Abstract
Vancomycin resistance has been reported in clinical isolates of both coagulase-negative staphylococci and Staphylococcus aureus. The emerging threat of widespread vancomycin resistance poses a serious public health concern given the fact that vancomycin has long been the preferred treatment of antibiotic-resistant gram-positive organisms. Though major efforts are now being focused on improving our understanding of vancomycin resistance, there is much that remains unknown at this time. This article reviews the major epidemiologic, microbiologic, and clinical characteristics of vancomycin resistance in both coagulase-negative staphylococci and S. aureus. The review begins with a discussion of issues common to both coagulase-negative staphylococci and S. aureus, such as definitions, laboratory detection of vancomycin resistance, and infection control issues related to vancomycin-resistant staphylococci. The rest of the article is then devoted to a discussion of issues unique to each organism, including epidemiology, risk factors for infection, mechanisms of resistance, and management options.
INTRODUCTION
The glycopeptide antibiotic vancomycin was introduced clinically in 1958 for the treatment of gram-positive bacteria. Use of this agent has increased dramatically in the last 20 years, in large part because of the increasing prevalence of methicillin resistance in both coagulase-negative staphylococci and Staphylococcus aureus (21). Data from the December 2000 report of the National Nosocomial Infection Surveillance (NNIS) System indicated that about 75% of coagulase-negative staphylococci and 47% of S. aureus isolates from intensive care units were resistant to methicillin (www.cdc.gov/ncidod/hip/NNIS/DEC2000sar.PDF). Vancomycin remains the drug of choice for these infections.
Vancomycin resistance among staphylococci was developed in laboratories even before the drug was in use clinically (27, 83). However, this resistance was so difficult to induce that many felt it would be unlikely to occur in a clinical setting (42). That no vancomycin-resistant staphylococci were reported in the first 20 years the drug was used only strengthened this assumption. Unfortunately, this confidence was shattered by the first reports of vancomycin resistance in coagulase-negative staphylococci in 1979 and 1983 (61, 74). Though a cause for concern, these reports did not generate a great deal of attention as coagulase-negative staphylococci are generally considered to be relatively avirulent organisms. A very different response greeted the first report of decreased susceptibility to vancomycin in S. aureus in 1997 (13, 35). This report was quickly followed by similar ones from other countries, including the United States (67). Given the well-known virulence of S. aureus, the isolation of these organisms generated enormous concern in the medical community and has prompted a flurry of activity aimed at limiting their emergence.
This review will serve as a general overview of vancomycin resistance in staphylococcal species. However, since vancomycin resistance in S. aureus is the major problem from both a clinical and public health standpoint, it will serve as the focus of the article.
Definition of Vancomycin Resistance
Unfortunately, confusion over the definitions of vancomycin resistance has been generated by recent literature. The source of this confusion seems to be the different breakpoints in vancomycin susceptibilities used in the various countries where vancomycin-resistant staphylococci have been reported. In the United States, the National Committee for Clinical Laboratory Standards (NCCLS) guidelines should be followed. NCCLS guidelines define staphylococci for which the MIC of vancomycin is ≤4 μg/ml to be susceptible, while isolates for which the MIC is 8 to 16 μg/ml are intermediate and those for which the MIC is ≥32 μg/ml are resistant (45). Japan, however, considers some isolates for which the MIC is 8 μg/ml to be resistant (68); as a result, some isolates reported as resistant in Japan have been reclassified as intermediate in the United States.
Confusion with respect to vancomycin resistance in staphylococci is also engendered by use of the term “heteroresistant staphylococci.” This phenomenon, which is seen in both coagulase-negative staphylococci and S. aureus, refers to the variability of vancomycin susceptibilities among subpopulations of single isolate. A heteroresistant isolate contains two populations of cells, a majority population that is susceptible to vancomycin and a minority population that is resistant. Heteroresistance is likely more common than pure resistance or diminished susceptibility, as evidenced by the fact that it was found in up to 20% of S. aureus isolates in one hospital in Japan (34). A similar study from the United States also found heteroresistant populations to be more common than homogenous populations with reduced susceptibilities; however, the overall incidence was much lower, with only 2 of 630 isolates (0.3%) demonstrating heteroresistance and none showing true reduced susceptibility to vancomycin (36).
The clinical significance of heteroresistance is not fully understood. Although one study (82) did show that patients who were infected with heteroresistant strains did have higher mortality rates than patients infected with sensitive isolates, it is difficult to conclusively determine impact based only on one small, retrospective study. Given the uncertain clinical significance and the difficulty and expense in detecting heteroresistance, there does not appear to be any role for screening outside of research studies (68). If screening is done and heteroresistant isolates are encountered, the MIC for the susceptible, parent strain and not that of the resistant subpopulation should be documented in the patient's record (36, 69).
Laboratory Detection of Vancomycin Resistance
Vancomycin resistance can be difficult to detect in the clinical microbiology laboratory. Disk diffusion sensitivity testing using the standard 30-μg vancomycin disk frequently misclassifies intermediately susceptible isolates as fully susceptible (70). In a recent study, 75% of microbiology laboratories from around the world misreported a glycopeptide-intermediate strain of Staphylococcus epidermidis as susceptible based on the results of disk diffusion testing (71).
Automated testing methods like MicroScan rapid panels (Dade Behring) and Vitek (version 7.07; bioMerieux) also have pitfalls. While conventional MicroScan panels performed well in detecting reduced susceptibilities to vancomycin, the rapid panels are less reliable as they do not allow for the recommended 24-h incubation (14, 70). Prior to 1999, Vitek software was not programmed to report vancomycin MICs above 4 μg/ml and would thus report the MICs of intermediately susceptible or resistant isolates as 4 μg/ml; a software upgrade in 1999 appears to have corrected this problem (14). At this time, however, nonautomated MIC determinations by broth or agar dilution or by E test are the “gold standard” for determining vancomycin susceptibility (69, 70).
Variation and irreproducibility among quantitative methods can be attributed in large part to difficulties encountered in attempting to detect resistance in a heterogeneous population of susceptible and resistant cells. Since vancomycin resistance has not been a homogenous characteristic of the majority of staphylococci that have been examined, agar-based susceptibility test methods, such as agar dilution and the agar diffusion E test, may be preferred, as they are more sensitive for detecting resistant subpopulations within a strain. However, this detection requires sufficient incubation time for expression of the resistance determinant and subsequent detectable growth. This usually translates into extended incubation times, i.e., a full 24 to 48 h, and precludes the use of the popular, rapid susceptibility methods (43). Another advantage of the agar methods is the fact that single colonies growing at higher drug concentrations can be visualized on solid media earlier and more readily than in broth-based systems. Whatever method is employed, it should be noted that inconsistencies have been reported even between gold standard testing methods for detecting vancomycin resistance in staphylococci (18, 20).
