Abstract
Mono-ADP-ribosylation is a post-translational modification that regulates the functions of target proteins or peptides by attaching an ADP-ribose moiety. Here we report the purification, molecular cloning, characterization and tissue-specific distribution of novel arginine-specific Arts (ADP-ribosyltransferases) from chicken. Arts were detected in various chicken tissues as GPI (glycosylphosphatidylinositol)-anchored forms, and purified from the lung membrane fraction. By molecular cloning based on the partial amino acid sequence using 5′- and 3′-RACE (rapid amplification of cDNA ends), two full-length cDNAs of chicken GPI-anchored Arts, cgArt1 (chicken GPI-anchored Art1) and cgArt2, were obtained. The cDNA of cgArt1 encoded a novel polypeptide of 298 amino acids which shows a high degree of identity with cgArt2 (82.9%), Art6.1 (50.2%) and rabbit Art1 (42.1%). In contrast, the nucleotide sequence of cgArt2 was identical with that of Art7 cloned previously from chicken erythroblasts. cgArt1 and cgArt2 proteins expressed in DT40 cells were shown to be GPI-anchored Arts with a molecular mass of 45 kDa, and these Arts showed different enzymatic properties from the soluble chicken Art, Art6.1. RNase protection assays and real-time quantitative PCR revealed distinct expression patterns of the two Arts; cgArt1 was expressed predominantly in the lung, spleen and bone marrow, followed by the heart, kidney and muscle, while cgArt2 was expressed only in the heart and skeletal muscle. Thus GPI-anchored Arts encoded by the genes cgArt1 and cgArt2 are expressed extensively in chicken tissues. It may be worthwhile determining the functional roles of ADP-ribosylation in each tissue.
Keywords: ADP-ribosyltransferase, DT40 cell, glycosylphosphatidylinositol (GPI) anchor, NAD+
Abbreviations: AP, adapter primer; Art, ADP-ribosyltransferase; cgArt, chicken glycosylphosphatidylinositol-anchored Art; ConA, concanavalin A; DTT, dithiothreitol; FAM, 6-carboxyfluorescein; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GPI, glycosylphosphatidylinositol; MGB, minor groove binding; PHA, phytohaemagglutinin; PI-PLC, phosphatidylinositol-specific phospholipase C; RACE, rapid amplification of cDNA ends; RPA, RNase protection assay; RT-PCR, reverse transcription–PCR
INTRODUCTION
Mono-ADP-ribosylation is a post-translational modification whereby a single ADP-ribose moiety of NAD+ is transferred to the amino acid residues of target proteins. Bacterial toxins, such as cholera, pertussis, diphtheria and clostridial toxins, are well known examples of mono-ADP-ribosyltransferases, and have been characterized with regard to their molecular structure, substrate specificity and functions [1]. ADP-ribosylation of eukaryotic proteins by these toxins, which alters the activity of proteins in critical metabolic or regulatory pathways, is a common mechanism in bacterial pathogenesis. For example, cholera toxin ADP-ribosylates an arginine residue of Gsα protein and inhibits its GTPase activity [2,3].
In eukaryotes, arginine-specific Arts (ADP-ribosyltransferases) have been shown to be present in GPI (glycosylphosphatidylinositol)-anchored or soluble forms in mammals and birds [4,5]. The family of vertebrate Arts was classified into seven members (Art1–Art7), based on their similarities in amino acid sequence and conservation of gene structures [6–8]. These Arts modify the arginine residue(s) of diverse target proteins, which could contribute to the regulation of various cellular functions [6–10]. For example, Art1 present in skeletal muscle and heart as a GPI-anchored form has been suggested to be involved in myogenesis [11–13], and Art2 (Rt6), localized predominantly in rodent T-cells with a GPI anchor, regulates the proliferation and apoptosis of lymphocytes [14,15].
Avian Art activities have been described in the turkey [16,17] and chicken [18–20]. Of these, Art6.1 is a soluble Art localized in the granules of chicken heterophils (polymorphonuclear leucocytes), and has been shown to be released into the extracellular space by various secretagogues, indicating possible functional roles in inflammatory sites [18–20]. Although Art7 was obtained from erythroblast cDNA and genomic DNA libraries of chicken using Art1 cDNA as a probe [21], the protein has not yet been fully characterized.
Previously we detected a novel GPI-anchored Art activity in chicken spleen membranes [22], indicating that chicken tissues contain not only soluble but also GPI-anchored Arts. Thus we attempted to identify the novel chicken Art. In the present study, we show that GPI-anchored Arts are widely distributed in chicken tissues. Here we describe the purification, molecular cloning, characterization and tissue-specific expression of novel GPI-anchored Arts in the chicken.
MATERIALS AND METHODS
Materials
Chickens were obtained from a local slaughterhouse. [adenylate-32P] NAD+ (29.6 TBq/mmol) and [α-32P]UTP (29.6 TBq/mmol) were purchased from New England Nuclear. [α-32P]dCTP (110 TBq/mmol), [carbonyl-14C] NAD+ (1.96 GBq/mmol) and ECL® Western blotting detection reagents were from Amersham Biosciences. PI-PLC (phosphatidylinositol-specific phospholipase C) from Bacillus cereus, L-arginine, poly(L-arginine), bathophenanthroline disulphonic acid and D-mannosamine were purchased from Sigma. Chicken B-cell lymphoma DT40 cells (JCRB 9130) were obtained from the Health Science Research Resource Bank (HSRRB), Japan.
Measurement of Art and NAD+ glycohydrolase activities
Each fresh chicken tissue (1 g) was cut and homogenized with 9 vol. of sucrose buffer (5 mM Tris/HCl, pH 8.0, 0.25 M sucrose, 3 mM CaCl2 and 1 mM EDTA), and centrifuged at 8000 g for 20 min. The supernatants were pelleted at 105000 g for 1 h, and suspended in 500 μl of buffer A (20 mM Tris/HCl, pH 7.5, 0.2 mM PMSF, 0.1 mM EDTA and 0.02% NaN3). Aliquots of the suspension were used as the membrane fraction. In some cases, an aliquot of 100 μl was incubated with 0.5 unit of PI-PLC at 30 °C for 30 min followed by centrifugation (105000 g for 1 h), and the supernatant was used as the PI-PLC-treated fraction.
Art activity was determined using L-arginine or poly(L-arginine) as the substrate. For the L-arginine assay, each membrane fraction (5 μl) was incubated with 50 mM Tris/HCl (pH 9.0), 5 mM NAD+ and 0.1 M L-arginine at 37 °C for 1 h in a total volume of 100 μl. After the reaction was terminated by adding 100 μl of 0.2% SDS, the sample was capillary electrophoresed and analysed as described previously [22]. For the poly(L-arginine) assay, each membrane fraction (5 μl) was incubated with 50 mM Tris/HCl (pH 9.0), 0.1 mM [32P]NAD+ (3.7 kBq/tube) and 0.1 mg/ml poly(L-arginine) at 25 °C for 20 min in a total volume of 200 μl, and the radioactivity in the acid-insoluble fraction was measured. In some cases, 5 μg of p33 purified from chicken heterophils [18], α-actin, casein or histone was used as substrate instead of poly(L-arginine). The optimal pH was determined using potassium phosphate (pH 5.5–7.5), Tris/HCl (pH 7.0–9.5) and glycine/NaOH (pH 8.5–11.0) buffers. The effects of the reagents were determined over concentration ranges 0–500 mM (NaCl), 0–1 mM (β-NMN) and 0.5–2.0 mM (novobiocin). In situ zymographic assays were carried out as described previously [19] using 10 μl of each membrane fraction.
