Abstract
Small non-coding bacterial RNAs (sRNAs) play important regulatory roles in a variety of cellular processes. Nearly all known sRNAs have been identified in Escherichia coli and most of these are not conserved in the majority of other bacterial species. Many of the E.coli sRNAs were initially predicted through bioinformatic approaches based on their common features, namely that they are encoded between annotated open reading frames and are flanked by predictable transcription signals. Because promoter consensus sequences are undetermined for most species, the successful use of bioinformatics to identify sRNAs in bacteria other than E.coli has been limited. We have created a program, sRNAPredict, which uses coordinate-based algorithms to integrate the respective positions of individual predictive features of sRNAs and rapidly identify putative intergenic sRNAs. Relying only on sequence conservation and predicted Rho-independent terminators, sRNAPredict was used to search for sRNAs in Vibrio cholerae. This search identified 9 of the 10 known or putative V.cholerae sRNAs and 32 candidates for novel sRNAs. Small transcripts for 6 out of 9 candidate sRNAs were observed by Northern analysis. Our findings suggest that sRNAPredict can be used to efficiently identify novel sRNAs even in bacteria for which promoter consensus sequences are not available.
INTRODUCTION
Numerous small, untranslated bacterial RNAs (sRNAs) that regulate myriad biological functions have been described within the last several years (1,2). Nearly all sRNA species identified to date are encoded in intergenic regions (IGRs) (3), suggesting that much remains to be discovered in portions of the genome once considered devoid of genetic information. Elucidation of the common features of sRNAs, along with advances in computational approaches used to predict these features on a genome-wide level, has recently led to a significant increase in the number of sRNAs identified (3). However, it is widely accepted that many more sRNAs remain undiscovered, particularly in less well-studied organisms.
In the search for bacterial sRNAs, no organism has been more rigorously examined than Escherichia coli. The first 10 E.coli sRNAs were found serendipitously, often owing to their high cellular abundance (2). Since these fortuitous discoveries, a number of additional E.coli sRNAs have been identified through physical and/or functional analyses (4–7), but the majority of new sRNAs have been identified through bioinformatic prediction followed by verification by Northern analysis (8–11). The predictive algorithms employed in many of these studies were based on the common features shared by the majority of known E.coli sRNAs: they are encoded in IGRs, are conserved in closely related species such as Salmonella typhi and Yersinia pestis, and are flanked by both a putative promoter and a predicted Rho-independent terminator (8–11).
Homologs of most E.coli sRNAs are not found in most bacterial species (3). Thus, relatively few sRNAs have been identified in other bacterial species based solely on sequence homology with known E.coli sRNAs (3). Moreover, directly applying the bioinformatic approaches used in E.coli to identify sRNAs in other bacterial species has had only limited success. The principal impediment to applying these approaches to other bacteria is that accurately predicting either promoters or transcription factor binding sites (TFBSs) requires reliable species-specific consensus sequences; few of these have been experimentally determined in bacterial species other than E.coli. Indeed, only two studies have used bioinformatics to predict sRNAs in bacteria other then E.coli. One used a consensus sequence for the Vibrio cholerae σ54 promoter to predict four functionally redundant sRNAs involved in quorum sensing (12). The other used a consensus sequence for the Pseudomonas aeruginosa Fur repressor binding site to identify two functionally redundant sRNAs involved in iron homeostasis (13).
A second significant limitation of the previously utilized approaches is computational rather than biological. Accurately predicting sRNAs requires not only identifying putative transcription signals and conserved sequences but also determining their locations in the genome relative to each other and to open reading frames (ORFs). For example, sequence conservation is suggestive of the presence of an sRNA only when found in an IGR, upstream of a putative terminator and/or downstream of a putative promoter. In our initial attempts to predict sRNAs in V.cholerae, the most time-consuming aspect of our search was determining the positional relationships of the nearly 10 000 relevant sequence elements. The time-intensive nature of this process severely limited the frequency at which searches incorporating different combinations of predictive parameters could be conducted. To overcome this computational limitation, we developed sRNAPredict, a program that uses the relative genomic locations of conserved sequences, transcription signals and ORFs, to rapidly identify putative sRNAs encoded in IGRs. sRNAPredict completes a genome-wide search for putative sRNAs in a matter of seconds, allowing searches using different parameters to be efficiently conducted until the desired stringency is achieved.
Recent findings strongly suggest that sRNAs regulate the virulence of the gram-negative diarrheal pathogen V.cholerae (12,14). Using sRNAPredict, we searched the IGRs of V.cholerae for sRNAs. Relying solely on putative terminators and regions of sequence conservation in IGRs, sRNAPredict predicted 9 of the 10 known or putative V.cholerae sRNAs as well as 32 candidates for novel sRNAs. Transcripts for 6 of 9 of the predicted novel sRNAs were detected by Northern analysis, suggesting that sRNAPredict is an effective tool for identifying sRNAs even in bacteria for which promoter consensus sequences are not available.