Further, even the most-sensitive techniques for determining vancomycin susceptibility are still vulnerable to the problems of inoculum size recently reported by Dunne et al. (20). NCCLS standards for susceptibility testing recommend an inoculum density of 5 × 105 CFU/ml for performance of standardized susceptibility testing, while vancomycin-intermediate subpopulations can occur at frequencies ranging from 10−6 to 10−7 CFU/ml. This raises the possibility that resistant subpopulations may be missed at the time of initial testing due to sampling error.
Though not helpful in MIC determinations, the use of selective agents or growth conditions, such as increased salt concentration or decreased temperature of incubation, to enhance the growth and expression of the resistant subpopulations appears to hold promise as a screening method for detecting vancomycin resistance. Screening isolates for growth on vancomycin-containing media appears to be a sensitive way to detect even low levels of vancomycin resistance. Commercially prepared agar media appear to be more specific, as susceptible isolates will occasionally grow on media prepared in-house, although in-house media appear to be equally sensitive (36, 70). Wong et al. have also described a sensitive system for detecting glycopeptide-intermediate or -resistant subpopulations of staphylococci, which utilizes increased NaCl concentrations (2 to 4%) and the monobactam aztreonam as an inducing agent (82).
The Centers for Disease Control and Prevention (CDC) has published recommendations to guide vancomycin susceptibility testing of S. aureus isolates (12, 15). These recommendations state that (i) primary testing of S. aureus requires at least 24 h of incubation, (ii) susceptibility determination with disk diffusion is not an acceptable method, and (iii) an MIC testing method should be used to confirm vancomycin susceptibility (15). Any S. aureus isolate for which the MIC is ≥4 μg/ml should be sent to the CDC for confirmatory testing.
Unfortunately, there are currently no official recommendations for screening isolates of coagulase-negative staphylococci for reduced susceptibility to vancomycin. At one point, a double zone of growth around an imipenem disk on a disk diffusion test was thought to indicate some degree of vancomycin resistance in coagulase-negative staphylococci (58). However, subsequent studies have revealed this method to be unreliable (33).
There is an obvious need for simple and accurate phenotypic screening and confirmatory tests for glycopeptide resistance in staphylococci. Unfortunately, such tests will require the elucidation of the molecular and biochemical mechanisms of resistance for validation. Until this occurs, clinical microbiologists should carefully pursue the evaluation of staphylococci for which the MICs of vancomycin initially appear to be elevated, particularly those from patients who are not responding to appropriate glycopeptide therapy or who have risk factors for the development of vancomycin-intermediate or -resistant staphylococci.
Infection Control Issues
The epidemiologic importance of vancomycin resistance has brought a great deal of attention to infection control issues regarding the spread of vancomycin-resistant staphylococci. The infection control community is understandably nervous, given the rapid dissemination of vancomycin-resistant enterococci that has occurred in hospitals around the world (11).
It has been established that S. aureus can be transmitted between patients by several routes, including environmental surfaces (9), hands of health care workers (44), and nasal shedding (8). Though less is known about the transmission of coagulase-negative staphylococci, there have been well-documented outbreaks of these organisms, and it is assumed that they are transmitted by many of the same routes as S. aureus (7, 41, 49). It is also assumed that vancomycin-resistant staphylococci can be transmitted by the same mechanisms as their sensitive counterparts. Indeed, there has been one reported outbreak of vancomycin-intermediate S. aureus (VISA) in a French hospital among patients who had not received any glycopeptide antibiotics and whose primary risk for acquiring the organism was environmental exposure (50).
Given the enormous public health concerns regarding the dissemination of these organisms, the Hospital Infection Control Practices Advisory Committee (HICPAC) of the CDC published infection control guidelines for all staphylococci for which the MIC of vancomycin is ≥8 μg/ml in 1997 (12). There were those, however, who felt the HICPAC guidelines did not go far enough, and a more elaborate set of infection control guidelines was published by a group from the Medical College of Virginia in 1998 (81). The similarities and differences between these two sets of guidelines are shown in Table 1. In addition to the various precautions listed in the table, both groups emphasize that vancomycin use must be limited to situations where it is deemed appropriate by current CDC guidelines (11).
TABLE 1.
Infection control guidelines for vancomycin-resistant staphylococcia
| Precaution | MCVb | HICPAC |
|---|---|---|
| Private room | Yes | Yes |
| Gloves | Yes | Yes |
| Gowns to enter room | Yes | No |
| Gowns for patient contact | Yes | Yes |
| Antibacterial hand washing agent | Yes | Yes |
| Record of all health care workers entering room | Yes | No |
| Mask and/or eye protection from aerosols | Yes | Yes |
| Mupirocin for nasal colonization | Yes | No |
| Limit number of health care workers caring for patient | Yes | Yes |
| Nares cultures for all health care workers who entered room | Yes | Yes |
| Health care workers at high risk for staphylococcal colonization should not care for patient | Yes | No |
| Isolation for duration of hospital stay | Yes | No |
| Environmental cultures after terminal room cleaning | Yes | Optional |
| Isolation on readmission until nares and previously infected, open sites are staphylococcus negative | Yes | No |
| Close unit to new admissions if nosocomial transmission occurs | Yes | No |
| All specimens carried to laboratory (no pneumatic tubes) | Yes | No |
| When possible, postpone tests that require patient to leave room | Yes | No |
| Post staff at doorway to ensure compliance with precautions | Yes | No |
Adapted from reference 81 with permission of the publisher.
MCV, Medical College of Virginia.
The results of contact tracing and culturing have been reported for three of the VISA cases in this country (67; L. Conway, T. Ross, M. O'Brien, J. Dick, and T. Perl, Abstr. 11th Annu. Meet. Soc. Healthcare Epidemiol. Am., abstr. 48, 2001). In each of these cases, the patients were already isolated on methicillin-resistant S. aureus (MRSA) precautions, and the HICPAC guidelines for VISA isolation were implemented as soon as the reduced vancomycin susceptibility was known. There was no transmission of VISA to any of the 285 contacts that were tested. However, in one case, a family member was colonized with the MRSA strain that later developed reduced vancomycin susceptibility in the patient (Conway et al., 11th Annu. Meet. Soc. Healthcare Epidemiol. Am.).
VANCOMYCIN RESISTANCE IN COAGULASE-NEGATIVE STAPHYLOCOCCI
Epidemiology of Vancomycin Resistance
Resistance to vancomycin among coagulase-negative staphylococci was first reported more than 20 years ago (61). However, the first report of a clinically significant isolate was in 1987 (59). Since that time, there have been at least five other case reports of clinically relevant coagulase-negative staphylococci that had diminished susceptibility to vancomycin (3, 20, 38, 57, 78).
