, the insulin promoters from the rat insulin 1 and 2 genes (there are two non-allelic insulin genes in the rat and mouse) and the human insulin gene have been characterized in detail. They all contain transcriptionally important E, A and C regulatory elements [2] as well as additional sequences that may have more subtle regulatory effects.
The E boxes bind proteins of the bHLH (basic helix–loop–helix) class of transcription factor. In β-cells, the ubiquitous bHLH protein E47 forms a heterodimer with NeuroD/BETA2 (β-cell E box transactivator 2; referred to in the remainder of the text as β2), a bHLH protein expressed in neuroendocrine cells [3] that also plays a role in islet cell development [4]. The rat insulin 1 promoter contains two E boxes at −104 (E1) and −228 (E2), whereas the rat insulin 2 and human insulin genes contain one E box at −91 and −103 respectively. Mutagenesis of the proximal E1 site in the rat insulin 1 and 2 genes completely abolished insulin promoter activity [5,6], whereas the relative importance of the equivalent E box site as well as a non-conserved E box at −225 in the human insulin gene is unclear [1].
There are up to five A box sequences in the insulin promoter. Their consensus TAAT sequence is a known binding site for homeodomain proteins [7], of which PDX-1 (pancreatic duodenal homeobox factor-1) is the most important [8–12]. PDX-1 is present in β-cells and some somatostatin-expressing islet cells as well as isolated endocrine cells of the intestine [13]. It interacts synergistically with E47/β2 [14–17] to activate the insulin promoter. PDX-1 also plays an important role in the development of the pancreas [18,19].
The C1 element of the rat insulin 2 gene binds a factor, RIPE3b1, which has recently been identified as the bZIP (basic leucine zipper) protein MafA [20–24]. MafA is a member of the large Maf family of transcription factors, which act as homodimers. Small Maf proteins lack a transactivation domain and activate transcription by forming heterodimers with other bZIP family members [22].
Previous studies using rat insulin 1 promoter constructs had shown that PDX-1, E47 and β2 could interact synergistically to stimulate promoter activity [16]. Given that there are a number of substantial differences in the regulation of the rodent and human insulin genes [1,25,26], the present studies were undertaken to establish the relative contribution of E47/β2, PDX-1 and MafA in regulating the human insulin promoter and to determine whether these factors interacted synergistically in the context of the human insulin promoter. The results show that both PDX-1 and MafA can activate the promoter and that their effects are additive. There was no evidence for any synergistic interactions between PDX-1, MafA, E47 or β2. PDX-1 had a powerful effect on promoter activity in non-islet cells, whereas MafA, E47 and β2 had weak effects. The results emphasize the difference in the regulation of the rodent and human insulin genes and may have important implications for inducing insulin expression in non-islet cells for the treatment of diabetes mellitus.
EXPERIMENTAL
Cell culture
The mouse islet glucagonoma αTC1.6 cell line [27] (American Type Culture Collection) was maintained in monolayer culture on plastic plates (Greiner) in DMEM (Dulbecco's modified Eagle's minimum essential medium) plus GlutaMAX™1, pyruvate and glucose (4.5 g/l) (Invitrogen) with the addition of 10% (v/v) FBS (fetal bovine serum; Invitrogen). Cells were cultivated at 37 °C in a humidified atmosphere of 5% CO2. HeLa cells were cultured in DMEM, glucose (4.5 g/l), 2 mM L-glutamine and 1 mM sodium pyruvate with addition of 10% (v/v) FBS. The rat insulinoma cell line INS-1 [28] was cultured in RPMI 1640 containing glucose (2.0 g/l) with 2 mM L-glutamine and supplemented with 10% (v/v) FBS, 1 mM sodium pyruvate, 10 mM Hepes balanced salt solution and 50 μM 2-mercaptoethanol. Cells were maintained at 37 °C without antibiotics in a humidified atmosphere containing 5% CO2.
Plasmids
Plasmids phINS260LUC and phINS171LUC contained fragments spanning the regions −260 to +148 and −171 to +148 of the human insulin gene [29] respectively. Mutagenesis of the C1 site within phINS171LUC to generate phINS171C1mLUC was achieved using the QuikChange Multi Site-Directed Mutagenesis kit (Stratagene) and the oligonucleotide 5′-CCGGAAATTGCAGCCTACTACCCCAGCCATCTGCCG-3′. The A1 site within phINS171LUC was mutated to generate phINS171A1mLUC using the QuikChange Site-Directed Mutagenesis kit (Stratagene) and the oligonucleotide 5′-ACCCCAGGCCCTCCTGGGCCAGGCG-3′ and its complementary reverse. The E1 site in phINS171LUC was mutated using the oligonucleotide 5′-CTCAGCCCCCAGCTCGGTGCCGACCCCCCCAC-3′ and its complementary reverse to create phINS171E1mLUC. The same technique was used to introduce extra bases into the human insulin promoter in phINS171LUC to create phINS171C+2EALUC and phINS171CE+2ALUC using the oligonucleotides 5′-AGCCTCAGCCCCTCTAGCCATCTGCCGAC-3′ and 5′-ATCTGCCGACCCACCCCCACCCCAGG-3′ respectively, and their reverse complements. The double mutant phINS171C+2E+2ALUC was created using both pairs of oligonucleotides.
Plasmid pcDNA3.1 was from Invitrogen, and the TK-Renilla Luciferase (phRL-TK) was from Promega. Plasmids pcDNA3-MafA, pcDNA3β2 and pJ3ΩE47 were from Dr Roland Stein (Vanderbilt University, Nashville, TN, U.S.A.). For the PDX-1/VP16 construct, the human PDX-1 cDNA was first mutated in pUC18 to generate a StuI recognition sequence (AGG/CCT). This was done in two stages, using the QuikChange Site-Directed Mutagenesis kit (Stratagene) and the primers 5′-CGCCTCGGCGGCCTCAGGAACCACG-3′ and 5′-CGTGGTTCCTGAGGCCGCCGAGGCG-3, which changed the sequence CGGCCG to CGGCCT. This sequence was then changed to AGGCCT using the primers 5′-GTCGCGCCTCGGAGGCCTCAGGAACC-3′, and 5′-GGTTCCTGAGGCCTCCGAGGCGCGAC-3′. A 384 bp (128 amino acid) fragment of the HSV-1 (herpes simplex virus-1) transcriptional activator VP16 was removed from the plasmid placI-4 with EcoRV and cloned into the newly created StuI site of PDX-1. The PDX-1/VP16 insert was then removed from pUC18 using EcoRI and cloned into the EcoRI site of pcDNA3.1.