MATERIALS AND METHODS
Summary of the sRNAPredict program
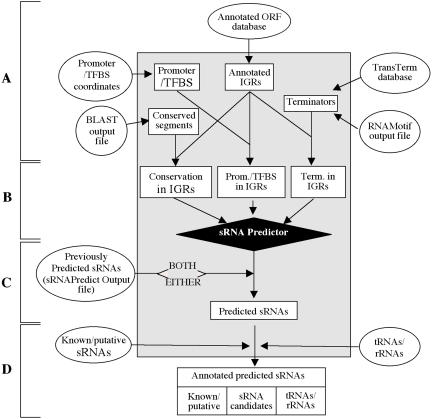
The general scheme of sRNAPredict is illustrated in Figure 1. The various algorithms used by sRNAPredict utilize only the coordinate locations and, when applicable, strand orientations of predictive elements; no sequence information is used. sRNAPredict is designed to automatically extract coordinates and strand orientations directly from certain published databases or from output files of particular programs (Figure 1A). These include databases of annotated ORFs available for download at NCBI and TIGR, databases of Rho-independent terminators predicted by TransTerm (15), output files of the predictive program RNAMotif (16,17) and output files of BLAST (18). sRNAPredict does not extract the coordinates and strand orientations of promoters or TFBSs from output files of predictive programs and thus these databases must be compiled by the user.
Figure 1.
General scheme of sRNAPredict. (A) The coordinates and, when applicable, strand orientations of transcription signals, conserved sequences and ORFs are extracted directly from output files of RNAMotif and BLAST, from published databases, or from a user-compiled database of predicted promoters/TFBSs. ORF coordinates and annotations are used to create a database of intergenic regions (IGRs) that includes the start and end positions of each IGR as well as the names and orientations of its flanking ORFs. (B) This database is used to produce databases of conserved sequences, terminators (Term.) and/or promoters (Prom.)/TFBSs located in IGRs, which are subsequently utilized in the prediction of sRNAs. (C) If the output file from a previous sRNAPredict search is provided, the program can be used to identify those sRNAs predicted in both searches or those predicted in either search. (D) If the appropriate databases are provided, predicted sRNAs corresponding to known or putative sRNAs or to tRNAs or rRNAs are identified.
Once the relevant information is extracted from the input files, redundant terminators, defined as those predicted in the same orientation as and within 15 bp of another terminator, are deleted. Redundant promoters/TFBSs, defined as those predicted in the same orientation and in the same location as another promoter/TFBS, are also deleted. The start and end coordinates of conserved genomic regions are parsed to remove redundancies and to resolve overlapping segments. sRNAPredict then uses the ORF database to create an annotated database of IGR coordinates. These IGR coordinates are compared with the coordinates extracted from other input files to identify the subsets of conserved sequences and/or transcription signals that are located in IGRs (Figure 1B). Next, sRNAPredict utilizes one of a number of algorithms to search for predictive elements co-localized in the proper relative orientations. sRNAPredict can search for regions encoding any combination of conserved sequences, predicted terminators and/or putative promoters/TFBSs; the specific algorithm employed in a given predictive search depends on the particular combination of parameters included in the initial input. For example, if only terminators and regions of sequence conservation are included, sRNAPredict will search for all instances in which a terminator is predicted inside or near the 3′ end of a region of conserved sequence. The predictive parameters included also influence the boundaries and strand orientations of predicted sRNAs. When putative transcription signals are included, the 5′ and/or 3′ ends of a putative sRNA are determined by the location of the predicted promoter/TFBS and/or terminator, respectively; the strand orientation of the predicted sRNAs is based on the strand orientations of its associated terminators and/or promoters/TFBSs. In the absence of predicted transcription signals, the boundaries of a putative sRNA are determined by the boundaries of its associated region of sequence conservation and sRNAs are predicted for both strands. The sRNAPredict output file, as shown in Figure 2, includes the coordinates, strand orientations and lengths of the predicted sRNAs, their distance from the flanking ORFs, and the locus names and orientations of those ORFs.
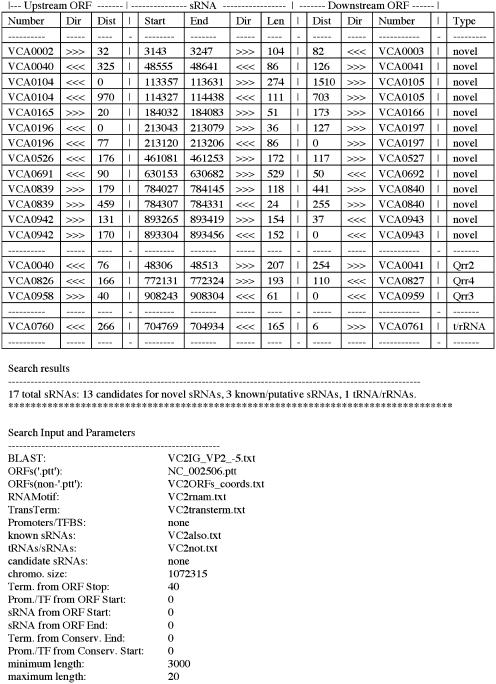
Figure 2.
Output file of sRNAPredict search. sRNAs were predicted using BLAST comparison of Vc chromosome II IGRs and Vp chromosome II (E < 1 × 10−5) as well as VcII terminators predicted by RNAMotif and TransTerm.
In addition to predicting sRNAs, sRNAPredict can utilize user-generated databases of tRNAs/rRNAs and known/putative sRNAs to separate those predicted sRNAs which correspond to previously identified transcripts from those which are potentially novel sRNAs (Figure 1D). Any overlap between the location of a predicted sRNA and a region encoding a tRNA or rRNA is sufficient to exclude that sRNA from the list of potential novel sRNAs, regardless of its strand orientation. To be classified as a known/putative sRNA, the location of the predicted sRNA must at least partially overlap the location of a known/putative sRNA. Furthermore, it must be encoded on the same strand as that of the known/putative sRNA. The respective numbers of predicted sRNAs falling into each category are reported at the end of the search, allowing the stringency of the search, as indicated both by the total number of sRNAs predicted and by the proportion of known sRNAs predicted, to be quickly and easily determined. If no single search yields the desired stringency, the sRNAPredict program can compare the results of two independent searches and report either those sRNAs predicted in both of the searches (to increase stringency) or those sRNAs predicted in either of the searches (to decrease stringency) (Figure 1C). In this comparison, two sRNAs are considered identical when they are predicted on the same strand and when their respective start or end coordinates are within 10 bp of each other.
