Abstract
The RET tyrosine kinase receptor and its ligand, glial cell line-derived neurotrophic factor (GDNF) are critical regulators of renal and neural development. It has been demonstrated that RET activates a variety of downstream signaling cascades, including the RAS/mitogen-activated protein kinase and phosphatidylinositol-3-kinase(PI3-K)/AKT pathways. However, nuclear targets specific to RET-triggered signaling still remain elusive. We have previously identified a novel zinc finger protein, GZF1, whose expression is induced during GDNF/RET signaling and may play a role in renal branching morphogenesis. Here, we report the DNA binding property of GZF1 and its potential target gene. Using the cyclic amplification and selection of targets technique, the consensus DNA sequence to which GZF1 binds was determined. This sequence was found in the 5′ regulatory region of the HOXA10 gene. Electrophoretic mobility shift assay revealed that GZF1 specifically binds to the determined consensus sequence and suppresses transcription of the luciferase gene from the HOXA10 gene regulatory element. These findings thus suggest that GZF1 may regulate the spatial and temporal expression of the HOXA10 gene which plays a role in morphogenesis.
INTRODUCTION
Glial cell line-derived neurotrophic factor (GDNF) was originally identified as a survival factor for midbrain dopaminergic neurons (1). Subsequently, it was shown that GDNF promotes the survival and/or differentiation of various central and peripheral neurons, such as spinal motoneurons and sympathetic, parasympathetic, sensory and enteric neurons (2). GDNF also plays an important role outside the nervous system. Based on studies of GDNF-transgenic mice, GDNF was shown to function as a morphogen in kidney development and to regulate spermatogonial differentiation (3–6). We and other investigators demonstrated that GDNF signals through a unique multicomponent receptor complex consisting of glycosyl-phosphatidylinositol (GPI)-anchored co-receptor as a ligand binding component and RET receptor tyrosine kinase as a signaling component (7,8).
Since GDNF was discovered as a RET ligand, the signaling pathways activated by GDNF have become well understood. Following GDNF stimulation, RET can activate a variety of intracellular signaling pathways, including the RAS/extracellular signal-regulated kinase (ERK), phosphatidylinositol 3-kinase (PI3-K)/AKT, p38 mitogen activated protein kinase (p38MAPK) and c-Jun N-terminal kinase (JNK) pathways (9,10). However, nuclear targets specific to RET-triggered signaling still remain elusive. By differential display analysis, we have recently identified a novel GDNF-inducible gene named GZF1 (GDNF-inducible zinc finger protein 1) (11). GZF1 encodes a protein with a BTB/POZ (Broad complex, Tramtrack, Bric á brac/Poxvirus and Zinc finger) domain at the N-terminus and 10 tandemly repeated C2H2 zinc finger motifs. The fact that antisense phosphorothioated oligodeoxynucleotides of the GZF1 gene markedly impair the ureteric bud branching in the metanephric organ culture suggests that GZF1 expression mediated by GDNF may play a role in renal branching morphogenesis (11).
The C2H2 zinc finger motif was initially found to be present in transcription factors. A number of transcription factors are known to utilize their C2H2 zinc fingers as DNA binding domains (12,13). The BTB/POZ domain is also a conserved structural motif found mainly in transcription factors and actin-binding proteins, and transcription factors with the BTB/POZ domain function as transcriptional repressors, activators or both (14,15). We reported that GZF1 has transcriptional repressive activity using the luciferase reporter gene assay (11), although no bona fide target genes for GZF1 have been elucidated.
In the present study, we determined a DNA binding sequence specific for GZF1, using the cyclic amplification and selection of targets (CASTing) technique (16,17). The C2H2 zinc fingers of GZF1 specifically recognized a 12 bp consensus sequence which was found in the regulatory regions of human and mouse HOXA10 genes. In addition, GZF1 was shown to function as a sequence-specific transcriptional repressor through its DNA binding activity.
MATERIALS AND METHODS
Plasmids
Full-length GZF1 and ΔBTB-GZF1 cDNAs were amplified by PCR and inserted into the pGEX5X-2 expression plasmid (Amersham Pharmacia) to produce glutathione S-transferase (GST) fusion proteins. For the expression of GFP-tagged protein, the ΔBTB-GZF1 cDNA was subcloned into the pEGFP-C3 vector (Clontech) to obtain pEGFP-ΔBTB-GZF1. The constructs pSRα-GZF1, pSRα-GAL4DBD-GZF1 and pSRα-GFP-GZF1 have been described previously (11).
CASTing
CASTing was performed as described previously (18). Oligonucleotides carrying defined ends and a 26-nt region of degeneracy [R76, 5′-CAGGTCAGTTCAGCGGATCCTGTCG-(N)26-GAGGCGAATTCAGTGCAACTGCAGC-3′] and PCR primers [forward (F) primer, 5′-GCTGCAGTTGCACTGAATTCGCCTC-3′; reverse (R) primer, 5′-CAGGTCAGTTCAGCGGATCCTGTCG-3′] were synthesized and used for the CASTing experiments. 32P-labeled double-stranded degenerate oligonucleotides were prepared by incubating 100 μM each of F and R76 oligonucleotides in 50 μl of reaction mixture containing 10 mM Tris–HCl (pH 9.0), 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100, 50 μM each of dATP, dGTP and dTTP, 20 μM [α-32P]dCTP (740 kBq) and 2 U of the Klenow fragment of DNA polymerase I at 37°C for 1 h. Binding reactions were performed by adding GST-ΔBTB-GZF1 (0.4 μg) to Buffer D [20 mM HEPES (pH 7.9), 120 mM KCl, 1 mM DTT, 1% NP-40 and 12% glycerol] containing 400 ng of poly(dI–dC), 400 ng of sonicated salmon sperm DNA, 50 μg of BSA and 32P-labeled double-stranded degenerate oligonucleotide (1.8 ng), and incubating the mixture at 4°C for 30 min. Then, glutathione-Sepharose beads (20 μl) were added and incubation was continued at 4°C for an additional 1 h. The DNA–protein complexes were pulled down by centrifugation, and pellets were washed six times with Buffer D. After elution with 100 μl of Buffer D containing 5 mM reduced glutathione, the oligonucleotides were recovered by phenol extraction and ethanol precipitation, and amplified by PCR in 20 μl of a mixture containing 300 ng of primers F and R, 20 μM [α-32P]dCTP (740 kBq), 50 μM each of dATP, dGTP and dTTP, 10 mM Tris–HCl (pH 8.0), 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100 and 0.5 U of Ex Taq polymerase (Takara). DNAs were amplified for 20 cycles of 1 min at 94°C, 30 s at 60°C and 1 min at 72°C, purified by passage through a Sephacryl-200 spin column, precipitated with ethanol, and dissolved in 50 μl of a buffer containing 10 mM Tris–HCl (pH 8.0) and 1 mM EDTA. The amount of amplified oligonucleotides was quantified, and 2 ng was used in subsequent CASTing cycles. After four cycles of CASTing, the radiolabeled oligonucleotides (1.5 × 105 c.p.m.) were used as probes in an electrophoretic mobility shift assay (EMSA). Oligonucleotides bound to GST-ΔBTB-GZF1 were excised from gels, eluted overnight in 0.2 ml of a solution containing 0.2 M NaCl, 20 mM EDTA and 0.1% SDS at 37°C, extracted once with phenol and precipitated with ethanol. DNAs were amplified by PCR as described above, and then subcloned into the pGEM-Teasy vector.
