Abstract
The pancreatic islet hormone glucagon stimulates hepatic glucose production and thus maintains blood glucose levels in the fasting state. Transcription factors of the Foxa [Fox (forkhead box) subclass A; also known as HNF-3 (hepatocyte nuclear factor-3)] family are required for cell-specific activation of the glucagon gene in pancreatic islet α-cells. However, their action on the glucagon gene is poorly understood. In the present study, comparative sequence analysis and molecular characterization using protein–DNA binding and transient transfection assays revealed that the well-characterized Foxa-binding site in the G2 enhancer element of the rat glucagon gene is not conserved in humans and that the human G2 sequence lacks basal enhancer activity. A novel Foxa site was identified that is conserved in rats, mice and humans. It mediates activation of the glucagon gene by Foxa proteins and confers cell-specific promoter activity in glucagon-producing pancreatic islet α-cell lines. In contrast with previously identified Foxa-binding sites in the glucagon promoter, which bind nuclear Foxa2, the novel Foxa site was found to bind preferentially Foxa1 in nuclear extracts of a glucagon-producing pancreatic islet α-cell line, offering a mechanism that explains the decrease in glucagon gene expression in Foxa1-deficient mice. This site is located just upstream of the TATA box (between −30 and −50), suggesting a role for Foxa proteins in addition to direct transcriptional activation, such as a role in opening the chromatin at the start site of transcription of the glucagon gene.
Keywords: Fox (forkhead box) subclass A protein 1 (Foxa1), G2, glucagon, hepatocyte nuclear factor (HNF), in silico, transcriptional regulation
Abbreviations: CRE, cAMP-response element; CREB, CRE-binding protein; DTT, dithiothreitol; EMSA, electrophoretic mobility-shift assay; Foxa, Fox (forkhead box) subclass A; GFP, green fluorescent protein; GST, glutathione S-transferase; HNF, hepatocyte nuclear factor; NFAT, nuclear factor of activated T-cells; PD, paired domain; TTR, transthyretin
INTRODUCTION
The peptide hormone glucagon raises blood glucose levels by acting on the liver to stimulate hepatic glycogenolysis and gluconeogenesis [1–3]. It thus serves as a functional antagonist of insulin and is of physiological importance in the maintenance of fasting blood glucose levels. Aberrant regulation of glucagon gene expression and relative hyperglucagonaemia in the background of insulin deficiency contribute significantly to the development of hyperglycaemia in diabetes mellitus [1–3]. Glucagon is synthesized in the α-cells of the pancreatic islets [1–3]. Pancreatic α-cell-specific activation of the glucagon gene is conferred by the 5′-flanking region of the glucagon gene [4–6]. Several cis elements, termed G1–G5 and CRE (cAMP-response element), have been identified within 300 bp 5′ of the transcription start site of the rat gene. This promoter fragment can efficiently support the expression of a reporter gene in islet α-cell lines [4–6]. Many transcription factors of distinct families have been shown to bind within the G1–G5 and CREs [6]. None of these transcription factors is, however, specific for pancreatic islet α-cells. These results thus support a model in which the distinct spatial arrangement of multiple binding sites and their combinatorial interaction with transcription factors confer pancreatic islet α-cell-specific activation of the glucagon gene in vivo. Of the transcription factors known to bind to the glucagon gene, the Foxa [Fox (forkhead box) subclass A] or HNF-3 (hepatocyte nuclear factor-3) proteins Foxa1, Foxa2 and Foxa3 (also called HNF-3α, HNF-3β and HNF-3γ respectively) seem to play an essential role [6–9].
The Foxa proteins belong to an evolutionarily conserved family of transcription factors [9]. They share a highly conserved wingedhelix motif comprising 100 amino acids responsible for monomeric recognition of specific DNA target sites with the consensus sequence VA(A/T)TRTT(G/T)RYTY (where R stands for purines, Y stands for pyrimidines and V stands for A, C or G) [9,10]. Foxa proteins are expressed in endoderm-derived tissues such as the liver, gut and endocrine pancreas [9]. Consistent with the importance of Foxa proteins for metabolic regulation, many genes involved in the regulation of glucose, lipid and amino acid metabolism have Foxa-binding sites in their promoters [9]. Foxa proteins determine glucose homoeostasis by dictating, among others, the expression of genes encoding the gluconeogenic enzyme phosphoenolpyruvate carboxykinase in the liver and the insulin secretion-regulating ion channel in β-cells of the pancreatic islets [9,11]. Studies in vivo support the view that Foxa proteins also regulate glucagon gene transcription in pancreatic islet α-cells (see [6,9] and references therein). Whereas mice with homozygous mutations in the Foxa2 gene die in utero, mice with targeted disruption of the Foxa1 gene develop normally but exhibit neonatal growth retardation and die post-natally before 4 weeks of age. Foxa1−/− mice exhibit hypoglycaemia associated with appropriately suppressed levels of insulin and show increased levels of cortisol and growth hormone but inappropriately low levels of circulating glucagon. Furthermore, the levels of islet proglucagon mRNA transcripts are reduced in hypoglycaemic Foxa1−/− mice, demonstrating an essential role for Foxa1 in proglucagon gene expression. Two binding sites for Foxa proteins have been described in the rat glucagon gene that can mediate the activation of glucagon gene transcription by Foxa1, Foxa2 or Foxa3 in cultured cell lines, one of them being located in the enhancer element G2 [7,8,12] and the other in the proximal promoter element G1 [13]. At these sites, Foxa2 is the predominant Foxa binding activity in α-cell lines in spite of the fact that Foxa1 and Foxa2 are concomitantly expressed and share identical consensus DNA-binding sites [7,8,10,13]. Thus, although Foxa proteins seem to be essential activators of the glucagon gene in pancreatic islets, their action on the glucagon gene is poorly understood.
The present study demonstrates that the Foxa-binding site in the rat G2 enhancer element is not conserved in the human glucagon gene. A novel Foxa site was identified that mediates the activation of glucagon gene transcription by Foxa proteins and that is conserved between rodents and humans. The preferential Foxa binding activity at this site in an α-cell line was found to be Foxa1, offering a likely explanation for the decrease in glucagon gene expression in Foxa1-deficient mice.
EXPERIMENTAL
Comparative sequence analysis
The 5′-flanking sequences 350 bp upstream of the transcription start site in rat, mouse and human orthologues of the glucagon gene were aligned using the multiple sequence alignment program DIALIGN [14]. The mouse gene 5′-flanking sequence was determined in the present study and has been deposited in GenBank® database (accession number AF356593). The rat and human glucagon gene promoter sequences have previously been reported [15]. Sequences were scanned for conserved potential transcription factor-binding sites against weight matrix descriptions of transcription factor-binding sites in the MatInspector library [16] with individually optimized matrix and core similarities thresholds, which determines matrix specificity, using the program MatInspector Professional (Genomatix Software, Munich, Germany). Similar and/or related matrices were grouped into matrix families to reduce match redundancy. MatInspector assigns a quality rating to a match (called matrix similarity), which is correlated with biological functionality [16].