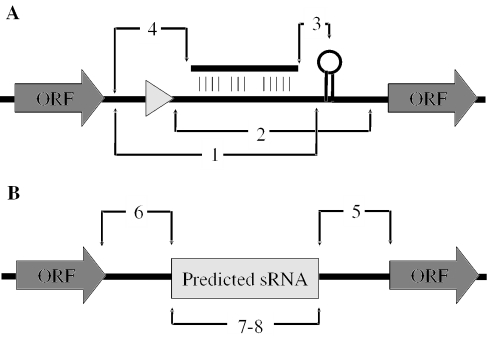
sRNAPredict allows a number of variables used in the sRNA predictive algorithms to be modified by the user (Figure 3A). These include the minimum distances of predicted promoters and terminators from the beginning and end of an ORF, respectively, and the maximum length of the gap allowed between a region of conservation and either a putative promoter or terminator. Several parameters pertaining to the predicted sRNAs can also be set by the user (Figure 3B). These include the maximum and minimum lengths of predicted sRNAs as well as their minimum distance from their flanking ORFs. The names of input files, the desired name of the output file and the values of adjustable search parameters are all extracted from a single file provided by the user. Thus, altering a few search parameters prior to conducting a new search can be accomplished simply by changing a few lines in the input file rather than reentering all input data de novo.
Figure 3.
sRNAPredict search parameters that can be set by the user. The gray triangle represents a putative promoter, hashed lines represent a region of sequence conservation, and the stem–loop represents a predicted terminator. (A) Adjustable search parameters applied prior to sRNA prediction. (1) The minimum distance of predicted terminator from end of an upstream ORF. (2) The minimum distance of promoter from a downstream ORF. (3) The maximum length of gap between a putative terminator and a region of sequence conservation. (4) The maximum length of gap between putative promoter and region of sequence conservation. (B) Adjustable search parameters applied following sRNA prediction. (5) The minimum distance of predicted sRNA from the beginning of a downstream ORF. (6) The minimum distance of predicted sRNA from the end of an upstream ORF. (7) The minimum and (8) maximum length of the predicted sRNA. Unless otherwise noted, all searches were conducted with parameters set to 0 with the exception of parameters 1, 7 and 8, which were set to 40, 20 and 3000, respectively.
ORF databases and genomic sequences
ORF databases were obtained from the NCBI and TIGR ftp bacterial databases (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/ and ftp://ftp.tigr.org/pub/data/Microbial_Genomes/). ORF databases available from NCBI contained a ‘.ptt’ extension. These databases include locus and product names for each ORF but do not include annotated genes with frame-shift mutations. The ORF databases obtained from TIGR contained a ‘.coords’ extension. These databases do not include locus and product names for each ORF but do include the coordinates of annotated genes with frame-shift mutations. Moreover, they can include annotated ORFs not found in the NCBI database. Genome databases were obtained from NCBI and contained an ‘.fna’ extension. All E.coli sRNA sequences and genomic coordinate positions were obtained from the EcoCyc database (19).
BLAST analysis
BLAST comparisons were conducted using BLASTN 2.0 (http://blast.wustl.edu). Unless otherwise noted, search parameters were set to default values.
Prediction of Rho-independent terminators
RNAMotif was used to predict Rho-independent terminators in chromosomes I and II of V.cholerae using a motif descriptor provided by D. J. Ecker. Rho-independent terminators predicted by TransTerm were obtained from the TransTerm website at TIGR (http://www.tigr.org/software/transterm.html). From this published database, terminators predicted in chromosomes I and II were separated into two databases.
Oligonucleotide sequences and probe preparation
The sequences of DNA oligonucleotides used in this study are provided in Supplementary Table S1. These oligonucleotides were designed to be complementary to the 30 nt directly 5′ of the predicted terminator. T4 polynucleotide kinase (New England Biolabs) was used to end-label ∼1 pmol of each synthetic DNA oligonucleotide with a 2-fold molar excess of [γ-32P]ATP (PerkinElmer). Radiolabled oligonucleotides were purified using Sephadex G-25 gel filtration columns (Amersham).
RNA isolation
Cultures of V.cholerae N16961 were grown in M63 media supplemented with 0.2% glucose and 1 μM MgSO4 or in Luria–Bertani (LB) either overnight or to an OD600 of ∼0.250. Total RNA was isolated with Trizol (Invitrogen) according to the manufacturer's protocol.
Northern analysis
An aliquot of 20 μg of total RNA per lane was fractionated on 10% polyacrylamide urea gels and transferred to BrightStar-Plus nylon membrane (Ambion). RNA was crosslinked to the membrane with UV light. Northern analysis was conducted according to the protocol accompanying the Ultrahyb-oligo buffer (Ambion). All hybridizations and washes were conducted at 40°C.
RESULTS
Testing the efficiency and sensitivity of sRNAPredict
To test the efficacy of sRNAPredict, we assessed whether it could accurately identify sRNAs encoded in IGRs of V.cholerae (Vc). Aside from our general interest in the role of sRNAs in Vc virulence, we chose Vc as the subject for our predictive search for three main reasons. First, at the time we began this study, the annotated genome sequences of two other Vibrio sp., Vibrio vulnificus (Vv) and Vibrio parahaemolyticus (Vp), were available for sequence comparison with Vc. Second, Vc had been shown to contain a relatively high number of predicted Rho-independent terminators (15). Finally, five Vc sRNAs had been functionally confirmed and could be used as positive controls in our predictions.