Electrophoretic mobility shift assay
Human embryonic kidney (HEK) 293T cell extracts or purified GST-ΔBTB-GZF1 were suspended in a reaction mixture containing 20 mM HEPES (pH 7.9), 150 mM KCl, 0.1 mM EDTA, 0.5 mM DTT, 10% glycerol and 1 μg of salmon sperm DNA. 32P-labeled probes (10 000 c.p.m.) were incubated in 15 μl of reaction mixture. When necessary, unlabeled DNA fragments were added as competitors at this step. Then, GST-ΔBTB-GZF1 fusion proteins (0.3 μg) or aliquots of the HEK293T cell extract (5 μg protein) were added, and the reaction mixture was incubated for 15 min on ice. In experiments with antibodies, the GST-fusion proteins were preincubated with the antibody for 1 h on ice. DNA–protein complexes were electrophoretically resolved on 4% polyacrylamide gels in 100 mM Tris–borate (pH 8.3) containing 2 mM EDTA and 2.5% glycerol at 25°C. The gels were dried and then autoradiographed.
Site-directed mutagenesis
Mutagenesis reactions were carried out on the GZF1 zinc finger domain subcloned into the pSRα or pGEX5X-2 (Amersham Pharmacia) vectors using the QuickChangeTM Site-directed Mutagenesis Kit (Stratagene). The reactions were set up essentially as recommended by the manufacturer. The mutations, as well as the fidelity of the rest of the DNA, were confirmed by sequencing.
Transfection and luciferase assay
HEK293T cells were grown at 60% confluency in 10 cm plates and transfected with the luciferase gene constructs using Lipofectamine 2000 (Invitrogen). Cells were harvested 48 h after transfection, and extracts were prepared using standard techniques. The transfection efficiency was normalized by cotransfection of the Renilla luciferase expression vector pRL-TK (Promega), and relative luciferase activity was determined as recommended by the manufacturer.
Chromatin immunoprecipitation (ChIP) assays
Cells (5 × 105) expressing GZF1 and GFP-GZF1 were fixed with 1% formaldehyde for 10 min. Crude cell lysates were sonicated to generate 200–1000 bp DNA fragments. After centrifugation, the supernatants were diluted 10-fold with dilution buffer (0.01% SDS, 1.1% TritonX-100, 1.2 mM EDTA, 16.7 mM Tris–HCl (pH 8.1) and 167 mM NaCl) and incubated with anti-GZF1 or anti-GFP antibody at 4°C overnight.
Immunocomplexes were collected with protein A agarose beads and eluted by adding 200 μl of 1% SDS in 0.1 M NaHCO3. Cross-links were reversed by heating at 65°C. Following protease K digestion, phenol–chloroform extraction and ethanol precipitation, the samples were subjected to 40 cycles of PCR amplification using HOXA10 promotor-specific primers 5′-AGAGTCTAGCCAGGAGGACTGCTCGCGGGC-3′ and 5′-TGGGCCGGAGGTTCCAGCCCCGAGCC-3′.
RESULTS
Identification of a consensus GZF1 binding sequence
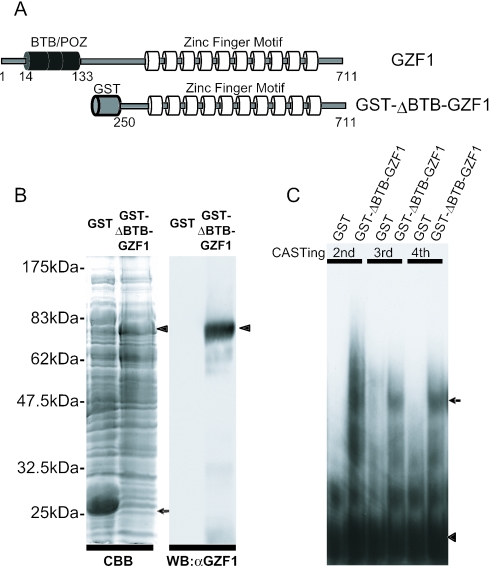
To investigate whether GZF1 binds to a specific regulatory sequence, we performed the CASTing technique using an unbiased set of degenerate oligonucleotides (16,17). We synthesized 76 bp oligonucleotides containing a random set of 26 bp flanked by PCR priming sequences. The products were expected to comprise a 426-fold degenerate oligonucleotide pool. After synthesis of the complementary strand by priming DNA synthesis with a 5′ PCR primer, the double-stranded oligomer was mixed with bacterially expressed GST-ΔBTB-GZF1 (amino acids 250–711 of GZF1) fusion protein (Figure 1A and B) to permit the formation of DNA–protein complexes. Expression of the GST-ΔBTB-GZF protein was confirmed by western blotting with anti-GZF1 antibody raised against the 19 C-terminal amino acids of GZF1 (Figure 1B).
Figure 1.
(A) Schematic illustration of full-length GZF1 (amino acids 1–711) and the GST-ΔBTB-GZF1 (amino acids 250–711) fusion protein. The BTB/POZ domain and GST are represented as black and gray cylinders, respectively. C2H2 zinc fingers are shown as white cylinders. (B) GST and GST-ΔBTB-GZF1 proteins expressed in bacteria were stained with Coomassie brilliant blue. An arrow and an arrowhead indicate GST and GST-ΔBTB-GZF1 proteins, respectively (left panel). The GST-ΔBTB-GZF1 fusion proteins were detected by western blotting with anti-GZF1 antibody raised against the 19 C-terminal amino acids of GZF1 (11) (right panel). (C) Progressive enrichment of oligonucleotide–GST-ΔBTB-GZF1 complex by the CASTing method. An arrow indicates an oligonucleotide–protein complex and an arrowhead indicates free probe.
The resulting complexes were pulled-down with glutathione-Sepharose beads and subjected to subsequent PCR amplification. The ratio of specific binding to random sequences was increased in subsequent rounds of CASTing as the highest affinity interactions were selected. As shown in Figure 1C, a specific signal was shown in the third cycle, which became a major fraction in the fourth cycle. Although we also used the GST-full-length GZF1 (amino acids 1–711) fusion protein for CASTing, no DNA–protein complexes were obtained, suggesting that the BTB-POZ domain may interfere with complex formation as has been observed for other BTB-POZ-containing proteins (19–23).