Transient transfections
Cells on six-well plates at approx. 80% confluence were transiently transfected with plasmids using Lipofectamine Reagent (Invitrogen), according to the manufacturer's protocol, modified by substituting serum-free DMEM (Invitrogen) for Opti-MEM 1 Reduced Serum Medium. Transcription efficiency has been determined previously using an EGFP (enhanced green fluorescent protein) reporter plasmid to be 8−10% for INS-1, αTC1.6 and HeLa cell lines. Cells were analysed 48 h post-transfection.
Reporter assays
Firefly and Renilla luciferase activities were determined in extracts of transfected cells using the Dual-Luciferase Reporter Assay System (Promega).
Insulin assay
Insulin secretion by transfected cells was measured by RIA of conditioned medium for total insulin-like immunoreactivity using an anti-(pro)insulin antibody (#1011; Linco Research) [30].
ChIP (chromatin immunoprecipitation) assays
ChIP assays were performed using reagents from Upstate. Formaldehyde (1%, v/v) was added to the cells in 15 cm plates and incubated for 10 min at 37 °C. Glycine was added to a final concentration of 0.125 M, after which the medium was removed and the cells washed twice in PBS containing 1 mM PMSF, 1 μg/ml aprotinin and 1 μg/ml pepstatin A. The cells were collected and resuspended in SDS lysis buffer containing 1% (w/v) SDS, 10 mM EDTA, 50 mM Tris/HCl, pH 8.1, and protease inhibitors. The samples were incubated on ice for 10 min and sonicated using 20×10 s pulses using a microtip at power setting 1.0 (Misonix S3000). Following centrifugation at 13000 g for 10 min at 4 °C, the supernatant was divided into three tubes and made up to 2 ml with ChIP dilution buffer containing 0.01% SDS, 1.1% (v/v) Triton X-100, 1.2 mM EDTA, 167 mM NaCl, 16.7 mM Tris/HCl, pH 8.1, and protease inhibitors. Salmon sperm DNA/Protein A slurry (80 μl; Upstate) was added and the samples mixed for 30 min at 4 °C on a rotating wheel. Normal rabbit serum (4 μl), anti-acetyl H3 antibody (4 μl) and anti-acetyl H4 antibody was added to each tube and the samples incubated at 4 °C overnight with rotation. Salmon sperm/Protein A slurry (20 μl) was added and the samples rotated at 4 °C for 1 h. The agarose was pelleted by centrifugation at 4 °C for 1 min at 1000 g and the supernatant was discarded. The agarose pellet was washed once in low-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris/HCl, pH 8.1, and 150 mM NaCl), once in high-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris/HCl, pH 8.1, and 500 mM NaCl), and once in LiCl wash buffer [0.25 M LiCl, 1% Igepal-CA630, 1% deoxycholic acid (sodium salt), 1 mM EDTA and 10 mM Tris/HCl, pH 8.1]. The pellet was then washed twice in 10 mM Tris/HCl and 1 mM EDTA, pH 8.0. The histone complex was eluted from the agarose following incubation with rotation at room temperature for 15 min in 1% SDS and 0.1 M NaHCO3. The agarose was pelleted, NaCl (0.2 M) was added and the samples were heated at 65 °C for 4 h. EDTA (10 mM), Tris/HCl (40 mM, pH 6.5) and proteinase K (40 μg/ml) were added and samples incubated at 45 °C for 1 h. The DNA was then extracted in phenol/chloroform and precipitated in ethanol.
PCR conditions were: 1 cycle at 94 °C for 2 min, followed by 38 cycles at 94 °C for 30 s, 62 °C for 30 s, 72 °C for 30 s, and finally 1 cycle at 72 °C for 5 min. The primers used were: insulin 1, 5′-TCAGCCAAAGATGAAGAAGGTCTC-3′ and 5′-TCCAAACACTTGCCTGGTGC-3′ [2]; insulin 2, 5′-AGGGCCCCTTGTTAAGACTCTAA-3′ and 5′-ACTGGGTCCCCACTACCTTTAT-3′ [31]; and GAPDH (glyceraldehyde-3-phosphate dehydrogenase), 5′-AGTGCCAGCCTCGTCCCGTAGACAAAATG-3′ and 5′-AAGTGGGCCCCGGCCTTCTCCAT-3′ [32].
RT (reverse transcriptase)-PCR
Total RNA was isolated from transfected cells using Trizol (Invitrogen) and 1 μg aliquots were treated with 1 unit of amplification grade DNase 1 (Invitrogen). cDNA was prepared by reverse transcription using 1 μg of DNA-free total RNA, 5 ng of oligo(dT) and 200 units of Superscript II RT(Invitrogen). Of the total RT reaction volume (21 μl), 1 μl was amplified using the following PCR conditions: denaturation for 2 min at 94 °C, followed by 30–38 cycles of 30 s at 94 °C, 30 s at the appropriate annealing temperature and then 30 s at 72 °C. Primer sequences along with product size and annealing temperatures were: mouse insulin 1, 5′-CCATCAGCAAGCAGGTCAT-3′ and 5′-CACTTGTGGGTCCTCCACTT-3′ (218 bp and 63 °C); mouse insulin 2, 5′-CAGCAAGCAGGAAGCCTATC and 5′-TTGTGCCACTTGTGGGTCCT-3′ (235 bp and 63 °C); mouse glucagon, 5′-CCAGATCATTCCCAGCTTCA-3′ and 5′-TGGTGCTCTCATCTGTCAGAG-3′ (384 bp and 60 °C); mouse somatostatin, 5′-CCACCGGGAAACAGGAACTG-3′ and 5′-GGGCCAGGAGTTAAGGAAGA-3′ (303 bp and 63 °C); and mouse GAPDH, 5′-AAGGGCTCATGACCACAGTC-3′ and 5′-CTTACTCCTTGGAGGCCATGT-3′ (495 bp and 68 °C).
Western blotting
The anti-PDX-1 antibody was from Dr Chris Wright (Vanderbilt University, Nashville, TN, U.S.A.), anti-MafA was from Dr Roland Stein (Vanderbilt University, Nashville, TN, U.S.A.) and anti-E47 and β2 were from Santa Cruz Biotechnology. Nuclear extracts (10 μg of protein) were fractionated by SDS/PAGE on a 10% (w/v) acrylamide gel and blotted on to Hybond-ECL nitrocellulose membrane (Amersham Biosciences). The membrane was incubated overnight in a buffer containing 20 mM Tris/HCl (pH 7.4), 0.2 M NaCl, 0.1% (v/v) Tween 20, 5% (w/v) non-fat milk powder and anti-PDX-1 (1:2000), anti-MafA (1:1000), anti-E47 (1:200) or anti-β2 (1:200) antibodies. The antigen–antibody complex was detected by incubating the membrane for 60 min in buffer containing a 1:5000 dilution of horseradish- peroxidase-conjugated anti-rabbit IgG secondary antibody.