Because a reliable consensus sequence for Vc σ70-dependent promoters is not available, all predictive searches relied only on sequence conservation and, in some cases, putative Rho-independent terminators. For each search, predicted sRNAs were compared with two databases of previously described Vc RNAs. One database held the coordinates of annotated tRNAs and rRNAs and of IGRs containing clusters of tRNA and rRNA operons. The second database contained the coordinates and strand orientations of 10 verified or putative Vc sRNAs. These included the four sRNAs identified by Lenz et al. (12) (Qrr1, Qrr2, Qrr3 and Qrr4) and RyhB, first predicted by Masse et al. (20) and subsequently characterized by Davis et al. (21). Furthermore, the second database included five putative Vc homologs of E.coli sRNAs—SsrA, RnpB, CsrB, Spf and Ffs—whose reported sequence conservation exceeded an E-value of 0.4 (3). The coordinate locations of these putative V.cholerae sRNA-encoding genes were based on BLAST alignment (E < 1) with their E.coli homologs.
The sRNAPredict program was first used to predict Vc sRNAs with sequence conservation as the only predictive parameter. To identify conserved sequences, chromosome I of Vc (VcI) was compared with chromosome I of Vv CMCP6 (VvI) by BLAST using very low stringency (E < 1 × 1010). Using this BLAST output file as input, sRNAPredict identified 1346 potential sRNAs encoded in VcI. These included only two of the seven known/putative VcI-encoded sRNAs, RnpB and Spf. These results were surprising, as one of the known sRNAs not identified in this search, Qrr1, is known to be conserved in Vv (12). Moreover, the remaining five known or putative sRNAs not predicted in the search are conserved in E.coli and thus are expected to be conserved in Vv as well.
We reasoned that the inability of BLAST to identify sRNA conservation when comparing entire chromosomes was due to the relatively short length of their conservation compared with the conservation associated with ORFs and postulated that removing the ORFs from the comparison would enhance the ability of BLAST to identify the short regions of sRNA conservation. To this end, we developed a program, IGRExtract, which extracts the sequences of IGRs from the sequence of the entire chromosome to produce a FASTA-formatted database of IGRs that can be directly entered into a BLAST search. Because the coordinates reported by this BLAST search correspond to the respective location of conserved sequences within each IGR rather than within the chromosome as a whole, they cannot be used by sRNAPredict to identify co-localization of sequence conservation and transcription signals. To overcome this technical problem, we modified both the IGRExtract and the sRNAPredict programs such that when databases created by IGRExtract are used as subject sequences in BLAST searches, the coordinates of conservation in the BLAST output file are automatically converted by sRNAPredict to ones corresponding to the location of the conservation within the chromosome.
sRNAPredict searches were performed using BLAST analysis (E < 1 × 1010) of either VcI IGRs versus VvI or VcII IGRs versus VvII as the only input. A total of 1118 sRNAs, including all 10 known/putative sRNAs, were predicted in these searches. We next performed searches to identify sRNAs using sequence conservation and Rho-independent terminators predicted by RNAMotif and/or TransTerm. Nine of the 10 known/putative Vc sRNAs were identified when both terminators predicted by RNAMotif and terminators predicted TransTerm were used; the only known/putative sRNA not identified was the putative Vc Ffs. Inclusion of both TransTerm and RNAMotif-predicted terminators in the search reduced the number of novel sRNAs predicted to 104. Based on the high proportion of known/putative sRNAs identified and the relatively low number of total sRNAs predicted, we concluded that identifying IGRs of conserved sequence co-localized with terminators predicted by RNAMotif or by TransTerm provided a sufficiently sensitive approach for the identification of putative sRNAs in Vc.
The searches described above also demonstrated the speed with which sRNAPredict can predict sRNAs on a genome-wide level. The search for VcI sRNAs using the VcI versus VvI BLAST with E < 1 × 10−5 included 2182 distinct IGRs, 1598 regions of conserved sequence and 7041 predicted terminators. When run on an Apple iBook with a 1.2 GHz PowerPC G4 processor, sRNAPredict completed this search in ∼20 s.
Increasing the stringency of the predictive search
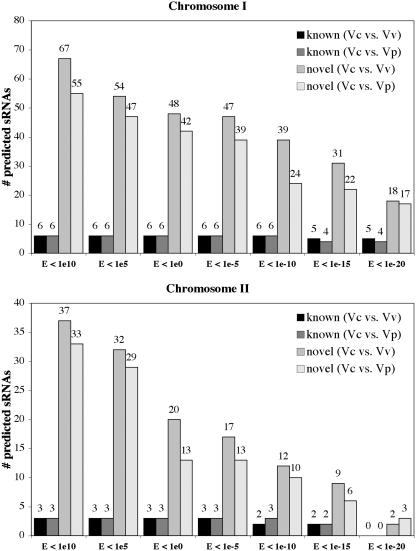
We next examined the effect of altering the stringency of BLAST analysis on the prediction of both previously identified and novel sRNAs. The VcI and VcII IGR databases were compared with the corresponding Vv and Vp chromosomes by BLAST with E set to values ranging from 1 × 1010 to 1 × 10−20. Conserved sequences identified by these comparisons were used by sRNAPredict in conjunction with predicted terminators to predict sRNAs. As shown in Figure 4, the number of novel sRNAs predicted decreased steadily as BLAST stringency increased, while the number of known/putative sRNAs predicted remained constant until the BLAST stringency exceeded a certain threshold. The lowest E-value tested at which all known/putative VcI-encoded sRNAs except Vc Ffs were predicted was 1 × 10−10; for the VcII-encoded known/putative sRNAs, this threshold was 1 × 10−5. To further increase the stringency of our search, sRNAPredict was used to identify the subset of sRNAs that was predicted both in the search using conservation in Vv and in the search using conservation in Vp. This reduced the total number of novel sRNAs predicted for VcI and VcII to 21 and 11, respectively.