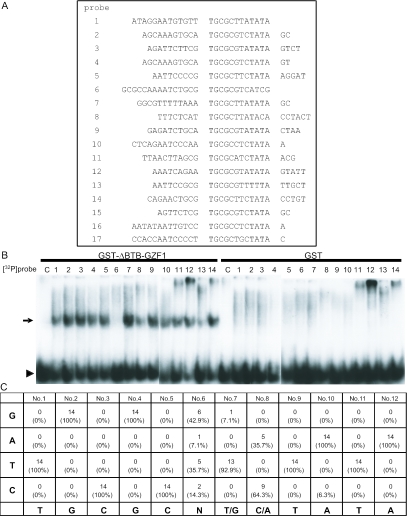
After the fourth round of selection, 17 different oligonucleotides were cloned from the specific band and sequenced (Figure 2A). Using these oligonucleotides, we performed EMSA to investigate their affinities for the GST-ΔBTB-GZF1 fusion protein. All 32P-labeled oligonucleotides except clones 6, 8 and 13 formed apparent DNA–protein complexes with identical mobility (Figure 2B). The oligonucleotide probes derived from clones 6, 8 and 13 showed very faint or no signal in repeated experiments (Figure 2B). As a result of these experiments, the consensus sequence TGCGCN (T/G)(C/A)TATA for DNA binding was obtained from the sequence alignment of 14 individual clones (the sequences from clones 6, 8 and 13 were excluded) (Figure 2A and C).
Figure 2.
Identification of the GZF1 binding consensus sequence. (A) Sequences of 17 cDNA clones recovered in the CASTing analysis. Putative core binding sequences from 17 clones were aligned. (B) EMSA to assess binding of the GST-ΔBTB-GZF1 to selected oligonucleotides. The arrow indicates complexes ofGST-ΔBTB-GZF1 with 32P-labeled probes (clones 1–14) and the arrowhead indicates free probe. The probes derived from clones 6, 8 and 13 showed very faint or no signals representing the DNA–protein complex. Core sequences of clones 15, 16 and 17 were the same as those of clones 2 (and 4), 10 and 5, respectively (A) and probes from clones 15–17 also showed a strong signal representing the DNA–protein complex (data not shown). (C) Consensus binding sequence of GZF1, as determined by CASTing. The frequency of nucleotides at each position is shown. The putative consensus sequence is 5′-TGCGCN(T/G)(C/A)TATA-3′.
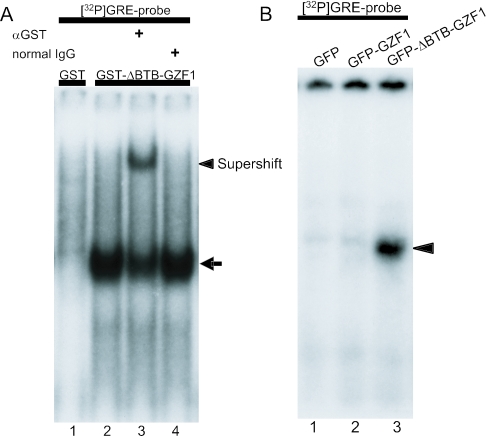
To confirm the binding specificity of GZF1 for this consensus sequence, we performed EMSA analysis using an oligonucleotide that represented the most frequently occurring sequence TGCGCGTCTATA (wild-type probe, wt), corresponding to clones 2, 4 and 15. This sequence was named the GZF1 responsive element (GRE). As shown in Figure 3A, the addition of anti-GST antibody supershifted the DNA–protein complex band, confirming the presence of the GST-ΔBTB-GZF1 protein in the retarded band (Figure 3A, lane 3). However, consistent with the finding that no specific DNA–protein complex was detected by CASTing using the GST-full-length GZF1 fusion protein, formation of the complex containing GST-full-length GZF1 and GRE was not induced (data not shown). To confirm our results, we transfected expression vectors carrying GFP-full-length GZF1 or GFP-ΔBTB-GZF1 into HEK293T cells. Whereas the DNA–protein complex was observed using extracts from cells transfected with GFP–ΔBTB–GZF1 (Figure 3B, lane 3), the use of the GFP-full-length GZF1 did not induce a specific complex (Figure 3B, lane 2). These results support the view that the BTB/POZ domain of GZF1 interferes with the DNA binding property of GZF1 in EMSA. A faint band detected in the GST and GFP lanes may represent a non-specific band or a band caused by endogenous GZF1.
Figure 3.
Complex formation of GRE with ΔBTB-GZF1. (A) 32P-labeled GRE was incubated with GST-ΔBTB-GZF1 in the absence or presence of anti-GST antibody or normal mouse IgG. The arrow indicates the GRE-GST-ΔBTB-GZF1 complex and the arrowhead indicates the supershifted complex in the presence of anti-GST antibody. (B) EMSA using GFP-full-length GZF1 or GFP-ΔBTB-GZF1 fusion proteins. Extracts from HEK293T cells transfected with empty pEGFP (lane 1), GFP-full-length GZF1 (lane 2) or GFP-ΔBTB-GZF1 (lane 3) plasmid were analyzed for binding with 32P-labeled GRE. An arrowhead indicates the DNA-GFP-ΔBTB-GZF1 complex (lane 3).
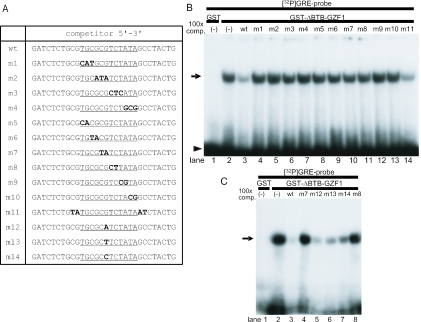
Mutation analysis of the GRE consensus sequence
To investigate the specificity of the GRE consensus sequence for GZF1 binding, a series of oligonucleotides with two or three base substitutions inside the GRE (m1–m10) and an oligonucleotide mutated just outside of the GRE (m11) were synthesized and used for EMSA as cold competitors (Figure 4A). As expected, the shifted band was markedly diminished by adding a 100-fold excess of wild-type unlabeled oligonucleotide (Figure 4B, lane 3). In contrast, oligonucleotides m1–m10 did not compete for binding at all (Figure 4B, lanes 4–13). The m11 oligonucleotide inhibited the binding of the GST-ΔBTB-GZF1 protein to labeled. GRE probe as efficiently as the wild-type cold competitor (Figure 4B, lane 14).
Figure 4.