Statistical analysis
Statistical analysis was performed using SPSS statistical software package, and statistical significance was measured by ANOVA.
RESULTS
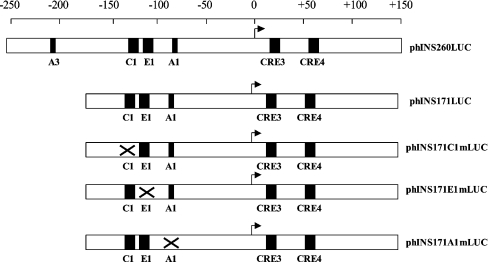
To investigate the role of MafA, PDX-1 and E47/β2 in regulating the human insulin gene, a reporter construct, phINS171LUC, was used (Figure 1). This construct contains: (i) a major PDX-1 binding site, the A1 box at −83 [8], and a recently identified weaker PDX-1 binding site (the GG2 box at −145) [26]; (ii) an E47/β2 binding site, the E1 box at −104; (iii) a MafA site, the C1 box at −128 [33]; and (iv) two CREs (cAMP-responsive elements) at +18 and +61 [34]. The CREs are important for basal promoter activity [35], and for this reason were included within the phINS171LUC construct.
Figure 1. Structure of the human insulin promoter constructs.
Schematic diagram showing the structure of the human insulin promoter constructs. The positions of the A1, A3, C1 and E1 sites are indicated along with the CREs (CRE3 and CRE4) that lie downstream of the transcription start site (arrow). The scale represents base pairs relative to the start site.
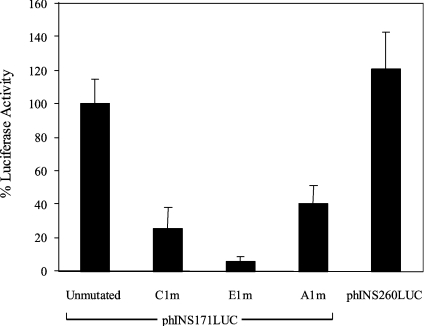
Initial experiments were carried out using the rat β-cell line INS-1, which we have found to respond to increased glucose levels by stimulating insulin transcription (results not shown). The phINS171LUC construct was found to be 22 times more active than the basal phRL-TK construct and was only slightly less active than phINS260LUC (Figures 1 and 2), a construct that has been shown previously to exhibit maximal insulin promoter activity [36]. Mutagenesis of the C1, E1 and A1 sites in phINS171LUC (Figure 1) reduced promoter activity by 75, 94 and 60% respectively (Figure 2). This indicates that all three sites are important for basal promoter activity, which is in keeping with a recent study [37]. The relatively weak effect of the A1m mutation may be due to the residual activity of the GG2 PDX-1 binding site [26].
Figure 2. Insulin promoter activity in INS-1 cells.
INS-1 cells were transfected with 250 ng of the indicated insulin promoter construct and 2.5 ng of phRL-TK with pcDNA3.1 added to a final total of 1 μg. After 48 h, the cells were harvested and assayed for firefly and Renilla luciferase activity. The data are expressed as percentage luciferase activity (firefly/Renilla) relative to cells transfected with unmutated phINS171LUC construct. Values are means±S.D. of between 3–9 separate experiments.
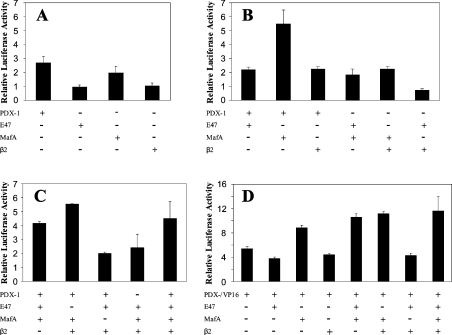
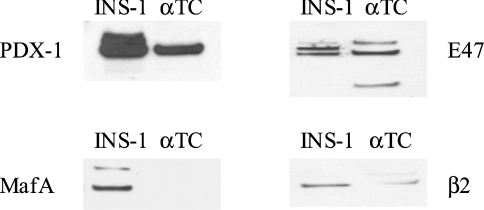
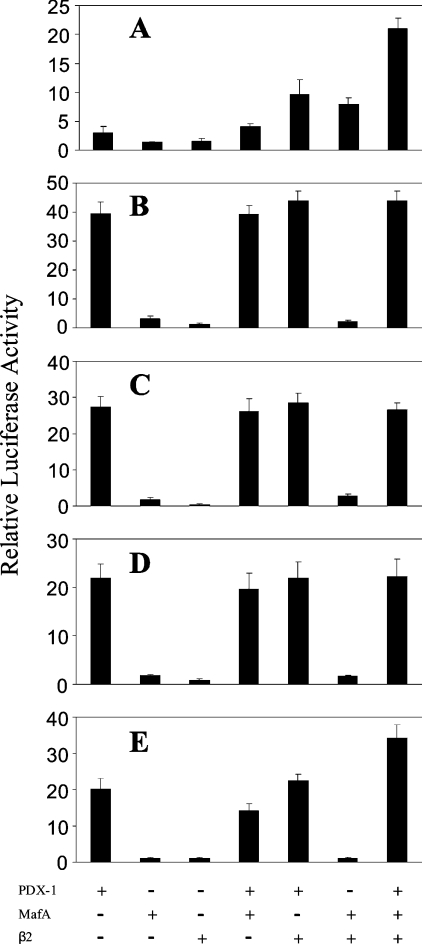
The activity of the reporter construct was also tested in the mouse glucagonoma cell line αTC1.6. The activity of the phINS171LUC construct was comparable with that in INS-1 cells with a firefly luciferase/Renilla luciferase ratio of 17. To determine whether PDX-1, MafA and E47/β2 could interact synergistically to activate the insulin promoter, plasmid phINS171LUC was transfected into αTC1.6 on its own or in combination with these factors. PDX-1 and MafA stimulated phINS171LUC activity 2.7- and 2-fold respectively, whereas β2 and E47 had no effect (Figure 3A). The effects of PDX-1, MafA, E47 and β2 were additive (Figures 3B and 3C). Statistical analysis revealed that, even at the 5% significance level, no combination of factors could interact synergistically to activate the phINS171LUC construct in αTC1.6. The inability of E47 to mediate an effect may be because αTC1.6 cells express relatively high concentrations of this factor as revealed by Western blotting (Figure 4). On the other hand, MafA was completely absent from αTC1.6 cells [22], whereas PDX-1 and β2 were expressed at low levels (<10%) compared with INS-1 cells.
Figure 3. Effect of PDX-1, PDX-1/VP-16, MafA, E47 and β2, individually and in combination, on phINS171LUC promoter activity in αTC1.6 cells.