Reporter plasmids and expression vectors
The expression plasmid pCMV-GFPtpz, encoding a GFP (green fluorescent protein) mutant, was purchased from Canberra Packard (Dreieich, Germany). Expression plasmids pCMV-Foxa1 and pCMV-Foxa2 encoding for Foxa1 and Foxa2 respectively were kindly provided by Dr R. H. Costa (University of Illinois, Chicago, IL, U.S.A.) [17,18]. The plasmids pMT2-Foxa1 and pMT2-Foxa2 were generated by subcloning the rat Foxa1 or Foxa2 cDNAs excised from plasmids pCMV-Foxa1 and pCMV-Foxa2 respectively, in a simian virus 40-based expression vector pMT2 (kindly provided by Dr K. S. Zaret, Cell and Development Biology Program, Fox Chase Cancer Center, Philadelphia, PA, U.S.A.). The plasmid pGEX2T-GST-Pax6-PD (where GST stands for glutathione S-transferase and PD stands for paired domain), which encodes the PD of Pax6 fused to GST, was kindly provided by Dr R. L. Maas (Division of Genetics, Department of Medicine and Howard Hughes Medical Institute, Brigham & Women's Hospital, Harvard Medical School, Boston, MA, U.S.A.) [19]. Construction of pT81Luc plasmid that contains the luciferase gene under the control of thymidine kinase minimal promoter [20], the rat glucagon promoter-reporter gene construct that contains the region −350 to +58, −350GluLuc [21], and the plasmids pSG5-Prep1, pSG5-Pbx1b and pLGPmNF-AT1b encoding the transcription factors Prep1, Pbx1b or NFATp [NFAT1B (nuclear factor of activated T-cells 1B)] respectively have been described previously [22,23]. The human glucagon gene promoter-reporter plasmid, H-350GluLuc, was generated by subcloning upstream of the luciferase gene a PCR-generated DNA fragment spanning the −350 to +58 region with respect to the transcription start site of the human glucagon gene [15] into the XhoI and BglII sites of pT81Luc. Plasmid constructs −350ΔG2GluLuc and H-350ΔG2GluLuc lacking the G2 element (rat G2, bp −165 to −201 and human G2, bp −172 to −208) in the context of the rat or human −350/+58 reporter construct respectively were generated by a PCR procedure, which replaced the G2 element with the NotI restriction site. The plasmid constructs −350hG2GluLuc and H-350rG2GluLuc, containing the human G2 element in the context of the rat −350/+58 reporter construct and vice versa, were generated by inserting the human or rat G2 into −350ΔG2GluLuc and H-350ΔG2GluLuc respectively. A schematic representation of chimaeric constructs is shown in Figures 3(B) and 3(C). The plasmid constructs H-350hG2GluLuc and −350rG2GluLuc were generated by reinserting the human or rat G2 element into the NotI site of plasmids H-350ΔG2GluLuc and −350ΔG2GluLuc respectively. A series of 5′- and 3′-deletion constructs of human glucagon −350/+58 reporter construct, depicted in Figures 4(A) and 4(B), were constructed by subcloning PCR-generated 5′-deletion fragments (bp −298/+58, −251/+58, −171/+58, −105/+58 or −30/+58) into the XhoI/BglII sites upstream of the luciferase gene in the plasmid pT81. The resulting constructs were termed H-298GluLuc (bp −298 to +58), H-251GluLuc (bp −251 to +58), H-171GluLuc (bp −171 to +58), H-105GluLuc (bp −105 to +58) and H-30GluLuc (bp −30/+58). The 3′-deletion fragments (bp −350/−299, −350/−225, −350/−161, −350/−103 or −350/−31) were ligated into the BamHI/XhoI sites upstream of the human glucagon gene minimal promoter (bp −30/+58) in the plasmid H-30GluLuc, as described above. The resulting constructs were termed H-350/-299GluLuc (bp −350 to −299), H-350/-225GluLuc (bp −350/−225), H-350/-161GluLuc (bp −350/−161), H-350/-103GluLuc (bp −350/−103) and H-350/-31GluLuc (bp −350/−31).
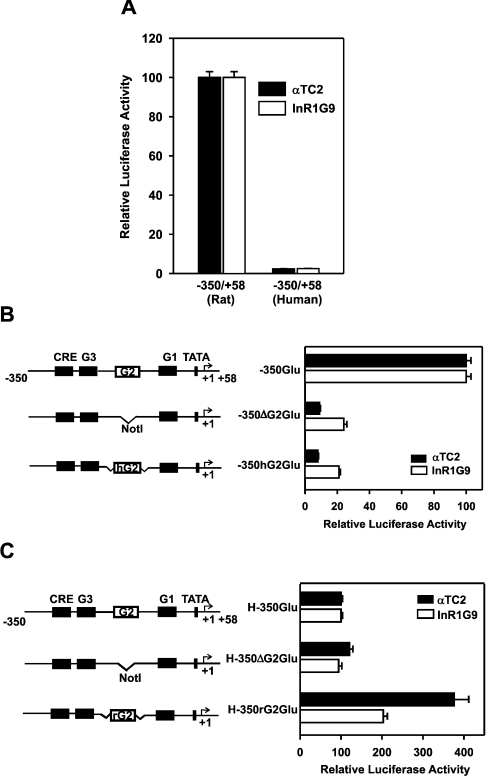
Figure 3. The G2 sequence of the human glucagon gene lacks enhancer activity.
(A) The −350/+58 region of the human glucagon gene shows no significant transcriptional activity in the glucagon-producing pancreatic islet cell lines αTC2 and InR1G9. The rat (−350GluLuc) or human (H-350GluLuc) glucagon gene fragment from −350 to +58 was fused to the coding region of the luciferase reporter gene. The reporter plasmids were transiently transfected into αTC2 or InR1G9 cells. In each cell line, luciferase activity of H-350GluLuc is expressed as a percentage of the mean value of the activity of −350GluLuc for each experiment. (B) Human G2 does not confer transcriptional activity to the rat glucagon promoter in glucagon-producing pancreatic islet α-cell lines. Left panel: a schematic representation of the wild-type and hybrid rat glucagon reporter constructs. From the wild-type rat glucagon gene promoter (−350GluLuc), the G2 element was deleted (−350ΔG2GluLuc); in the newly created NotI restriction site, an oligonucleotide containing the human G2 sequence was inserted to obtain a hybrid plasmid, −350hG2GluLuc. The reporter plasmids were transiently transfected into αTC2 and InR1G9 cells. In each cell line, luciferase activity is expressed as a percentage of the mean value of the activity measured in the respective −350GluLuc group for each experiment. (C) The rat G2 element confers weak transcriptional activity to the human glucagon promoter in glucagon-producing pancreatic islet α-cell lines. Left panel: a schematic representation of the wild-type and hybrid human glucagon reporter gene constructs. From the wild-type human glucagon gene promoter (H-350GluLuc), the G2 sequence was deleted (H-350ΔG2GluLuc); into the newly created NotI restriction site, an oligonucleotide containing the rat G2 element was inserted to generate a hybrid plasmid, H-350rG2GluLuc. The plasmids were transiently transfected into αTC2 and InR1G9 cells. In each cell line, luciferase activity is expressed as a percentage of the mean value of the activity measured in the respective H-350GluLuc group for each experiment. Values are the means±S.E.M. for at least four independent experiments, each performed in duplicate.
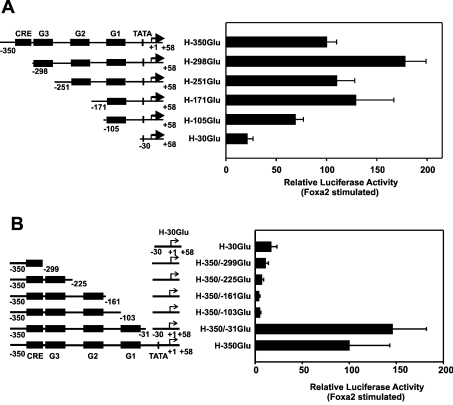
Figure 4. Mapping by 5′- and 3′-deletion analysis of the region imparting transactivation by Foxa2 of the human glucagon gene promoter.
(A) 5′-Deletion analysis. (B) 3′-Deletion analysis. The indicated constructs were transfected into HepG2 cells, both with and without co-transfection of the expression vector encoding Foxa2 (450 ng/dish). The Foxa2-induced luciferase activity is expressed as a percentage of the mean value of the activity measured with H-350GluLuc for each experiment. Values are means±S.E.M. for at least four independent experiments, each performed in duplicate. Potential control elements in the 5′-flanking region of the human glucagon gene are indicated, referring to characterized elements in the rat glucagon gene (see the text for an explanation).
