Abstract
Functional genomics methods are used to investigate the huge amount of information contained in genomes. Numerous experimental methods rely on the use of oligo- or polynucleotides. Nucleotide strand hybridization forms the underlying principle for these methods. For all these techniques, the probes should be unique for analyzed genes. In addition to being unique for the studied genes, the probes should fulfill a large number of criteria to be usable and valid. The criteria include for example, avoidance of self-annealing, suitable melting temperature and nucleotide composition. We developed a method for searching unique and valid oligonucleotides or probes for genes so that there is not even a similar (approximate) occurrence in any other location of the whole genome. By using probe size 25, we analyzed 17 complete genomes representing a wide range of both prokaryotic and eukaryotic organisms. More than 92% of all the genes in the investigated genomes contained valid oligonucleotides. Extensive statistical tests were performed to characterize the properties of unique and valid oligonucleotides. Unique and valid oligonucleotides were relatively evenly distributed in genes except for the beginning and end, which were somewhat overrepresented. The flanking regions in eukaryotes were clearly underrepresented among suitable oligonucleotides. In addition to distributions within genes, the effects on codon and amino acid usage were also studied.
INTRODUCTION
The complete genome of a large number of organisms including bacteria, archaea and eukaryotes have been determined along with the human genome. Currently, there are some 230 bacterial and archaeal, and 34 finished eukaryal genomes. Genomes contain overwhelming amount of information, which can be investigated with numerous experimental and computational techniques. Many experimental methods rely on the use of oligo- or polynucleotides. Nucleotide strand hybridization—preferably with unique probes—forms the underlying principle for these methods. PCR technology, the workhorse of molecular biology, utilizes oligonucleotides as primers to copy and amplify genetic material. Gene expression studies such as Southern and northern blotting and more advanced SAGE and microarrays also utilize oligonucleotides. Gene function can be modulated by short oligonucleotides either by antisense technology or by RNA interference (RNAi). For all these techniques, the probes ought to be unique for analyzed genes.
Oligonucleotides for an organism can be identified from complete genomes. If working with mRNA only, genes and flanking regions have to be analyzed, whereas if genomic DNA is the target, the probes should be unique for the whole genome. In addition to being unique for the studied genes, the probes have to fulfill a large number of criteria, which vary due to the use of probes. These criteria include, for example, the avoidance of self-annealing, suitable melting temperature (Tm) and nucleotide composition. A number of methods have been developed for primer design [e.g. (1–13), http://www-genome.wi.mit.edu/genome_software/other/primer3.html; for a review see (14)]. MEDUSA shows visually the location of the primer pairs (15). Simulated annealing and Lagrangian relaxation algorithms have been used to design oligonucleotides to study microbial communities (16). Organisms can be identified with proper oligos (17). Methods for oligonucleotide selection and probe production for microarrays have also been developed (18–31). Numerous methods have been developed to predict antisense oligonucleotides (32–36) and RNAi (37–41). Probes for full gene synthesis have to be specially designed (42). Oligos can be designed also for protein interaction studies (43). When degenerate oligonucleotides are used for cloning orthologues and paralogues, primers have to be specially designed. The properties of genome-wide unique oligonucleotide studies have not been published, although some methods for the search of such strings have been presented (44).
Our aim was to develop a method for searching unique and valid oligonucleotides or probes for genes so that there is not even a similar (approximate) occurrence in any other location of the whole genome. Thus, for unique oligonucleotides there are no matches present in a genome within certain edit distance. All other oligonucleotides are called redundant. Not all the unique probes are suitable for practical experiments, therefore valid probes have to be distinguished from unique sequences. The Levenshtein edit distance (45) was used as the measure of similarity between two oligonucleotides. Let ed(x,y) denote the Levenshtein edit distance between the strings x and y. Then ed(x,y) is defined as the minimum number of edit operations needed to convert x to y or vice versa, where a single edit operation can either replace, delete or insert a single character. Given an oligonucleotide x and an error threshold k, we deem x to be unique if there is no such other oligonucleotide y that ed(x,y) ≤ k and some occurrence of y does not overlap x.
By using probe size 25, we analyzed 17 complete genomes representing a wide range of both prokaryotic and eukaryotic organisms. It was possible to find a large number of unique oligonucleotides for all the genomes. To avoid cross-hybridization when using the probes, edit distance of four was used, i.e. only such sequences were accepted for which related sequences with at most 20 matches were present. We define unique sequences as those, which do not have matches within allowed edit distance. As valid oligos are called when they are unique and they in addition meet a number of criteria for avoiding adverse effects of self-annealing and have high enough Tm. More than 92% of all the genes in the investigated genomes contained valid oligonucleotides, and thus were probeable. Extensive statistical tests were performed to characterize the properties of oligonucleotides. These segments were relatively evenly distributed in genes except for the beginning and end, which were somewhat overrepresented. In addition to distributions within genes, also the effects on codon and amino acid usage were tested. Although the majority of codons and residues had expected distributions in majority of the genomes some interesting trends were apparent.
MATERIALS AND METHODS
Genomes
The 17 genomes used in the tests were taken from the NCBI database (Table 1). When analyzing coding sequences (CDSs) in eukaryotes, the coding areas were concatenated with 100 nt extensions on both sides. Oligos used in laboratories are often directed to 5′ regions of genes. Many genes in genomes of prokaryotes are spaced so closely that the downstream and upstream regions of adjacent genes overlap. Therefore, it was possible to analyze the extensions only in the larger eukaryotic genomes.
Table 1.