αTC1.6 cells were transfected with 250 ng of phINS171LUC, 2.5 ng of phRL-TK and 250 ng of expression plasmids for the indicated transcription factors with pcDNA3.1 added to a final total of 1 μg. After 48 h, the cells were harvested and assayed for firefly and Renilla luciferase activity. (A) The effect of transcription factors on their own was assayed. (B) Transcription factors were assayed in pairs. (C) Multiple combinations of transcription factors were assayed. (D) PDX-1/VP-16 was used in place of PDX-1 in a series of experiments in combination with the indicated transcription factors. The data are expressed as luciferase activity (firefly/Renilla) relative to that measured in the absence of the indicated transcription factors. The data shown (means±S.D.) are representative of up to seven separate experiments.
Figure 4. Western blot showing the level of expression of exogenous gene products in transfected cells.
Nuclear extracts were prepared from INS-1 or αTC1.6 cells and Western blots were performed using antibodies against PDX-1, MafA, E47 and β2 as indicated.
Given the pre-eminent role of PDX-1 in stimulating the human insulin promoter, we were interested to learn whether PDX-1/VP16, a construct containing the powerful transactivation domain of the HSV-1 VP16 fused to the C-terminus of PDX-1, could overcome requirements for other transcription factors. PDX-1/VP16 was observed to stimulate phINS171LUC activity in αTC1.6 cells by 5.4-fold (Figure 3D). In combination with MafA, this was increased to 8.9-fold, whereas the further presence of E47 or β2 in combination elicited 10.6- and 11.3-fold stimulatory effects respectively, that were more than additive (P<0.05 in both cases).
We next investigated the stimulatory effect of PDX-1, MafA and E47/β2 on the human insulin promoter in HeLa cells, since it had been shown previously that E47 and β2 interacted synergistically with PDX-1 to activate the rat 1 insulin promoter in this cell line [16]. HeLa cells were transfected with either the pFOX410LUC or phINS171LUC reporter constructs that contained the rat insulin 1 or human insulin promoter respectively. The transcription factor E47 had no effect either on its own or in any combination with either promoter, so the study concentrated on PDX-1, MafA and β2. As shown in Figure 5(A), PDX-1 stimulated the rat insulin 1 promoter 3-fold, whereas MafA and β2 stimulated by less than 2-fold. Significant synergy was detected between PDX-1 and β2 (P<0.001), β2 and MafA (P<0.001) and all three transcription factors in combination (P<0.001), which resulted in stimulation of 21-fold. These results showed that our assay system could readily detect synergy. The human insulin promoter in phINS171LUC was weakly active in HeLa cells with a firefly/Renilla luciferase activity of 2. PDX-1 stimulated the human promoter to a much greater degree than the rodent form with a 40-fold increase (Figure 5B), whereas MafA and β2 exhibited a relatively weak, less than 3-fold, stimulatory effect. In combination, the effects of PDX-1, MafA and β2 were additive (Figure 5B) and statistical analysis could uncover no evidence that MafA or β2 significantly potentiated the effect of PDX-1 on the promoter. Also, unlike the rat insulin 1 promoter, no synergy was detected between MafA and β2 in the human promoter (Figures 5A and 5B).
Figure 5. Effect of PDX-1, MafA and β2, individually and in combination, on rat insulin 1 and human insulin promoter activity in HeLa cells.
HeLa cells were transfected with 250 ng of reporter plasmid indicated below, 0.25 ng of phRL-TK and 250 ng of expression plasmids for the indicated transcription factors with pcDNA3.1 added to a final total of 1 μg. The plasmids used were (A) pFOX410LUC containing the rat insulin 1 promoter, (B) phINS171LUC containing the human insulin promoter, (C) phINS171C+2EALUC, (D) phINS171CE+2ALUC, and (E) phINS171C+2E+2ALUC. After 48 h, the cells were harvested and assayed for firefly and Renilla luciferase activity. The data are expressed as luciferase activity (firefly/Renilla) relative to that measured in the absence of the indicated transcription factors for each reporter plasmid used. The data shown (means±S.D.) are representative of three separate experiments performed in triplicate.
Given the major dissimilarities in stimulation observed between the rat insulin 1 and human promoters, a comparison of their sequences in the region of the C1, E1 and A1 sites was carried out (Figure 6A). Although the consensus sequences are highly conserved, there is divergence in the separation of these regulatory elements from each other. The human insulin promoter lacks 2 bp in both of the intervening regions, which would in turn lead to alterations in the rotational alignment of transcription factors along the DNA double helix (Figure 6B), radically altering the potential for protein–protein interactions. It can be argued that using single base pairs at the centres of regulatory consensus sequences to denote transcription factor binding could be unrepresentative due to variations in the size of footprints and in the volume of the assembled proteins, e.g. PDX-1 monomer and E47/β2 heterodimer; however, the schematic diagrams do clearly show the major consequences of apparently minor spacing differences on transcription factor spatial arrangements. The possible bearing of these modifications was explored using a series of mutated versions of phINS171LUC (Figure 6), which contained the human promoter with additional bases to mimic the rat insulin 1 promoter. PDX-1 was the dominant transcription factor in all three mutant plasmids Figures 5C, 5D and 5E) in keeping with the human promoter; however, the degree of stimulation was halved in the rat-like double mutant phINS171C+2E+2ALUC. Importantly, this construct displayed synergistic stimulation with a combination of PDX-1, MafA and β2 (P<0.01).
Figure 6. Comparison of the rat insulin 1 and human insulin promoters.
(A) Alignment of the DNA sequences of the relevant region of the rat insulin 1 promoter (−133 to −74) and the human insulin promoter (−130 to −75). The MafA, β2 and PDX-1 consensus sequences are underlined and bases inserted into the mutated forms of phINS171LUC are shown in bold. (B) Schematic diagram showing the effect of spacing between the regulatory elements on potential transcription factor alignments. The insulin promoter is depicted looking down the DNA helical axis and is represented as a circle. The transcription factors are denoted as differently shaded ovals and are displayed binding to the centres of their respective regulatory elements at the bottom of (A). Note that the human insulin promoter contains an additional PDX-1 binding sequence (GG2) starting 9 bp upstream of the region shown in (A). The potential differences in spatial orientation of MafA, β2 and PDX-1 relative to each other within the various promoter constructs are portrayed by showing the transcription factors coming in contact with the DNA at the centre of their respective consensus binding sequences. Each schematic diagram illustrates the effects of regulatory element spacing in the promoter to the immediate left in (A).