The NAD+ glycohydrolase assay was performed as described previously [23]. Briefly, the enzyme fractions were incubated with 50 mM Tris/HCl (pH 8.0), 0.1 mM NAD+ and [carbonyl-14C]NAD+ (0.1 μCi/assay) in the presence (Art activity) or absence (NAD+ glycohydrolase activity) of 0.1 M L-arginine at 30 °C for 2 h in a total volume of 50 μl. The samples (20 μl) were separated on a reverse-phase Cosmosil 5C-18MS column (4.6 mm×150 mm; Nacalai Tesque) with 0.05% (v/v) trifluoroacetic acid as the mobile phase, and the radioactivity of reacted nicotinamide was measured. cgArt1 (chicken GPI-anchored Art1) and cgArt2 enzyme fractions were obtained from COS-7 cells transfected with each cDNA by partial purification using ConA (concanavalin A)–Sepharose chromatography. Basal activity found in the preparation from pcDNA3-transfected cells was subtracted from the NAD+ glycohydrolase activity of each cgArt.
Purification of Art from chicken lung, and amino acid sequencing
The membrane fraction obtained from chicken lung (250 g) was suspended in 500 ml of buffer A, incubated with 20 units of PI-PLC overnight at room temperature, and centrifuged at 105000 g for 1 h. The supernatant was diluted 5-fold and loaded on to a CM52 column equilibrated with buffer B (20 mM Tris/HCl, pH 7.5, 150 mM NaCl, 0.2 mM PMSF, 0.1 mM EDTA and 0.02% NaN3). The flow-through fraction was applied directly to a ConA–Sepharose column equilibrated with buffer A, and eluted with 0.5 M methyl α-D-glucoside in buffer A. The enzyme-rich fraction was subjected to sequential column chromatography on Sephadex G-100 (equilibrated with buffer B), MonoQ (HPLC; equilibrated with buffer A and eluted with 0–0.5 M NaCl in buffer A), ConA–Sepharose (equilibrated with buffer A, and eluted with 0.5 M methyl α-D-glucoside in buffer A) and TSK-3000SW (HPLC; equilibrated with buffer B). Finally, 80 μg of the nearly homogeneous protein was obtained.
N-terminal sequencing of the purified transferase (2 μg) was performed by automated Edman degradation using a model 477 or 492 peptide sequencer (Applied Biosystems, Foster City, CA, U.S.A.).
Detection of glycoprotein
To detect glycoproteins, electroblotted Art was reacted with the lectin PHA (phytohaemagglutinin)-E4 labelled with biotin, and then with avidin-labelled horseradish peroxidase (Honen Lectin Sensor plus C kit, Seikagaku Co., Tokyo, Japan).
RT-PCR (reverse transcription–PCR)
Chicken lung mRNA was prepared using a QuickPrep Micro mRNA purification kit (Amersham Biosciences), and reverse-transcribed with Moloney murine leukaemia virus reverse transcriptase and random primers. The first strand was amplified with the degenerate primers corresponding to the N-terminal amino acid sequence (KKGPIKEV) of the purified lung Art (AARAARGGNCCNATHAARGARGT), and to the catalytic region amino acid sequence (EVLIPPFE) that is conserved between vertebrate Arts (TCRAANGGNGGDATNAGNACYTC) (see Figures 3A and 3B).
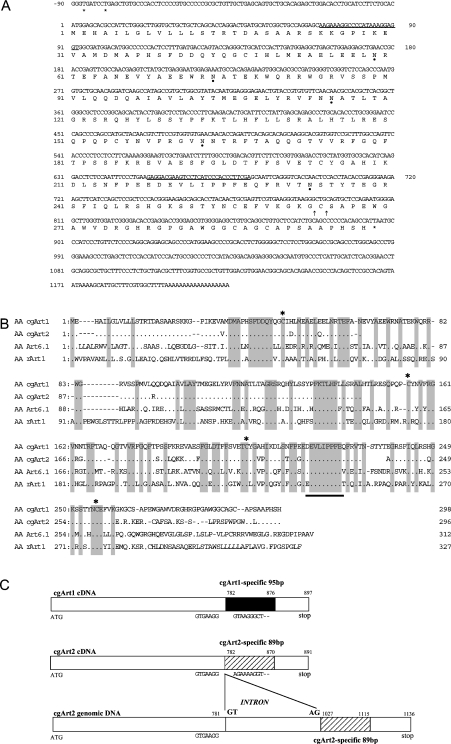
Figure 3. Nucleotide and deduced amino acid sequences of cgArt1, and structures of cgArts.
(A) The nucleotide sequence (cgArt1; GenBank® accession number AY772192) is numbered relative to the initiation codon. Termination codons are indicated by asterisks. The degenerate oligonucleotides used as primers for RT-PCR are underlined. Possible sites for addition of the GPI anchor are indicated by arrows. Predicted sites for N-glycosylation are indicated by closed circles. (B) Comparison of the amino acid (AA) sequences of representative vertebrate Arts. Catalytic regions and four cysteine residues conserved between the Arts are underlined and indicated by asterisks respectively. Amino acid sequences of cgArts were compared with those of Art6.1 (GenBank® accession no. D31864) and rabbit Art1 (rArt1; M98764) using Genetyx-Mac 10 software. (C) Schematic representations of the structures of cgArt1 and cgArt2. Each cDNA encodes a consensus catalytic region, which is common between vertebrate Arts, and a distinct region that lies at the 3′-end (cgArt1- or cgArt2-specific region). The nucleotide sequences excluding the cgArt-specific regions show striking identity of 94% between cgArt1 and cgArt2. The cgArt2 sequence obtained from genomic DNA (GenBank® accession no. AY772194) included a 245 bp intron between positions 781 and 782 of cgArt2 cDNA.
5′- and 3′-RACE (rapid amplification of cDNA ends)
The reverse-transcribed cDNA was prepared and ligated with cDNA adaptor primer AP-1 using a Marathon cDNA amplification kit (Clontech). 5′-RACE was performed with gene-specific primers CGATR1 (5′-CACCGTGCCTTGCTGTGCTG-3′) and CGATR2 (5′-CATCGCCACCTCCTTTATGG-3′), and adaptor primers AP-1 and AP-2. 3′-RACE was performed with the adaptor-ligated oligo(dT)-primed double-stranded cDNA prepared using a cDNA amplification kit, with the gene-specific primer CGATF1 (5′-ACTGAGCAGAGCCCTGCACACCC-3′) and AP-1, and then with nested PCR primers CGATF2 (5′-AGCTTCATCCAGCTCCGCTCCC-3′) and AP-2. Both 5′- and 3′-RACE products were inserted into pGEM-T or pGEM 3zf(+) vectors (Promega, Madison, WI, U.S.A.).