Properties of studied genomes and oligonucleotides
| Organism | Classa | Genome size (106) | C+G (%) | CDS | Genome | CDS | |||
|---|---|---|---|---|---|---|---|---|---|
| Unique oligos (106) | Valid oligos (106) | Unique oligos (106) | Valid oligos (106) | Unique oligos/gene | Valid oligos/gene | ||||
| Buchnera sp. | Prγ | 0.64 | 27.5 | 0.4 | 0.296 | 0.343 | 0.265 | 708 | 525 |
| B.burgdorferi | S | 0.91 | 28.9 | 0.586 | 0.416 | 0.5 | 0.367 | 689 | 489 |
| C.acetobutylicum | F | 3.94 | 31.7 | 1.812 | 1.428 | 1.411 | 1.118 | 494 | 389 |
| S.solfataricus | A | 2.99 | 36.6 | 1.696 | 1.317 | 1.452 | 1.128 | 571 | 444 |
| H.pylori | Prδ/ε | 1.67 | 39.8 | 1.107 | 0.715 | 0.963 | 0.626 | 707 | 456 |
| B.subtilis | F | 4.21 | 44.5 | 2.945 | 2.121 | 2.6 | 1.873 | 718 | 517 |
| A.aeolicus | Bh | 1.55 | 43.8 | 1.183 | 0.741 | 1.115 | 0.699 | 778 | 487 |
| Thermotoga maritime | Bh | 1.86 | 46.5 | 1.44 | 0.958 | 1.341 | 0.889 | 780 | 519 |
| A.fulgidus | A | 2.17 | 49.6 | 1.619 | 0.944 | 1.53 | 0.893 | 673 | 392 |
| E.coli | Prγ | 4.63 | 52.1 | 3.182 | 2.058 | 2.82 | 1.824 | 742 | 480 |
| N.meningitidis | Prβ | 2.27 | 53.4 | 1.31 | 0.739 | 1.15 | 0.659 | 647 | 365 |
| S.typhimurium | Prβ | 4.81 | 53.5 | 3.202 | 1.986 | 2.788 | 1.741 | 697 | 432 |
| A.tumefaciens | Prα | 2.84 | 60.4 | 1.81 | 0.944 | 1.512 | 0.806 | 665 | 347 |
| M.tuberculosis | Ac | 4.4 | 66.0 | 2.116 | 0.936 | 1.584 | 0.749 | 505 | 223 |
| C.elegans | E | 95.2 | 41.0 | 8.53 | 6.36 | 2.128 | 1.391 | 516 | 385 |
| A.thaliana | E | 116.7 | 42.9 | 9.61 | 6.912 | 2.232 | 1.315 | 374 | 269 |
| S.cerevisiae | E | 12.1 | 38.9 | 6.105 | 4.672 | 4.466 | 3.387 | 968 | 741 |
aA, Archae; Ac, Actinobacteria; Bh, hyperthermophilic bacterium; E, eukaryote; F, Firmicute; Pr, Probacteria; S, Spirochete.
Overview of the search method
The method for locating unique oligonucleotides is a modification of a central pattern partitioning principle in approximate string matching. We will use the notation x ∪ y to denote the concatenation of x and y, and the notation x ⊆ y means that x is a substring of y.
The best current methods for indexed approximate string matching (46,47) are essentially based on the following pattern partitioning principle:
If ed(x,y) ≤ k and x = x1 ∪ x2 ∪ ⋯ ∪ xj, then for some index i, where 1 ≤ i ≤ j, there exists string z, such that ed(z,xi) ≤ ⌊k/j⌋ and z ⊆ y.
A direct consequence of the above principle is that if the oligonucleotide x is partitioned into j pieces x1, x2, …, xj, then, for any oligonucleotide y, ed(x,y) ≤ k only if the oligonucleotide y contains at least one of the pieces x1, x2, …, xj with at most ⌊k/j⌋ errors. This permits using the following steps to check whether a given oligonucleotide x is unique:
Partition x into j pieces x1, x2, …, xj.
Find all locations in the genome where one of the pieces x1, x2, …, xj occurs with at most ⌊k/j⌋ errors.
Check the surroundings of each pattern piece occurrence for a k-match of the complete oligonucleotide x.
If no such k-match of x is found that does not overlap with x itself, x is unique.
This basic approach can be improved in certain circumstances. Let di denote the number of errors permitted when searching for the piece xi. Previous methods typically assign di = ⌊k/j⌋ for each piece xi, as discussed above. But we note that it is possible to set di = ⌊k/j⌋ for (k mod j) + 1 pieces and di = ⌊k/j⌋ − 1 for the rest, if any left, without missing a single k-match of x. This is because if no piece xi is found inside y with at most di errors, then the total number of errors needed in converting y into x is at least (d1 + 1) + (d2 + 1) + ⋯ + (dj + 1) = j + d1 + d2 + ⋯ + dj = j + [(k,mod,j) + 1] × ⌊k/j⌋ + [j − (k mod j) − 1] × (⌊k/j⌋ − 1) = j + j × ⌊k/j⌋ − j + (k mod j) + 1 = j × ⌊k/j⌋ + (k mod j) + 1 = k + 1 and thus ed(x, y) > k. Our method is equal to the basic method when (k mod j) + 1 = j and leads into an improvement in all other cases. The algorithm was implemented on C++ and run either in a normal PC with sufficient RAM or in a Linux cluster of 10 virtual parallel computers.
Selection criteria for oligonucleotides
Primers can be utilized for many purposes and therefore in addition to uniqueness they have to meet other criteria depending on the intended use. The oligonucleotides designed here were primarily aimed for gene expression studies in microarrays. The typical length of such oligonucleotides is 25, which has been used also on commercial chips by Affymetrix. The valid oligonucleotides were defined by the following conditions. They may include at most 12 A, 12 T, 10 C or 10 G nucleotides, and no window of 8 nt includes more than 6 A, 6 T, 4 C or 4 G nucleotides. Further, the oligonucleotides include at most 6 successive A, 6 successive T, 5 successive C or 5 successive G nucleotides. An inverse complementary oligonucleotide of an oligonucleotide can match at most six symbols from the beginning of an oligonucleotide. These criteria were used to avoid self-annealing, self-end annealing and to provide high enough Tm. The distance threshold was four edit operations, i.e. no more than four errors were allowed.
RESULTS AND DISCUSSION
There is a great demand for functional oligonucleotides for a large spectrum of techniques. The oligonucleotides should be unique to allow specific and reliable binding. Genome-wide analyses are routine in many fields and therefore the probes utilized should not hybridize with any other genes or parts of genome. A method to determine, analyze and identify unique oligonucleotides from complete genomes was developed.
The method was applied to the analysis of 17 complete genomes (Table 1). The Archaeoglobus fulgidus and Sulfolobus solfataricus represented Archae, Aquifex aeolicus and Thermotoga maritima hyperthermophilic bacteria, Escherichia coli, Salmonella typhimurium and Buchnera sp. for Probacteria gamma subdivision, Agrobacterium tumefaciens for alpha subdivision, Neisseria meningitidis for beta subdivision and Helicobacter pylori for delta/epsilon subdivision. Of the Firmicutes included were Bacillus subtilis and Clostridium acetobutylicum, and of Actinobacteria, Mycobacterium tuberculosis was included. Borrelia burgdorferi exemplified Spirochete. The Eukaryotes included were Caenorhabditis elegans, a nematode, Saccharomyces cerevisiae, baker's yeast for fungi, and Arabidopsis thaliana for plants.
The genomes contained 564–25 694 genes and spanned 0.6–117 Mb. Some general properties of the genomes and oligonucleotides are in Tables 1 and 2. The organisms are listed in the order of their ascending C+G content and the eukaryotes are in the end. The C+G content affects many functional properties of DNA and genes. Therefore, this intrinsic property of genomes was taken into account and used to organize the genomes in analyses and for visualization of results. The lowest C+G content, 26.2%, was for Buchnera sp., and the highest, 65.6%, for M.tuberculosis. The organisms analyzed were chosen to represent different genuses and large variation of environmental growth conditions. The number of genes increases linearly along with genome size, however there are less genes than expected in eukaryotes due to the presence of mosaic genes (i.e. those having exons and introns) that make individual genes larger. In addition, the coding regions of eukaryotes were few hundred bases longer on average than for prokaryotes. All the analyzed small genomes are for intronless prokaryotes.