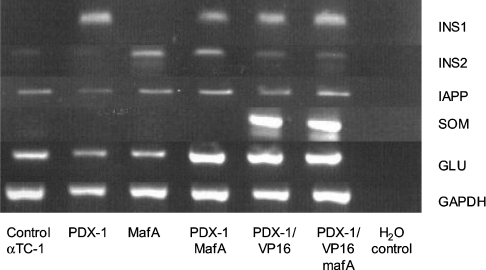
Collectively, the results indicated that PDX-1 was a powerful regulator of the human insulin promoter in non-islet cells and that MafA and E47/β2 were primarily involved in basal promoter activity with a modest effect on promoter activity in non-islet cells. It was important to determine what effect, if any, PDX-1 and MafA might have on the endogenous insulin gene in non-expressing cells. In initial experiments, there was no detectable insulin mRNA in HeLa cells transiently transfected with PDX-1 and MafA either individually or in combination (results not shown). In the absence of a suitable human islet cell line, we therefore studied the effects of overexpressing PDX-1 and MafA in αTC1.6 cells. These cells express a small amount of mouse insulin 2 gene, but the insulin 1 gene is completely suppressed (Figure 7). From Figure 7 it can also be seen that, in keeping with previous findings [33], expression of MafA switched on the rat insulin 2 gene with no effect on the rat insulin 1 gene. In contrast, PDX-1 activated the insulin 1 gene with little effect on the insulin 2 gene. In combination, MafA and PDX-1 activated both insulin genes. Interestingly, the PDX-1/VP16 construct activated the insulin 1 gene and, to a lesser degree, the insulin 2 gene as well as the somatostatin gene [38]. The somatostatin gene was unaffected by PDX-1, MafA or combinations thereof. Cells transfected with PDX-1/VP16 plus MafA were found to secrete detectable amounts of insulin; 32±18 pg/ml (n=3) of insulin-like immunoreactivity was found to have accumulated in conditioned medium 48 h post-transfection, whereas no insulin could be measured in media from untransfected cells.
Figure 7. Effect of PDX-1 and MafA on endogenous gene expression in αTC1.6 cells.
αTC1.6 cells were transfected with 250 ng of expression plasmids for PDX-1, MafA and PDX-1/VP16 as indicated, with pcDNA3.1 added to a final total of 2 μg. After 48 h, the cells were harvested and assayed by RT-PCR for mRNA encoding insulin 1 (INS1), insulin 2 (INS2), IAPP, somatostatin (SOM), glucagon (GLU) and GAPDH, as indicated. The data are representative of three separate experiments.
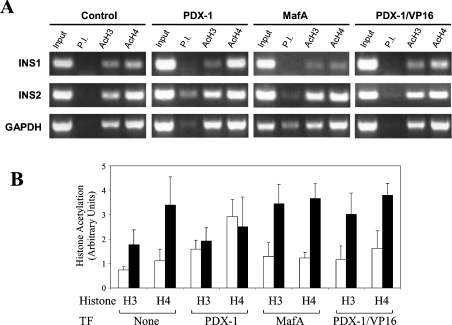
These results were validated further by ChiP assays that examined the effects of the different transcription factors on the recruitment of histone acetylation (Figure 8). In untransfected αTC1.6 cells, the insulin 2 promoter contained acetylated histone H4 and to a lesser extent acetylated histone H3. This is in keeping with our findings that the insulin 2 gene is expressed at low levels in αTC1.6 cells and with previous studies showing that the mouse insulin genes in β-cells, but not in non-β-cells, are associated with hyperacetylated histones H3 and H4 [39]. The insulin 1 promoter exhibited minor association of acetylated histones H3 and H4. Transfection with PDX-1 increased the degree of acetylation of histones H3 and H4 in the insulin 1 promoter (P<0.05 in both instances) with no increase in histone acetylation in the insulin 2 promoter. MafA had no effect on the acetylation of histones H3 and H4 in the insulin 1 promoter, but increased the amount of acetylated histone H3 in the insulin 2 promoter (P<0.05). Use of antibodies against MafA, E47 and PDX-1 in further ChIP assays showed that, in keeping with its pre-eminent role in stimulation, PDX-1 is permanently bound to the mouse insulin promoters, whereas E47 and MafA bind transiently (results not shown). Although expression of PDX1/VP16 activated both insulin 1 and 2 genes (Figure 7), only the insulin 2 promoter showed a gain in histone acetylation, namely H3 (P<0.05).
Figure 8. ChiP of the insulin 1 and 2 genes in αTC1.6 cells transfected with PDX-1 or MafA.
αTC1.6 cells transfected with expression vectors for transcription factors (TF) PDX-1, MafA, PDX-1/VP16 or empty vector (control) were subjected to ChiP assays using anti-acetylated H3 (AcH3) or anti-acetylated H4 (AcH4) antibodies and PCR primers specific for insulin 1 (INS1), insulin 2 (INS2) and GAPDH. (A) PCR results representative of four separate experiments. (B) Quantification of the PCR products with the amounts of DNA in each band calculated relative to the pre-immune (P.I.) signal. The data represent the means±S.D. of the four separate experiments. Insulin 1, open bars; insulin 2, closed bars.
DISCUSSION
The results of the present study show that (i) PDX-1, MafA and E47/β2 acting through the promoter proximal A1, C1 and E1 sites play an important role in maintaining basal promoter activity of the human insulin gene; (ii) the effects of PDX-1, MafA and E47/β2 are additive in αTC1.6 cells with no evidence for synergistic interactions on the human promoter; (iii) PDX-1 has the strongest stimulatory effect on the promoter in non-islet cells, especially in the human form, whereas MafA, E47 and β2 have very weak effects; and (iv) mutating the human insulin promoter to more closely resemble the rat insulin 1 promoter generates synergy between the various factors in HeLa cells. Together, these results emphasize that the regulation of the human insulin promoter is substantially different from the well-characterized rodent insulin promoters.
Of the three binding sites examined, the E1 site in the human insulin promoter was shown to be particularly important, since mutagenesis almost completely abolished promoter activity. Mutagenesis of the C1 and A1 sites partially inhibited promoter activity, probably due to the presence of additional weak MafA and PDX-1 binding sites within the proximal region of the promoter [21,26]. Our finding that the C1 MafA binding site plays an important role in the basal activity of the human insulin promoter is in keeping with previous studies in transfected αTC1.6 [33] and MIN6 [37] cells. In non-islet cells, PDX-1 had a powerful effect on the weakly active promoter, whereas MafA, E47 and β2 individually or in combinations had marginal effects. This suggests that PDX-1 may be less dependent than MafA and E47/β2 on additional factors present in the β-cell.