Transfection and expression of cgArts in COS-7 and DT40 cells
The entire coding regions of the cgArts were inserted into pcDNA3 at the BamHI site (Invitrogen). The plasmids pcDNA3-cgArt1 and pcDNA3-cgArt2 were transfected into COS-7 cells with PolyFect Transfection Reagents (Qiagen, Hilden, Germany). In some cases, COS-7 cells were treated simultaneously with Dulbecco's modified Eagle's medium containing 2.5 mM D-mannosamine (Sigma), 0.2 mg/ml glucose and 10% (v/v) fetal calf serum to inhibit the incorporation of mannose residues into the glycan portion of the GPI anchor and thus reduce synthesis of the anchor [24,25]. DT40 cells (5×107 cells) suspended in 0.5 ml of PBS were incubated on ice for 10 min with 30 μg of each plasmid linearized with BglII, and subjected to electroporation (250 V, 500 μF; Bio-Rad Gene Pulser). The cells were plated and incubated for 1 day in RPMI 1640 medium, and selected with 2 mg/ml G418 for 2–3 weeks.
Northern blot analysis
Northern blot analysis was carried out with 20 μg of total RNA as described previously [19,26]. The blots were prehybridized at 43 °C for 2 h in hybridization buffer [5×SSPE (1×SSPE=0.15 M NaCl, 10 mM sodium dihydrogen phosphate, 1.25 mM EDTA), 50% (v/v) formamide, 5×Denhardt's solution (0.02% Ficoll 400/0.02% polyvinylpyrrolidene/0.02% BSA), 0.5% SDS and 50 μg/ml salmon sperm carrier DNA]. The oligonucleotide probe [897 bp entire coding region for cgArt1 and 456 bp 3′-region for chicken GAPDH (glyceraldehyde-3-phosphate dehydrogenase)] labelled with [α-32P]dCTP was added and incubated for a further 16 h. After hybridization, the blots were washed twice with 2×SSPE/0.1% SDS at room temperature, and twice with 1×SSPE/0.1% SDS at 55 °C.
RPA (RNase protection assay)
The templates for cgArt1 and cgArt2 were produced from pcDNA3-cgArt1/2 by PCR (the forward primers for cgArt1 and cgArt2 were 5′-GTAAGGGCTGCAGTGCTCC-3′ and 5′-AGAAAAGGTGCAAGGAGCGG-3′ respectively, and the reverse primer behind the SP6 promoter in pcDNA3 was 5′-TGATCAGCGAGCTCTAGC-3′). The GAPDH template was prepared by linearization of pGEM-cGAPDH using EcoRI. Antisense riboprobes were transcribed in vitro with SP6 RNA polymerase and [α-32P]UTP according to the manufacturer's protocol (MAXI-script In Vitro Transcription SP6/T7 kit; Ambion, Austin, TX, U.S.A.). After DNase I digestion, the radiolabelled probes were purified on denaturing gel containing 5% (w/v) acrylamide and 8 M urea. Using a Ribonuclease Protection Assay III kit (Ambion), chicken RNA was hybridized with the radiolabelled probe (5×105 c.p.m. per tube) at 60 °C overnight, and then treated with RNase A/T1 at 37 °C for 30 min. Protected fragments were analysed on a denaturing gel followed by autoradiography.
Real-time quantitative PCR: TaqMan assay
Specific primers and probes for analysis of cgArt1 and cgArt2 expression were designed using Primer Express software (ABI Model 7000 sequence detector; Applied Biosystems). For cgArt1, the forward and reverse primers were 5′-C382GGTGTCCCCGATCCA397-3′ and 5′-C450TGCGAGTTCGTGAAGGGTAAG429-3′ respectively, and the internal probe was 5′-(FAM)-C399CAAGCTCCCCATTC413-(MGB)-3′, where FAM is 6-carboxyfluorescein and MGB is minor groove binding. For cgArt2, the forward and reverse primers were 5′-G405GGACTGCTCTTATCTGCACT-GA427-3′ and 5′-C474TGCGAGTTTGTGAAGGAGAAAA452-3′ respectively, and the internal probe was 5′-(FAM)-C431GCATGGCCGCTG443-(MGB)-3′. Chicken total RNA (2 μg; from chickens ∼2 months old for bone marrow cells; and from 6-month-old chickens for all others) was reverse transcribed, and aliquots of the products were used as templates. Each sample contained 1×TaqMan Universal PCR Master Mix, 250 nM TaqMan probe, 900 nM gene-specific primers and 2 μl of reverse-transcribed cDNA or sequentially diluted plasmids (pcDNA3-cgArt1/2) as standard in a total volume of 50 μl in triplicate. The reaction conditions were 2 min at 50 °C, 10 min at 95 °C, then 40 cycles each of 15 s at 95 °C and 1 min at 60 °C on MicroAmpOptical 96-well plates. The results are expressed as relative copy numbers compared with standards.
RESULTS
Detection of Art activities in the membrane fractions of chicken tissues
Previously, we detected Art activity in the membrane fraction of chicken spleen [22]. To determine whether Art activities are present in other chicken tissues, each membrane fraction was subjected to a poly(L-arginine) assay. Spleen and lung membrane fractions demonstrated high Art activities, followed by heart, liver and kidney. The spleen membrane fraction exhibited the highest total activity (1140 nmol/h per g of tissue), whereas the lung membrane fraction showed the highest specific activity (224 nmol/h per mg of protein) in the poly(L-arginine) assay. Similar results were obtained with the L-arginine assay. The Art activities in the nuclear, mitochondrial and supernatant fractions in each tissue were below the level of detection (results not shown).
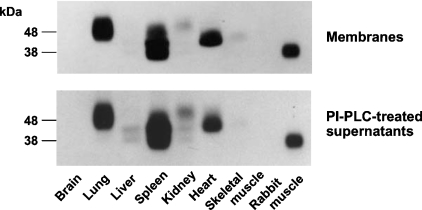
Next, we carried out in situ zymographic assays to determine the molecular size of the Art in each tissue. As shown in Figure 1 (upper panel), lung, spleen and heart membrane fractions demonstrated strongly radiolabelled spots, followed by kidney and skeletal muscle. These Arts exhibited higher molecular masses (between 40 and 50 kDa) as compared with the size of Art6.1 (27.5 kDa) [18,19] and rabbit Art1 (38 kDa) [11,12]. Thus the enzyme activities and in situ zymographic assays indicated that Arts with different molecular sizes are widely distributed in chicken tissues.
Figure 1. In situ zymographic analysis of chicken Arts.
The membrane fraction (upper panel) prepared from each chicken tissue and the PI-PLC-treated supernatant (lower panel) of the membrane fraction were subjected to in situ zymographic analysis. The labels on the left indicate the migration of protein size markers (in kDa). The data are representative of four independent experiments.