Since the initial report of reduced susceptibility to vancomycin in coagulase-negative staphylococci, there have been at least 20 studies that have screened large numbers of isolates in an attempt to define the prevalence of this problem. Eleven of these studies did not find any isolates of coagulase-negative staphylococci with reduced vancomycin susceptibility (4, 5, 22, 31, 39, 52, 65, 72, 75, 77, 79), and the nine that did are described in more detail in Table 2. From the studies, it appears that the incidence of these organisms is very low. The notable exception is the study by Froggatt et al., in which 42% of Staphylococcus haemolyticus isolates were intermediately resistant (MIC ≥ 6.25 μg/ml) to vancomycin. It is unclear from the study why this institution had such a high prevalence of vancomycin resistance. Potential explanations include high vancomycin use in the months preceding the survey, the fact that these isolates were all part of an outbreak, or the possibility that the investigators used a more sensitive technique for detecting vancomycin resistance. Unfortunately, none of these factors was addressed in the study. Nonetheless, the report is a cause for alarm, as it indicates that these organisms can become endemic in a health care setting.
TABLE 2.
Summary of in vitro studies that examined reduced susceptibility to vancomycin among coagulase-negative staphylococci
| Reference | Location | Study design | No. of isolates tested | % of isolates for which MIC of vancomycin was >4 μg/ml (resistant species [if reported]) | Susceptibility testing method(s) |
|---|---|---|---|---|---|
| Henwood et al. (32) | England | Multicenter | 769 | 0.5 | E test |
| Santos Sanches et al. (56) | Worldwide | Multicenter | 1,351 | 0.07 | |
| Luh et al. (40) | Taiwan | Single center | 405 | 5.0 (S. epidermidis, S. haemolyticus, S. simulans, and others) | Agar dilution |
| de Neeling et al. (19) | The Netherlands | Multicenter | 7,334 | 0.4% | Agar dilution |
| Felmingham et al. (23) | Europe | Multicenter | 1,444 | 0.2 (S. haemolyticus) | Agar dilution |
| Gruneberg et al. (29) | Europe | Multicenter | 1,480 | 0.03 | Agar dilution |
| Herwaldt et al. (33) | Iowa | Single center | 28 | 0.04 (S. haemolyticus) | Agar and broth dilution |
| Goldstein et al. (28) | France | Single center | 362 | 0.03 | Agar dilution |
| Froggatt et al. (25) | Virginia | Single center | 70 | 42 (S. haemolyticus) | Agar dilution |
Risk Factors for Vancomycin Resistance
Because of the small number of cases, determining risk factors for the development of reduced vancomycin susceptibility among coagulase-negative staphylococci is difficult. Thus far, all resistant strains have been recovered from patients in acute-care hospitals. There was no common underlying illness in all the cases, although two of the five patients were on peritoneal dialysis. Exposure to a glycopeptide antibiotic would certainly appear to play an important role, as four of the five reported patients received at least 30 days of vancomycin before a less-susceptible isolate was recovered. The fifth patient had received a prolonged course of teicoplanin (a glycopeptide antibiotic not available in the United States) before the S. haemolyticus developed intermediate resistance to vancomycin. Though not a risk factor for developing vancomycin resistance, it should be noted that all of these isolates were resistant to multiple other antibiotics, including methicillin, quinolones, cephalosporins, and macrolides.
Species Variation in Vancomycin Susceptibility
There may be species differences in the coagulase-negative staphylococci with respect to vancomycin susceptibility. Despite a few case reports of intermediate susceptibility (26, 38, 57), almost all S. epidermidis isolates, which represent 60 to 90% of clinical isolates (47), remain sensitive to vancomycin. This relatively uniform susceptibility has been demonstrated in the in vitro prevalence studies in Table 2, in which only one group reported S. epidermidis isolates with reduced susceptibility to vancomycin (40). This finding seems to be supported in the laboratory, where some investigators have reported that vancomycin resistance in S. epidermidis has been difficult to induce (1, 59).
Although still unusual, reduced susceptibility to vancomycin may be more common in S. haemolyticus, as indicated in the prevalence studies in Table 2. Initially, this higher incidence of reduced susceptibility in S. haemolyticus was supported by in vitro studies in which researchers attempted to induce vancomycin resistance in several coagulase-negative staphylococcal species but were successful only with isolates of S. haemolyticus (58, 59). However, in another study, vancomycin resistance was induced with similar frequencies in both S. haemolyticus and S. epidermidis (33). Given the mixed results of these in vitro studies and the small numbers of resistant isolates detected in the prevalence studies, it is impossible to say if the increased frequency of reduced susceptibility in S. haemolyticus is truly a species phenomenon. Only one prevalence study (40) has reported reduced vancomycin susceptibility among species of coagulase-negative staphylococci other than S. epidermidis and S. haemolyticus. Thus, commenting on other potential species differences is simply not possible at this time.
Mechanisms of Vancomycin Resistance
The true mechanism of vancomycin resistance in these species, though currently unknown, is a topic of active investigation. Vancomycin acts by binding irreversibly to the terminal d-alanyl-d-alanine of cell wall disaccharide-pentapeptide precursors, thereby inhibiting production of the bacterial cell wall. Enterococci have developed vancomycin resistance by replacing the terminal alanine with a lactate, which has a greatly reduced affinity for vancomycin (10). Studies of vancomycin-resistant, coagulase-negative staphylococci have shown that these altered cell wall precursors are produced, but in amounts that are likely too small to account for the degree of resistance observed (6). Analysis of the cell wall peptidoglycans from highly resistant coagulase-negative staphylococci has demonstrated the presence of altered cross-links compared to susceptible strains (6). It has been suggested that these altered cross-links may inhibit vancomycin binding to target peptides, but this hypothesis has yet to be proven. Vancomycin resistance in coagulase-negative staphylococci is likely multifactorial, but the exact mechanisms await elucidation.
Treatment Options for Coagulase-Negative Staphylococci with Reduced Susceptibility to Vancomycin
Given the paucity of cases, there are no formal recommendations regarding the treatment of infections with coagulase-negative staphylococci with reduced susceptibility to vancomycin. In the cases reported to date, the infections were successfully treated by the addition of rifampin to vancomycin in one case (59) and by changes in therapy to rifampin and fusidic acid in one case (3) and to erythromycin in another (57).