Table 2.
General properties of studied genomes and oligonucleotides
| Organism | Classa | Genome | Number of genes | Average gene length | Probeable genes | Probeable genes (%) | ||
|---|---|---|---|---|---|---|---|---|
| Unique oligos/gene | Valid oligos/gene | Invalid oligos/gene | ||||||
| Buchnera sp. | Prγ | 609 | 469 | 495 | 564 | 987.3 | 564 | 100 |
| B.burgdorferi | S | 588 | 432 | 547 | 850 | 1002.9 | 847 | 99.6 |
| C.acetobutylicum | F | 384 | 304 | 593 | 3672 | 921.0 | 3659 | 99.6 |
| S.solfataricus | A | 489 | 380 | 449 | 2968 | 852.4 | 2648 | 89.2 |
| H.pylori | Prδ/ɛ | 615 | 400 | 531 | 1566 | 954.5 | 1529 | 97.6 |
| B.subtilis | F | 634 | 457 | 413 | 4100 | 893.5 | 4095 | 99.9 |
| A.aeolicus | Bh | 733 | 460 | 471 | 1521 | 954.7 | 1515 | 99.6 |
| T.maritima | Bh | 727 | 481 | 443 | 1846 | 948.6 | 1832 | 99.2 |
| A.fulgidus | A | 635 | 371 | 434 | 2407 | 829.2 | 2379 | 98.8 |
| E.coli | Prγ | 658 | 425 | 504 | 4289 | 953.6 | 4218 | 98.3 |
| N.meningitidis | Prβ | 568 | 326 | 523 | 2025 | 872.7 | 1866 | 92.1 |
| S.typhimurium | Prβ | 607 | 379 | 514 | 4595 | 917.1 | 4509 | 98.1 |
| A.tumefaciens | Prα | 556 | 296 | 613 | 2722 | 933.1 | 2714 | 99.7 |
| M.tuberculosis | Ac | 378 | 179 | 750 | 4187 | 952.9 | 4132 | 98.7 |
| C.elegans | E | 129 | 84 | 1380 | 16522 | 1288 | 15673 | 94.9 |
| A.thaliana | E | 87 | 51 | 1424 | 25694 | 1499.5 | 24392 | 94.9 |
| S.cerevisiae | E | 708 | 537 | 1054 | 6306 | 1414.7 | 6013 | 95.34 |
aA, Archae; Ac, Actinobacteria; Bh, hyperthermophilic bacterium; E, eukaryote; F, Firmicute; Pr, Probacteria; S, Spirochete.
Search for unique and valid oligonucleotides
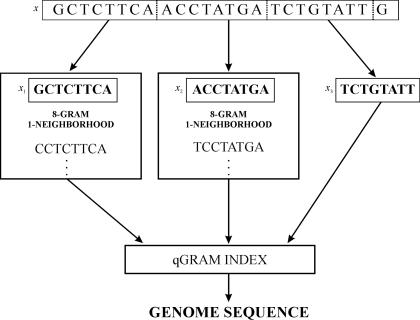
We were looking for unique 25mers with the error threshold k = 4. First, the oligonucleotides were partitioned into three pieces of length 8, which under our partitioning principle leads into locating the occurrences of (3 mod 1) + 1 = 2 of the pieces with at most ⌊k/j⌋ = ⌊4/3⌋ = 1 error, and the single remaining piece with ⌊k/j⌋ − 1 = 0 errors (Figure 1). Then, these occurrences were located by using a method reminiscent of the d-neighborhood generation (46). An 8-gram 1-neighborhood was generated for each piece xi by enumerating a sufficient set of 8-grams that will contain or be contained in any string z such that ed(xi, z) ≤ 1. An index containing all the locations of all 48 = 65 536 different oligonucleotides with length 8 in the genome was used in finding fast the occurrences of the generated 8-grams. Next, a two-phase filtering method (44) was applied. The surroundings of a given 8-gram occurrence was checked for a complete k-match of x, only if the 8-gram matched xi exactly or if the surroundings contained also an occurrence of an 8-gram belonging to the 1-neighborhood of some other pattern piece. Fast bit-parallel approximate string matching algorithm (48) was used in the final stage of checking for a k-match of x. If a match was found, then x was non-unique and the checking process was terminated. If no match was found, x was unique.
Figure 1.
Principle of the oligonucleotide analysis program. The oligos are searched by sliding a window of 25 positions along the analyzed sequence. The 25mer is partitioned to three 8mers and a single nucleotide. 1-neighborhoods (difference of one character allowed) are constructed for each piece and compared to the precomputed index of the locations of all 8mers in the investigated data (coding regions or complete genome). Two-phase filtering program and fast bit-parallel approximate string matching algorithm are used to identify the uniqueness of the 25mers.
Different computer setups were used for calculations and analysis. The genomes of prokaryotes as well as of S.cerevisiae were processed on a single PC with 1 GB RAM. The use of large enough memory facilitated storage of the complete genome and avoidance of excessive I/O operations. The two eukaryotes with larger genomes, C.elegans and A.thaliana, were processed in parallel on a Linux cluster of 10 PCs. The processing time was ∼3 days for A.thaliana and somewhat <2 days for C.elegans. It is thus feasible to search unique and valid probes for any organism.
Unique and valid probes
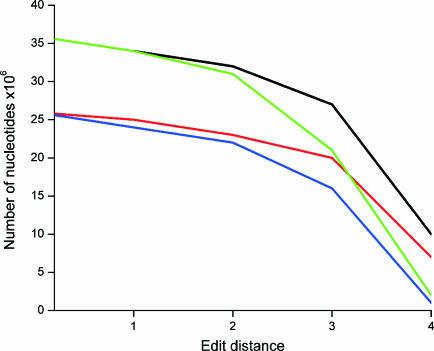
The analysis was divided into two parts to obtain full picture of the properties and distribution of unique and valid oligos in a single strand and in both strands. We determined unique and valid oligos both for CDS regions (in eukaryotes together with 5′ and 3′ extensions of 100 bp) and for the complete genome. If not otherwise stated, the results refer to genome-wide analysis. The proportion of unique oligonucleotides varied between 18.2 and 59.4% (25.3 and 83.6%) depending on the organism being smaller for the larger genomes. The corresponding values for valid oligos are 3.5 and 52.5 (18.2 and 52.3%). The numbers in parentheses are for CDS regions. Unique and valid probes were found for at least 92% of the genes, which is in agreement with the theoretical calculations based on the size of the genome and density of the genes (data not shown). The number of redundant probes exceeded significantly the number of valid oligos for all the eukaryotes analyzed as well as A.tumefaciens, N.meningitidis and M.tuberculosis. The total number of valid oligos per gene was high, the average varying from 51 to 537. The use of annealing and composition criteria clearly reduced the number of valid oligos compared to unique ones. Naturally, the use of more stringent edit distance has similar effect (Figure 2). C+G content has no direct effect on the number or ratios of valid and redundant oligos.