To determine whether these Arts are GPI-anchored proteins, as is the case for the spleen Art [22], we treated each membrane fraction with PI-PLC. The results indicated that up to 85% of the lung Art activity in the membrane fraction was released by PI-PLC into the supernatant. The PI-PLC-treated supernatant of each membrane fraction was subjected to in situ zymographic assay, and the cleaved Arts migrated slightly more slowly than those in the membrane fractions (Figure 1, lower panel), indicating that each Art was attached to the membrane via a GPI anchor. Extraction by 1 M NaCl or 1% Triton X-100 failed to release the enzymes from the membrane fractions (results not shown).
Purification of GPI-anchored Art from chicken lung and its N-linked glycosylation
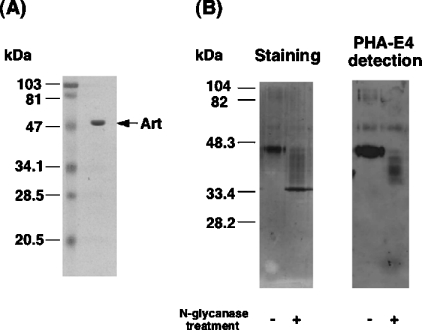
To characterize the structure and properties of a possible cgArt, we purified the transferase from chicken lung, as described in the Materials and methods section, more than 6700-fold with a yield of 14.8%. The specific activity of the transferase was 1.3 mmol/h per mg, comparable with that of purified Art6.1 (0.4 mmol/h per mg) [18]. SDS/PAGE analysis of the purified lung Art revealed a single band with an apparent molecular mass of 48 kDa (Figure 2A).
Figure 2. SDS/PAGE analysis of purified chicken lung Art and effects of N-glycanase treatment.
(A) The final preparation (3 μg) following purification of lung Art was analysed by SDS/PAGE, and stained with Coomassie Brilliant Blue R-250. The arrow indicates the position of purified Art. (B) Effects of N-glycanase treatment on molecular size and reactivity to lectin PHA-E4 of purified Art. Purified Art (8 μg) was treated or not with N-glycanase (0.4 unit) at 37 °C for 18 h and subjected to SDS/PAGE under reducing conditions. Staining of the protein (left) and PHA-E4-reacted protein bands (right) is shown. The data are representative of three independent experiments. Protein sizes are indicated in kDa.
When the purified Art was treated with N-glycanase, which cleaves the amide bonds between GlcNAc and asparagine residues of N-linked glycoproteins, the apparent molecular size on SDS/PAGE was decreased to 34 kDa (Figure 2B). The N-glycanase-treated enzyme, blotted on nitrocellulose from SDS/PAGE, was reacted with several lectins conjugated with biotin and stained using an avidin-conjugated peroxidase reaction. PHA-E4, which binds specifically to some bisected complex-type oligosaccharides [27], recognized the purified enzyme, but did not recognize it after cleavage of glycosyl moieties by N-glycanase (Figure 2B). These results indicate the presence of asparagine-linked glycosyl moieties on the enzyme.
Molecular cloning of chicken lung Art
The N-terminal amino acid sequence of the purified lung Art was determined to be SKKGPIKEVAMDMAPHSFDDQY. Degenerate oligonucleotide primers corresponding to a stretch of the N-terminal sequence (K24KGPIKEV31) and a region that is highly conserved among vertebrate Arts (E220VLIPPFE227) were used for RT-PCR using chicken lung mRNA. Based on the cDNA sequence from the obtained RT-PCR product (approx. 610 bp), we performed 5′- and 3′-RACE, and obtained two full-length cDNAs, designated cgArt1 and cgArt2 respectively (GenBank® accession numbers AY772192 and AY772193 respectively; Figure 3A). Both cgArt cDNAs have two in-frame termination codons (TGA) at 81 and 87 bp upstream of the ATG initiation codon in the 5′-untranslated region. In the 3′-untranslated region, an AATAA polyadenylation signal is present approx. 20 bp upstream of the polyadenylated tail. On the 5′-side of the coding region, a possible leader sequence of 22 amino acids is followed by the sequence SKKGPIKEVAMDMAPHSFDDQY, which was determined using the purified Art. On the 3′-side of the coding region, possible signal sequences and sites for the GPI anchor are present (see Figure 3A). There are five possible N-glycosylation sites, which could explain the higher molecular mass of the purified Art as compared with the estimated value. The entire coding region of cgArt1 shows 88.8% and 82.9% similarity with cgArt2 at the nucleotide and amino acid sequence levels respectively, and consists of 897 bp nucleotides (298 amino acids) with a predicted molecular mass of 33619 Da. Moreover, as shown in Figure 3(B), the predicted amino acid sequence of the entire coding region of cgArt1 shows a high degree of sequence identity with those of chicken Art6.1 (50.2%), human Art1 (44.1%) and rabbit Art1 (42.1%), retaining the conserved catalytic region, including a glutamate residue critical for the transferase activity, and four cysteine residues that may be involved in disulphide bond formation [28,29]. The cgArt2 nucleotide sequence is identical with that of Art7 [21]. cgArt1 and cgArt2 contain distinct sequences as long as 95 bp and 89 bp respectively in the 3′-region (cgArt1- and cgArt2-specific regions), which show 44.7% and 22.5% identity at the nucleotide and amino acid sequence level respectively (Figure 3C). The nucleotide sequences show 94% identity outside these specific regions.
PCR analysis of chicken genomic DNA demonstrated a possible intron of 245 bp after position 781 in the sequence of cgArt2, but not cgArt1 (cgArt2 genomic DNA; GenBank® accession number AY772194). Interestingly, the sequence of a 91 bp stretch at the 5′-end of this intron resembles the cgArt1-specific region (see Figure 3C), although the sequence did not appear in the cgArt2 cDNA. In contrast with cgArt2, the cgArt1 sequence obtained by PCR amplification from the genomic DNA was identical with that obtained by RT-PCR, indicating that cgArt1 had no intron. However, the position of the intron in cgArt2 is quite different from that of Art7, in which an intron was reported to be located close to the 5′-end [21].