Mutation analysis of the GRE consensus sequence. (A) Sequences of oligonucleotides used for competitive EMSAs m1–m4 contain triple base substitutions in the wild-type (wt) GRE sequence, and m5–m10 contain double base substitutions. m11 was mutated just outside of GRE, and m12–m14 contains a single base mutation at the sixth position of the GRE. (B) 100-fold molar excess of unlabeled probes was added to the binding reaction as cold competitors. The wild-type and m11 oligonucleotides efficiently competed for GST-ΔBTB-GZF1 binding to 32P-labeled GRE, whereas m1–m10 cold competitors did not compete at all. An arrow indicates the DNA–protein complex and an arrowhead indicates free probe. (C) All oligonucleotides with single base substitutions at the sixth position of the GRE (m12–m14) competed efficiently for GST-ΔBTB-GZF1 binding to the 32P-labeled GRE.
Because the sixth position of the defined consensus sequence was not specific (Figure 2C), oligonucleotides with a series of single base substitutions at the sixth position of the GRE (m12–m14) were synthesized as cold competitors (Figure 4A). All of these mutants competed efficiently for binding of GST-ΔBTB-GZF1 to the labeled GRE probe (Figure 4C).
Analysis of the zinc finger motifs in GZF1 required for DNA binding
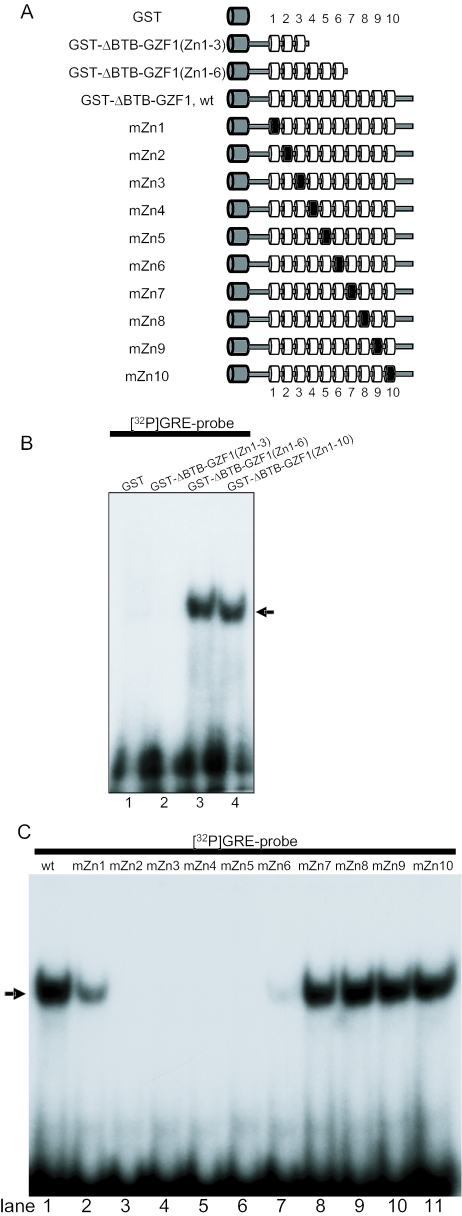
We next investigated which zinc fingers in GZF1 are involved in the interaction with DNA. Two mutants of GZF1, in which zinc finger motifs were deleted, were generated and subjected to EMSA using 32P-labeled GRE (Figure 5A). The GST-fusion protein with only the first three zinc fingers (zinc fingers 1–3) did not induce a DNA–protein complex (Figure 5B, lane 2). In contrast, the mutant consisting of zinc fingers 1–6 provided a specific complex band (Figure 5B, lane 3), suggesting that one or more of these zinc finger motifs are necessary for sequence-specific DNA binding.
Figure 5.
Analysis of zinc finger motifs in GZF1 required for DNA binding. (A) Schematic representation of GZF1 zinc finger motif mutants used for EMSA. Two deletion zinc finger mutants are shown. Two cysteine residues in the C2H2 motifs of each finger were replaced with arginines. The wild-type and mutated fingers are shown as a white and a black cylinder, respectively. GST is shown as a grey cylinder. (B) Two deletion mutants of zinc finger motifs in GZF1 were subjected to EMSA. The arrow indicates the DNA–protein complex. (C) EMSA with GZF1 constructs mutated in the individual zinc finger motifs.
To further define which fingers in GZF are required for DNA binding, two cysteine residues in the C2H2 motif of each finger were replaced with arginines (Figure 5A). As shown in Figure 5C, mutation of any one of zinc finger motifs 2–5 completely abolished binding to the wild-type probe (lanes 3–6). In addition, mutation of zinc finger 1 or 6 markedly reduced binding (lanes 2 and 7) whereas mutation of zinc fingers 7–10 had no effect (lanes 8–11). These results demonstrate that zinc fingers 1–6 are necessary for full DNA binding ability of GZF1.
Transcriptional repression by GZF1
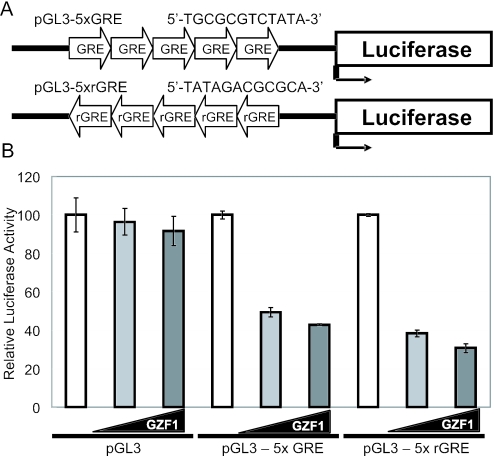
To address the question of whether GZF1 can regulate the expression of a reporter gene containing the GRE, five tandem repeats of GRE were cloned upstream of the luciferase gene in a pGL3-basic reporter vector (pGL3-5×GRE) (Figure 6A). When pGL3-5×GRE and an expression plasmid carrying full-length GZF1 (pSRα-GZF1) were transfected into HEK293T cells, expression of the luciferase gene decreased by 50–60% compared to co-transfection with pGL3 and pSRα-GZF1 (Figure 6B).
Figure 6.
Transcriptional repressive activity of GZF1. (A) Schematic representation of the reporter vectors. Five tandem repeats of GRE were cloned upstream of the luciferase gene in a pGL3-basic reporter vector (designated pGL3-5 × GRE). The reporter vector containing five GREs in the antisense direction was also constructed (pGL3-5 × rGRE). (B) Luciferase assay using the extracts from HEK293T cells transiently co-transfected with increasing amounts (50 and 100 ng) of an expression vector carrying full-length GZF1 cDNA (pSRα-GZF1) and pGL3, pGL3-5 × GRE or pGL3-5 × rGRE.
Because it has been established that transcription factors regulate the expression of target genes regardless of the orientation of their binding sequences, a reporter gene containing GRE in the antisense direction was constructed to examine the effect of GRE orientation on transcription (Figure 6A). As in the case of the reporter construct containing GRE in the sense direction, GZF1 efficiently repressed the expression of this antisense reporter gene (Figure 6B).