Foxa sites were selectively mutated by the oligonucleotide-directed mutagenesis strategy [24]. In the context of human glucagon gene reporter construct, H-350GluLuc, the Foxa sites A and B were replaced with the unrelated EcoRI/NotI and XbaI/NotI sequences respectively. The resulting plasmid constructs were termed H-350GluLuc-mtA, H-350GluLuc-mtB and H-350GluLuc-mtAB, carrying mutations at the Foxa site A (−48 to −33 bp), site B (−94 to −81 bp) or the sites A and B respectively. A similar strategy was used to mutate Foxa sites in the context of the rat glucagon promoter-reporter construct, −350GluLuc, which replaced the Foxa sites A, B and C with the unrelated EcoRI/NotI, XbaI/NotI or Pst1/CGGC sequence respectively. The resulting plasmid constructs were termed −350GluLuc-mtA, −350GluLuc-mtB, −350GluLuc-mtC, −350GluLuc-mtAB and −350GluLuc-mtABC carrying mutations at the Foxa site A (bp −47 to −33), site B (bp −93 to −80), site C (bp −194 to −185), sites A and B or sites A–C respectively. All constructs were sequenced by the dideoxy-chain termination approach to confirm their identity.
Cell culture and transient transfection assays
Human hepatoma cells (HepG2 cells) and glucagon-producing pancreatic islet α-cell lines (the hamster InR1G9 [25] and mouse αTC2 [26]) were maintained in culture as described previously [8]. COS-1 cells (African green monkey kidney cells) were grown in Dulbecco's modified Eagle's medium (4.5 g of glucose/l) supplemented with 10% (v/v) fetal calf serum, 100 units/ml penicillin and 100 μg/ml streptomycin (Gibco BRL/Invitrogen). Transient transfections were performed using 0.5 μg (COS-1), 2 μg (αTC2, InR1G9) or 3 μg (HepG2) of the appropriate reporter plasmid per 6 cm dish, either by the DEAE–dextran method (InR1G9, αTC2 and COS-1 cells) or the calcium phosphate co-precipitation method (HepG2) as described previously [8,27,28]. pCMV-GFPtpz (0.5 μg/6 cm dish) was co-transfected for monitoring transfection efficiency. Where indicated, cells were co-transfected with the expression plasmid for Foxa1 or Foxa2. The amounts used are indicated in the Figure legends. The total amount of DNA transfected was kept constant by supplementing sample DNAs with appropriate amounts of the plasmid vector Bluescript (Stratagene Heidelberg, Germany). At 48 h post-transfection, cells were harvested, lysed by three cycles of freeze–thawing and luciferase activity was measured using an AutoLumat LB 953 luminometer (Berthold Technologies, Bad 1 Wilbad, Germany) as described in [21]. Fluorescence signal (GFP) was measured in cell extracts using the Fusion™ microplate fluorimeter (Packard, Zürich, Switzerland).
In vitro translation and preparation of nuclear and whole cell extracts
Nuclear protein extracts from InR1G9 cells were prepared by the procedure described by Schreiber et al. [29]. Whole cell lysates from COS-1 cells that were transfected with expression plasmids for Foxa1 or Foxa2 proteins, namely pMT2-Foxa1 and pMT2-Foxa2 (0.5 μg) respectively, were prepared at 48 h post-transfection as described in [30]. Prep1/Pbx1b and NFAT1B proteins were synthesized using the plasmid templates pSG5-Prep1/pSG5-Pbx1b or pLGPmNFAT1b respectively in conjunction with the TNT-coupled in vitro transcription/translation system according to the manufacturer's instructions (Promega, Mannheim, Germany). His-tagged CREB-327 (CRE-binding protein-327) and Foxa2 fusion proteins and GST-tagged Pax6-PD were bacterially (Escherichia coli) expressed from plasmid templates pRSET-CREB-327, pRSET-Foxa2 or pGEX2T-GST-Pax6-PD respectively and affinity-purified as described in [31,32].
Oligonucleotides and probes
Oligonucleotides were purchased from MWG-Biotech (Ebersberg, Germany). The DNA sequences of the sense strand of oligonucleotides directed to the 5′-flanking region of human glucagon gene are as follows [15]: G1(H), 5′-AAAAACTCATTATTTACAGATGAGAAATTTATATTGT-3′; G2(H), 5′-AGATACAAGAGTGCATAAAAAGTTTCCAGGTCTCTAA-3′; G3(H), 5′-TTAAGTTGATTTTCATGCGTGATTGAAAGTAGAAGGTGGA-3′; GluCRE(H), 5′-TTTGTCTTCAACGTCAAAATTCACTTT-3′. The DNA sequences of oligonucleotides corresponding to G1–G3 regulatory elements of the rat glucagon gene [denoted G1(R), G2(R) and G3(R) respectively in the present study] [4,5], GluCRE and mGluCRE, representing wild-type and mutated rat glucagon promoter CRE respectively, and SomCRE, spanning the rat somatostatin gene CRE, as well as TTR (transthyretin) and TTR-HNF-4, oligonucleotides representing TTR gene promoter high-affinity Foxa- and HNF-4-binding sites respectively, have been described previously [5,10,31–33]. The coding strand sequences of oligonucleotides encompassing the novel Foxa site A in the rat and human glucagon promoters are as follows: rat Foxa site A, 5′-GTAATATCTGCAAGGCTAAACAGCCTG-3′ and human Foxa site A, 5′-CGTAATATCTGTGAGGCTAAACAGAGCTG-3′. Oligonucleotides were incorporated with 5′-overhangs for labelling purposes. Double-stranded oligonucleotides were labelled by filling in with [α-32P]dCTP (Amersham Biosciences) using the Klenow fragment of DNA polymerase (Fermentas, Munich, Germany).
EMSA (electrophoretic mobility-shift assay)
The procedure used for EMSA was adapted from Knepel et al. [27] and Klemsz et al. [34]. All steps were performed at 4 °C unless otherwise noted. Nuclear extracts, whole cell lysates (∼15 μg of protein each) or in vitro-synthesized NFAT1B were preincubated on ice for 20 min with 2 μg of poly(dI-dC)·(dI-dC) (Sigma) in a 20 μl reaction containing Klemsz binding buffer [20 mM Hepes, pH 7.9, 75 mM NaCl, 0.5 mM EDTA, 1 mM DTT (dithiothreitol) and 5%, v/v, glycerol]. CREB-327, Pax6(PD) or Prep1/Pbx1b were incubated in a buffer (20 mM Hepes, pH 7.9, 140 mM KCl, 1 mM EDTA, pH 8.0, 0.5 mM DTT and 5%, v/v, glycerol) containing either 0.2 μg (for bacterially expressed proteins) or 2.5 μg (for in vitro translated proteins) of poly(dI-dC)·(dI-dC). Recombinant Foxa2 protein was incubated for 15 min under buffer conditions (50 mM NaCl, 1 mM PMSF, 1 mM MgCl2, 1 mM EGTA, 5 mM DTT and 10% glycerol in 10 mM Tris/HCl, pH 7.5). After preincubation, an appropriate labelled oligonucleotide probe (20000 c.p.m.) was added and incubation was continued for a further 20 min. In competition assays, 200-fold molar excess of the specific unlabelled oligonucleotide was added to the binding reaction before the addition of a labelled probe. In antibody supershift studies, nuclear or whole cell extracts were preincubated for 20 min at room temperature (22 °C) with either 0.5 μg of anti-Foxa1 IgG, 2 μg of anti-Foxa2 antibody or control preimmune goat IgG before the initiation of the binding reaction as described above. The polyclonal Foxa2 antibody (sc-6554X) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, U.S.A.); it recognizes both Foxa1 and Foxa2 (see Figure 7C) and is hereinafter referred to as anti-Foxa1/2. The polyclonal rabbit anti-Foxa1 IgG was a gift from R. H. Costa's group [17]. Bound and free probes were resolved on a non-denaturing polyacrylamide gel, blotted on to Whatman 3 MM filter paper, dried for 2 h under vacuum at 75 °C and exposed to an X-ray film at −80 °C with intensifying screens as described in [27].
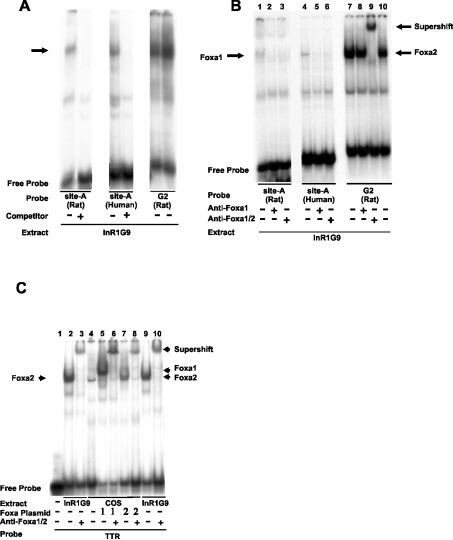
Figure 7. The novel Foxa site A in both the rat and human glucagon promoters binds preferentially to Foxa1 in nuclear extracts from a pancreatic islet α-cell line as indicated by the EMSA.