Recently, two new agents to treat gram-positive infections have been approved for use in this country, the streptogramin quinupristin-dalfopristin and the oxazolidinone linezolid. Unfortunately, there are few in vitro and no in vivo data on the effectiveness of these new agents in treating infections caused by coagulase-negative staphylococci with reduced susceptibility to vancomycin. A study from Taiwan (40) found high rates of resistance (16%) to quinupristin-dalfopristin among coagulase-negative staphylococci, suggesting that it would not be a good option for vancomycin-resistant isolates in that country. However, the long-standing use of the streptogramin virginiamycin in animal husbandry in Taiwan is likely to have contributed significantly to the quinupristin-dalfopristin resistance seen in this study. Indeed, studies in other countries (2, 79) have reported excellent activity of quinupristin-dalfopristin against coagulase-negative staphylococci, although no vancomycin-resistant strains were tested. Likewise, linezolid has also shown excellent activity against coagulase-negative staphylococci, with no resistance reported to date (32, 37, 54). One of these studies (32) did test a few isolates that had reduced susceptibility to vancomycin.
VANCOMYCIN RESISTANCE IN S. AUREUS
Introduction
Since first being reported in 1997, the threat of vancomycin resistance in S. aureus has been the topic of intense research and discussion. Although vancomycin resistance in S. aureus remains extremely rare and is less common than vancomycin resistance in enterococci or even coagulase-negative staphylococci, there is widespread concern that vancomycin-resistant S. aureus (VRSA) poses, by far, the greatest risk to patients, given the virulence of the organism. Though there have been only a few reports of S. aureus isolates with reduced susceptibility to vancomycin, the high prevalence of MRSA and vancomycin use, both thought to be risk factors for VRSA, make the widespread dissemination of these organisms an alarmingly realistic possibility (48). Furthermore, there is the equally alarming threat of the risk of transmission of these organisms between patients.
Epidemiology of Vancomycin Resistance in S. aureus
To date, there have been no verified clinical isolates of S. aureus that were truly resistant to vancomycin by the NCCLS standards. Instead, the organisms have had intermediate susceptibility, which has led to the term “vancomycin intermediate S. aureus” or “VISA.” The term “glycopeptide intermediate S. aureus” or “GISA” is synonymous, but because vancomycin is the only glycopeptide used in this country, most American physicians are more familiar with the acronym VISA.
VISA isolates were first found in nature more than 15 years ago while investigators were screening isolates for vancomycin susceptibility (80). However, it was not until 1995 that the first clinical isolate was reported, which was from a French child who had been receiving vancomycin for an MRSA line infection (51). In 1996, a wound infection caused by VISA was reported in Japan in a child receiving vancomycin for an MRSA wound infection (35). The following year, the first VISA isolate was reported in the United States from Michigan. Since then, there have been at least seven confirmed cases of VISA from around the country (Table 3).
TABLE 3.
VISA cases in the United States
| State and yr (reference) | Source | Underlying illness(es) | Vancomycin exposure (wk) |
|---|---|---|---|
| Michigan, 1997 (67) | Peritoneal fluid | Renal failure, MRSA peritonitis, cancer | 18 |
| New Jersey, 1997 (67) | Blood | Acute renal failure, MRSA bacteremia | 18 |
| New York, 1998 (53) | Blood | Renal failure, MRSA bacteremia | 6 |
| Illinois, 1999 (15) | Blood | Renal failure, MRSA endocarditis | 3.5 |
| Minnesota, 2000 (24) | Blood | Renal failure, MRSA osteomyelitis | 18 |
| Nevada, 2000 (24) | Abscess fluid | Complicated cholecystectomy with polymicrobial intrahepatic abscess (including MRSA) | 10 |
| Maryland, 2000 | Blood | MRSA endocarditis, psoriasis, sleep apnea | 14 |
Interestingly, unlike the case for coagulase-negative staphylococci, it does not appear that there are undetected isolates of VISA in hospital microbiology laboratories. Several studies that have screened S. aureus isolates for reduced susceptibility to vancomycin have found none (19, 22, 23, 29, 31, 35, 39, 72, 75).
Risk Factors for Vancomycin Resistance
As is the case with coagulase-negative staphylococci, the relative rarity of decreased vancomycin susceptibility in S. aureus makes risk factors difficult to ascertain. Exposure to vancomycin (or other glycopeptide antibiotics) again stands out as a strong risk as every patient in this country who developed a VISA isolate had been on vancomycin therapy for some period of time, though the duration did vary widely, from just a few weeks to several months. Prior infection caused by MRSA would also appear to be a strong risk, as no known cases of VISA have developed from methicillin-susceptible strains. The relative risk posed individually by vancomycin exposure and MRSA infection is difficult to determine, as they tend to go hand-in-hand in most cases. Renal failure appears to be a significant risk factor, as it was present in five of the seven cases from the United States. Again, whether renal failure itself is a risk or merely serves to increase the risk of MRSA infection and vancomycin exposure is unknown.
Mechanisms of Vancomycin Resistance
The true mechanism of vancomycin resistance in S. aureus is not known. It was initially feared that S. aureus would acquire the van genes that code for vancomycin resistance in Enterococcus species, especially after this transfer was successfully accomplished in the laboratory (46). Further, vancomycin-resistant Enterococcus faecalis emits a sex pheromone that promotes plasmid transfer, and it has been recently demonstrated that this same pheromone is produced by S. aureus. Emission of this pheromone by S. aureus organisms that are in proximity to vancomycin-resistant enterococci that contain plasmids encoding van genes could result in transfer of these resistance genes (60). However, thus far, neither the van genes nor their altered peptidoglycan products have been recovered in vancomycin-intermediate or resistant S. aureus isolates. Instead, it appears that vancomycin resistance in S. aureus is conferred by other alterations in the bacterial cell wall.
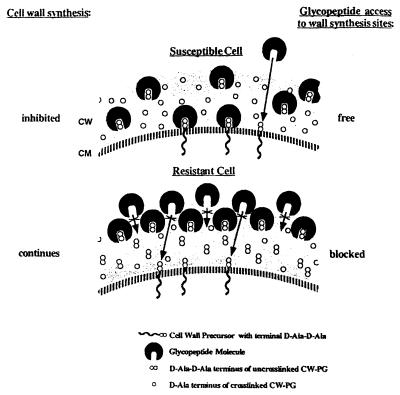
Several years prior to the first clinical VISA isolate being reported, Daum et al. produced laboratory strains of VISA and VRSA that had much thicker cell walls than the sensitive parent strains (17). Subsequent investigators have demonstrated that cell wall synthesis and turnover are upregulated in VRSA isolates, leading to thicker and more-disorganized cell walls (30). Further, it appears that resistant isolates have significantly less cross-linking in the peptidoglycan component of the cell wall (64). In order to exert an effect, vancomycin must reach the cytoplasmic membrane and bind with nascent cell wall precursors, thereby inhibiting their incorporation into the growing cell wall. It has been proposed that the thicker, disorganized cell walls can actually trap vancomycin at the periphery of the cell, thereby blocking its action (Fig. 1) (62). In fact, it has been shown that vancomycin can be recovered intact from the cell walls of VISA and VRSA isolates (63), indicating that the antibiotic is not being inactivated but merely sequestered by the bacteria. Furthermore, the altered cell walls appear to have a reduced affinity for vancomycin as soluble targets are able to bind more antibiotic in the presence of vancomycin-resistant isolates (6).