HOXA10 as a potential target gene for GZF1
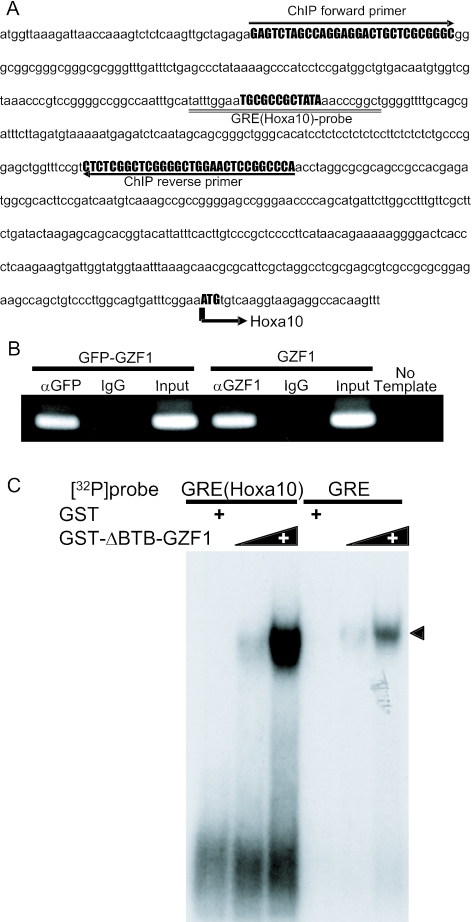
We searched a database of transcriptional start sites for potential target genes that may be regulated by GZF1, and found that several genes, including HOXA10, have the GZF1 binding sequence in their 5′-upstream regulatory regions (24). We focused on the HOXA10 gene because both human and mouse HOXA10 genes contain the GZF1 binding site within their regulatory regions. In several other genes, the GZF1 binding sequences were detected in either human or mouse genes but not in both. The GZF1 recognition sequence 5′-TGCGCCGCTATA-3′ observed in the regulatory region of HOXA10 was named GRE(HOXA10).
To investigate whether GZF1 is targeted to GRE(HOXA10) in vivo, we performed chromatin immunoprecipitation. Cross-linked chromatin fragments from HEK293T cells expressing GZF1 or GFP-GZF1 were immunoprecipitated with anti-GZF1 or anti-GFP antibody. DNA from the resulting immunoprecipitates was subjected to PCR in order to amplify a target sequence of 300 bp corresponding to the human HOXA10 regulatory region (Figure 7A). Chromatin fragments containing this region were specifically precipitated with anti-GZF1 antibody or anti-GFP antibody but not with normal IgG (Figure 7B). These results reveal that GZF1 binds to the recognition sequence in the HOXA10 regulatory region in vivo.
Figure 7.
(A) Genomic sequence of the human HOXA10 regulatory region. The GRE(HOXA10) sequence used for EMSA is indicated by a double underline. The primer sets used for the ChIP assays are also shown. (B) Binding of GZF1 to the HOXA10 regulatory region as assessed by ChIP. Crosslinked chromatin fragments from HEK293T cells expressing GZF1 or GFP-GZF1 were immunoprecipitated with anti-GZF1 antibody or anti-GFP antibody. DNA from the resulting immunoprecipitates was subjected to PCR amplification using the primers shown in (A). As a positive control (input), 10 ng of genomic DNA was amplified in parallel. (C) Binding of GZF1 to GRE(HOXA10) as assessed by EMSA. EMSA was performed using a 32P-labeled GRE(HOXA10) probe and GST-ΔBTB-GZF1 protein (0.1 and 0.3 μg). The GRE(HOXA10) probe and GST-ΔBTB-GZF1 protein showed a single complex band, which was identical to that shifted with the GRE probe.
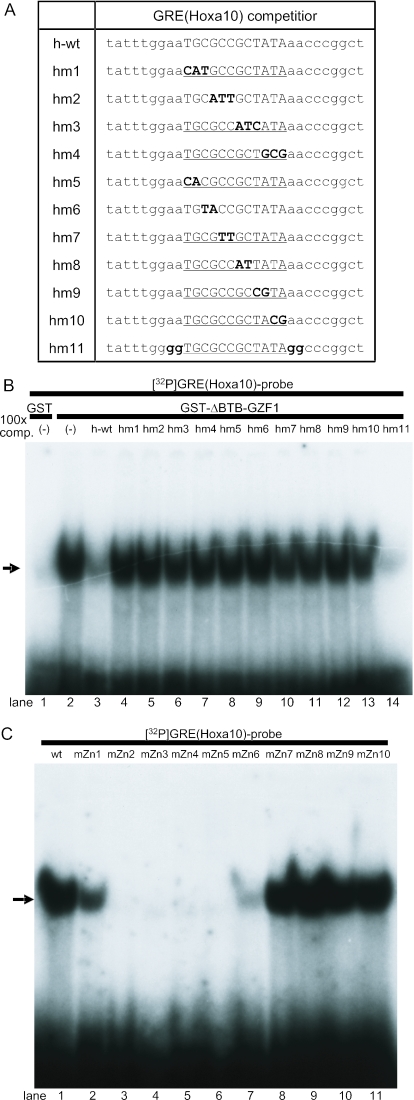
To confirm that GZF1 binds to GRE(HOXA10), we performed EMSA with a probe containing the GRE(HOXA10) sequence (Figure 7A). Incubation of the 32P-labeled GRE (HOXA10) probe with the GST-ΔBTB-GZF1 protein resulted in a single complex band, which was identical to the band shifted with the GRE probe (Figure 7C). The shifted band was markedly diminished by adding a 100-fold excess amount of unlabeled oligonucleotides of GRE(HOXA10) wild-type (h-wt) or a mutant (hm11) (Figure 8A and B) in which 4 nt just outside of GRE(HOXA10) were substituted. In contrast, none of the mutants with two or three base substitutions inside the GRE (hm1–hm10) competed with the labeled GRE(HOXA10) probe for binding to GST-ΔBTB-GZF1 (Figure 8A and B).
Figure 8.
Mutation analysis of the GRE(HOXA10) sequence and zinc finger motifs in GZF1 required for DNA–protein complex formation. (A) Oligonucleotide sequences used for the competitive EMSAs. Hm1–hm4 contain triple base substitutions in the wild-type (h-wt) GRE(HOXA10) sequence, andhm5–hm10 contain double base substitutions. Hm11 mutated outside ofGRE(HOXA10) is also used in this assay. (B) 100-fold molar excess of unlabeled probes was added to the binding reaction as cold competitors. The wild-type and hm11 oligonucleotides efficiently competed for GST-ΔBTB-GZF1 binding to 32P-labeled GRE(HOXA10), whereas hm1–hm10 cold competitors did not compete at all. An arrow indicates the DNA–protein complex. (C) EMSA using 32P-labeled GRE(HOXA10) and GST-ΔBTB-GZF1 mutated in the individual zinc finger motifs. The arrow indicates the DNA–protein complex.