Figure 2.
Effects of edit distance and the use of criteria on the number of unique and valid oligonucleotides in A.thaliana data. The analysis was done for unique (black) and valid (red) oligos on coding region as well as for unique (green) and valid (blue) oligos in the whole genome.
Nucleotide distribution
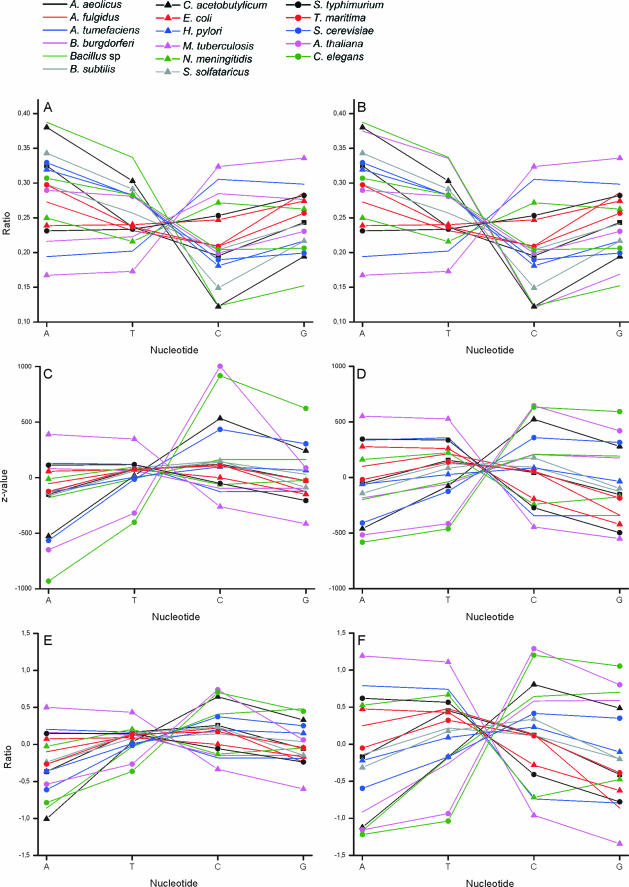
To analyze the properties of the oligos, the distribution of nucleotides within the unique and valid oligonucleotides were analyzed. The significance of the observations was estimated by calculating the Z-values based on normal distribution. The Z-values indicate the statistical bias in each position for the proportion of each base type. The nucleotide distributions follow well the Chargaff's first parity rule for duplex DNA (%A = %T and %C = %G) (49) and the Chargaff's second parity rule for single-stranded DNA (50,51) (Figure 3A–D). It is of interest that the curves pass through almost a single point when traversing from T to C ratio. The major differences to the parity rules is the genome of A.fulgidus, which is the only one where the ratios for A and T, and C and G are not close to each other.
Figure 3.
Nucleotide distribution within oligonucleotides. The ratio of nucleotides in (A) unique oligos in coding region and (B) valid oligos in genome. Z-values for the distribution of nucleotides in (C) unique oligos in coding region and (D) valid oligos in genome. The difference between the nucleotide usage and (E) unique oligos in coding regions and (F) all oligos in genome data.
The use of criteria to choose for valid oligos biases the distribution in B.burgdorferi, which has quite low C+G content. This seems to be related to nucleotide composition because the distribution of Buchera sp., which has the lowest C+G content among the analyzed genomes, has also slightly biased U-shaped distribution. When looking at the actual differences compared to normal distributions, the biggest change can be seen in C.acetobutylicum and the other genomes with extreme C+G values (Figure 3E and F). The criteria for valid oligos significantly reduce the number of oligos (Table 2). Valid/unique oligo ratio is from 0.47 to 0.79. Valid/invalid oligo ratio for genome data is from 0.036 to 1.11.
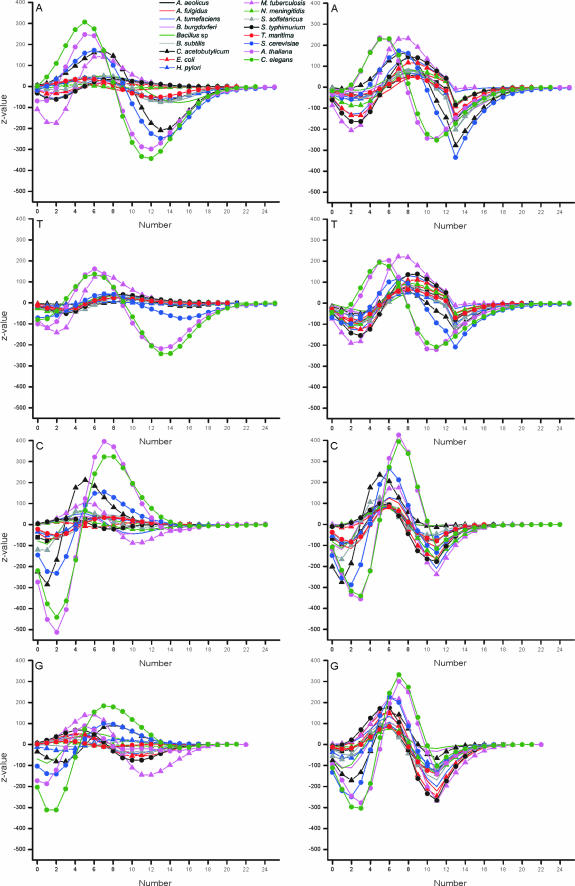
Further analysis of the nucleotide numbers in oligonucleotides indicated that the distribution in the majority of bacterial and archaeal genomes was as expected (Figure 4). The major exceptions were M.tuberculosis and C.acetobutylicum. Of these, M.tuberculosis has the highest C+G content among the analyzed genomes. It has more than expected number of oligonucleotides with 4–11 A, or 5–10 T, or 6–7 C, or 5–8 G bases among the valid oligos for genome data. On the other hand, oligonucleotides with large numbers of C or G are in fact underrepresented.
Figure 4.
Distribution of nucleotide numbers in unique oligonucleotides in coding region (panels on left) and in valid oligos in genome data (panels to the right).
The distributions of yeast, C.elegans and A.thaliana are all very biased. Common to all these is the underrepresentation of small numbers of nucleotides in oligos, overrepresentation usually in the range 3–8 and again underrepresentation in the range 9–19 nt. The actual borders of these patterns vary between nucleotide types and organisms. Interestingly, the location of peaks is shifted towards smaller base counts for A and T, and towards higher counts for C and G when compared to bacteria and archaea. The Z-values are very high for the eukaryotic organisms.