PDX-1 has been shown previously to interact synergistically with E47 and β2 to activate the rat insulin 1 gene [14,16,17,40]. In the present study, we observed synergistic interactions between PDX-1 and β2 in the rat insulin 1 promoter; however, there was no evidence that any of the factors acted synergistically with PDX-1 to activate the human insulin promoter. The results also show that expression of MafA in αTC1.6 stimulates the human insulin promoter approx. 2-fold and that co-expression with PDX-1 and β2 either on their own or together results in a stimulatory effect that is the sum of the individual effects of the factors. Thus, although MafA has been shown to act synergistically with PAX6 to activate the glucagon promoter [41], in the context of the proximal region of the human insulin promoter there was no evidence for any synergistic interactions between MafA and other major regulatory factors such as PDX-1, E47 and β2. Interestingly, synergistic interactions between MafA and β2 were recorded in the rat insulin 1 promoter, providing another example of differences in the operation of the human and rodent insulin genes.
Collectively these findings emphasize, as pointed out previously [1,25,26], that although there are similarities in the arrangement of cis-acting elements in the rodent and human insulin genes, there are major differences in their regulation. Recent studies have shown that promoters may well accumulate mutations at greater rates than coding sequences [42], and comparison of over 50 genes in mammals has revealed that approximately one-third of regulatory binding sites in humans are probably not functional in rodents [43]. Minor base changes within regulatory elements can greatly alter the efficiency and outcome of transcription factor binding and this has important implications when attempting to extrapolate data using promoters from different species. For example, binding of MafA at the C1 site in the rat 1, rat 2 and human insulin promoters elicits quite different responses due to specific single nucleotide substitutions [22,23]. Regions outwith the more highly conserved consensus sequences can also have a bearing on transcription factor binding, as exemplified in the C1 site of the rat insulin 2 promoter, where upstream regions have been shown to be critical for the binding of MafA [44].
Comparison of the rat 1 and human insulin promoters reveals not only extensive nucleotide substitutions, but also subtle and significant 2 bp disparities in the spacing along the DNA of the C1, E1 and A1 regulatory elements. These small differences affect the relative distances of the regulatory elements along the helical axis and, more importantly, the rotational alignment of bound transcription factors. Insertion of base pairs into the human insulin promoter to replicate the relative spacing of the C1, E1 and A1 regulatory elements in the rat insulin 1 promoter resulted in synergistic interactions between MafA, β2 and PDX-1 in a manner similar to the rat insulin 1 promoter. A further example of the effect of promoter arrangement on insulin gene regulation is provided by the recent findings that, within the comparable region of the rat insulin 2 promoter, MafA (binding to the C1 site) can interact synergistically with β2 and PDX-1 at the E1 and A1 sites respectively [45]. Our experiments using the rat insulin 1 promoter, which has the same spacing between the C1 and E1 sites as the rat insulin 2 promoter, also showed synergy between MafA and β2. Significantly, the human and rat insulin 1 promoters, which contain additions of 8 and 12 bp respectively, between the C1 and A1 sites compared with the rat insulin 2 promoter, did not display synergy between MafA and PDX-1. When multiple binding sites are less than 35 bp apart, the DNA-turn is the most important factor for permitting co-operative protein–protein interactions. This has been demonstrated in experiments employing the rat tryptophan oxidase gene where the CACCC box and glucocorticoid receptor elements were separated by various lengths of DNA in a series of reporter plasmids. Stimulation of the reporter plasmids showed a 10 bp periodicity and changes of only 2 or 3 bp could drastically reduce the degree of co-operativity [46]. DNA-turn-dependent reductions in rat prolactin promoter activity have also been reported for the two proximal elements [47]. In addition, the region of spacing divergence between the human and rat insulin 1 promoters begins only 2 bp upstream of the 6 bp E1 consensus sequence and would, therefore, be within the binding footprint of E47/β2. These sequence differences could affect both the binding of proteins and their ability to form synergistic interactions with other transcription factors. This may also explain why β2 plays a more important role in stimulating rat 1 [16] than the human insulin promoter.
The necessity of cell- or species-specific cofactors for co-operative binding between MafA, E47/β2 and PDX-1 cannot be absolutely ruled out; however, it is an unlikely explanation for the lack of synergy in the human insulin promoter as no cofactor requirements have ever been reported for these transcription factors in β-cells. Also, the experiments were carried out using three different cell types from three different species. Possible saturating levels of the expressed transcription factors could theoretically mask synergy; however, this is unlikely as PDX-1/VP16, which stimulated the human insulin promoter on its own to a greater degree than the wild-type protein, was found to exhibit synergistic interactions, whereas the wild-type factor did not.
It has been shown previously that exogenous PDX-1 can induce expression of IAPP (islet amyloid polypeptide) in αTC1 cells [48]. We extend this to show that PDX-1 activates the mouse insulin 1 but not the insulin 2 gene. We show further that PDX-1/VP16 can activate both insulin genes. A similar PDX-1/VP16 construct has been shown to induce liver cells to transdifferentiate into insulin-secreting β-like cells in Xenopus embryos [49] and has been proposed as a route for long-term treatment of diabetes. However, our observation that PDX-1/VP16, but not PDX-1, strongly activated the somatostatin gene suggests that the PDX-1/VP16 construct may have limited applications in gene therapy of diabetes mellitus.
The results of the present study also confirm that MafA can activate the endogenous insulin 2, but not the insulin 1, gene in αTC cells [33], and show that both insulin genes can be activated in the presence of MafA and PDX-1, which specifically activates the insulin 1 gene. We show further that this differential effect may be due to the ability of PDX-1 and MafA to recruit histone-modifying enzymes, resulting in acetylated forms of both histones H3 and H4, and histone H3 alone in the mouse insulin 1 and 2 gene promoters respectively.
Acknowledgments
This work was supported by the Wellcome Trust.