Figure 4.
Effect of increased BLAST stringency on the numbers of known/putative sRNAs and novel sRNAs predicted by sRNAPredict in Vc. Vc IGRs in chromosomes I and II were compared by BLAST with the corresponding chromosomes of Vv and Vp with the threshold for reporting set to the indicated values. The regions of conservation determined by these BLAST analyses, along with predicted terminators, were utilized by sRNAPredict to identify putative sRNAs and to determine how many of these corresponded to known/putative Vc sRNAs and how many correspond to previously unidentified sRNAs.
While sequence in an IGR may be conserved because it encodes an sRNA, it may alternatively be conserved because it encodes an untranslated regulatory region of an messenger RNA (mRNA). Indeed, a significant proportion of the sRNAs predicted in E.coli, when tested by Northern analysis, proved to be 5′-untranslated portions of mRNAs (11). We postulated that the closer a predicted sRNA was to the start codon of an ORF, the greater the likelihood that it was a conserved regulatory region of an mRNA rather than a functional sRNA. The search parameters of sRNAPredict were adjusted to report only those candidates for novel sRNAs in VcI and VcII that were farther than 50 bp from the start codon of an ORF. In addition, the search was modified so that only sRNAs between 30 and 450 nt in length were reported. These modifications reduced the number of novel sRNAs predicted to 10 in VcI and 7 in VcII.
Functional verification of sRNA candidates by Northern analysis
From this group of 17 predicted novel sRNAs, 9 were selected at random and subjected to Northern analysis (Table 1). In this analysis, RyhB was used as a positive control. As shown in Figure 5, transcripts <350 nt were detected for six candidates. For one of the three candidates for which no small transcript was detected, a large transcript (>800 nt) was observed (data not shown), suggesting this predicted sRNA may correspond to an untranslated region (UTR) of an mRNA. For the other two, distinct transcripts were not detected, suggesting that these predicted sRNAs were either unstable, not expressed under the conditions tested, or represent false positives.
Table 1.
Annotation of the nine Vc sRNAs predicted by sRNAPredict that were subjected to Northern analysis
| Chromosomes | sRNA | Upstream ORF | Downstream ORF | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Name | Length | Direction | Name | Direction | Distancea | Name | Direction | Distanceb | |
| VcI | B2 | 386 | <<< | VC0142 | >>> | 47 | VC0143 | >>> | 68 |
| B3 | 163 | <<< | VC0168 | <<< | 935 | VC0169 | <<< | 235 | |
| A4 | 44 | >>> | VC0331 | >>> | 6 | VC0332 | >>> | 191 | |
| B4 | 72 | >>> | VC1131.1 | >>> | 0 | VC1132 | >>> | 76 | |
| B1 | 203 | <<< | VC1844 | <<< | 56 | VC1845 | <<< | 159 | |
| C1 | 201 | >>> | VC2489 | <<< | 83 | VC2490 | >>> | 97 | |
| VcII | A7 | 172 | >>> | VCA0526 | <<< | 176 | VCA0527 | >>> | 117 |
| A9 | 154 | >>> | VCA0942 | >>> | 131 | VCA0943 | <<< | 37 | |
| A10 | 153 | <<< | VCA0942 | >>> | 170 | VCA0943 | <<< | 0 | |
aDistance between 3′ boundary of upstream ORF and 5′ boundary of predicted sRNA.
bDistance between 3′ boundary of predicted sRNA and 5′ boundary of downstream ORF.
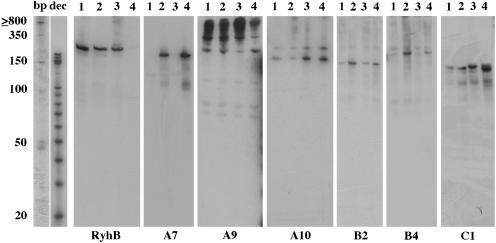
Figure 5.
Detection of novel sRNAs by Northern analysis. Total RNA was extracted from V.cholerae N16961 cells grown in M63-glucose media to early exponential (1) or stationary (2) phase, or in LB media to early exponential (3) or stationary phase (4). Blots were hybridized to radiolabeled DNA oligonucleotide probes and then exposed to film for varying times; thus the relative intensities of the signals do not correspond to the relative abundance of each sRNA. Each individual gel included radiolabeled Decade (dec) RNA markers (Ambion) and 50 bp (bp) DNA markers (Invitrogen). One representative lane of each marker is shown for reference. Additional Northern analysis of candidate A9 suggested that those signals corresponding to transcripts >350 nt were due to non-specific probe hybridization to rRNA (data not shown).