Valid and redundant oligonucleotides
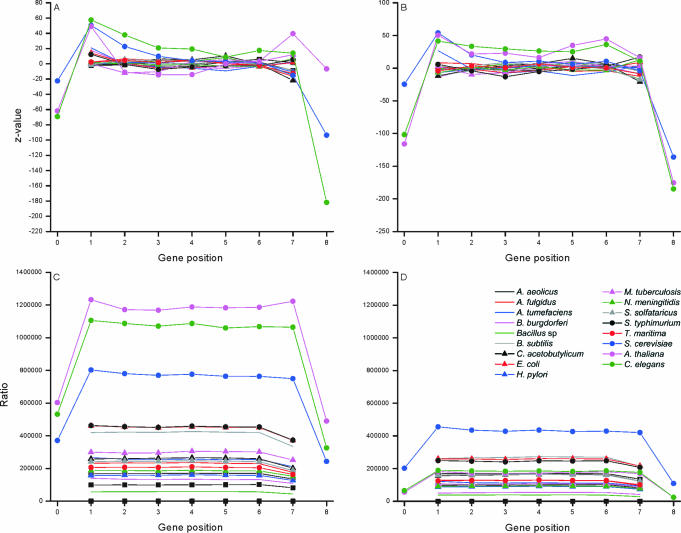
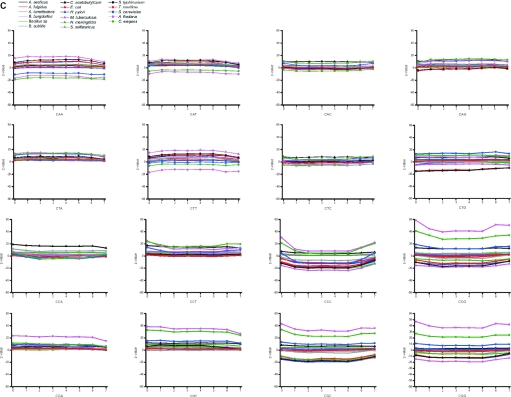
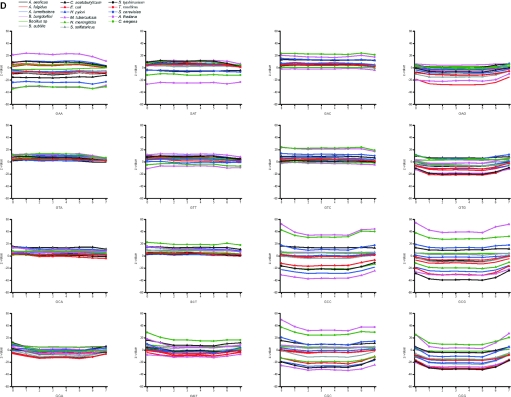
The distribution of the unique, valid and redundant oligonucleotides within the genes and flanking regions were estimated by calculating Z-values. The flanking regions of eukaryotic genes are numbered as 0 and 8 in Figure 5A, where coding regions have been divided into seven equal partitions. If there was uneven number of nucleotides, the middlemost (4th) partition was shorter. Both the 5′ and 3′ flanking sequences are highly underrepresented among the valid oligonucleotides. The reason is that these regions contain common and therefore conserved patterns involved in transcription and translation start and stop. In all these genomes, the last section contains slightly reduced proportion of valid oligos. The bacterial and arhaeal genomes have quite unbiased distribution throughout the genes. It has been a general trend to select probes, for example for antisense and microarray applications from the beginning of genes. The first and last sections in eukaryal genomes are somewhat surprisingly overrepresented among the unique and valid oligos. As a conclusion, oligonucleotides can be selected almost equally well from all the sections within coding region whereas in eukaryotes the flanking regions contain much less than expected of valid and unique oligonucleotides.
Figure 5.
Distribution of oligonucleotides in different sections of genes for (A) unique oligos in CDS regions and (B) valid oligos on genome. The ratio of (C) unique versus invalid oligos in coding regions and (D) valid versus invalid oligos in genome data.
When looking at the ratio of the valid and invalid oligos (Figure 5C and D), the same trends are apparent. In all the organisms, the graphs have remarkably flat distribution except for sections 0 and 8. The section 7 is universally somewhat decreased in all the prokaryal genomes. Sections 1 and 7 contain only slightly higher ratios than sections 2–6 for eukaryotes.
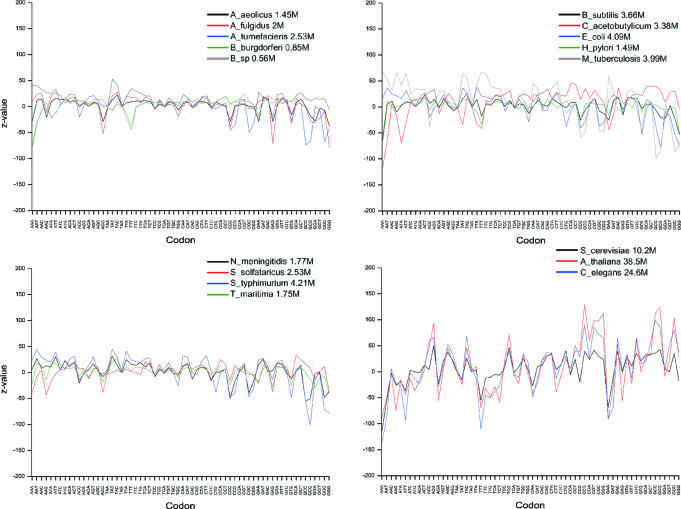
Effects on codon usage
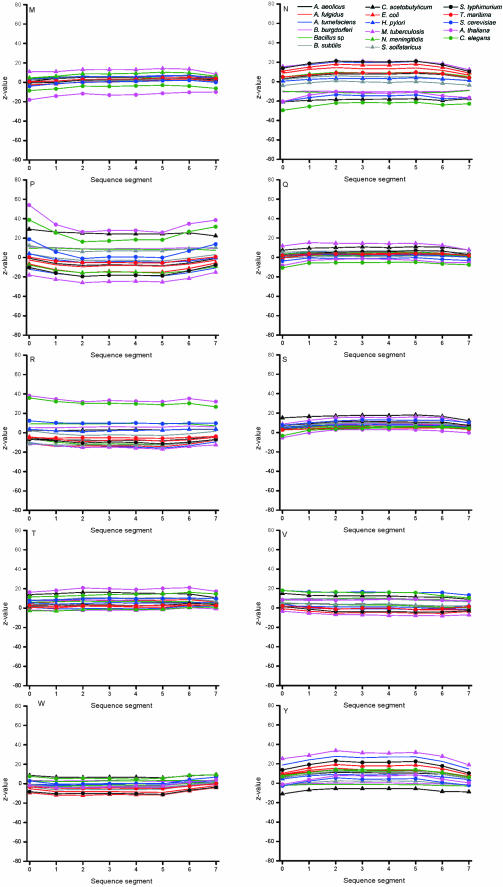
The effect on the coding properties of the valid oligonucleotides was studied by calculating the Z-values for the distribution of each codon (Figure 6). The expected codon frequencies were calculated based on the nucleotide content. The codons were analyzed according to the gene, i.e. the coding region within oligos started either from the first, second or third position dependent on the match with the gene sequence. In prokaryotes, the distributions are rather normal for all the codons except for in C.acetobutylicum, S.solfataricus and M.tuberculosis, which show large deviations for most of the codons. Even higher deviations are apparent in the eukaryal genomes. It is intriguing, that in most instances all the eukaryotes have similar trends for a large number of codons although the extent of the bias varies. This is of notion because these organisms have different codon preferences.