(A) Nuclear extracts from InR1G9 cells were probed with labelled oligonucleotides containing the Foxa site A of the rat glucagon promoter [site A (rat)], the Foxa site A of the human glucagon promoter [site A (human)] or, as a control, the G2 element of the rat glucagon promoter [G2 (rat)]. Where indicated, a 200-fold molar excess of unlabelled oligonucleotide (the same as probe) was added as a specific competitor. The band corresponding to specific binding is indicated by an arrow. (B) The Foxa site A-binding protein is recognized by a specific anti-Foxa1 antibody. Nuclear extracts from InR1G9 cells were incubated with labelled Foxa site A of the rat glucagon promoter [site A (rat)], Foxa site A of the human glucagon promoter [site A (human)] or, as a control, G2 of the rat glucagon promoter [G2 (rat)]. Preimmune IgG (2 μg) or antibodies recognizing specifically Foxa1 (anti-Foxa1) (0.5 μg) or antibodies recognizing Foxa1 and Foxa2 (anti-Foxa1/2) (2 μg) were added to the binding reaction as indicated. Note that the specific anti-Foxa1 antibody recognizes nuclear protein binding to the Foxa site A (both rat and human) but not nuclear protein binding to rat G2 (containing the Foxa site C). This shows the specificity of the anti-Foxa1 antibody and, at the same time, demonstrates that distinct nuclear proteins bind to the Foxa site A (Foxa1) and Foxa site C in G2 (Foxa2). (C) Binding specificity of the anti-Foxa1/2 antibody. Whole cell lysates from COS-1 cells transfected with expression vectors encoding Foxa1, Foxa2 or the empty vector were incubated with a labelled oligonucleotide (TTR) that contains a high-affinity Foxa-binding site from the mouse TTR gene promoter, which can be bound by each of the three major Foxa isoforms 1–3 [10,35]. Nuclear extracts of InR1G9 cells were included as controls. The anti-Foxa1/2 antibody was added to the binding reaction as indicated. Note that the complex formed by labelled TTR with COS-expressed Foxa1 as well as the complex formed by labelled TTR with COS-expressed Foxa2 is abolished and supershifted by the anti-Foxa1/2 antibody, indicating that this antibody (named anti-Foxa2 antibody by the manufacturer) recognizes both Foxa1 and Foxa2 under the conditions used.
RESULTS
Sequence analysis of the 5′-flanking regions of the rat, murine and human glucagon genes
The 5′-flanking region sequences of the rat, murine and human orthologues of the glucagon gene were aligned and searched for potential transcription factor-binding sites using a weight matrix description for transcription factor families. The analysis revealed a novel match to Foxa/forkhead matrix (matrix similarity score, 0.882), termed Foxa site A, downstream of the G1 element and proximal to the TATA box in all three sequences (Figure 1). The novel Foxa site A matches, at eight out of 12 nucleotide positions, the consensus Foxa/forkhead box recognition sequence [VA(A/T)TRTT(G/T)RYTY] [10] and is in reverse orientation relative to the previously characterized Foxa site in the G1 regulatory region, denoted here as Foxa site B (matrix similarity score, 0.923). A match to Foxa/forkhead matrix in the G2 element, denoted here as Foxa site C, is only detected in rodent sequences. No match to the Foxa/forkhead matrix was detected in the human G2 element.
Figure 1. Comparative sequence analysis.
Nucleotide sequences of the 5′-flanking regions in rat, mouse and human orthologues of the glucagon gene were aligned using the program DIALIGN [14] and scanned for potential transcription factor-binding sites using weight matrix description of binding sites and the program MatInspector Professional. Upper-case letters denote aligned regions. The positions of a novel Foxa/forkhead site (the Foxa site A), which is located downstream of the G1 element, and the previously described Foxa sites in the G1 and G2 elements as well as the TATA box and CREs are illustrated. Note that the Foxa/forkhead site in the G2 element is detected only in rodent sequences, not in human sequences, and that the nucleotides at Foxa sites A–C are identical between the rat and mouse sequences. Nucleotides matching the Foxa/forkhead consensus recognition sequence (VAWTRTTKRYTY; V=A, C or G; W=A or T; R=A or G; K=G or T; Y=C or T) are highlighted in boldface. The sequences are numbered relative to the transcription start site (+1). G1–G3 regulatory regions are marked by overbars above the sequence. The mouse sequence has been deposited in GenBank® database under the accession number AF356593. ATF, activating transcription factor.
The human G2 element lacks interaction with recombinant Foxa2 and enhancer activity
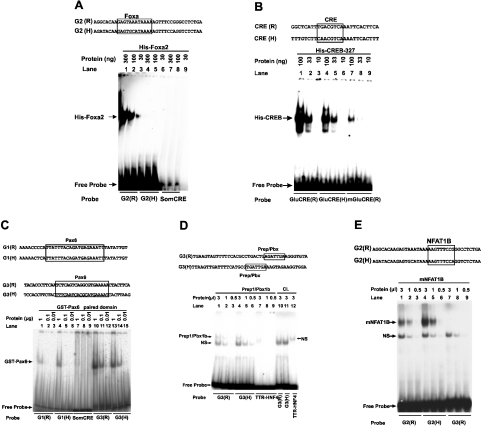
To validate the in silico prediction of the lack of a Foxa/forkhead site in human G2, the ability of human G2 to bind bacterially expressed and purified Foxa2 protein was evaluated using EMSA. The rat G2 element that harbours a high-affinity Foxa-binding site (Foxa site C) [7,8] was employed as a positive control. The gel mobility-shift profiles of the rat and human G2 with Foxa2 are shown in Figure 2(A). A specific complex with the rat G2 probe (lanes 1–3), but not with the human G2 probe (lanes 4–6), was seen. No complex was observed with the unrelated somatostatin CRE probe (lanes 7–9), demonstrating the specificity of Foxa2 interaction with rat G2. These results demonstrate that human G2 does not interact significantly with Foxa2. EMSA analysis of the binding ability of potential regulatory elements in the 5′-flanking region of the human gene to transcription factors CREB, Pax6, Prep1/Pbx1b and NFATp, previously shown to bind and regulate the expression of the rat glucagon gene [6], showed that each of the transcription factors binds specifically to respective sequences in the human glucagon gene (Figures 2B–2E). DNA–protein complexes of similar mobility were observed with rat and human sequences.
Figure 2. Analysis of the interaction between potential regulatory sequences in the 5′-flanking region of the human glucagon gene and recombinant Foxa2, CREB, Pax6, Prep1/Pbx1b and NFAT1B by EMSA.
A comparison of rat and human regulatory sequences. The locations of the putative binding site in the human sequence and the experimentally characterized binding site in the rat promoter are boxed. (A) Binding of the indicated amounts of recombinant Foxa2 to labelled oligonucleotide probes, either the rat G2 [G2(R), lanes 1–3], human G2 [G2(H), lanes 4–6] or somatostatin CRE (SomCRE, lanes 7–9). (B) Binding of the indicated amounts of purified-bacterially expressed CREB-327 to labelled oligonucleotide probes, either the CRE of the rat glucagon gene [GluCRE(R), lanes 1–3], the CRE of the human glucagon gene [GluCRE(H), lanes 4–6] or the rat glucagon gene mutant CRE [mGluCRE(R), lanes 7–9]. (C) Binding of the indicated amounts of purified bacterially expressed Pax6-PD to either the rat G1 [G1(R), lanes 1–3], human G1 [G1(H), lanes 4–6], rat G3 [G3(R), lanes 10–12], human G3 [G3(H), lanes 13–15] or the somatostatin CRE (SomCRE, lanes 7–9) as probes. (D) Binding of the indicated amounts of in vitro co-transcribed/translated heterodimer complex consisting of the homeodomain transcription factors Prep1/Pbx1b or control lysate (CL). The rat G3, G3(R), human G3, G3(H) or TTR gene HNF-4 recognition sequence (TTR-HNF-4) probes were incubated with either Prep1/Pbx1b (lanes 1–3, 4–6 and 7–9) or control lysate (lanes 10–12). (E) Binding of the indicated amounts of in vitro-transcribed/translated NFAT1B to either the rat G2 [G2(R)], human G2 [G2(H)] or rat G3 [G3(R)]. The upper arrow indicates the specific complex; NS, a complex formed with non-specific DNA-binding activity; R, sequence from the rat glucagon gene; H, sequence from the human gene.