References
- 1.Melloul D., Marshak S., Cerasi E. Regulation of insulin gene transcription. Diabetologia. 2002;45:309–326. doi: 10.1007/s00125-001-0728-y. [DOI] [PubMed] [Google Scholar]
- 2.German M., Ashcroft S., Docherty K., Edlund H., Edlund T., Goodison S., Imura H., Kennedy G., Madsen O., Melloul D. The insulin gene promoter. A simplified nomenclature. Diabetes. 1995;44:1002–1004. doi: 10.2337/diab.44.8.1002. [DOI] [PubMed] [Google Scholar]
- 3.Naya F. J., Stellrecht C. M., Tsai M. J. Tissue-specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev. 1995;9:1009–1019. doi: 10.1101/gad.9.8.1009. [DOI] [PubMed] [Google Scholar]
- 4.Naya F. J., Huang H. P., Qiu Y., Mutoh H., DeMayo F. J., Leiter A. B., Tsai M. J. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes Dev. 1997;11:2323–2334. doi: 10.1101/gad.11.18.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karlsson O., Edlund T., Moss J. B., Rutter W. J., Walker M. D. A mutational analysis of the insulin gene transcription control region: expression in β cells is dependent on two related sequences within the enhancer. Proc. Natl. Acad. Sci. U.S.A. 1987;84:8819–8823. doi: 10.1073/pnas.84.24.8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whelan J., Poon D., Weil P. A., Stein R. Pancreatic β-cell-type-specific expression of the rat insulin II gene is controlled by positive and negative cellular transcriptional elements. Mol. Cell. Biol. 1989;9:3253–3259. doi: 10.1128/mcb.9.8.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rudnik A., Ling T. Y., Odagiri H., Rutter W. J., German M. S. Pancreatic β cells express a diverse set of homeobox genes. Proc. Natl. Acad. Sci. U.S.A. 1994;91:12203–12207. doi: 10.1073/pnas.91.25.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boam D. S., Docherty K. A tissue-specific nuclear factor binds to multiple sites in the human insulin-gene enhancer. Biochem. J. 1989;264:233–239. doi: 10.1042/bj2640233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohlsson H., Karlsson K., Edlund T. IPF1, a homeodomain-containing transactivator of the insulin gene. EMBO J. 1993;12:4251–4259. doi: 10.1002/j.1460-2075.1993.tb06109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller C. P., McGehee R. E., Jr, Habener J. F. IDX-1: a new homeodomain transcription factor expressed in rat pancreatic islets and duodenum that transactivates the somatostatin gene. EMBO J. 1994;13:1145–1156. doi: 10.1002/j.1460-2075.1994.tb06363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leonard J., Peers B., Johnson T., Ferreri K., Lee S., Montminy M. R. Characterization of somatostatin transactivating factor-1, a novel homeobox factor that stimulates somatostatin expression in pancreatic islet cells. Mol. Endocrinol. 1993;7:1275–1283. doi: 10.1210/mend.7.10.7505393. [DOI] [PubMed] [Google Scholar]
- 12.Marshak S., Totary H., Cerasi E., Melloul D. Purification of the β-cell glucose-sensitive factor that transactivates the insulin gene differentially in normal and transformed islet cells. Proc. Natl. Acad. Sci. U.S.A. 1996;93:15057–15062. doi: 10.1073/pnas.93.26.15057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKinnon C. M., Docherty K. Pancreatic duodenal homeobox-1, PDX-1, a major regulator of β cell function and identity. Diabetologia. 2001;44:1203–1214. doi: 10.1007/s001250100628. [DOI] [PubMed] [Google Scholar]
- 14.Peers B., Leonard J., Sharma S., Teitelman G., Montminy M. R. Insulin expression in pancreatic islet cells relies on cooperative interactions between the helix loop helix factor E47 and the homeobox factor STF-1. Mol. Endocrinol. 1994;8:1798–1806. doi: 10.1210/mend.8.12.7708065. [DOI] [PubMed] [Google Scholar]
- 15.Peshavaria M., Henderson E., Sharma A., Wright C. V., Stein R. Functional characterization of the transactivation properties of the PDX-1 homeodomain protein. Mol. Cell. Biol. 1997;17:3987–3996. doi: 10.1128/mcb.17.7.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glick E., Leshkowitz D., Walker M. D. Transcription factor BETA2 acts cooperatively with E2A and PDX1 to activate the insulin gene promoter. J. Biol. Chem. 2000;275:2199–2204. doi: 10.1074/jbc.275.3.2199. [DOI] [PubMed] [Google Scholar]
- 17.Ohneda K., Mirmira R. G., Wang J., Johnson J. D., German M. S. The homeodomain of PDX-1 mediates multiple protein–protein interactions in the formation of a transcriptional activation complex on the insulin promoter. Mol. Cell. Biol. 2000;20:900–911. doi: 10.1128/mcb.20.3.900-911.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jonsson J., Carlsson L., Edlund T., Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature (London) 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 19.Offield M. F., Jetton T. L., Labosky P. A., Ray M., Stein R. W., Magnuson M. A., Hogan B. L., Wright C. V. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- 20.Olbrot M., Rud J., Moss L. G., Sharma A. Identification of β-cell-specific insulin gene transcription factor RIPE3b1 as mammalian MafA. Proc. Natl. Acad. Sci. U.S.A. 2002;99:6737–6742. doi: 10.1073/pnas.102168499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuoka T. A., Zhao L., Artner I., Jarrett H. W., Friedman D., Means A., Stein R. Members of the large Maf transcription family regulate insulin gene transcription in islet β-cells. Mol. Cell. Biol. 2003;23:6049–6062. doi: 10.1128/MCB.23.17.6049-6062.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kataoka K., Han S. I., Shioda S., Hirai M., Nishizawa M., Handa H. MafA is a glucose-regulated and pancreatic β-cell-specific transcriptional activator for the insulin gene. J. Biol. Chem. 2002;277:49903–49910. doi: 10.1074/jbc.M206796200. [DOI] [PubMed] [Google Scholar]
- 23.Kajihara M., Sone H., Amemiya M., Katoh Y., Isogai M., Shimano H., Yamada N., Takahashi S. Mouse MafA, homologue of zebrafish somite Maf 1, contributes to the specific transcriptional activity through the insulin promoter. Biochem. Biophys. Res. Commun. 2003;312:831–842. doi: 10.1016/j.bbrc.2003.10.196. [DOI] [PubMed] [Google Scholar]
- 24.Kataoka K., Shioda S., Ando K., Sakagami K., Handa H., Yasuda K. Differentially expressed Maf family transcription factors, c-Maf and MafA, activate glucagon and insulin gene expression in pancreatic islet α- and β-cells. J. Mol. Endocrinol. 2004;32:9–20. doi: 10.1677/jme.0.0320009. [DOI] [PubMed] [Google Scholar]
- 25.Clark A. R., Docherty K. The insulin gene. In: Ashcroft F. M., Ashcroft S. J., editors. Insulin: Molecular Biology to Pathology. Oxford: IRL Press; 1992. pp. 37–63. [Google Scholar]