Expression of recombinant cgArt1 and cgArt2 proteins
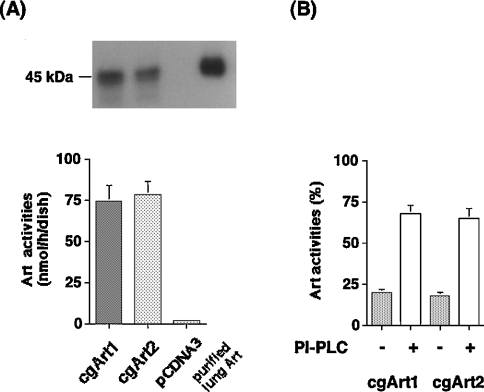
To determine whether these cDNAs encode catalytically active GPI-anchored Arts, the coding regions of cgArt1 and cgArt2 inserted into the mammalian expression vector pcDNA3 were stably transfected into DT40 cells. As shown in Figure 4(A), apparent enzyme activities were detected in cell lysates transfected with both cDNAs, but not with pcDNA3. In situ zymographic analysis of each cell lysate revealed bands corresponding to 44–45 kDa, somewhat lower than the molecular mass of the purified lung Art (Figure 4A, upper panel). On subcellular fractionation analysis of the enzyme activity in the Art-expressing cells, approx. 60% of the transferase activity was detected in the membrane fraction in each cell lysate. As shown in Figure 4(B), PI-PLC treatment of the membrane caused significant release (up to 70%) of transferase activities into the supernatant. Furthermore, treatment of the transfected COS-7 cells with 2.5 mM D-mannosamine to inhibit the incorporation of mannose residues into the glycan portion of the GPI anchor [24,25] significantly decreased the membrane-bound Art activities of cgArt1 and cgArt2 to 45.8% and 40.2% respectively (compared with the corresponding activity in COS-7 cells without treatment with D-mannosamine; results not shown). Thus both cgArt1 and cgArt2 proteins are proved to be GPI-anchor-type Arts.
Figure 4. Detection of GPI-anchored Arts in pcDNA3-cgArt1/2-transfected DT40 cells.
Stable transfection of the pcDNA3-cgArt1 or pcDNA3-cgArt2 construct into DT40 cells was performed. (A) Art assay and in situ zymographic analysis were performed with the lysates of pcDNA3-cgArt1/2 or pcDNA3 (empty vector)-transfected cells and purified lung Art. (B) Release by PI-PLC treatment of recombinant Arts from the membrane fractions of the transfected DT40 cells. The data are expressed as means±S.D. for four independent experiments.
Expression levels of cgArt mRNAs
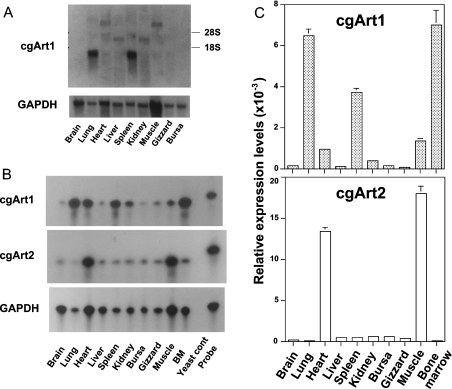
Northern blot analysis of chicken tissues using a probe spanning the entire coding region of cgArt1 revealed prominent bands of 1.3 kb in both lung and spleen, weak bands of approx. 9 kb in heart and muscle, and faint bands of 3.2 kb in liver and kidney (Figure 5A). The transcripts were barely detected in brain, gizzard or bursa. To discriminate between the expression levels of the two Art genes, we performed RPA and real-time quantitative PCR with specific probes for cgArt1 and cgArt2. As shown in Figure 5(B), RPA for cgArt1 revealed strong radiolabelled bands corresponding to the 116-base transcript in RNA from the lung, spleen, heart and bone marrow cells, whereas RPA for cgArt2 demonstrated prominent bands of 110 bases in RNA from the heart and muscle. Both probes failed to hybridize with a yeast RNA control. Using real-time quantitative PCR, marked expression of cgArt1 was demonstrated in the lung, spleen and bone marrow, while cgArt2 expression was detected in heart and muscle (Figure 5C), showing a good correlation with the results of RPA. Thus the distribution of cgArt1 transcripts in tissues was shown to be distinct from that of cgArt2.
Figure 5. Levels of expression of cgArt1 and cgArt2.
(A) Northern blot analysis of chicken GPI-anchored Arts. Total RNA (20 μg) from each chicken tissue was hybridized with the cgArt1 or chicken GAPDH probe. Labels on the right show the positions of 28 S and 18 S rRNAs. The upper and lower panels show the cgArt1 and chicken GAPDH Northern blot analyses respectively. The data are representative of four independent experiments. (B) RPA for the expression of cgArt1/2. Total RNA (25 μg) from each chicken tissue was hybridized with cgArt1, cgArt2 and chicken GAPDH probes and assayed. Bands corresponding to 116, 110 and 450 nucleotides for cgArt1 (top panel), cgArt2 (middle panel) and chicken GAPDH (bottom panel) respectively were detected. The protected band after hybridization with yeast RNA (5 μg) or the band without treatment of RNase (probe band) is shown as a control on the right of each panel. The data are representative of four independent experiments. BM, bone marrow. (C) Quantitative expression analysis of cgArt1 and cgArt2 mRNAs in chicken tissues. Total RNA (2 μg) from each tissue was reverse-transcribed, and the level of expression of each transferase was measured by real-time quantitative PCR. The data are expressed as means±S.D. for three independent experiments performed in triplicate.
Enzymatic properties of cgArts
Previously, we reported that Arts from spleen membrane and bone marrow cells respond differently to thiol agents and NaCl [19,22]. To characterize the enzymatic properties of cgArts, we examined the effects of DTT (dithiothreitol), NaCl, β-NMN and novobiocin, a potent inhibitor of Art [30], on the prepared transferases (Table 1). Thiol agents such as DTT or β-mercaptoethanol were prerequisites for expression of the maximum enzyme activity of Art6.1, but this was not the case for GPI-anchored Arts (results not shown). NaCl significantly decreased the transferase activity of cgArt1 and Art6.1, but not that of cgArt2. β-NMN significantly lowered the enzyme activity of cgArt2, although it had less effect on that of cgArt1, and did not affect Art6.1 activity. ADP-ribose, ADP, AMP and adenosine were less effective than β-NMN (results not shown). Novobiocin at 1 mM eliminated the activities of all transferases examined. We also examined the substrate specificity of chicken Arts. As shown in Table 1, the degree of ADP-ribosylation of α-actin and casein by cgArt1/2 was markedly lower than that catalysed by Art6.1, although the extent of modification of p33 and histone was nearly comparable. Thus the properties of the cgArts are different from those of Art6.1.
Table 1. Properties of chicken Arts.
The activities of chicken Arts were measured by poly(L-arginine) assay. The PI-PLC-treated supernatants of the membrane fractions from DT40 cells transfected with the pcDNA3-cgArt1/2 constructs were used as the enzyme fractions. Molecular masses were determined by SDS/PAGE and in situ zymographic assay. For measurement of substrate specificity, incorporation of radioactivity into the substrates by purified Art6.1 was set as 100%; incorporation by cgArt1 or cgArt2 is shown as relative to this.
| cgArt1 | cgArt2 | Art6.1 | |
|---|---|---|---|
| Molecular mass (kDa) | 45 | 45 | 27.5 |
| IC50 for: | |||
| NaCl (mM) | 125 | 278 | 102 |
| β-NMN (μM) | 350 | 21 | >1000 |
| Substrate specificity (%) | |||
| Actin | 3 | 2 | 100 |
| p33 | 97 | 100 | 100 |
| Casein | 8 | 8 | 100 |
| Histone | 65 | 130 | 100 |
To investigate whether cgArt1 and cgArt2 possess NAD+ glycohydrolase activity, as in the case of Art1 and Art2 [23,31], we prepared a partially purified enzyme fraction from COS-7 cells transfected with each cgArt cDNA, and carried out an NAD+ glycohydrolase assay. Both cgArt1 and cgArt2 demonstrated apparent NAD+ glycohydrolase activity (2.49 and 2.34 nmol/h per dish respectively). Art activity relative to NAD+ glycohydrolase activity was calculated as 20.7 and 17.7 for cgArt1 and cgArt2 respectively. Both of these values are higher than that for Art1 [31], indicating that NAD+ glycohydrolase activity of cgArts is less important.