To study the transcriptional activity of the human G2 sequence, transient transfection experiments were performed in glucagon-producing pancreatic islet α-cell lines. It has been shown previously that the Foxa site in the rat G2 element is essential for its enhancer activity and for rat glucagon promoter activity [6–8]. As shown in Figure 3(A), the reporter gene construct driven by the −350/+58 fragment of the human glucagon gene (H-350GluLuc) showed very low transcriptional activity in two glucagon-producing pancreatic islet α-cell lines (αTC2 and InR1G9), confirming the reported lack of transcriptional activity of the human glucagon gene 5′-flanking region (up to 3355 bp) in InR1G9 cells [15]. To study the transcriptional activity of the human G2 sequence in the context of the rat glucagon promoter, a construct was prepared (−350hG2GluLuc) in which human G2 replaces rat G2 within the rat glucagon promoter. As shown in Figure 3(B), swapping of the rat G2 element with human G2 resulted in a marked attenuation of basal transcriptional activity to levels comparable with the activity of the rat glucagon promoter lacking the G2 element (−350ΔG2GluLuc). The reinsertion of the rat G2 element into the rat promoter (construct −350rG2GluLuc) resulted in a somewhat higher transcriptional activity than that of the wild-type promoter (construct −350GluLuc; results not shown), indicating that the conversion of G2-flanking sequences into an NotI restriction site, as performed in the constructs −350hG2GluLuc and −350rG2GluLuc, does somewhat enhance but does not disrupt G2 transcriptional activity and, thus, cannot explain the inability of the human G2 sequence to confer transcriptional activity. On the other hand, swapping of the human G2 with the rat G2 element enhanced human glucagon promoter activity (Figure 3C). The rat G2 element augmented the basal activity of the human glucagon promoter construct approx. 2- and 4-fold in InR1G9 and αTC2 cells respectively (Figure 3C). However, the level of activity reached was approx. 8% of the activity of the wild-type rat glucagon promoter (compare Figure 3C with Figure 3A). These results indicate that the human G2 element, in marked contrast with the rat G2 element, lacks enhancer activity in islet cell lines. Furthermore, the low basal transcriptional activity of the human glucagon promoter is in part due to the lack of a functional Foxa site in the human G2 element.
The predicted novel Foxa site A contributes to transactivation of the glucagon promoter by exogenously expressed Foxa proteins
Exogenously expressed Foxa1, Foxa2 or Foxa3 proteins have been shown to activate the rat glucagon gene promoter in the heterologous cell lines HepG2 or BHK (baby hamster kidney) in transient transfection assays [7,8,12,13]. To investigate whether the human glucagon gene is transactivated by Foxa proteins like the rat gene, HepG2 cells were transfected with the human glucagon-reporter gene construct H-350GluLuc together with an expression plasmid for Foxa1 or Foxa2. The human glucagon gene promoter responded to Foxa1 and Foxa2 similarly to the rat promoter, also after internal deletion of the G2 element (results not shown), suggesting that response elements outside G2 are sufficient to activate the human glucagon promoter in response to exogenously expressed Foxa proteins.
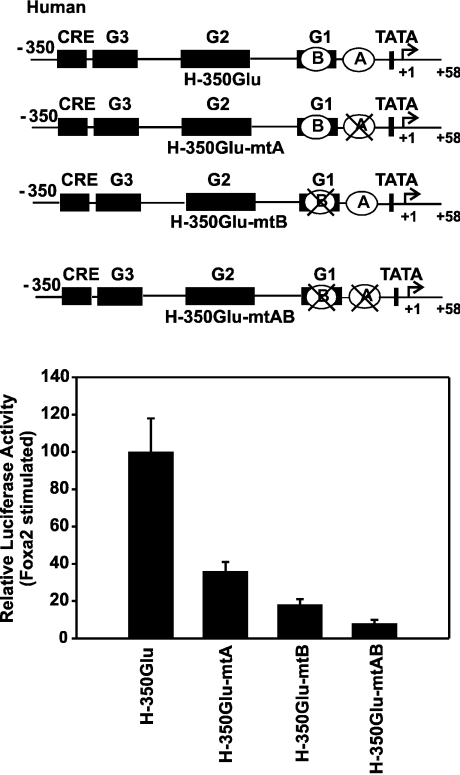
To localize the cis-acting DNA sequence(s) of the human glucagon gene that mediate transcriptional activation by Foxa proteins, a 5′-, 3′- and internal-deletion analysis was performed. The results of the 5′-deletion analysis are shown in Figure 4(A). Deletion from bp −350 to −105 resulted in no significant decrease in Foxa2-dependent activity (loss of ∼30% activity), whereas further deletion up to bp −30 caused approx. 80% loss of Foxa2-dependent transcriptional activity compared with that of the wild-type construct (H-350GluLuc; Figure 4A). The results of the 3′-deletion analysis are shown in Figure 4(B). Fragments of the human glucagon promoter with deletions at their 3′-end were linked to the minimal human glucagon promoter (H-30GlucLuc). This minimal promoter shows almost no response to Foxa2 (Figures 4A and 4B). The human glucagon gene 5′-flanking DNA from −350 to −31 conferred full Foxa2 responsiveness (Figure 4B). When only sequences from −350 to −103 were fused to the promoter, Foxa2 no longer activated gene transcription (Figure 4B). The combined results obtained with the 5′- and 3′-deletion analyses suggest that transactivation by Foxa2 of the human glucagon gene is conferred by element(s) located within the region encompassing bp −105 to −31. This region includes the predicted novel Foxa site A and the previously described Foxa site B in the G1 element. To examine their role more directly, the Foxa site A, Foxa site B or both were mutated in the context of the human glucagon gene promoter (Figure 5). As shown in Figure 5, the mutation of site A markedly reduced the activation of the human glucagon gene promoter by exogenously expressed Foxa2. The reduction was by approx. 65% (Figure 5). A reduction was also seen after mutation of site B (reduction by ∼80%), and Foxa2 responsiveness was virtually lost after mutation of both sites in combination (Figure 5). The results show that the Foxa site A is essential for transactivation by Foxa2 of the human glucagon gene.
Figure 5. Effect of mutations in Foxa site A and Foxa site B on the transactivation by Foxa2 of the human glucagon gene promoter.
A schematic representation of wild-type (H-350GluLuc) and mutated constructs shows the location of the Foxa site A (A) and the Foxa site B (B). The mutated sites are marked by crosses. HepG2 cells were co-transfected with either H-350GluLuc or mutated constructs (H-350GluLuc-mtA, H-350GluLuc-mtB or H-350GluLuc-mtAB) and the expression vector encoding Foxa2 (450 ng/dish). The Foxa2-induced luciferase activity is expressed as a percentage of the mean value of the activity measured with H-350GluLuc for each experiment. Values are means±S.E.M. for at least four independent experiments, each performed in duplicate.