Figure 6.
Distribution of codons in oligonucleotides. Data is shown only for valid oligonucleotides in genome data. Note that yeast, C.elegans and A.thaliana data contain also the flanking 5′ and 3′ regions.
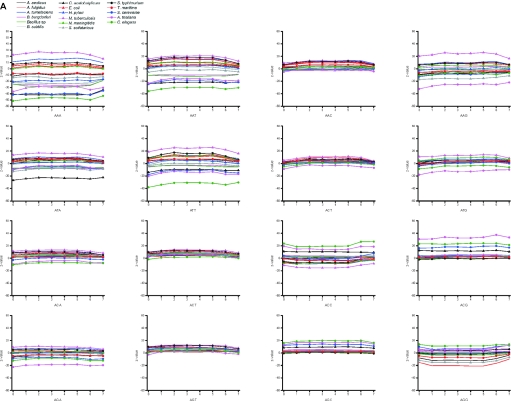
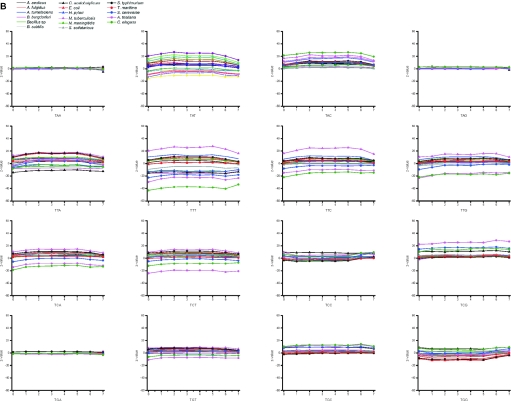
We compared further the Z-values for all codons (Figure 7A–D). Synonymous codons are known to have strong bias. Codon usage has effect, for example, on the translation. Highly expressed genes contain mainly those codons for which there are abundant tRNAs. The codon usage varies between organisms. There were no general trends for the usage of codons.
Figure 7.
Distribution of codons in different sections of genes. The figures (A–D) are for valid oligos in genome data.
When looking at the codon usage within the seven sections of genes, the majority of the codons in the majority of organisms have a normal distribution. The distribution is almost equal for the majority of codons in each section. However, certain patterns are visible. The largest, eukaryal genomes have the highest Z-scores, especially A.thaliana, which had significant bias in many places. Also C.elegans and S.cerevisiae have biased distribution to sections, but not that often and generally the Z-values are smaller than for A.thaliana, which is clearly the largest of the studied genomes. Clear examples of C+G-rich codons for alanine and glycine are A.tumefaciens and M.tuberculosis, which have significantly less codons containing C or G in the third position, although these organisms have the highest overall C+G content. Also S.typhimurium is biased towards not having G at the third position in codon for alanine. C.acetobutylicum has strong bias in a number of codons, and S.solfataricus and A.fulgidus in some individual cases. The genomes of prokaryotes have less biased distribution. Usually, the eukaryotes clearly favor certain triplets when synonymous codons appear.
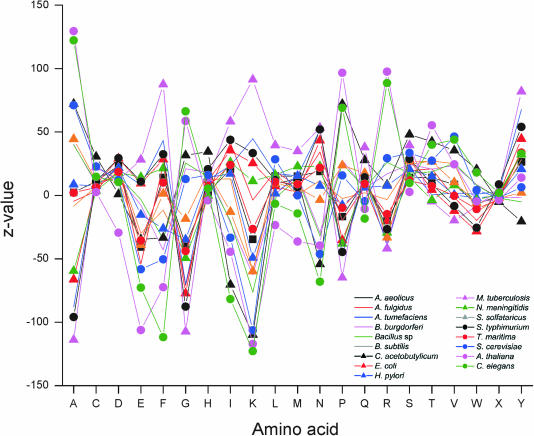
Effects on amino acid level
The 25mers from coding regions were further studied on amino acid level. Depending on the location within codons, the oligo-encoded sequence matched with either 7 or 8 amino acids in the protein sequence. The encoded amino acids of the oligonucleotides were compared to general amino acid compositions in each organism. Z-values (Figure 8) indicate strong bias from general pattern. As already seen in the codon usage, C.acetobutylicum and M.tuberculosis along with B.burgdorferi have the largest Z-values. Otherwise, the bacterial genomes have rather even distribution. The yeast, nematode and plant genomes have the largest variation and there are in fact only a few residue types that have normal distribution in these organisms.
Figure 8.
Distribution of amino acids within the valid oligonucleotides in genome data.
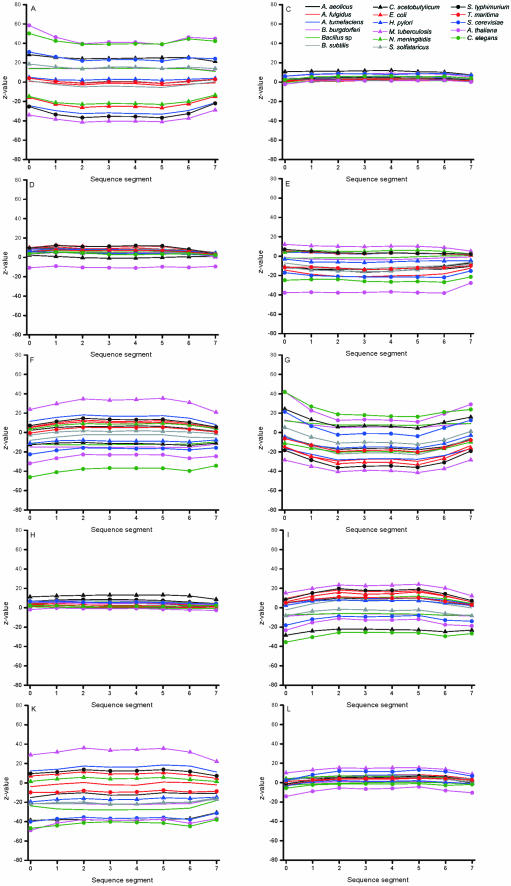
When looking at the effect of C+G content on the amino acid bias, it is evident that most of the residues have normal distribution. However, there is quite linear correlation between the decrease in alanine and glycine along increased C+G content and opposite effect in lysine. The distribution within the genes was further investigated by calculating the distribution within eight equally sized gene segments (Figure 9).
Figure 9.
Distribution of the amino acids within eight sections of proteins. Data is for valid oligonucleotides in genome data.