DISCUSSION
Arts are widely distributed in diverse species, including rabbit, mouse, human and chicken. The tissue distribution also varies among species; Arts are found in muscle, heart, spleen, brain, T- cells and polymorphonuclear leucocytes [4–7,11–13,18,19]. The putative roles of ADP-ribosylation include functional changes via modification of many target proteins, including guanine- nucleotide-binding protein, actin, integrin, fibronectin and some physiologically active peptides [9,10,32–39]. There are at least two forms of Arts, i.e. GPI-anchored and soluble, and we reported previously the presence of not only the soluble form (Art6), but also GPI-anchored Arts, in the chicken [22].
In the present study, we demonstrate that GPI-anchored Arts are distributed extensively in chicken tissues. We purified GPI-anchored Art from the lung, and obtained two Art clones, designated cgArt1 and cgArt2; the former has a novel sequence, while the sequence of the latter is identical with that of Art7 [21]. Since Art7 cDNA is a composite sequence derived from an immature cDNA sequence filled with its genomic sequence at the 5′-terminus, the position of the deduced initiation codon was not confirmed by its cDNA sequence [21]. In the present paper, we successfully obtained a full-length cDNA and determined the initiation codon and in-frame stop codon (cgArt2 cDNA; GenBank® accession no. AY772193). The position of this initiation codon of cgArt2 is different from that of Art7. A sequence of approx. 90 bp that is distinct between cgArt1 and cgArt2 was detected on the 3′-side of the coding region, showing 44.7% and 22.5% identity at the nucleotide and amino acid sequence levels respectively (see Figure 3C). Sequence analysis of genomic DNA revealed that cgArt2 genomic DNA has a novel intron of 245 bp adjacent to its 3′-end, whereas cgArt1 has no intron. This intron contains a sequence that is similar to, but not identical with, the cgArt1-specific region, although the sequence was not observed in cgArt2 cDNA. The position of the intron in the present cgArt2 sequence was quite different from that reported previously, in which the intron was determined to be close to the 5′-end [21]. The nucleotide sequences of the cgArt1 and cgArt2 coding regions demonstrate striking identity of 88.8%; the coding region without each cgArt-specific region showed an even higher degree of identity of 94%. Moreover, when we searched for the cgArt1 and cgArt2 cDNA sequences in a chicken genome database (NCBI chicken genome resources), each sequence corresponded to the independent genome sequence with highest consistency. Thus cgArt2 and cgArt1 should be regarded as Art7 itself and its homologue respectively.
We expressed each cDNA in DT40 cells, and confirmed that both cgArt1 and cgArt2 proteins were expressed in the membrane fraction as GPI-anchored forms. Art7 was not recognized as a GPI-anchored transferase, as its C-terminus had a less typical hydrophobic signal sequence for a GPI anchor compared with Art1. However, the results of the present study confirmed that Art7 and its homologue cgArt1 are both GPI-anchored transferases, consistent with the observation that the purified transferases are GPI-anchored proteins. The proteins encoded by cgArt1 and cgArt2 without the N- and C-terminal signal sequences had predicted molecular mass of approx. 28 kDa, although the apparent molecular masses of the recombinant or purified proteins were 45–48 kDa (Figures 2A and 4). These differences may be due to the addition of sugar chains, i.e. aspargine-linked glycosyl moieties as shown in the purified enzyme, via post-translational modification. In fact, there were five and four predicted sites for N-glycosylation in the amino acid sequences of cgArt1 and cgArt2 respectively (see Figure 3A).
The tissue distributions of cgArt1 and cgArt2 determined by RPA and quantitative real-time PCR analysis revealed distinct expression patterns. The expression of cgArt1 mRNA was highest in the lung, spleen and bone marrow, followed by the heart, muscle and kidney. On the other hand, the expression of cgArt2 mRNA was limited to heart and muscle, which is reminiscent of the expression pattern of Art1 [12,13]. Northern blot analysis using full-length probes for cgArt1 and cgArt2 demonstrated transcripts of 9 kb in the heart and muscle, 3.2 kb in the liver and kidney, and 1.3 kb in the lung and spleen (see Figure 5). This, together with the data on tissue distribution from RPA and quantitative real-time PCR analysis, indicates that the 1.3 kb transcript apparently corresponds to cgArt1. Although there may be another gene encoding a GPI-anchored Art, at least two types of GPI-anchored transferases are confirmed to be expressed independently in chicken tissues.
In the present study, the activities of cgArt1 and Art6.1 were inhibited by increasing concentrations of NaCl, while the activity of cgArt2 was less sensitive. Four types of turkey erythrocyte Arts (A, B, C and A′) have been reported; types A and B were from the cytosol, and types C and A′ were from the plasma membrane and nucleus respectively [16,17]. The activities of transferases A and A′ were markedly enhanced by NaCl in a concentration-dependent manner, although that of transferase B was inhibited only slightly, and that of transferase C was enhanced 2-fold by 300 mM NaCl. These observations, taken together with salt sensitivity, molecular size and subcellular distribution, demonstrate that cgArt1 and cgArt2 are clearly distinct transferases from the turkey Arts.
Arts have been shown to be expressed in mammalian lung tissue. Art1 was demonstrated to be expressed weakly in mouse lung by Northern blot analysis [40]. Both Art1 mRNA and the protein itself have been detected in human bronchial epithelial cells. Furthermore, Art3 and Art4 mRNA expression has been observed by in situ hybridization and RT-PCR analysis [41]. Human neutrophil peptide-1, which possesses antimicrobial and cytotoxic activities, was also shown to be modified by Art1 localized in airway epithelial cells, resulting in a decrease in its biological activity [42]. Tuftsin, a phagocytosis-activating peptide, was demonstrated to be modified by Art6, resulting in loss of its activity [38]. These reports suggested that ADP-ribosylation of the immunomodulatory peptides by Arts in the extracellular milieu may be involved in the physiological functions of the lung. Taken together with our observation that cgArt1 is expressed predominantly in the lung, this indicates that these lung Arts may contribute to airway inflammatory responses through the modification of physiologically active peptides or cell surface proteins, although their precise roles remain to be elucidated.