Using the zinc finger mutants of GST-ΔBTB-GZF1 shown in Figure 5A, we performed EMSA to examine whether the GRE(HOXA10) probe binds specific zinc finger motifs of GZF1. As observed for the GRE probe (Figure 5C), zinc finger motifs 2–5 are essential for binding of GZF1 toGRE(HOXA10) and zinc fingers 1 and 6 are also required for the full-binding activity of GZF1 (Figure 8C).
Transcriptional repression by GZF1 via GRE(HOXA10)
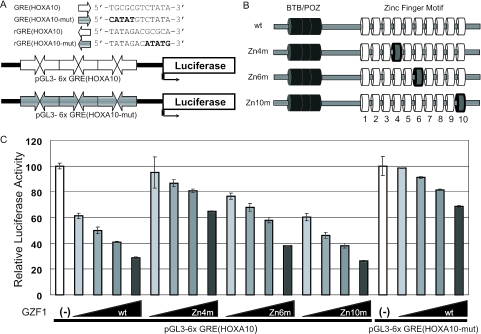
We synthesized an oligonucleotide containing six copies of GRE(HOXA10) closely packed in both orientations and inserted the oligonucleotide upstream of the luciferase gene by utilizing a pGL3-basic reporter vector (Figure 9A). We also synthesized 6×GRE(HOXA10-mut), in which the conserved TGCGC (sense orientation) or GCGCA (antisense orientation) motif in 6×GRE(HOXA10) was changed to CATAT or ATATG (Figure 9A). Co-transfection of pSRα-GZF1 with the 6×GRE(HOXA10) vector decreased the expression of the luciferase gene in a dose-dependent manner (Figure 9C). Transfection of the 6×GRE(HOXA10-mut) vector markedly impaired the repressive activity of GZF1 (Figure 9C).
Figure 9.
(A) Schematic representation of a pGL3-basic reporter vector containing six copies of GRE(HOXA10) in both orientations upstream of the luciferase gene. We also synthesized 6×GRE(HOXA10-mut) in which the conserved TGCGC (sense orientation) or GCGCA (antisense orientation) motifs in 6×GRE(HOXA10) were changed to CATAT or ATATG, respectively. (B) Schematic representation of mutant GZF1 (Zn4m, Zn6m and Zn10m) in which two cysteines in zinc fingers 4, 6 or 10 were replaced with arginines. The wild-type and mutated fingers are shown as white and gray cylinders, respectively. The BTB/POZ domain is shown as a black cylinder. (C) Transcriptional repressive activity of GZF1 was markedly impaired by mutation of zinc finger 4 (Zn4m). Mutation of zinc finger 6 (Zn6m) moderately reduced the repressive activity of GZF1 but mutation of zinc finger 10 (Zn10m) showed no effect. Increased amounts of wild-type or mutant GZF1 (50, 100, 200 and 500 ng) were co-transfected with pGL3-6×GRE(HOXA10) or pGL3-6×GRE(HOXA10-mut).
We next used GZF1 mutants (Zn4m, Zn6m and Zn10m) in which two cysteines in zinc finger motif 4, 6 or 10 were replaced with arginines (Figure 9B). Consistent with the results of EMSA (Figure 8C), transcriptional repressive activity of GZF1 was strongly reduced by the mutation of zinc finger 4. The mutation of zinc finger 6 moderately reduced GZF1 repressive activity but mutation of zinc finger 10 had no effect. These results suggest that GZF1 regulates HOXA10 expression through binding to GRE(HOXA10).
DISCUSSION
GZF1 is a sequence-specific transcriptional repressor
GZF1 is a C2H2 zinc finger protein that possesses a BTB/POZ domain, which is generally known to bind DNA directly and regulate the expression of target genes. C2H2 zinc finger proteins comprise the largest family of transcription factors in eukaryotes, and play important roles in multiple cellular processes, including development, cell growth and differentiation (15). Structurally, the C2H2 zinc finger motif is composed of a beta-hairpin followed by an alpha-helix that folds around a single zinc ion (13). Sequence-specific recognition of DNA is mediated primarily by interactions between the variable amino acids within and around the alpha-helix and nucleotides within the major groove of DNA. The present study provides direct evidence that GZF1 is a sequence-specific DNA binding protein, and represses transcription of a reporter gene in a manner that depends on the presence of the GZF1 responsive element (GRE). We determined that the GRE consists of a 12 bp consensus sequence, TGCGCN(T/G)(C/A)TATA.
The BTB/POZ domain is an evolutionarily conserved protein–protein interaction motif found in many transcription factors. It is estimated that the BTB/POZ domain is found in 0.6 and 0.8% of total genes in the human and mouse genomes, respectively (25). Previously, it was shown that the BTB/POZ domains of ZID, ZF5, GAGA, KAISO and HIC1 act to inhibit the interaction of the proteins' zinc finger regions with DNA by direct interaction with the zinc finger motifs (19–23). Deletion of their BTB/POZ domains resulted in strong binding to target DNA sequences. These results reveal that the BTB/POZ domain negatively regulates DNA binding of full-length zinc finger proteins. Consistent with these reports, the BTB/POZ domain of GZF1 interfered with the sequence-specific DNA binding of the zinc finger domains of GZF1, as observed by EMSA, although the BTB/POZ domain itself appears to play a role in the transcriptional repressive activity of GZF1 (11).
As has been observed in other BTB/POZ domain-containing proteins such as PLZF and BCL6, the BTB/POZ domain in GZF1 may have the ability to interact with other key regulatory proteins, including co-repressors, CtBP and histone deacetylases (26). The CtBP family has been reported to mediate the repressor function of several transcription factors by binding to the consensus sites, PXDSLXK (27). However, because GZF1 lacks this CtBP binding motif, CtBP does not appear to mediate the repressor function of GZF1. In addition, GZF1-mediated transcriptional repression was not inhibited by trichostatin A (TSA) (data not shown), suggesting that transcriptional repression mediated by GZF1 through the GRE does not involve the recruitment of TSA-sensitive class I and II histone deacetylases as observed for HIC1 (28). Alternative mechanisms that include the recruitment of members of the TSA-insensitive histone deacetylases may underlie GZF1-mediated transcriptional repression in mammalian cells. Identification of the proteins associated with GZF1 will be necessary to elucidate the precise mechanisms of transcriptional repression by GZF1.
GZF1 represses transcription via a binding sequence found in the HOXA10 regulatory region
The optimal GZF1 binding sequence was found in the human and murine HOXA10 gene regulatory region. We demonstrated that GZF1 binds specifically to the identified sequence and represses the expression of the reporter gene linked to this sequence. In addition, ChIP assay revealed that GZF1 interacts with the genomic fragment containing GRE(HOXA10), suggesting that HOXA10 is a physiological target gene of GZF1.