Certain residues such as C, D, H, M, Q and S have very equal distribution in all sections. G has generally somewhat pronounced underrepresentation in the middle of the sequence when compared to the termini. Also in this data, the organisms that have greatest bias in the other features are biased, namely the three eukaryotes and C.acetobutylicum, and M.tuberculosis of prokaryotes.
CONCLUSIONS
A new method for searching unique and valid oligonucleotides from complete genomes was developed. Despite the exhaustive analysis approach, genomes of any size could be analyzed. The presented method can be modified to permit other probe lengths and/or error thresholds. This may affect the run time, which is dependent on the ratio between the permitted number of errors and the probe length. When processing even larger genomes, such as the human genome, one should take into account the fact that using a too large error threshold may lead to a situation where practically all oligonucleotides are non-unique. Unique and valid oligos can clearly be found from any part of the gene, however the termini are overrepresented. The use of the criteria for valid oligos changes the overall properties of the oligonucleotides. By changing the criteria, the method could be easily modified for different purposes, e.g. to search for oligos functional in RNAi technology (37).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
Tampere Graduate School in Information Science and Engineering, the Academy of Finland, and the Medical Research Fund of Tampere University Hospital are acknowledged for financial support. Funding to pay the Open Access publication charges for this article was provided by the Academy of Finland.
Conflict of interest statement. None declared.
REFERENCES
- 1.Rychlik W., Rhoads R.E. A computer program for choosing optimal oligonucleotides for filter hybridization, sequencing and in vitro amplification of DNA. Nucleic Acids Res. 1989;17:8543–8551. doi: 10.1093/nar/17.21.8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hillier L., Green P. OSP: a computer program for choosing PCR and DNA sequencing primers. PCR Methods Appl. 1991;1:124–128. doi: 10.1101/gr.1.2.124. [DOI] [PubMed] [Google Scholar]
- 3.Cutichia A., Arnold J., Timberlake W.E. PCAP: probe choice and analysis package—a set of programs to aid in choosing synthetic oligomers for contig mapping. Comput. Appl. Biosci. 1993;9:201–203. doi: 10.1093/bioinformatics/9.2.201. [DOI] [PubMed] [Google Scholar]
- 4.Li P., Kupfer K.C., Davies C.J., Burbee D., Evans G.A., Garner H.R. PRIMO: a primer design program that applies base quality statistics for automated large-scale DNA sequencing. Genomics. 1997;40:476–485. doi: 10.1006/geno.1996.4560. [DOI] [PubMed] [Google Scholar]
- 5.Mecklenburg M. Design of high-annealing-temperature primers for PCR and development of a versatile low-copy-number amplification protocol. Adv. Mol. Cell Biol. 1997;15B:473–490. [Google Scholar]
- 6.Haas S., Vingron M., Poutska A., Wiemann S. Primer design for large scale sequencing. Nucleic Acids Res. 1998;26:3006–3012. doi: 10.1093/nar/26.12.3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rozen S., Skaletsky H. 1998. Primer3 code.
- 8.Herwig R., Schmitt A.O., Steinfath M., O'Brian J., Seidel H., Meier-Ewert S., Lehrach H., Radelof U. Information theoretical probe selection for hybridisation experiments. Bioinformatics. 2000;10:890–898. doi: 10.1093/bioinformatics/16.10.890. [DOI] [PubMed] [Google Scholar]
- 9.Emrich S.J., Love M., Delcher A.L. PROBEmer: a web-based software tool for selecting optimal DNA oligos. Nucleic Acids Res. 2003;31:3746–3750. doi: 10.1093/nar/gkg569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen S.H., Lin C.Y., Cho C.S., Lo C.Z., Hsiung C.A. Primer Design Assistant (PDA): a web-based primer design tool. Nucleic Acids Res. 2003;31:3751–3754. doi: 10.1093/nar/gkg560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Baren M.J., Heutink P. The PCR suite. Bioinformatics. 2004;20:591–593. doi: 10.1093/bioinformatics/btg473. [DOI] [PubMed] [Google Scholar]
- 12.Jarman S.N. Amplicon: software for designing PCR primers on aligned DNA sequences. Bioinformatics. 2004;20:1644–1645. doi: 10.1093/bioinformatics/bth121. [DOI] [PubMed] [Google Scholar]
- 13.Wu J.S., Lee C., Wu C.C., Shiue Y.L. Primer design using genetic algorithm. Bioinformatics. 2004;20:1710–1717. doi: 10.1093/bioinformatics/bth147. [DOI] [PubMed] [Google Scholar]
- 14.Kämpke T., Kieninger M., Mecklenburg M. Efficient primer design algorithms. Bioinformatics. 2001;17:214–225. doi: 10.1093/bioinformatics/17.3.214. [DOI] [PubMed] [Google Scholar]
- 15.Podowski R.M., Sonnhammer E.L. MEDUSA: large scale automatic selection and visual assessment of PCR primer pairs. Bioinformatics. 2001;17:656–657. doi: 10.1093/bioinformatics/17.7.656. [DOI] [PubMed] [Google Scholar]
- 16.Borneman J., Chrobak M., Della Vedova G., Figueroa A., Jiang T. Probe selection algorithms with applications in the analysis of microbial communities. Bioinformatics. 2001;17:S39–S48. doi: 10.1093/bioinformatics/17.suppl_1.s39. [DOI] [PubMed] [Google Scholar]
- 17.Kaderali L., Schliep A. Selecting signature oligonucleotides to identify organisms using DNA arrays. Bioinformatics. 2002;18:1340–1349. doi: 10.1093/bioinformatics/18.10.1340. [DOI] [PubMed] [Google Scholar]
- 18.Talaat A.M., Hunter P., Johnston S.A. Genome-directed primers for selective labeling of bacterial transcripts for DNA microarray analysis. Nat. Biotechnol. 2000;18:679–682. doi: 10.1038/76543. [DOI] [PubMed] [Google Scholar]
- 19.Li F., Stormo G. Selection of optimal DNA oligos for gene expression arrays. Bioinformatics. 2001;17:1067–1076. doi: 10.1093/bioinformatics/17.11.1067. [DOI] [PubMed] [Google Scholar]
- 20.Raddatz G., Dehio M., Meyer F.T., Dehio C. PrimeArray: genome-scale primer design for DNA-microarray construction. Bioinformatics. 2001;17:98–99. doi: 10.1093/bioinformatics/17.1.98. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen H.B., Knudsen S. Avoiding cross hybridization by choosing nonredundant targets on cDNA arrays. Bioinformatics. 2002;18:321–322. doi: 10.1093/bioinformatics/18.2.321. [DOI] [PubMed] [Google Scholar]
- 22.Rouillard J.M., Herbert C.J., Zuker M. OligoArray: genome-scale oligonucleotide design for microarrays. Bioinformatics. 2002;18:486–487. doi: 10.1093/bioinformatics/18.3.486. [DOI] [PubMed] [Google Scholar]