In summary, the results of the present study have revealed that: (1) Arts are distributed widely in chicken tissues as GPI-anchored forms with different molecular sizes; (2) the nucleotide sequence of cgArt2 is identical with that of Art7, except for the ATG position at the 5′-end, and shows a high degree of identity with that of cgArt1, indicating that cgArt1 is an Art7 homologue; (3) recombinant cgArt1 and cgArt2 are GPI-anchored transferases; (4) cgArts show distinct responses to some reagents and different substrate specificities; and (5) the expression patterns of cgArt1 and cgArt2 are quite distinct. Thus we have demonstrated the existence of chicken GPI-anchored Arts and characterized them by molecular cloning and expression experiments. Both soluble (Art6.1 and Art6.2) and GPI-anchored (cgArt1 and cgArt2) Arts are expressed in chicken tissues. Further studies should be performed to explore the functional roles of ADP-ribosylation in the different tissues.
Acknowledgments
We thank M. Takahashi for excellent technical assistance.
References
- 1.Krueger K. M., Barbieri J. T. The family of bacterial ADP-ribosylating exotoxins. Clin. Microbiol. Rev. 1995;8:34–37. doi: 10.1128/cmr.8.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cassel D., Pfeuffer T. Mechanism of cholera toxin action: covalent modification of the guanyl nucleotide-binding protein of the adenylate cyclase system. Proc. Natl. Acad. Sci. U.S.A. 1978;75:2669–2673. doi: 10.1073/pnas.75.6.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gill D. M., Meren R. ADP-ribosylation of membrane proteins catalysed by cholera toxin: basis of the activation of adenylate cyclase. Proc. Natl. Acad. Sci. U.S.A. 1978;75:3050–3054. doi: 10.1073/pnas.75.7.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zolkiewska A., Okazaki I. J., Moss J. Vertebrate mono-ADP-ribosyltransferases. Mol. Cell. Biochem. 1994;138:107–112. doi: 10.1007/BF00928450. [DOI] [PubMed] [Google Scholar]
- 5.Okazaki I. J., Moss J. Mono-ADP-ribosylation: a reversible posttranslational modification of proteins. Adv. Pharmacol. 1996;35:247–280. doi: 10.1016/s1054-3589(08)60277-x. [DOI] [PubMed] [Google Scholar]
- 6.Okazaki I. J., Moss J. Glycosylphosphatidylinositol-anchored and secretory isoforms of mono-ADP-ribosyltransferases. J. Biol. Chem. 1998;273:23617–23620. doi: 10.1074/jbc.273.37.23617. [DOI] [PubMed] [Google Scholar]
- 7.Saxty B. A., Kefalas P., Yodollahi-Farsani M., MacDermot J. Arginine-specific ADP-ribosyltransferases in leukocytes. J. Leukocyte Biol. 1998;63:15–21. doi: 10.1002/jlb.63.1.15. [DOI] [PubMed] [Google Scholar]
- 8.Corda D., Di Girolamo M. Functional aspects of protein mono-ADP-ribosylation. EMBO J. 2003;22:1953–1958. doi: 10.1093/emboj/cdg209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsuchiya M., Shimoyama M. Target protein for eukaryotic arginine-specific ADP-ribosyltransferase. Mol. Cell. Biochem. 1994;138:113–118. doi: 10.1007/BF00928451. [DOI] [PubMed] [Google Scholar]
- 10.Donnelly L. E., Boyd R. S., Clifford C. P., Olmos G., Allport J. R., Lo G., MacDermot J. Endogenous substrates and functional role of eukaryotic mono(ADP-ribosyl)transferases. Biochem. Pharmacol. 1994;48:1669–1675. doi: 10.1016/0006-2952(94)90450-2. [DOI] [PubMed] [Google Scholar]
- 11.Soman G., Mickelson J. R., Louis C. F., Graves D. J. NAD:guanidino group specific mono ADP-ribosyltransferase activity in skeletal muscle. Biochem. Biophys. Res. Commun. 1984;120:973–980. doi: 10.1016/s0006-291x(84)80202-8. [DOI] [PubMed] [Google Scholar]
- 12.Zolkiewska A., Nightingale M. S., Moss J. Molecular characterization of NAD:arginine ADP-ribosyltransferase from rabbit skeletal muscle. Proc. Natl. Acad. Sci. U.S.A. 1992;89:11352–11356. doi: 10.1073/pnas.89.23.11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okazaki I. J., Zolkiewska A., Nightingale M. S., Moss J. Immunological and structural conservation of mammalian skeletal muscle glycosylphosphatidylinositol-linked ADP-ribosyltransferases. Biochemistry. 1994;33:12828–12836. doi: 10.1021/bi00209a014. [DOI] [PubMed] [Google Scholar]
- 14.Koch-Nolte F., Petersen D., Balasubramanian S., Haag F., Kahlke D., Willer T., Kastelein R., Bazan F., Thiele H.-G. Mouse T cell membrane proteins Rt6-1 and Rt6-2 are arginine/protein mono(ADPribosyl)transferases and share secondary structure motifs with ADP-ribosylating bacterial toxins. J. Biol. Chem. 1996;271:7686–7693. doi: 10.1074/jbc.271.13.7686. [DOI] [PubMed] [Google Scholar]
- 15.Moss J., Stevens L. A., Cavanaugh E., Okazaki I. J., Bortell R., Kanaitsuka T., Mordes J. P., Greiner D. L., Rossini A. A. Characterization of mouse Rt6.1 NAD:arginine ADP-ribosyltransferase. J. Biol. Chem. 1997;272:4342–4346. doi: 10.1074/jbc.272.7.4342. [DOI] [PubMed] [Google Scholar]
- 16.Yost D. A., Moss J. Amino acid-specific ADP-ribosylation. J. Biol. Chem. 1983;258:4926–4929. [PubMed] [Google Scholar]
- 17.West R. E., Jr, Moss J. Amino acid specific ADP-ribosylation: specific NAD:arginine mono-ADP-ribosyltransferases associated with turkey erythrocyte nuclei and plasma membrane. Biochemistry. 1986;25:8057–8062. doi: 10.1021/bi00372a039. [DOI] [PubMed] [Google Scholar]
- 18.Mishima K., Terashima M., Obara S., Yamada K., Imai K., Shimoyama M. Arginine-specific ADP-ribosyltransferase and its acceptor protein p33 in chicken polymorphonuclear cells: co-localization in the cell granules, partial characterization, and in situ mono(ADP-ribosyl)ation. J. Biochem. (Tokyo) 1991;110:388–394. doi: 10.1093/oxfordjournals.jbchem.a123591. [DOI] [PubMed] [Google Scholar]
- 19.Tsuchiya M., Hara N., Yamada K., Osago H., Shimoyama M. Cloning and expression of cDNA for arginine-specific ADP-ribosyltransferase from chicken bone marrow cells. J. Biol. Chem. 1994;269:27451–27454. [PubMed] [Google Scholar]