HOX proteins regulate the morphogenesis of specific organs indirectly by activating networks of transcription factors and signaling molecules; they also influence cell behavior, thereby affecting tissue and organ shape (29). It is well-known that some mouse Hox genes are involved in renal development (30,31). Mutant mice homozygous for either Hoxa11 or Hoxd11 have normal kidneys; however, the kidneys of double homozygous mutants of these genes are absent or rudimentary (32). In the triple mutant of the Hox11 paralogous group (Hoxa11, Hoxc11 and Hoxd11), no GDNF expression can be detected, and there is complete failure of ureteric bud formation (33). These findings suggest that functional redundancy occurs between paralogous HOX11 genes in kidney development.
Functional redundancy has also been found between non-paralogous Hox genes as has been described in the limbs in the case of Hoxa10 and Hoxd11, as well as in the case of the contiguous genes Hoxa10 and Hoxa11 (34,35). In this regard, it is interesting to note that paralogous genes of Hox10 and Hox11 are expressed predominantly within the metanephric mesenchyme in the developing kidney (36). Hoxa10-deficient mice are sexually sterile but have normal kidneys (37). Interestingly, mice doubly mutant for Hoxa10 and Hoxd10 show altered kidney placement and size (38). These findings suggest that the Hox10 paralogous group may have a role in branching morphogenesis of the ureteric bud in the developing kidney; the same can be said of the Hox11 group. The results of gene targeting of the entire Hox10 paralogous group are anxiously awaited.
Emx2 is a homeobox gene located outside the Hox cluster. It has also been shown that Emx2 is essential for urogenital development. In Emx2 mutant mice, Ret expression in the ureteric bud and Gdnf expression in the mesenchyme are greatly reduced, resulting in ureteric bud branching failure (39). Troy et al. (40) recently reported that constitutive expression of Hoxa10 diminished Emx2 mRNA, and Hoxa10 bound to the Emx2 5′ regulatory region. Because GZF1 is expressed in the branching ureteric bud, it is interesting to speculate that GZF1 suppresses Hoxa10 expression, thereby maintaining Emx2 and Ret expression in the ureteric buds for normal kidney development. We are currently generating conditional knockout mice in order to elucidate the in vivo role of GZF1 in morphogenesis.
Acknowledgments
We thank Mr Y. Imaizumi, Mr K. Uchiyama and Ms M. Kawai for technical assistance. This study was supported in part by Grants-in-aid for the 21st Century COE (Center Of Excellence) Research, Scientific Research (A) and Scientific Research on Priority Areas ‘Cancer’ from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and by a Grant from Uehara Memorial Foundation. Funding to pay the Open Access publication charges for this article was provided by the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lin L.F., Doherty D.H., Lile J.D., Bektesh S., Collins F. GDNF: a glial cell line-derived neurotrophic factor for midbrain dopaminergic neurons. Science. 1993;260:1130–1132. doi: 10.1126/science.8493557. [DOI] [PubMed] [Google Scholar]
- 2.Airaksinen M.S., Saarma M. The GDNF family: signalling, biological functions and therapeutic value. Nat. Rev. Neurosci. 2002;3:383–394. doi: 10.1038/nrn812. [DOI] [PubMed] [Google Scholar]
- 3.Sanchez M.P., Silos-Santiago I., Frisen J., He B., Lira S.A., Barbacid M. Renal agenesis and the absence of enteric neurons in mice lacking GDNF. Nature. 1996;382:70–73. doi: 10.1038/382070a0. [DOI] [PubMed] [Google Scholar]
- 4.Pichel J.G., Shen L., Sheng H.Z., Granholm A.C., Drago J., Grinberg A., Lee E.J., Huang S.P., Saarma M., Hoffer B.J., et al. Defects in enteric innervation and kidney development in mice lacking GDNF. Nature. 1996;382:73–76. doi: 10.1038/382073a0. [DOI] [PubMed] [Google Scholar]
- 5.Moore M.W., Klein R.D., Farinas I., Sauer H., Armanini M., Phillips H., Reichardt L.F., Ryan A.M., Carver-Moore K., Rosenthal A. Renal and neuronal abnormalities in mice lacking GDNF. Nature. 1996;382:76–79. doi: 10.1038/382076a0. [DOI] [PubMed] [Google Scholar]
- 6.Meng X., Lindahl M., Hyvonen M.E., Parvinen M., de Rooij D.G., Hess M.W., Raatikainen-Ahokas A., Sainio K., Rauvala H., Lakso M., et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science. 2000;287:1489–1493. doi: 10.1126/science.287.5457.1489. [DOI] [PubMed] [Google Scholar]
- 7.Treanor J.J., Goodman L., de Sauvage F., Stone D.M., Poulsen K.T., Beck C.D., Gray C., Armanini M.P., Pollock R.A., Hefti F., et al. Characterization of a multicomponent receptor for GDNF. Nature. 1996;382:80–83. doi: 10.1038/382080a0. [DOI] [PubMed] [Google Scholar]
- 8.Jing S., Wen D., Yu Y., Holst P.L., Luo Y., Fang M., Tamir R., Antonio L., Hu Z., Cupples R., et al. GDNF-induced activation of the ret protein tyrosine kinase is mediated by GDNFR-alpha, a novel receptor for GDNF. Cell. 1996;85:1113–1124. doi: 10.1016/s0092-8674(00)81311-2. [DOI] [PubMed] [Google Scholar]
- 9.Takahashi M. The GDNF/RET signaling pathway and human diseases. Cytokine Growth Factor Rev. 2001;12:361–373. doi: 10.1016/s1359-6101(01)00012-0. [DOI] [PubMed] [Google Scholar]
- 10.Ichihara M., Murakumo Y., Takahashi M. RET and neuroendocrine tumors. Cancer Lett. 2004;204:197–211. doi: 10.1016/S0304-3835(03)00456-7. [DOI] [PubMed] [Google Scholar]
- 11.Fukuda N., Ichihara M., Morinaga T., Kawai K., Hayashi H., Murakumo Y., Matsuo S., Takahashi M. Identification of a novel glial cell line-derived neurotrophic factor-inducible gene required for renal branching morphogenesis. J. Biol. Chem. 2003;278:50386–50392. doi: 10.1074/jbc.M309629200. [DOI] [PubMed] [Google Scholar]
- 12.Klug A., Schwabe J.W. Protein motifs 5. Zinc fingers. FASEB J. 1995;9:597–604. [PubMed] [Google Scholar]
- 13.Pabo C.O., Peisach E., Grant R.A. Design and selection of novel Cys2His2 zinc finger proteins. Annu. Rev. Biochem. 2001;70:313–340. doi: 10.1146/annurev.biochem.70.1.313. [DOI] [PubMed] [Google Scholar]