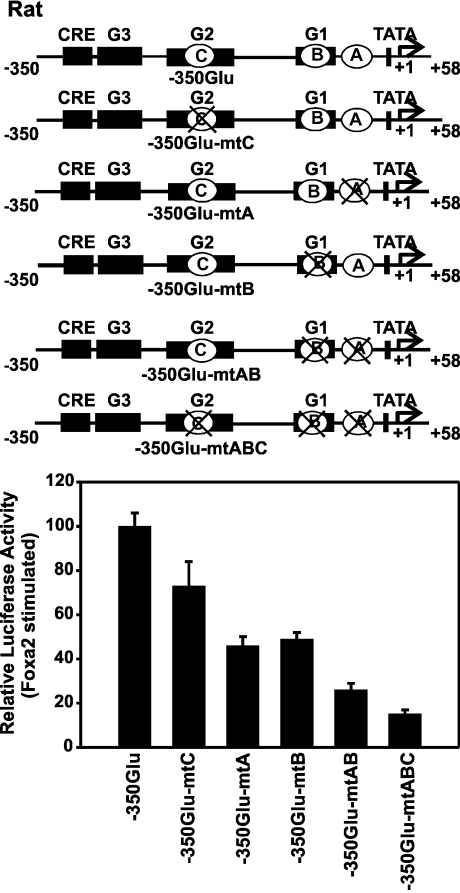
A comparative sequence analysis suggested that the Foxa site A is present also in the rat glucagon gene (Figure 1). Furthermore, a 5′- and 3′-deletion analysis of the rat glucagon promoter indicated that sequences within −136 bp are sufficient and required for transcriptional activation of the rat promoter by exogenously expressed Foxa2 in HepG2 cells (results not shown), similar to the results obtained with the human glucagon promoter (Figure 4). Therefore a role for the Foxa site A in the rat glucagon promoter was further investigated by internal mutations (Figure 6). Mutations in either site A or site B repressed transactivation by Foxa2 to a similar extent (decreased by ∼50%; Figure 6). In contrast, mutating the Foxa site C alone had no significant effect (Figure 6), indicating that a combination of site A and site B, but not a combination of site C and either site A or site B, is sufficient to confer almost full responsiveness to Foxa2. Simultaneous mutations in Foxa site A and site B or in all three sites (site A, site B and site C) nearly completely abolished transactivation by Foxa2 (decreased by ∼80%; Figure 6). These results demonstrate the functionality of the Foxa site A. For both the rat and the human genes, the activation of the glucagon promoter by Foxa2 in a heterologous cell line depends primarily on intact Foxa sites A and B.
Figure 6. Effect of mutations in the Foxa sites A, B and C on the transactivation by Foxa2 of the rat glucagon gene promoter.
A schematic representation of wild-type (−350GluLuc) and mutated constructs shows the location of the Foxa site A (A), Foxa site B (B) and Foxa site C (C). The mutated site is marked by a cross. HepG2 cells were co-transfected with either −350GluLuc or mutated constructs (−350GluLuc-mtC, −350GluLuc-mtA, −350GluLuc-mtB, −350GluLuc-mtAB or −350GluLuc-mtABC) and the Foxa2 expression plasmid (450 ng/dish). The Foxa2-induced luciferase activity is expressed as a percentage of the mean value of the activity measured with −350GluLuc for each experiment. Values are means±S.E.M. for at least four independent experiments, each performed in duplicate.
The novel Foxa site A binds preferentially Foxa1 in nuclear extracts from pancreatic islet α-cells
To determine which protein factors, expressed in an α-cell line, bind to the Foxa site A, EMSAs were performed. Nuclear protein extract prepared from InR1G9 cells, which endogenously express the Foxa1 and Foxa2 isoforms [13], was probed with labelled oligonucleotides containing Foxa site A from either the rat or human glucagon promoter (Figure 7A). The rat G2 (Foxa site C) probe, which binds Foxa2 in InR1G9 extracts [8,13], was included as a positive control. A retarded band with similar mobility was detected with all three probes (Figure 7A, indicated by an arrow). This complex with the rat or human site A probe was competed out by an excess of an unlabelled ‘self’ oligonucleotide (Figure 7A), suggesting that the retarded band represents specific interaction. To identify the protein that binds the Foxa site A, EMSAs were performed in the presence of preimmune goat IgG, antibodies recognizing specifically Foxa1 (anti-Foxa1) [17] or antibodies recognizing Foxa1 and Foxa2 (anti-Foxa1/2) (see Figure 7C). The results are shown in Figure 7(B). The rat G2 probe (Foxa site C) was again included as a positive control. The retarded complex of the rat G2 binding activity in InR1G9 extracts was not affected by the preimmune IgG or the anti-Foxa1 antibody (Figure 7B, lanes 10 and 8) but was abolished and supershifted by the anti-Foxa1/2 antibody (Figure 7B, lane 9), consistent with previous reports that rat G2 (Foxa site C) binds Foxa2 in InR1G9 extracts [8,13]. In contrast, the addition of the antibody specific for Foxa1 as well as the antibody recognizing Foxa1 and Foxa2 to the binding reaction inhibited the binding of the retarded complex to the rat and human Foxa site A (Figure 7B, lanes 2 and 3 and lanes 5 and 6 respectively). It is worth noting that a complex of higher mobility, observed with all three probes, was not influenced by the antibodies used (Figure 7B). Collectively, data from Figure 7 point to Foxa1 as the nuclear protein in InR1G9 extract that preferentially binds to the Foxa site A.
The novel Foxa site A is required for cell-specific transcriptional activity of the glucagon promoter
To study the functional significance of the novel Foxa site A in cell-specific transcriptional activation of the glucagon gene in pancreatic islet cells, the activity of the rat glucagon promoter, both wild-type and mutant, was measured in two glucagon-producing pancreatic islet α-cell lines (InR1G9, αTC2). Because the human glucagon gene region from −350 to +58 exhibits very low basal transcriptional activity in these cell lines (Figure 3A), only the rat promoter was studied. As a control, the rat glucagon promoter with a mutated Foxa site C in the G2 element was included (construct −350GluLuc-mtC). Consistent with previous reports [6–8], mutation of the Foxa site C markedly decreased glucagon promoter-reporter gene activity (Figure 8), confirming that Foxa2 binding to the G2 element is required for islet-specific rat glucagon promoter activity. Mutation of the Foxa site A (construct −350GluLuc-mtA) decreased reporter gene expression by approx. 80% in both the pancreatic islet α-cell lines (Figure 8). Mutation of the Foxa site A, which is close to the TATA box (Figure 1), does not cause a general disruption of initiation of transcription at the glucagon promoter, since it does not inhibit the activation of glucagon promoter activity by transcription factors like CREB in JEG cells (results not shown). Thus, when taken together, results of the present study suggest that the novel Foxa site A binds preferentially Foxa1 in pancreatic islet α-cells and confers pancreatic islet α-cell-specific activation of the glucagon gene.
Figure 8. Mutation of the novel Foxa site A markedly decreases cell-specific transcriptional activity of the glucagon promoter in glucagon-producing pancreatic islet α-cell lines.
The rat glucagon promoter, either wild-type (−350GluLuc) or mutated at the Foxa site A (construct −350GluLuc-mtA) or, as a control, mutated at the Foxa site C in G2 (construct −350GluLuc-mtC), was transfected into αTC2 and InR1G9 cells. pXP2, empty vector. In each cell line, luciferase activity is expressed as a percentage of the mean value of the activity of −350GluLuc wild-type in each experiment. Values are means±S.E.M. for at least three independent experiments, each performed in quadruplicate.
DISCUSSION
Considering cross-species homologies between the rat, mouse and human glucagon gene 5′-flanking sequences and the identical pattern of glucagon gene expression [5,6,15], it was surmised that the rat and human glucagon genes probably share key regulatory elements and molecular mechanisms. However, the findings of a previous study suggest that transcriptional regulatory mechanisms governing pancreatic α-cell-specific activation of the glucagon gene have diverged between rats and humans [15], but the underlying molecular determinants have not yet been defined. The G2 sequence is remarkably conserved in rodents and humans except for an AA to GC substitution in the human G2 sequence. These bases correspond to core positions in the characterized Foxa site in the rat element (Figure 1). The sequence analysis correctly predicted the lack of a Foxa site in the human G2 sequence, demonstrating that the weight matrix description of Foxa/forkhead site is able to distinguish between sequences that show high sequence similarity but differ in binding/function because of crucial mismatches in an otherwise highly conserved site. The human G2 sequence did not exhibit enhancer activity in islet cell lines, suggesting that the lack of a functional Foxa site renders human G2 non-functional. This could have been sufficient to explain the low transcriptional activity of the human glucagon promoter in the islet α-cell lines. However, although the rat G2 element augmented the transcriptional activity of the −350/+58 human glucagon gene fragment in islet cell lines, the difference in transcriptional activities of the rat and human glucagon reporter gene constructs is not accounted for by the increase in activity seen. The results therefore clearly indicate that further differences exist between the −350/+58 region in the rat and human glucagon genes apart from the lack of a functional Foxa site in human G2. These remain to be defined, although each of the transcription factors CREB, Pax6, Prep1/Pbx1b and NFATp, previously shown to bind and regulate the expression of the rat glucagon gene [6], were found by EMSA in the present study to interact specifically with respective sequences in the human gene.