- 26.Le Lay J., Matsuoka T. A., Henderson E., Stein R. Identification of a novel PDX-1 binding site in the human insulin gene enhancer. J. Biol. Chem. 2004;279:22228–22235. doi: 10.1074/jbc.M312673200. [DOI] [PubMed] [Google Scholar]
- 27.Powers A. C., Efrat S., Mojsov S., Spector D., Habener J. F., Hanahan D. Proglucagon processing similar to normal islets in pancreatic α-like cell line derived from transgenic mouse tumor. Diabetes. 1990;39:406–414. doi: 10.2337/diab.39.4.406. [DOI] [PubMed] [Google Scholar]
- 28.Asfari M., Janjic D., Meda P., Li G., Halban P. A., Wollheim C. B. Establishment of 2-mercaptoethanol-dependent differentiated insulin-secreting cell lines. Endocrinology. 1992;130:167–178. doi: 10.1210/endo.130.1.1370150. [DOI] [PubMed] [Google Scholar]
- 29.Bell G. I., Pictet R. L., Rutter W. J., Cordell B., Tischer E., Goodman H. M. Sequence of the human insulin gene. Nature (London) 1980;284:26–32. doi: 10.1038/284026a0. [DOI] [PubMed] [Google Scholar]
- 30.Shaw J. A., Delday M. I., Hart A. W., Docherty H. M., Maltin C. A., Docherty K. Secretion of bioactive human insulin following plasmid-mediated gene transfer to non-neuroendocrine cell lines, primary cultures and rat skeletal muscle in vivo. J. Endocrinol. 2002;172:653–672. doi: 10.1677/joe.0.1720653. [DOI] [PubMed] [Google Scholar]
- 31.Parrizas M., Maestro M. A., Boj S. F., Paniagua A., Casamitjana R., Gomis R., Rivera F., Ferrer J. Hepatic nuclear factor 1-α directs nucleosomal hyperacetylation to its tissue-specific transcriptional targets. Mol. Cell Biol. 2001;21:3234–3243. doi: 10.1128/MCB.21.9.3234-3243.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Annicotte J. S., Fayard E., Swift G. H., Selander L., Edlund H., Tanaka T., Kodama T., Schoonjans K., Auwerx J. Pancreatic-duodenal homeobox 1 regulates expression of liver receptor homolog 1 during pancreas development. Mol. Cell Biol. 2003;23:6713–6724. doi: 10.1128/MCB.23.19.6713-6724.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuoka T. A., Artner I., Henderson E., Means A., Sander M., Stein R. The MafA transcription factor appears to be responsible for tissue-specific expression of insulin. Proc. Natl. Acad. Sci. U.S.A. 2004;101:2930–2933. doi: 10.1073/pnas.0306233101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inagaki N., Maekawa T., Sudo T., Ishii S., Seino Y., Imura H. c-Jun represses the human insulin promoter activity that depends on multiple cAMP response elements. Proc. Natl. Acad. Sci. U.S.A. 1992;89:1045–1049. doi: 10.1073/pnas.89.3.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Inada A., Someya Y., Yamada Y., Ihara Y., Kubota A., Ban N., Watanabe R., Tsuda K., Seino Y. The cyclic AMP response element modulator family regulates the insulin gene transcription by interacting with transcription factor IID. J. Biol. Chem. 1999;274:21095–21103. doi: 10.1074/jbc.274.30.21095. [DOI] [PubMed] [Google Scholar]
- 36.Boam D. S., Clark A. R., Docherty K. Positive and negative regulation of the human insulin gene by multiple trans-acting factors. J. Biol. Chem. 1990;265:8285–8296. [PubMed] [Google Scholar]
- 37.da Silva G., Rutter J., Rutter G. A. Involvement of Per-Arnt-Sim (PAS) kinase in the stimulation of preproinsulin and pancreatic duodenum homeobox 1 gene expression by glucose. Proc. Natl. Acad. Sci. U.S.A. 2004;101:8319–8324. doi: 10.1073/pnas.0307737101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.German M. S., Moss L. G., Wang J., Rutter W. J. The insulin and islet amyloid polypeptide genes contain similar cell-specific promoter elements that bind identical β-cell nuclear complexes. Mol. Cell. Biol. 1992;12:1777–1788. doi: 10.1128/mcb.12.4.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chakrabarti S. K., Francis J., Ziesmann S. M., Garmey J. C., Mirmira R. G. Covalent histone modifications underlie the developmental regulation of insulin gene transcription in pancreatic β cells. J. Biol. Chem. 2003;278:23617–23623. doi: 10.1074/jbc.M303423200. [DOI] [PubMed] [Google Scholar]
- 40.Qiu Y., Guo M., Huang S., Stein R. Insulin gene transcription is mediated by interactions between the p300 coactivator and PDX-1, BETA2, and E47. Mol. Cell. Biol. 2002;22:412–420. doi: 10.1128/MCB.22.2.412-420.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Planque N., Leconte L., Coquelle F. M., Benkhelifa S., Martin P., Felder-Schmittbuhl M. P., Saule S. Interaction of Maf transcription factors with Pax-6 results in synergistic activation of the glucagon promoter. J. Biol. Chem. 2001;276:35751–35760. doi: 10.1074/jbc.M104523200. [DOI] [PubMed] [Google Scholar]
- 42.Wray G. A., Hahn M. W., Abouheif E., Balhoff J. P., Pizer M., Rockman M. V., Romano L. A. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- 43.Dermitzakis E. T., Clark A. G. Evolution of transcription factor binding sites in Mammalian gene regulatory regions: conservation and turnover. Mol. Biol. Evol. 2002;19:1114–1121. doi: 10.1093/oxfordjournals.molbev.a004169. [DOI] [PubMed] [Google Scholar]
- 44.Harrington R. H., Sharma A. Transcription factors recognizing overlapping C1-A2 binding sites positively regulate insulin gene expression. J. Biol. Chem. 2001;276:104–113. doi: 10.1074/jbc.M008415200. [DOI] [PubMed] [Google Scholar]
- 45.Zhao L., Guo M., Matsuoka T. A., Hagman D. K., Parazzoli S. D., Poitout V., Stein R. The islet β-cell-enriched MafA activator is a key regulator of insulin gene transcription. J. Biol. Chem. 2005;280:11887–11894. doi: 10.1074/jbc.M409475200. [DOI] [PubMed] [Google Scholar]
- 46.Schule R., Muller M., Otsuka-Murakami H., Renkawitz R. Cooperativity of the glucocorticoid receptor and the CACCC-box binding factor. Nature (London) 1988;332:87–90. doi: 10.1038/332087a0. [DOI] [PubMed] [Google Scholar]
- 47.Harvey C., Jackson S. M., Siddiqui S. K., Gutierrez-Hartmann A. Structure-function analysis of the rat prolactin promoter: phasing requirements of proximal cell-specific elements. Mol. Endocrinol. 1991;5:836–843. doi: 10.1210/mend-5-6-836. [DOI] [PubMed] [Google Scholar]
- 48.Serup P., Jensen J., Andersen F. G., Jorgensen M. C., Blume N., Holst J. J., Madsen O. D. Induction of insulin and islet amyloid polypeptide production in pancreatic islet glucagonoma cells by insulin promoter factor 1. Proc. Natl. Acad. Sci. U.S.A. 1996;93:9015–9020. doi: 10.1073/pnas.93.17.9015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Horb M. E., Shen C. N., Tosh D., Slack J. M. Experimental conversion of liver to pancreas. Curr. Biol. 2003;13:105–115. doi: 10.1016/s0960-9822(02)01434-3. [DOI] [PubMed] [Google Scholar]