Based on the difference between its predicted and observed length (Table 1 and Figure 5), candidate B4 may be a false positive, corresponding to the 3′-UTR of the 45 bp upstream gene. For candidates A9, A10 and C1, two distinct transcripts <350 nt were observed. Recent findings in E.coli suggest that this may be due to the presence of two overlapping sRNAs whose transcription initiates from adjacent promoters and ends at a shared terminator (22). Alternatively, one transcript may be produced by post-transcriptional processing of the other. For A7 and B2, transcript levels were found to be significantly higher in stationary phase than in exponential phase. Northern analysis of RNA isolated from an rpoS− V.cholerae strain suggests that the expression of A7 and B2 is regulated by RpoS (data not shown). Moreover, preliminary findings suggest that the stability of A7, A9 and A10 is dependent on Hfq (data not shown).
Comparisons of sRNA predictions based on conservation between Vc and several related species
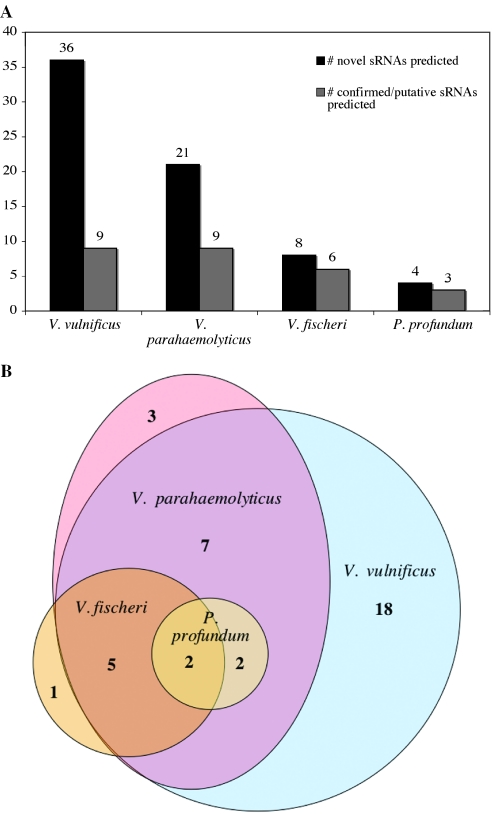
To examine how the prediction of sRNAs is affected by the degree of evolutionary divergence between the species compared by BLAST, we conducted sRNAPredict searches using BLAST comparisons of VcI versus the corresponding chromosomes of Vibrio fischeri and Photobacterium profundum, whose complete genome sequences were published while this study was in progress (23,24). Phylogenetic analysis suggests that V.fischeri and P.profundum belong to families that are closely related to but distinct from the Vibrionaceae family that includes Vc, Vv and Vp, with the V.fischeri group located between Vibrionaceae and Photobacteriaceae in the phylogenetic tree (25). Furthermore, within Vibrionaceae Vc appears to be more closely related to Vv than to Vp. The results of the sRNAPredict searches using BLAST (E < 1 × 10−10) of VcI versus chromosome I of these various species (Figure 6A) are consistent with the evolutionary relationships suggested by the phylogenetic study. Both the number of novel sRNAs and the number of confirmed/putative sRNAs tend to decline as the degree of evolutionary divergence between Vc and its BLAST partner increases. In total, 38 distinct novel Vc sRNAs were predicted using sequence conserved between VcI and each of the 4 related species (E < 1 × 10−10) (Figure 6B). A total of 16 (42%) of these sRNAs were predicted using conservation between VcI and at least two other species; 9 (24%) were predicted using conservation between VcI and at least three species; 2 (5%) were predicted using conservation between VcI and all four species. Because they are conserved in all four species, the predicted sRNAs in this latter group likely represent the strongest candidates for novel sRNAs. Taken together, these findings suggest that the stringency of the sRNAPredict search can be altered by varying the degree of relatedness between the species of interest and its BLAST partner. Thus, our general approach should be especially effective in predicting sRNAs in those species for which the genome sequences of several other species in the same or closely related families are available, such as Pseudomonas sp. and Yersinia sp.
Figure 6.
Comparative analysis of sRNAs predicted by sRNAPredict for VcI based on its homology to chromosome I of V.vulnificus, V.parahaemolyticus, V.fischeri and P.profundum, respectively. (A) BLAST comparisons were conducted with E < 10−10. Confirmed/putative sRNAs include the three VcI sRNAs confirmed in this study. (B) Venn diagram of novel VcI sRNAs predicted using different species in BLAST comparisons.
Identification of E.coli sRNAs using sRNAPredict
To further test the utility of our predictive approach, we used sRNAPredict to identify putative sRNAs in E.coli K12 and compared the results of these searches with a database of 55 experimentally verified sRNAs compiled by Hershberg et al. (3). Using sequence conservation between E.coli IGRs and Shigella flexneri (E < 1 × 10−10; B = 1 × 104, V = 1 × 104) as the only predictive parameter, sRNAPredict identified 50 (91%) of the confirmed E.coli sRNAs. When putative E.coli Rho-independent terminators predicted by either RNAMotif or TransTerm were incorporated into the search, 30 (55%) of the confirmed sRNAs were identified. Of the 55 confirmed sRNAs in the database, 20 were initially identified by algorithms that relied only on sequence conservation and/or putative transcription signals as predictive parameters (8,9); 15 (75%) of these were identified in our search. In addition, sRNAPredict was able to identify 11 (58%) of the 19 sRNAs identified by non-bioinformatic approaches, including at least four sRNAs that had not been predicted by any of the previously utilized bioinformatic approaches. Overall, these observations validate the utility of our predictive approach.
DISCUSSION
To make the search for sRNAs more accessible and efficient, we developed a program, sRNAPredict, that, using a coordinate-based algorithm, integrates several combinations of predictive parameters and, in a matter of seconds, identifies the location and strand orientation of putative sRNAs encoded in IGRs. The stringency of each sRNAPredict search can be adjusted both by modifying the primary searches used to identify individual predictive features and by modifying a number of variables in the sRNAPredict search itself. This flexibility, along with the speed at which the search is completed, allows numerous searches to be efficiently conducted until the desired stringency is achieved.