FIG. 1.
Proposed model for the capture of vancomycin in the cell wall of VISA organisms. Reprinted from reference 62 with permission of the publisher.
The role of penicillin-binding proteins (PBPs) in vancomycin resistance remains unclear. PBPs are a group of enzymes that catalyze various steps in cell wall synthesis and are the targets of beta-lactam antibiotics. It is a mutation in one of these enzymes, PBP2A, that confers methicillin resistance in MRSA. While some studies have shown an increase in the production of PBPs in VRSA (30, 43), others have shown that these enzymes are down regulated (66). That the MIC of oxacillin for some of the VISA isolates has decreased as the vancomycin MIC goes up (64; Conway et al., 11th Annu. Meet. Soc. Healthcare Epidemiol. Am.) supports the in vitro finding that expression of the mutated PBP2A is down regulated in these isolates (66). What role, if any, this altered expression plays in vancomycin resistance is unknown.
Clearly, more research is needed to further elucidate the exact mechanism of vancomycin resistance. However, given what is currently known, it seems safe to assume that it will be multifactorial.
Microbiology of VISA and VRSA
The cell wall alterations in VRSA can cause difficulties with laboratory identification. Morphologically, colonies of VISA and VRSA isolates often look smaller than their susceptible counterparts, which can lead some to confuse them with coagulase-negative staphylococci (64). Furthermore, vancomycin-resistant strains may require more incubation time for coagulase detection. If the coagulase reactions are incubated for less than 4 h, the result may be falsely negative and the isolate may be misclassified as coagulase-negative staphylococci (43). Thus, for any staphylococcus with suspected vancomycin resistance, the coagulase test should be incubated for more than 4 h before being interpreted (43).
Treatment of Infections Caused by VISA and VRSA
The susceptibilities and treatments of the VISA isolates that have been reported in this country are summarized in Table 4. It is interesting that all isolates have been sensitive to trimethoprim-sulfamethoxazole and tetracycline. Investigators and clinicians have also attempted to exploit the decreased resistance to oxacillin of some of the VISA isolates. In the laboratory, the combination of nafcillin and vancomycin was synergistic in the treatment of VISA endocarditis in rabbits (16). Beta-lactam antibiotics have been used clinically in the treatment of two of the VISA cases, once in combination with an aminoglycoside (35) and once in combination with an aminoglycoside and vancomycin (24). In both cases, the infection was cleared, although only one of the patients survived.
TABLE 4.
Susceptibilities and treatments of U.S. VISA isolates
| State and yr (reference) | Drugs to which isolate was susceptible | Treatment |
|---|---|---|
| Michigan, 1997 (67) | Trimethoprim-sulfamethoxazole, chloramphenicol, rifampin, tetracycline | Vancomycin, tobramycin then trimethoprim-sulfamethoxazole, rifampin |
| New Jersey, 1997 (76, 77) | Trimethoprim-sulfamethoxazole, chloramphenicol, tetracycline, gentamicin | Gentamicin, rifampin |
| New York, 1998 (53) | Trimethoprim-sulfamethoxazole, chloramphenicol, tetracycline, gentamicin, clindamycin | Vancomycin, tobramycin |
| Illinois, 1999 (15) | Trimethoprim-sulfamethoxazole, tetracycline, gentamicin | Vancomycin, tobramycin, rifampin |
| Minnesota, 2000 (24) | Not available | Vancomycin, nafcillin, gentamicin |
| Nevada, 2000 (24) | Trimethoprim-sulfamethoxazole, tetracycline, gentamicin | Linezolid, doxycycline, trimethoprim-sulfamethoxazole |
| Maryland, 2000 | Trimethoprim-sulfamethoxazole, chloramphenicol, tetracycline, linezolid | Linezolid |
Given the rarity of these infections, it is impossible to say what role the recently approved antibiotics quinupristin-dalfopristin and linezolid will play in their management. One study did show that both agents had good activity against three separate VISA strains (55); however, at least one of the clinical isolates was resistant to quinupristin-dalfopristin (Conway et al., 11th Annu. Meet. Soc. Healthcare Epidemiol. Am.). Only linezolid has been used in reported clinical cases, being used once in conjunction with trimethoprim-sulfamethoxazole and doxycycline (24) and once as a single agent (Conway et al., 11th Annu. Meet. Soc. Healthcare Epidemiol. Am.). Again, though there was a microbiologic cure in both cases, only one of the patients survived. Though VISA isolates thus far have all been susceptible to linezolid, the recent report of linezolid resistance in an isolate of MRSA, combined with growing use of this agent, raises real concern over how long this uniform susceptibility will hold (73).
Thus, there do appear to be at least a few treatment options for VISA and VRSA infections. However, until more experience exists, the best treatment strategy would appear to be to tailor therapy to the susceptibilities of the isolate in each case.
CONCLUSION
Vancomycin resistance in staphylococcal species is only beginning to emerge as a clinical issue, yet the attention it has already received serves to underscore the seriousness of the problem. Although much work has been and is being done on these organisms, much is also yet to be done, especially with respect to determining the true risk factors for infection and the mechanisms of vancomycin resistance. A better understanding of these issues will be key to helping prevent and treat these infections in the future. If past experience with multidrug resistant organisms is any indicator, the problem of vancomycin-resistant staphylococci will only grow in the future. However, heightened awareness of the issues and strict adherence to current guidelines for vancomycin use and infection control practices may help limit the impact of these organisms.