- 20.Terashima M., Badruzzaman M., Tsuchiya M., Shimoyama M. Exocytosis of arginine-specific ADP-ribosyltransferase and p33 induced by A23187 and calcium or serum-opsonized zymosan in chicken polymorphonuclear leukocytes. J. Biochem. (Tokyo) 1996;120:1209–1215. doi: 10.1093/oxfordjournals.jbchem.a021543. [DOI] [PubMed] [Google Scholar]
- 21.Davis T., Shall S. Sequence of chicken erythroblast mono(ADP-ribosyl)transferase-encoding gene and its upstream region. Gene. 1995;164:371–372. doi: 10.1016/0378-1119(95)00504-y. [DOI] [PubMed] [Google Scholar]
- 22.Tsuchiya M., Osago H., Shimoyama M. A newly identified GPI-anchored arginine-specific ADP-ribosyltransferase activity in chicken spleen. Biochem. Biophys. Res. Commun. 1994;214:760–764. doi: 10.1006/bbrc.1995.2351. [DOI] [PubMed] [Google Scholar]
- 23.Hara N., Terashima M., Shimoyama M., Tsuchiya M. Mouse T-cell antigen Rt6.1 has thiol-dependent NAD glycohydrolase activity. J. Biochem. (Tokyo) 2000;128:601–607. doi: 10.1093/oxfordjournals.jbchem.a022792. [DOI] [PubMed] [Google Scholar]
- 24.Pan Y.-T., Kamitani T., Bhuvaneswaran C., Hallaq Y., Warren C. D., Yeh E. T. H., Elbein A. D. Inhibition of glycosylphosphatidylinositol anchor formation by mannosamine. J. Biol. Chem. 1992;267:21250–21255. [PubMed] [Google Scholar]
- 25.Lisanti M. P., Field M. C., Caras I. W., Menon A. K., Rodriguez-Boulan E. Mannosamine, a novel inhibitor of glycosylphosphatidylinositol incorporation into proteins. EMBO J. 1991;10:1969–1971. doi: 10.1002/j.1460-2075.1991.tb07726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chomczynski P., Sacci N. Single-step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1986;162:11447–11453. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 27.Kobata A., Yamashita Y. Affinity chromatography of oligosaccharides on E4-phytohemoagglutinin-agarose column. Methods Enzymol. 1989;179:46–54. doi: 10.1016/0076-6879(89)79112-6. [DOI] [PubMed] [Google Scholar]
- 28.Takada T., Iida K., Moss J. Conservation of a common motif in enzymes catalyzing ADP-ribose transfer. J. Biol. Chem. 1995;270:541–544. doi: 10.1074/jbc.270.2.541. [DOI] [PubMed] [Google Scholar]
- 29.Glowacki G., Braren R., Firner K., Nissen M., Kühl M., Reche P., Bazan F., Cetkovic-Cvrlje M., Leiter E., Haag F., Koch-Nolte F. The family of toxin-related ecto-ADP-ribosyltransferases in humans and the mouse. Protein Sci. 2002;11:1657–1670. doi: 10.1110/ps.0200602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banasik M., Komura H., Shimoyama M., Ueda K. Specific inhibitors of poly(ADP-ribose) synthetase and mono(ADP-ribosyl)transferase. J. Biol. Chem. 1992;267:1569–1575. [PubMed] [Google Scholar]
- 31.Bourgeois C., Okazaki I., Cavanaugh E., Nightingale M., Moss J. Identification of regulatory domains in ADP-ribosyltransferase-1 that determine transferase and NAD glycohydrolase activities. J. Biol. Chem. 2003;278:26351–26355. doi: 10.1074/jbc.M303193200. [DOI] [PubMed] [Google Scholar]
- 32.Terashima M., Mishima M., Yamada K., Tsuchiya M., Wakutani T., Shimoyama M. ADP-ribosylation of actins by arginine-specific ADP-ribosyltransferase purified from chicken heterophils. Eur. J. Biochem. 1992;204:305–311. doi: 10.1111/j.1432-1033.1992.tb16638.x. [DOI] [PubMed] [Google Scholar]
- 33.Terashima M., Yamamori C., Shimoyama M. ADP-ribosylation of Arg28 and Arg206 on the actin molecule by chicken arginine-specific ADP-ribosyltransferase. Eur. J. Biochem. 1995;231:242–249. [PubMed] [Google Scholar]
- 34.Zolkiewska A., Moss J. Integrin α7 as substrate for a glycosylphosphatidylinositol-anchored ADP-ribosyltransferase on the surface of skeletal muscle cells. J. Biol. Chem. 1993;268:25273–25276. [PubMed] [Google Scholar]
- 35.Zolkiewska A., Moss J. Processing of ADP-ribosylated integrin α7 in skeletal muscle myotubes. J. Biol. Chem. 1995;270:9227–9233. doi: 10.1074/jbc.270.16.9227. [DOI] [PubMed] [Google Scholar]
- 36.Terashima M., Yamamori C., Shimoyama M., Tsuchiya M. Suppression of cell adhesion and spreading activities of fibronectin by arginine-specific ADP-ribosyltransferase from chicken polymorphonuclear leukocytes. Biochim. Biophys. Acta. 1998;1404:299–304. doi: 10.1016/s0167-4889(98)00067-6. [DOI] [PubMed] [Google Scholar]
- 37.Jones E. M., Baird A. Cell-surface ADP-ribosylation of fibroblast growth factor-2 by an arginine-specific ADP-ribosyltransferase. Biochem. J. 1997;323:173–177. doi: 10.1042/bj3230173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Terashima M., Hara N., Badruzzaman M., Shimoyama M., Tsuchiya M. ADP-ribosylation of tuftsin suppresses its receptor-binding capacity and phagosytosis-stimulating activity to murine peritoneal macrophages. FEBS Lett. 1997;412:227–232. doi: 10.1016/s0014-5793(97)00784-9. [DOI] [PubMed] [Google Scholar]
- 39.Terashima M., Yamamori C., Tsuchiya M., Shimoyama M. ADP-ribosylation of tubulin by chicken NAD-arginine ADP-ribosyltransferase suppresses microtubule formation. J. Nutr. Sci. Vitaminol. 1999;45:393–400. doi: 10.3177/jnsv.45.393. [DOI] [PubMed] [Google Scholar]
- 40.Okazaki I. J., Kim H.-J., Mcelvaey G., Lesma E., Moss J. Molecular characterization of a glycosylphosphatidylinositol-linked ADP-ribosyltransferase from lymphocytes. Blood. 1996;88:915–921. [PubMed] [Google Scholar]
- 41.Balducci E., Horiba K., Usuki J., Park M., Ferrans V. J., Moss J. Selective expression of RT6 superfamily in human bronchial epithelial cells. Am. J. Respir. Cell Mol. Biol. 1999;21:337–346. doi: 10.1165/ajrcmb.21.3.3638. [DOI] [PubMed] [Google Scholar]
- 42.Paone G., Wada A., Stevens L. A., Matin A., Hirayama T., Levine R. L., Moss J. ADP ribosylation of human neutrophil peptide-1 regulates its biological properties. Proc. Natl. Acad. Sci. U.S.A. 2002;99:8231–8235. doi: 10.1073/pnas.122238899. [DOI] [PMC free article] [PubMed] [Google Scholar]