Using sequence conservation and putative Rho-independent terminators as predictive parameters, 32 previously unidentified putative Vc sRNAs were predicted by sRNAPredict. Of these, 17 were selected as the subset of predicted sRNAs least likely to be 5′-UTR of mRNAs. Nine of these were subjected to Northern analysis and transcripts <350 nt were detected for 6, suggesting that sRNAPredict is a fairly accurate predictive tool. It is important to note that simply demonstrating that an sRNA is transcribed does not necessarily mean it possesses a biological function. In future studies, we will further characterize these transcripts by determining their 5′ and 3′ boundaries and test their biological function by microarray analysis; we will refrain from assigning gene names to the confirmed sRNAs until their biological functions are ascertained.
Prior to the work described here, only one algorithm used to identify bacterial sRNAs had not relied at least in part on putative promoters or TFBSs, utilizing instead predicted secondary structure conservation in IGRs as its only predictive parameter (10). The accuracy of this approach was significantly lower than that of ours, with small transcripts detected for only 11 (22%) of the 49 predicted sRNAs subjected to Northern analysis. Indeed, even in the three studies in which σ70 promoters were used as predictive parameters, the average accuracy of the predictions as determined by Northern analysis was 59% (2,8,9). Thus, the reliability of our approach appears to be at least comparable with those of algorithms previously tested in E.coli. Furthermore, since it does not require promoter or TFBS consensus sequences, this general approach should be applicable to a much wider variety of bacterial species.
While generally successful in predicting sRNAs, our approach does have limitations. The main limitation is due to its reliance on sequence conservation as a predictive parameter. First, this restricts our search to IGRs, as sequence conservation found in a coding region would probably indicate conservation of the encoded protein rather than the presence of a conserved functional sRNA. Though nearly all sRNAs identified to date are encoded in IGRs, this likely reflects the fact that most were discovered using predictive searches limited to IGRs rather than a biological bias against the presence of sRNAs in coding regions. Indeed, recent cloning-based screens for E.coli sRNAs have identified a number of sRNAs that are partially encoded on the non-coding strands of ORFs (4,5). Second, predicting sRNAs by sequence conservation requires that genomic sequences of bacterial species that are appropriately diverged from the species of interest are available. If the two species being compared are too similar, distinguishing functional RNAs from the high background of overall sequence homology may be difficult. Alternatively, if they are too evolutionary distant, the sRNA sequences may no longer be conserved. For example, when P.profundum was used as the Vc BLAST partner, several known sRNAs were not identified by sRNA predict (Figure 6A), including two confirmed in this study.
Another limitation of our approach is its reliance on Rho-independent terminators. Rho-dependent termination is thought to occur in most bacterial species (26), and in some bacterial species with few predicted Rho-independent terminators [e.g. Helicobacter pylori, Mycoplasma genitalium and Treponema pallidum (15)] it may be the principal mechanism of transcriptional termination. Some sRNAs may be associated with a Rho-dependent terminator and thus would not be identified in our search. Other classes of sRNAs would not be detectable by our approach. Recently, three sRNAs identified by a shot-gun cloning approach were found to be processed derivatives of mRNAs (5), suggesting that post-transcriptional processing of mRNAs may be a relatively common mechanism by which sRNAs are produced. sRNAs processed from the 5′ ends of mRNAs would presumably not require a downstream terminator and thus would be missed in our predictions. This limitation of our approach accounts for our failure to identify the putative Vc Ffs in our predictive searches. Functional homologs of the E.coli Ffs, the 4.5S RNA component of the signal recognition particle, exist in numerous prokaryotes (27–29). The putative Vc ffs, though very well conserved both in Vv and in Vp (E < 10−14), is not associated with a predicted Rho-independent terminator and thus was not identified in our search.
Taken together, our findings suggest that searching for sequence conservation associated with predicted Rho-independent terminators in IGRs is an effective approach for identifying a particular class of sRNAs. Successfully predicting more elusive classes of sRNAs will require the development and efficient execution of new algorithms utilizing newly identified predictive parameters. The sRNAPredict program, because it utilizes a generic coordinate-based algorithm, can be easily modified to incorporate any new predictive approach that includes combinations of novel and/or established predictive parameters. By making this version and future versions of sRNAPredict publicly available, we hope to facilitate the extension of bioinformatic searches for bacterial sRNAs to a greater number of species and to more intractable types of sRNAs.
PROGRAM AVAILIBILITY
sRNAPredict and IGRExtract are written in C++. The source codes, user instructions and Mac OS X-compatible executables are available for download at http://www.tufts.edu/sackler/waldorlab/sRNAPredict/.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
The authors are grateful for support from HHMI and NIAID. The authors thank D. Ecker for his help with RNAMotif. Funding to pay the Open Access publication charges for this article was provided by HHMI.
Conflict of interest statement. None declared.