REFERENCES
- 1.Archer, G. L. 1978. Antimicrobial susceptibility and selection of resistance among Staphylococcus epidermidis isolates recovered from patients with infections of indwelling foreign devices. Antimicrob. Agents Chemother. 14:353-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archer, G. L., P. Auger, G. V. Doern, M. J. Ferraro, P. C. Fuchs, J. H. Jorgensen, D. E. Low, P. R. Murray, L. B. Reller, C. W. Stratton, et al. 1993. RP 59500, a new streptogramin highly active against recent isolates of North American staphylococci. Diagn. Microbiol. Infect. Dis. 16:223-226. [DOI] [PubMed] [Google Scholar]
- 3.Aubert, G., S. Passot, F. Lucht, and G. Dorche. 1990. Selection of vancomycin- and teicoplanin-resistant Staphylococcus haemolyticus during teicoplanin treatment of S. epidermidis infection. J. Antimicrob. Chemother. 25:491-493. [DOI] [PubMed] [Google Scholar]
- 4.Barelli, C., E. C. Minto, R. Martinez, and A. L. Darini. 1999. Evaluation of the antimicrobial susceptibilities of coagulase-negative staphylococci by E-test. Rev. Latinoam. Microbiol. 41(2):67-72. [PubMed] [Google Scholar]
- 5.Biavasco, F., C. Vignaroli, and P. E. Varaldo. 2000. Glycopeptide resistance in coagulase-negative staphylococci. Eur. J. Clin. Microbiol. Infect. Dis. 19:403-417. [DOI] [PubMed] [Google Scholar]
- 6.Billot-Klein, D., L. Gutmann, D. Bryant, D. Bell, J. Van Heijenoort, J. Grewal, and D. M. Shlaes. 1996. Peptidoglycan synthesis and structure in Staphylococcus haemolyticus expressing increasing levels of resistance to glycopeptide antibiotics. J. Bacteriol. 178:4696-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyce, J. M., S. M. Opal, J. W. Chow, M. J. Zervos, G. Potter-Bynoe, C. B. Sherman, R. L. Romulo, S. Fortna, and A. A. Medeiros. 1994. Outbreak of multidrug-resistant Enterococcus faecium with transferable vanB class vancomycin resistance. J. Clin. Microbiol. 32:1148-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyce, J. M., S. M. Opal, G. Potter-Bynoe, and A. A. Medeiros. 1993. Spread of methicillin-resistant Staphylococcus aureus in a hospital after exposure to a health care worker with chronic sinusitis. Clin. Infect. Dis. 17:496-504. [DOI] [PubMed] [Google Scholar]
- 9.Boyce, J. M., G. Potter-Bynoe, C. Chenevert, and T. King. 1997. Environmental contamination due to methicillin-resistant Staphylococcus aureus: possible infection control implications. Infect. Control Hosp. Epidemiol. 18:622-627. [PubMed] [Google Scholar]
- 10.Bugg, T. D., G. D. Wright, S. Dutka-Malen, M. Arthur, P. Courvalin, and C. T. Walsh. 1991. Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry 30:10408-10415. [DOI] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. 1995. Recommendations for preventing the spread of vancomycin resistance. Recommendations of the Hospital Infection Control Practices Advisory Committee (HICPAC). Morb. Mortal. Wkly. Rep. 44(RR-12):1-13. [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention. 1997. Interim guidelines for prevention and control of staphylococcal infection associated with reduced susceptibility to vancomycin. Morb. Mortal. Wkly. Rep. 46:626-628, 635. [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention. 1997. Reduced susceptibility of Staphylococcus aureus to vancomycin—Japan, 1996. Morb. Mortal. Wkly. Rep. 46:624-626. [PubMed] [Google Scholar]
- 14.Centers for Disease Control and Prevention. 2000. Laboratory capacity to detect antimicrobial resistance, 1998. Morb. Mortal. Wkly. Rep. 48:1167-1171. [PubMed] [Google Scholar]
- 15.Centers for Disease Control and Prevention. 2000. Staphylococcus aureus with reduced susceptibility to vancomycin—Illinois, 1999. Morb. Mortal. Wkly. Rep. 48:1165-1167. [PubMed] [Google Scholar]
- 16.Climo, M. W., R. L. Patron, and G. L. Archer. 1999. Combinations of vancomycin and beta-lactams are synergistic against staphylococci with reduced susceptibilities to vancomycin. Antimicrob. Agents Chemother. 43:1747-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daum, R. S., S. Gupta, R. Sabbagh, and W. M. Milewski. 1992. Characterization of Staphylococcus aureus isolates with decreased susceptibility to vancomycin and teicoplanin: isolation and purification of a constitutively produced protein associated with decreased susceptibility. J. Infect. Dis. 166:1066-1072. [DOI] [PubMed] [Google Scholar]
- 18.Del' Alamo, L., R. F. Cereda, I. Tosin, E. A. Miranda, and H. S. Sader. 1999. Antimicrobial susceptibility of coagulase-negative staphylococci and characterization of isolates with reduced susceptibility to glycopeptides. Diagn. Microbiol. Infect. Dis. 34:185-191. [DOI] [PubMed] [Google Scholar]
- 19.de Neeling, A. J., W. J. van Leeuwen, L. M. Schouls, C. S. Schot, A. van Veen-Rutgers, A. J. Beunders, A. G. Buiting, C. Hol, E. E. Ligtvoet, P. L. Petit, L. J. Sabbe, A.J. van Griethuysen, and J. D. van Embden. 1998. Resistance of staphylococci in The Netherlands: surveillance by an electronic network during 1989-1995. J. Antimicrob. Chemother. 41:93-101. [DOI] [PubMed] [Google Scholar]
- 20.Dunne, W. M., Jr., H. Qureshi, H. Pervez, and D. A. Nafziger. 2001. Staphylococcus epidermidis with intermediate resistance to vancomycin: elusive phenotype or laboratory artifact? Clin. Infect. Dis. 33:135-137. [DOI] [PubMed] [Google Scholar]
- 21.Ena, J., R. W. Dick, R. N. Jones, and R. P. Wenzel. 1993. The epidemiology of intravenous vancomycin usage in a university hospital. A 10-year study. JAMA 269:598-602. [PubMed] [Google Scholar]
- 22.Ena, J., A. Houston, R. P. Wenzel, and R. N. Jones. 1993. Trends in gram-positive bloodstream organism resistance: a seven-year audit of five glycopeptides and other drugs at a large university hospital. J. Chemother. 5:17-21. [DOI] [PubMed] [Google Scholar]
- 23.Felmingham, D., D. F. Brown, C. J. Soussy, et al. 1998. European Glycopeptide Susceptibility Survey of gram-positive bacteria for 1995. Diagn. Microbiol. Infect. Dis. 31:563-571. [DOI] [PubMed] [Google Scholar]