Notably, mutating the Foxa site C in G2 had virtually no effect on the activation of the rat glucagon gene by the expressed Foxa2 in a heterologous cell line, whereas it markedly decreased rat glucagon promoter activity in pancreatic islet α-cell lines. This suggests that Foxa binding to the G2 element is dispensable for activation of the rat glucagon gene by Foxa proteins alone, but is required for its islet-specific activation, probably due to synergistic interactions between Foxa proteins and other islet-enriched transcription factors. In addition to islet-specific enhancer activity, the G2 element in the rat glucagon gene has also been shown to confer protein kinase C [8] and depolarization responsiveness [33]. The binding of Foxa proteins is required for both of these activities, synergizing with Ets-like transcription factors [8] and NFATp [33] respectively. Thus the lack of Foxa binding to the human G2 sequence may also affect the regulation of the human glucagon gene by these signalling pathways.
In contrast with Foxa binding to the G2 element, the novel Foxa site in the glucagon gene (site A, between −30 and −50), identified by the present study, is conserved in rats, mice and humans. This interspecies conservation of site A suggests functional importance. Indeed, mutation of site A decreased by 50–80% both the activation of the rat or human glucagon promoter by the expressed Foxa2 in a heterologous cell line and the cell-specific activity of the rat glucagon promoter in glucagon-producing pancreatic islet α-cell lines. These results suggest that the novel Foxa site A, together with site B (and, in the rat, site C), mediates the activation of the glucagon gene by Foxa proteins, being an essential component of the synergistic interaction of a unique combination of multiple transcription factors that confers cell-specific activation of the glucagon gene in pancreatic islet α-cells.
Although two Foxa isoforms, Foxa1 and Foxa2, are concomitantly expressed in glucagon-producing pancreatic islet α-cell lines [13] and share identical consensus DNA-binding sites [10], previous studies have shown that both the Foxa site B in G1 and the Foxa site C in rat G2 bind Foxa2 from α-cell lines [8,13]. In contrast, the present study shows that the novel Foxa site A binds preferentially Foxa1 in nuclear extracts from an α-cell line. The reason for these distinct binding specificities is unknown. Assuming a similar occupancy in vivo, the unique binding specificity of the novel Foxa site A may shed light on the regulation of glucagon gene transcription by Foxa1. Two independent studies have demonstrated that, in Foxa1-deficient mice, glucagon-producing pancreatic islet α-cells develop normally but glucagon mRNA levels are reduced by 50–70%, followed by hypoglycaemia and death [9]. These findings imply a role for Foxa1 in the regulation of the glucagon gene, which cannot be substituted by Foxa2. However, how Foxa1 fulfils this role remained unclear, since both the Foxa site B and Foxa site C bind Foxa2 in α-cell lines. The results of the present study now suggest that the ability of islet α-cell Foxa1 to bind to the novel Foxa site A distinguishes it from islet α-cell Foxa2. The results thereby suggest a mechanism for the essential and unique role played by Foxa1 in the regulation of the glucagon gene, according to which islet α-cell Foxa1 directly binds to the novel Foxa site A, conferring transcriptional activity that is required for pancreatic islet α-cell-specific activity of the glucagon gene, both in rodents and humans.
The Foxa site A has limited identity to the consensus Foxa/forkhead box recognition sequence (at eight of 12 nucleotide positions). The Foxa site A (rat, GGCTGTTTAGCC; human, CTCTGTTTAGCC) in the glucagon promoter falls into the TGT3 subclass (nucleotides underlined) of Foxa-binding sites from which Foxa dissociates significantly faster than from the high-affinity Foxa recognition sequence ATTATTGACTTAG defined in the mouse TTR gene promoter [35–37]. This subclass of Foxa sites is often found in close association with binding sites for other transcription factors (see [37] and references therein). The Foxa site A in the glucagon gene may thus provide another example of a functional framework of transcription factor binding sites that involves a combination of high- and low-affinity sites for the fine tuning of transcriptional activity and/or to reinforce possible synergy with neighbouring transcription factors [35–37].
The proximal rat glucagon gene promoter up to bp −136 is essential for G2 and G3 enhancer function, but has itself low intrinsic transcriptional activity [6]. The novel Foxa site A is located inside this proximal promoter between bp −30 and −50, immediately adjacent to the TATA box, suggesting that Foxa1 at this site may serve additional functions besides direct transcriptional activation. Foxa proteins have been shown to decompact DNA [38]. Chromatin opening by Foxa requires only DNA binding and direct protein–protein interactions between the C-terminus of Foxa and histones H3 and H4, which may disrupt the internucleosomal interactions between H3/H4 molecules in neighbouring nucleosomes [38]. Considering the intrinsic chromatin opening property of Foxa proteins, it is tempting to speculate that Foxa proteins binding to site A might impart to the glucagon promoter competence for transcription by placing the transcription start site of the glucagon gene in a permissive chromatin structure and, in so doing, help the general transcription machinery to assemble at the start site of transcription of the glucagon gene.
Acknowledgments
We greatly appreciate the gifts of unique reagents from the following individuals: R. H. Costa (anti-Foxa1 antibody, pCMV-Foxa1 and pCMV-Foxa2), Dr R. Maas (pGEX2T-GST-Pax6-PD) and K. S. Zaret (pMT2). We thank A. Matsiulka for excellent proofreading of this paper prior to submission. This work was funded by the Deutsche Forschungsgemeinschaft (SFB402/A3).