- 23.Xu D., Li G., Wu L., Zhou J., Xu Y. PRIMEGENS: robust and efficient design of gene-specific probes for microarray analysis. Bioinformatics. 2002;18:1432–1437. doi: 10.1093/bioinformatics/18.11.1432. [DOI] [PubMed] [Google Scholar]
- 24.Blick R.J., Revel A.T., Hansen E.J. FindGDPs: identification of primers for labeling microbial transcriptomes for DNA microarray analysis. Bioinformatics. 2003;19:1718–1719. doi: 10.1093/bioinformatics/btg218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mei R., Hubbell E., Bekiranov S., Mittmann M., Christians F.C., Shen M.M., Lu G., Fang J., Liu W.M., Ryder T., et al. Probe selection for high-density oligonucleotide arrays. Proc. Natl Acad. Sci. USA. 2003;100:11237–11242. doi: 10.1073/pnas.1534744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thareau V., Dehais P., Serizet C., Hilson P., Rouze P., Aubourg S. Automatic design of gene-specific sequence tags for genome-wide functional studies. Bioinformatics. 2003;19:2191–2198. doi: 10.1093/bioinformatics/btg286. [DOI] [PubMed] [Google Scholar]
- 27.Tolstrup N., Nielsen P.S., Kolberg J.G., Frankel A.M., Vissing H., Kauppinen S. OligoDesign: optimal design of LNA (locked nucleic acid) oligonucleotide capture probes for gene expression profiling. Nucleic Acids Res. 2003;31:3758–3762. doi: 10.1093/nar/gkg580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X., Seed B. Selection of oligonucleotide probes for protein coding sequences. Bioinformatics. 2003;19:796–802. doi: 10.1093/bioinformatics/btg086. [DOI] [PubMed] [Google Scholar]
- 29.Chou H.H., Hsia A.P., Mooney D.L., Schnable P.S. Picky: oligo microarray design for large genomes. Bioinformatics. 2004;20:2893–2902. doi: 10.1093/bioinformatics/bth347. [DOI] [PubMed] [Google Scholar]
- 30.Hornshøj H., Stengaard H., Panitz F., Bendixen C. SEPON, a selection and evaluation pipeline for oligonucleotides based on ESTs with a non-target Tm algorithm for reducing cross-hybridization in microarray gene expression experiments. Bioinformatics. 2004;20:428–429. doi: 10.1093/bioinformatics/btg434. [DOI] [PubMed] [Google Scholar]
- 31.Reymond N., Charles H., Duret L., Calevro F., Beslon G., Fayard J.M. ROSO: optimizing oligonucleotide probes for microarrays. Bioinformatics. 2004;20:271–273. doi: 10.1093/bioinformatics/btg401. [DOI] [PubMed] [Google Scholar]
- 32.Sczakiel G. Theoretical and experimental approaches to design effective antisense oligonucleotides. Front. Biosci. 2000;5:D194–D201. doi: 10.2741/sczakiel. [DOI] [PubMed] [Google Scholar]
- 33.Toschi N. Influence of mRNA self-structure of hybridisation: computational tools for antisense sequence selection. Methods. 2000;22:261–269. doi: 10.1006/meth.2000.1078. [DOI] [PubMed] [Google Scholar]
- 34.Vickers T.A., Wyatt J.R., Freier S.M. Effects of RNA secondary structure on cellular antisense activity. Nucleic Acids Res. 2000;28:1340–1347. doi: 10.1093/nar/28.6.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ding Y., Lawrence C.E. Statistical prediction of single-stranded regions in RNA secondary structure and application to predicting effective antisense target sites and beyond. Nucleic Acids Res. 2001;29:1034–1046. doi: 10.1093/nar/29.5.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Far R.K., Nedbal W., Sczakiel G. Concepts to automate the theoretical design of effective antisense oligonucleotides. Bioinformatics. 2001;17:1058–1061. doi: 10.1093/bioinformatics/17.11.1058. [DOI] [PubMed] [Google Scholar]
- 37.Chalk A.M., Wahlestedt C., Sonnhammer E.L. Improved and automated prediction of effective siRNA. Biochem. Biophys. Res. Commun. 2004;319:264–274. doi: 10.1016/j.bbrc.2004.04.181. [DOI] [PubMed] [Google Scholar]
- 38.Levenkova N., Gu Q., Rux J.J. Gene specific siRNA selector. Bioinformatics. 2004;20:430–432. doi: 10.1093/bioinformatics/btg437. [DOI] [PubMed] [Google Scholar]
- 39.Pancoska P., Moravek Z., Moll U.M. Efficient RNA interference depends on global context of the target sequence: quantitative analysis of silencing efficiency using Eulerian graph representation of siRNA. Nucleic Acids Res. 2004;32:1469–1479. doi: 10.1093/nar/gkh314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reynolds A., Leake D., Boese Q., Scaringe S., Marshall W.S., Khvorova A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 41.Wang L., Mu F.Y. A web-based design center for vector-based siRNA and siRNA cassette. Bioinformatics. 2004;20:1818–1820. doi: 10.1093/bioinformatics/bth164. [DOI] [PubMed] [Google Scholar]
- 42.Hoover D.M., Lubkowski J. DNAWorks: an automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic Acids Res. 2002;30:e43. doi: 10.1093/nar/30.10.e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu G., Hallett M., Pollock S., Thomas D. DePIE: designing primers for protein interaction experiments. Nucleic Acids Res. 2003;31:3755–3757. doi: 10.1093/nar/gkg577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hyyrö H. On using two-phase filtering in indexed approximate string matching with application to searching unique oligonucleotides. Proceedings of String Processing and Information Retrieval (SPIRE 2001); November 13–15; Laguna de San Rafael, Chile. IEEE Press; 2001. pp. 84–95. [Google Scholar]
- 45.Levenshtein V. Binary coded capable of correcting deletions, insertions and reversals. Soviet Phys. Doklady. 1966;10:707–710. [Google Scholar]
- 46.Myers E. A sublinear algorithm for approximative keyword searching. Algorithmica. 1994;12:345–374. [Google Scholar]
- 47.Navarro G., Baeza-Yates R. A hybrid indexing method for approximate string matching. J. Discrete Algorithms. 2000;1:205–239. [Google Scholar]
- 48.Myers G. A fast bit-vector algorithm for approximate string matching based on dynamic programming. J. ACM. 1999;46:539–553. [Google Scholar]
- 49.Chargaff E. Structure and function of nucleic acids as cell constituents. Fed. Proc. 1951;10:654–659. [PubMed] [Google Scholar]
- 50.Karkas J.D., Rudner R., Chargaff E. Separation of B.subtilis DNA into complementary strands. II. Template functions and composition as determined by transcription with RNA polymerase. Proc. Natl Acad. Sci. USA. 1968;60:915–920. doi: 10.1073/pnas.60.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rudner R., Karkas J.D., Chargaff E. Separation of B.subtilis DNA into complementary strands. 3. Direct analysis. Proc. Natl Acad. Sci. USA. 1968;60:921–922. doi: 10.1073/pnas.60.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.