- 24.Fridkin, S. K. 2001. Vancomycin-intermediate and -resistant Staphylococcus aureus: what the infectious disease specialist needs to know. Clin. Infect. Dis. 32:108-115. [DOI] [PubMed] [Google Scholar]
- 25.Froggatt, J. W., J. L. Johnston, D. W. Galetto, and G. L. Archer. 1989. Antimicrobial resistance in nosocomial isolates of Staphylococcus haemolyticus. Antimicrob. Agents Chemother. 33:460-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garrett, D. O., E. Jochimsen, K. Murfitt, B. Hill, S. McAllister, P. Nelson, R. V. Spera, R. K. Sall, F. C. Tenover, J. Johnston, B. Zimmer, and W. R. Jarvis. 1999. The emergence of decreased susceptibility to vancomycin in Staphylococcus epidermidis. Infect. Control Hosp. Epidemiol. 20:167-170. [DOI] [PubMed] [Google Scholar]
- 27.Geraci, J. E. 1956. Some laboratory and clinical experiences with a new antibiotic, vancomycin. p. 90-106. In Antibiotic annual 1955-1956. Medical Encyclopedia, New York, N.Y. [PubMed]
- 28.Goldstein, F. W., A. Coutrot, A. Sieffer, and J. F. Acar. 1990. Percentages and distributions of teicoplanin- and vancomycin-resistant strains among coagulase-negative staphylococci. Antimicrob. Agents Chemother. 34:899-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gruneberg, R. N., and W. Hryniewicz. 1998. Clinical relevance of a European collaborative study on comparative susceptibility of gram-positive clinical isolates to teicoplanin and vancomycin. Int. J. Antimicrob. Agents 10:271-277. [DOI] [PubMed] [Google Scholar]
- 30.Hanaki, H., K. Kuwahara-Arai, S. Boyle-Vavra, R. S. Daum, H. Labischinski, and K. Hiramatsu. 1998. Activated cell-wall synthesis is associated with vancomycin resistance in methicillin-resistant Staphylococcus aureus clinical strains Mu3 and Mu50. J. Antimicrob. Chemother. 42:199-209. [DOI] [PubMed] [Google Scholar]
- 31.Hanberger, H., M. Hoffmann, S. Lindgren, and L. E. Nilsson. 1997. High incidence of antibiotic resistance among bacteria in 4 intensive care units at a university hospital in Sweden. Scand. J. Infect. Dis. 29:607-614. [DOI] [PubMed] [Google Scholar]
- 32.Henwood, C. J., D. M. Livermore, A. P. Johnson, D. James, M. Warner, A. Gardiner, and The Linezolid Study. 2000. Susceptibility of Gram-positive cocci from 25 UK hospitals to antimicrobial agents including linezolid. J. Antimicrob. Chemother. 46:931-940. [DOI] [PubMed] [Google Scholar]
- 33.Herwaldt, L., L. Boyken, and M. Pfaller. 1991. In vitro selection of resistance to vancomycin in bloodstream isolates of Staphylococcus haemolyticus and Staphylococcus epidermidis. Eur. J. Clin. Microbiol. Infect. Dis. 10:1007-1012. [DOI] [PubMed] [Google Scholar]
- 34.Hiramatsu, K., N. Aritaka, H. Hanaki, S. Kawasaki, Y. Hosoda, S. Hori, Y. Fukuchi, and I. Kobayashi. 1997. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet 350:1670-1673. [DOI] [PubMed] [Google Scholar]
- 35.Hiramatsu, K., H. Hanaki, T. Ino, K. Yabuta, T. Oguri, and F. C. Tenover. 1997. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40:135-136. [DOI] [PubMed] [Google Scholar]
- 36.Hubert, S. K., J. M. Mohammed, S. K. Fridkin, R. P. Gaynes, J. E. McGowan, Jr., and F. C. Tenover. 1999. Glycopeptide-intermediate Staphylococcus aureus: evaluation of a novel screening method and results of a survey of selected U.S. hospitals. J. Clin. Microbiol. 37:3590-3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jorgensen, J. H., M. L. McElmeel, and C. W. Trippy. 1997. In vitro activities of the oxazolidinone antibiotics U-100592 and U-100766 against Staphylococcus aureus and coagulase-negative Staphylococcus species. Antimicrob. Agents Chemother. 41:465-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krcmery, V., Jr., J. Trupl, L. Drgona, J. Lacka, E. Kukuckova, and E. Oravcova. 1996. Nosocomial bacteremia due to vancomycin-resistant Staphylococcus epidermidis in four patients with cancer, neutropenia, and previous treatment with vancomycin. Eur. J. Clin. Microbiol. Infect. Dis. 15:259-261. [DOI] [PubMed] [Google Scholar]
- 39.Laverdiere, M., K. Weiss, R. Rivest, and J. Delorme. 1998. Trends in antibiotic resistance of staphylococci over an eight-year period: differences in the emergence of resistance between coagulase positive and coagulase-negative staphylococci. Microb. Drug Resist. 4:119-122. [DOI] [PubMed] [Google Scholar]
- 40.Luh, K. T., P. R. Hsueh, L. J. Teng, H. J. Pan, Y. C. Chen, J. J. Lu, J. J. Wu, and S. W. Ho. 2000. Quinupristin-dalfopristin resistance among gram-positive bacteria in Taiwan. Antimicrob. Agents Chemother. 44:3374-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lyytikainen, O., H. Saxen, R. Ryhanen, M. Vaara, and J. Vuopio-Varkila. 1995. Persistence of a multiresistant clone of Staphylococcus epidermidis in a neonatal intensive-care unit for a four-year period. Clin. Infect. Dis. 20:24-29. [DOI] [PubMed] [Google Scholar]
- 42.Moellering, R. C. 1998. The specter of glycopeptide resistance: current trends and future considerations. Am. J. Med. 104(Suppl. 5A):3S-6S. [DOI] [PubMed] [Google Scholar]
- 43.Moreira, B., S. Boyle-Vavra, B. L. deJonge, and R. S. Daum. 1997. Increased production of penicillin-binding protein 2, increased detection of other penicillin-binding proteins, and decreased coagulase activity associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 41:1788-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mulligan, M. E., K. A. Murray-Leisure, B. S. Ribner, H. C. Standiford, J. F. John, J. A. Korvick, C. A. Kauffman, and V. L. Yu. 1993. Methicillin-resistant Staphylococcus aureus: a consensus review of the microbiology, pathogenesis, and epidemiology with implications for prevention and management. Am. J. Med. 94:313-328. [DOI] [PubMed] [Google Scholar]
- 45.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. NCCLS approved standard M7-A5. National Committee for Laboratory Standards, Wayne, Pa.