REFERENCES
- 1.Storz G. An expanding universe of noncoding RNAs. Science. 2002;296:1260–1263. doi: 10.1126/science.1072249. [DOI] [PubMed] [Google Scholar]
- 2.Wassarman K.M., Zhang A., Storz G. Small RNAs in Escherichia coli. Trends Microbiol. 1999;7:37–45. doi: 10.1016/s0966-842x(98)01379-1. [DOI] [PubMed] [Google Scholar]
- 3.Hershberg R., Altuvia S., Margalit H. A survey of small RNA-encoding genes in Escherichia coli. Nucleic Acids Res. 2003;31:1813–1820. doi: 10.1093/nar/gkg297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawano M., Reynolds A.A., Miranda-Rios J., Storz G. Detection of 5′- and 3′-UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acids Res. 2005;33:1040–1050. doi: 10.1093/nar/gki256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vogel J., Bartels V., Tang T.H., Churakov G., Slagter-Jager J.G., Huttenhofer A., Wagner E.G. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 2003;31:6435–6443. doi: 10.1093/nar/gkg867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tjaden B., Saxena R.M., Stolyar S., Haynor D.R., Kolker E., Rosenow C. Transcriptome analysis of Escherichia coli using high-density oligonucleotide probe arrays. Nucleic Acids Res. 2002;30:3732–3738. doi: 10.1093/nar/gkf505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang A., Wassarman K.M., Rosenow C., Tjaden B.C., Storz G., Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- 8.Argaman L., Hershberg R., Vogel J., Bejerano G., Wagner E.G., Margalit H., Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 9.Chen S., Lesnik E.A., Hall T.A., Sampath R., Griffey R.H., Ecker D.J., Blyn L.B. A bioinformatics based approach to discover small RNA genes in the Escherichia coli genome. Biosystems. 2002;65:157–177. doi: 10.1016/s0303-2647(02)00013-8. [DOI] [PubMed] [Google Scholar]
- 10.Rivas E., Eddy S.R. Noncoding RNA gene detection using comparative sequence analysis. BMC Bioinformatics. 2001;2:8. doi: 10.1186/1471-2105-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wassarman K.M., Repoila F., Rosenow C., Storz G., Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lenz D.H., Mok K.C., Lilley B.N., Kulkarni R.V., Wingreen N.S., Bassler B.L. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 13.Wilderman P.J., Sowa N.A., FitzGerald D.J., FitzGerald P.C., Gottesman S., Ochsner U.A., Vasil M.L. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl Acad. Sci. USA. 2004;101:9792–9797. doi: 10.1073/pnas.0403423101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding Y., Davis B.M., Waldor M.K. Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol. Microbiol. 2004;53:345–354. doi: 10.1111/j.1365-2958.2004.04142.x. [DOI] [PubMed] [Google Scholar]
- 15.Ermolaeva M.D., Khalak H.G., White O., Smith H.O., Salzberg S.L. Prediction of transcription terminators in bacterial genomes. J. Mol. Biol. 2000;301:27–33. doi: 10.1006/jmbi.2000.3836. [DOI] [PubMed] [Google Scholar]
- 16.Lesnik E.A., Sampath R., Levene H.B., Henderson T.J., McNeil J.A., Ecker D.J. Prediction of rho-independent transcriptional terminators in Escherichia coli. Nucleic Acids Res. 2001;29:3583–3594. doi: 10.1093/nar/29.17.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macke T.J., Ecker D.J., Gutell R.R., Gautheret D., Case D.A., Sampath R. RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res. 2001;29:4724–4735. doi: 10.1093/nar/29.22.4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keseler I.M., Collado-Vides J., Gama-Castro S., Ingraham J., Paley S., Paulsen I.T., Peralta-Gil M., Karp P.D. EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res. 2005;33:D334–D337. doi: 10.1093/nar/gki108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masse E., Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl Acad. Sci. USA. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis B.M., Quinones M., Pratt J., Ding Y., Waldor M.K. Characterization of the small untranslated RNA RyhB and its regulon in Vibrio cholerae. J. Bacteriol. 2005;187:4005–4014. doi: 10.1128/JB.187.12.4005-4014.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vogel J., Argaman L., Wagner E.G., Altuvia S. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr. Biol. 2004;14:2271–2276. doi: 10.1016/j.cub.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Ruby E.G., Urbanowski M., Campbell J., Dunn A., Faini M., Gunsalus R., Lostroh P., Lupp C., McCann J., Millikan D., et al. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc. Natl Acad. Sci. USA. 2005;102:3004–3009. doi: 10.1073/pnas.0409900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vezzi A., Campanaro S., D'Angelo M., Simonato F., Vitulo N., Lauro F.M., Cestaro A., Malacrida G., Simionati B., Cannata N., et al. Life at depth: Photobacterium profundum genome sequence and expression analysis. Science. 2005;307:1459–1461. doi: 10.1126/science.1103341. [DOI] [PubMed] [Google Scholar]
- 25.Thompson F.L., Iida T., Swings J. Biodiversity of Vibrios. Microbiol. Mol. Biol. Rev. 2004;68:403–431. doi: 10.1128/MMBR.68.3.403-431.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richardson J.P. Rho-dependent termination and ATPases in transcript termination. Biochim. Biophys. Acta. 2002;1577:251–260. doi: 10.1016/s0167-4781(02)00456-6. [DOI] [PubMed] [Google Scholar]
- 27.Brown S. 4.5S RNA: does form predict function? New Biol. 1991;3:430–438. [PubMed] [Google Scholar]
- 28.Jenkins G.S., Chandler M.S., Fink P.S. Functional characterization of the Haemophilus influenzae 4.5S RNA. Can. J. Microbiol. 1998;44:91–94. [PubMed] [Google Scholar]
- 29.Struck J.C., Lempicki R.A., Toschka H.Y., Erdmann V.A., Fournier M.J. Escherichia coli 4.5S RNA gene function can be complemented by heterologous bacterial RNA genes. J. Bacteriol. 1990;172:1284–1288. doi: 10.1128/jb.172.3.1284-1288.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]