References
- 1.Unger R. H., Orci L. Glucagon and the A cell: physiology and pathophysiology. N. Engl. J. Med. 1981;304:1518–1524. doi: 10.1056/NEJM198106183042504. [DOI] [PubMed] [Google Scholar]
- 2.Unger R. H., Orci L. Glucagon and the A cell: physiology and pathophysiology. N. Engl. J. Med. 1981;304:1575–1580. doi: 10.1056/NEJM198106253042604. [DOI] [PubMed] [Google Scholar]
- 3.Lefebvre P. J. Glucagon and its family revisited. Diabetes Care. 1995;18:715–730. doi: 10.2337/diacare.18.5.715. [DOI] [PubMed] [Google Scholar]
- 4.Drucker D. J., Philippe J., Jepeal L., Habener J. F. Glucagon gene 5′-flanking sequences promote islet cell-specific gene transcription. J. Biol. Chem. 1987;262:15659–15665. [PubMed] [Google Scholar]
- 5.Philippe J., Drucker D. J., Knepel W., Jepeal L., Misulovin Z., Habener J. F. Alpha-cell-specific expression of the glucagon gene is conferred to the glucagon promoter element by the interactions of DNA-binding proteins. Mol. Cell. Biol. 1988;8:4877–4888. doi: 10.1128/mcb.8.11.4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knepel W. The α-cell and regulation of glucagon gene transcription. In: Habener J. F., Hussain M. A., editors. Molecular Basis of Endocrine Pancreas Development and Function. Boston, MA: Kluwer Academic Publishers; 2001. pp. 67–89. [Google Scholar]
- 7.Philippe J., Morel C., Prezioso V. R. Glucagon gene expression is negatively regulated by hepatocyte nuclear factor 3 beta. Mol. Cell. Biol. 1994;14:3514–3523. doi: 10.1128/mcb.14.5.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fürstenau U., Schwaninger M., Blume R., Kennerknecht I., Knepel W. Characterisation of a novel protein kinase C response element in the glucagon gene. Mol. Cell. Biol. 1997;17:1805–1816. doi: 10.1128/mcb.17.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lantz K. A., Kaestner K. H. Winged-helix transcription factors and pancreatic development. Clin. Sci. 2005;108:195–204. doi: 10.1042/CS20040309. [DOI] [PubMed] [Google Scholar]
- 10.Overdier D. G., Porcella A., Costa R. H. The DNA-binding specificity of the hepatocyte nuclear factor 3/forkhead domain is influenced by amino-acid residues adjacent to the recognition helix. Mol. Cell. Biol. 1994;14:2755–2766. doi: 10.1128/mcb.14.4.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J. C., Stafford J. M., Scott D. K., Sutherland C., Granner D. K. The molecular physiology of hepatic nuclear factor 3 in the regulation of gluconeogenesis. J. Biol. Chem. 2000;275:14717–14721. doi: 10.1074/jbc.275.19.14717. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y., Shen W., Brubaker P. L., Kaestner K. H., Drucker D. J. Foxa3 (HNF-3gamma) binds to and activates the rat proglucagon gene promoter but is not essential for proglucagon gene expression. Biochem. J. 2002;366:633–641. doi: 10.1042/BJ20020095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gauthier B. R., Schwitzgebel V. M., Zaiko M., Mamin A., Ritz-Laser B., Philippe J. Hepatic nuclear factor-3 (HNF-3 or Foxa2) regulates glucagon gene transcription by binding to the G1 and G2 promoter elements. Mol. Endocrinol. 2002;16:170–183. doi: 10.1210/mend.16.1.0752. [DOI] [PubMed] [Google Scholar]
- 14.Morgenstern B., Frech K., Dress A., Werner T. DIALIGN: finding local similarities by multiple sequence alignment. Bioinformatics. 1998;14:290–294. doi: 10.1093/bioinformatics/14.3.290. [DOI] [PubMed] [Google Scholar]
- 15.Nian M., Drucker D. J., Irwin D. Divergent regulation of human and rat proglucagon gene promoters in vivo. Am. J. Physiol. Gastrointest. Liver Physiol. 1999;277:G829–G837. doi: 10.1152/ajpgi.1999.277.4.G829. [DOI] [PubMed] [Google Scholar]
- 16.Quandt K., Frech K., Karas H., Wingender E., Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai E., Prezioso V. R., Tao W., Chen W. S., Darnell J. E., Jr Hepatocyte nuclear factor 3 alpha belongs to a gene family in mammals that is homologous to the Drosophila homeotic gene fork head. Genes Dev. 1991;5:416–427. doi: 10.1101/gad.5.3.416. [DOI] [PubMed] [Google Scholar]
- 18.Pani L., Overdier D. G., Porcella A., Qian X., Lai E., Costa R. H. Hepatocyte nuclear factor 3 beta contains two transcriptional activation domains, one of which is novel and conserved with the Drosophila fork head protein. Mol. Cell. Biol. 1992;12:3723–3732. doi: 10.1128/mcb.12.9.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Epstein J., Cai J., Glaser T., Jepeal L., Maas R. Identification of a Pax paired domain recognition sequence and evidence for DNA-dependent conformational changes. J. Biol. Chem. 1994;269:8355–8361. [PubMed] [Google Scholar]
- 20.Nordeen S. K. Luciferase reporter gene vectors for analysis of promoters and enhancers. BioTechnique. 1988;6:454–457. [PubMed] [Google Scholar]
- 21.Schwaninger M., Lux G., Blume R., Oetjen E., Hidaka H., Knepel W. Membrane depolarization and calcium influx induce glucagon gene transcription in pancreatic islet cells through the cyclic AMP-responsive element. J. Biol. Chem. 1993;268:5168–5177. [PubMed] [Google Scholar]
- 22.Luo C., Burgen E., Carew J. A., McCaffrey P. G., Badalian T. M., Lane W. S., Hogan P. G., Rao A. Recombinant NFAT1 (NFATp) is regulated by calcineurin in T cells and mediates transcription of several cytokine genes. Mol. Cell. Biol. 1996;16:3955–3966. doi: 10.1128/mcb.16.7.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bertheisen J., Zappavigna V., Mavillo F., Biasi F. Prep1, a novel functional partner of Pbx proteins. EMBO J. 1998;17:1423–1433. doi: 10.1093/emboj/17.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaret K. S., Liu J. K., DiPersio C. M. Site-directed mutagenesis reveals a liver transcription factor essential for the albumin transcriptional enhancer. Proc. Natl. Acad. Sci. U.S.A. 1990;87:5469–5473. doi: 10.1073/pnas.87.14.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takaki R., Ono J., Nakamura M., Yokogawa Y., Kumae S., Hiraoka T., Yamaguchi K., Hamaguchi K., Uchida S. Isolation of glucagon-secreting cell lines by cloning insulinoma cells. In Vitro Cell Dev. Biol. 1986;22:120–126. doi: 10.1007/BF02623498. [DOI] [PubMed] [Google Scholar]
- 26.Powers A. C., Efrat S., Mojsov S., Spector D., Habener J. F., Hanahan D. Proglucagon processing similar to normal islets in pancreatic alpha-like cell line derived from transgenic mouse tumor. Diabetes. 1990;39:406–414. doi: 10.2337/diab.39.4.406. [DOI] [PubMed] [Google Scholar]
- 27.Knepel W., Jepeal L., Habener J. F. A pancreatic islet cell-specific enhancer-like element in the glucagon gene contains two domains binding distinct cellular proteins. J. Biol. Chem. 1990;265:8725–8735. [PubMed] [Google Scholar]
- 28.Knepel W., Vallejo M., Chafitz J. A., Habener J. F. The pancreatic islet-specific glucagon G3 transcription factors recognize control elements in the rat somatostatin and insulin-I genes. Mol. Endocrinol. 1991;5:1457–1466. doi: 10.1210/mend-5-10-1457. [DOI] [PubMed] [Google Scholar]
- 29.Schreiber E., Matthias P., Muller M. M., Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paulweber B., Sandhofer F., Wilson B. E. The mechanism by which the human apolipoprotein B gene reducer operates involves blocking of transcriptional activation by hepatocyte nuclear factor 3. Mol. Cell. Biol. 1993;13:1534–1546. doi: 10.1128/mcb.13.3.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knepel W., Chafitz J., Habener J. F. Transcriptional activation of the rat glucagon gene by the cyclic AMP-responsive element in pancreatic islet cells. Mol. Cell. Biol. 1990;10:6799–6804. doi: 10.1128/mcb.10.12.6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zaret K. S., Stevens K. Expression of a highly unstable and insoluble transcription factor in Escherichia coli: purification and characterisation of the fork head homolog HNF3 alpha. Protein Exp. Purif. 1995;6:821–825. doi: 10.1006/prep.1995.0014. [DOI] [PubMed] [Google Scholar]
- 33.Fürstenau U., Schwaninger M., Blume R., Jendrusch E. M., Knepel W. Characterisation of a novel calcium response element in the glucagon gene. J. Biol. Chem. 1999;274:5851–5860. doi: 10.1074/jbc.274.9.5851. [DOI] [PubMed] [Google Scholar]
- 34.Klemsz M. J., McKercher S. R., Celada A., van Beveren C., Maki R. A. The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell (Cambridge, Mass.) 1990;61:113–124. doi: 10.1016/0092-8674(90)90219-5. [DOI] [PubMed] [Google Scholar]
- 35.Costa R. H., Grayson D. R., Darnell J. E., Jr Multiple hepatocyte-enriched nuclear factors function in the regulation of transthyretin and alpha 1-antitrypsin genes. Mol. Cell. Biol. 1989;9:1415–1426. doi: 10.1128/mcb.9.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jackson D. A., Rowader K. E., Stevens K., Jiang C., Milos P., Zaret K. S. Modulation of liver-specific transcription by interactions between hepatocyte nuclear factor 3 and nuclear factor 1 binding DNA in close apposition. Mol. Cell. Biol. 1993;13:2401–2410. doi: 10.1128/mcb.13.4.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaudet J., Mango S. E. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4A. Science. 2002;295:821–825. doi: 10.1126/science.1065175. [DOI] [PubMed] [Google Scholar]
- 38.Cirillo L. A., Lin F. R., Cuesta I., Friedman D., Jarnik M., Zaret K. S. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]