- 14.Albagli O., Dhordain P., Deweindt C., Lecocq G., Leprince D. The BTB/POZ domain: a new protein–protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Differ. 1995;6:1193–1198. [PubMed] [Google Scholar]
- 15.Collins T., Stone J.R., Williams A.J. All in the family: the BTB/POZ, KRAB, and SCAN domains. Mol. Cell. Biol. 2001;21:3609–3615. doi: 10.1128/MCB.21.11.3609-3615.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pollock R., Treisman R. A sensitive method for the determination of protein–DNA binding specificities. Nucleic Acids Res. 1990;18:6197–6204. doi: 10.1093/nar/18.21.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wright W.E., Binder M., Funk W. Cyclic amplification and selection of targets (CASTing) for the myogenin consensus bindingsite. Mol. Cell. Biol. 1991;11:4104–4110. doi: 10.1128/mcb.11.8.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohshima N., Takahashi M., Hirose F. Identification of a human homologue of the DREF transcription factor with a potential role in regulation of the histone H1 gene. J. Biol. Chem. 2003;278:22928–22938. doi: 10.1074/jbc.M303109200. [DOI] [PubMed] [Google Scholar]
- 19.Bardwell V.J., Treisman R. The POZ domain: a conserved protein–protein interaction motif. Genes Dev. 1994;8:1664–1677. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- 20.Kaplan J., Calame K. The ZiN/POZ domain of ZF5 is required for both transcriptional activation and repression. Nucleic Acids Res. 1997;25:1108–1116. doi: 10.1093/nar/25.6.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katsani K.R., Hajibagheri M.A., Verrijzer C.P. Co-operative DNA binding by GAGA transcription factor requires the conserved BTB/POZ domain and reorganizes promoter topology. EMBO J. 1999;18:698–708. doi: 10.1093/emboj/18.3.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daniel J.M., Spring C.M., Crawford H.C., Reynolds A.B., Baig A. The p120(ctn)-binding partner Kaiso is a bi-modal DNA-binding protein that recognizes both a sequence-specific consensus and methylated CpG dinucleotides. Nucleic Acids Res. 2002;30:2911–2919. doi: 10.1093/nar/gkf398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinte S., Stankovic-Valentin N., Deltour S., Rood B.R., Guerardel C., Leprince D. The tumor suppressor gene HIC1 (hypermethylated in cancer 1) is a sequence-specific transcriptional repressor: definition of its consensus binding sequence and analysis of its DNA binding and repressive properties. J. Biol. Chem. 2004;279:38313–38324. doi: 10.1074/jbc.M401610200. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki Y., Yamashita R., Sugano S., Nakai K. DBTSS, DataBase of Transcriptional Start Sites: progress report 2004. Nucleic Acids Res. 2004;32:D78–D81. doi: 10.1093/nar/gkh076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 26.David G., Alland L., Hong S.H., Wong C.W., DePinho R.A., Dejean A. Histone deacetylase associated with mSin3A mediates repression by the acute promyelocytic leukemia-associated PLZF protein. Oncogene. 1998;16:2549–2556. doi: 10.1038/sj.onc.1202043. [DOI] [PubMed] [Google Scholar]
- 27.Chinnadurai G. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol. Cell. 2002;9:213–224. doi: 10.1016/s1097-2765(02)00443-4. [DOI] [PubMed] [Google Scholar]
- 28.Deltour S., Guerardel C., Leprince D. Recruitment of SMRT/N-CoR-mSin3A-HDAC-repressing complexes is not a general mechanism for BTB/POZ transcriptional repressors: the case of HIC-1 and gammaFBP-B. Proc. Natl Acad. Sci. USA. 1999;96:14831–14836. doi: 10.1073/pnas.96.26.14831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hombria J.C., Lovegrove B. Beyond homeosis–HOX function in morphogenesis and organogenesis. Differentiation. 2003;71:461–476. doi: 10.1046/j.1432-0436.2003.7108004.x. [DOI] [PubMed] [Google Scholar]
- 30.Shah M.M., Sampogna R.V., Sakurai H., Bush K.T., Nigam S.K. Branching morphogenesis and kidney disease. Development. 2004;131:1449–1462. doi: 10.1242/dev.01089. [DOI] [PubMed] [Google Scholar]
- 31.Patterson L.T., Potter S.S. Hox genes and kidney patterning. Curr. Opin. Nephrol. Hypertens. 2003;12:19–23. doi: 10.1097/00041552-200301000-00004. [DOI] [PubMed] [Google Scholar]
- 32.Patterson L.T., Pembaur M., Potter S.S. Hoxa11 and Hoxd11 regulate branching morphogenesis of the ureteric bud in the developing kidney. Development. 2001;128:2153–2161. doi: 10.1242/dev.128.11.2153. [DOI] [PubMed] [Google Scholar]
- 33.Wellik D.M., Hawkes P.J., Capecchi M.R. Hox11 paralogous genes are essential for metanephric kidney induction. Genes Dev. 2002;16:1423–1432. doi: 10.1101/gad.993302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Favier B., Rijli F.M., Fromental-Ramain C., Fraulob V., Chambon P., Dolle P. Functional cooperation between the non-paralogous genes Hoxa-10 and Hoxd-11 in the developing forelimb and axial skeleton. Development. 1996;122:449–460. doi: 10.1242/dev.122.2.449. [DOI] [PubMed] [Google Scholar]
- 35.Branford W.W., Benson G.V., Ma L., Maas R.L., Potter S.S. Characterization of Hoxa-10/Hoxa-11 transheterozygotes reveals functional redundancy and regulatory interactions. Dev. Biol. 2000;224:373–387. doi: 10.1006/dbio.2000.9809. [DOI] [PubMed] [Google Scholar]
- 36.Patterson L.T., Potter S.S. Atlas of Hox gene expression in the developing kidney. Dev. Dyn. 2004;229:771–779. doi: 10.1002/dvdy.10474. [DOI] [PubMed] [Google Scholar]
- 37.Satokata I., Benson G., Maas R. Sexually dimorphic sterility phenotypes in Hoxa10-deficient mice. Nature. 1995;374:460–463. doi: 10.1038/374460a0. [DOI] [PubMed] [Google Scholar]
- 38.Lin A.W., Carpenter E.M. Hoxa10 and Hoxd10 coordinately regulate lumbar motor neuron patterning. J. Neurobiol. 2003;56:328–337. doi: 10.1002/neu.10239. [DOI] [PubMed] [Google Scholar]
- 39.Miyamoto N., Yoshida M., Kuratani S., Matsuo I., Aizawa S. Defects of urogenital development in mice lacking Emx2. Development. 1997;124:1653–1664. doi: 10.1242/dev.124.9.1653. [DOI] [PubMed] [Google Scholar]
- 40.Troy P.J., Daftary G.S., Bagot C.N., Taylor H.S. Transcriptional repression of peri-implantation EMX2 expression in mammalian reproduction by HOXA10. Mol. Cell. Biol. 2003;23:1–13. doi: 10.1128/MCB.23.1.1-13.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]