- 46.Noble, W. C., Z. Virani, and R. G. Cree. 1992. Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC 12201 to Staphylococcus aureus. FEMS Microbiol. Lett. 72:195-198. [DOI] [PubMed] [Google Scholar]
- 47.Paradisi, F., G. Corti, and D. Messeri. 2001. Antistaphylococcal (MSSA, MRSA, MSSE, MRSE) antibiotics. Med. Clin. N. Am. 85:1-17. [DOI] [PubMed] [Google Scholar]
- 48.Perl, T. M. 1999. The threat of vancomycin resistance. Am. J. Med. 106(Suppl. 5A):26S-37S, 48S-52S. [DOI] [PubMed] [Google Scholar]
- 49.Perl, T. M., W. A. Kruger, A. Houston, L. D. Boyken, M. A. Pfaller, and L. A. Herwaldt. 1999. Investigation of suspected nosocomial clusters of Staphylococcus haemolyticus infections. Infect. Control Hosp. Epidemiol. 20:128-131. [DOI] [PubMed] [Google Scholar]
- 50.Pina, P., C. Marliere, F. Vandenesch, J. P. Bedos, J. Etienne, and P. Y. Allouch. 2000. An outbreak of Staphylococcus aureus strains with reduced susceptibility to glycopeptides in a French general hospital. Clin. Infect. Dis. 31:1306-1308. [DOI] [PubMed] [Google Scholar]
- 51.Ploy, M. C., C. Grelaud, C. Martin, L. de Lumley, and F. Denis. 1998. First clinical isolate of vancomycin-intermediate Staphylococcus aureus in a French hospital. Lancet 351:1212. [DOI] [PubMed] [Google Scholar]
- 52.Refsahl, K., and B. M. Andersen. 1992. Clinically significant coagulase-negative staphylococci: identification and resistance patterns. J. Hosp. Infect. 22:19-31. [DOI] [PubMed] [Google Scholar]
- 53.Rotun, S. S., V. McMath, D. J. Schoonmaker, P. S. Maupin, F. C. Tenover, B. C. Hill, and D. M. Ackman. 1999. Staphylococcus aureus with reduced susceptibility to vancomycin isolated from a patient with fatal bacteremia. Emerg. Infect. Dis. 5:147-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rybak, M. J., D. M. Cappelletty, T. Moldovan, J. R. Aeschlimann, and G. W. Kaatz. 1998. Comparative in vitro activities and postantibiotic effects of the oxazolidinone compounds eperezolid (PNU-100592) and linezolid (PNU-100766) versus vancomycin against Staphylococcus aureus, coagulase-negative staphylococci, Enterococcus faecalis, and Enterococcus faecium. Antimicrob. Agents Chemother. 42:721-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rybak, M. J., E. Hershberger, T. Moldovan, and R. G. Grucz. 2000. In vitro activities of daptomycin, vancomycin, linezolid, and quinupristin-dalfopristin against staphylococci and enterococci, including vancomycin-intermediate and -resistant strains. Antimicrob. Agents Chemother. 44:1062-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Santos Sanches, I., R. Mato, H. de Lencastre, and A. Tomasz. 2000. Patterns of multidrug resistance among methicillin-resistant hospital isolates of coagulase-positive and coagulase-negative staphylococci collected in the international multicenter study RESIST in 1997 and 1998. CEM/NET Collaborators and the International Collaborators. Microb. Drug Resist. 6:199-211. [DOI] [PubMed] [Google Scholar]
- 57.Sanyal, D., A. P. Johnson, R. C. George, B. D. Cookson, and A. J. Williams. 1991. Peritonitis due to vancomycin-resistant Staphylococcus epidermidis. Lancet 337:54. [DOI] [PubMed] [Google Scholar]
- 58.Schwalbe, R. S., W. J. Ritz, P. R. Verma, E. A. Barranco, and P. H. Gilligan. 1990. Selection for vancomycin resistance in clinical isolates of Staphylococcus haemolyticus. J. Infect. Dis. 161:45-51. [DOI] [PubMed] [Google Scholar]
- 59.Schwalbe, R. S., J. T. Stapleton, and P. H. Gilligan. 1987. Emergence of vancomycin resistance in coagulase-negative staphylococci. N. Engl. J. Med. 316:927-931. [DOI] [PubMed] [Google Scholar]
- 60.Showsh, S. A., E. H. De Boever, and D. B. Clewell. 2001. Vancomycin resistance plasmid in Enterococcus faecalis that encodes sensitivity to a sex pheromone also produced by Staphylococcus aureus. Antimicrob. Agents Chemother. 45:2177-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Siebert, W. T., N. Moreland, and T. W. Williams, Jr. 1979. Synergy of vancomycin plus cefazolin or cephalothin against methicillin-resistance Staphylococcus epidermidis. J. Infect. Dis. 139:452-457. [DOI] [PubMed] [Google Scholar]
- 62.Sieradzki, K., M. G. Pinho, and A. Tomasz. 1999. Inactivated pbp4 in highly glycopeptide-resistant laboratory mutants of Staphylococcus aureus. J. Biol. Chem. 274:18942-18946. [DOI] [PubMed] [Google Scholar]
- 63.Sieradzki, K., and A. Tomasz. 1997. Inhibition of cell wall turnover and autolysis by vancomycin in a highly vancomycin-resistant mutant of Staphylococcus aureus. J. Bacteriol. 179:2557-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sieradzki, K., and A. Tomasz. 1999. Gradual alterations in cell wall structure and metabolism in vancomycin-resistant mutants of Staphylococcus aureus. J. Bacteriol. 181:7566-7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sieradzki, K., P. Villari, and A. Tomasz. 1998. Decreased susceptibilities to teicoplanin and vancomycin among coagulase-negative methicillin-resistant clinical isolates of staphylococci. Antimicrob. Agents Chemother. 42:100-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sieradzki, K., S. W. Wu, and A. Tomasz. 1999. Inactivation of the methicillin resistance gene mecA in vancomycin-resistant Staphylococcus aureus. Microb. Drug Resist. 5:253-257. [DOI] [PubMed] [Google Scholar]
- 67.Smith, T. L., M. L. Pearson, K. R. Wilcox, C. Cruz, M. V. Lancaster, B. Robinson-Dunn, F. C. Tenover, M. J. Zervos, J. D. Band, E. White, and W. R. Jarvis. 1999. Emergence of vancomycin resistance in Staphylococcus aureus. Glycopeptide-Intermediate Staphylococcus aureus Working Group. N. Engl. J. Med. 340:493-501. [DOI] [PubMed] [Google Scholar]
- 68.Tenover, F. C. 1999. Implications of vancomycin-resistant Staphylococcus aureus. J Hosp. Infect. 43(Suppl.):S3-S7. [DOI] [PubMed] [Google Scholar]
- 69.Tenover, F. C., J. W. Biddle, and M. V. Lancaster. 2001. Increasing resistance to vancomycin and other glycopeptides in Staphylococcus aureus. Emerg. Infect. Dis. 7:327-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tenover, F. C., M. V. Lancaster, B. C. Hill, C. D. Steward, S. A. Stocker, G. A. Hancock, C.M. O'Hara, S. K. McAllister, N. C. Clark, and K. Hiramatsu. 1998. Characterization of staphylococci with reduced susceptibilities to vancomycin and other glycopeptides. J. Clin. Microbiol. 36:1020-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tenover, F. C., M. J. Mohammed, J. Stelling, T. O'Brien, and R. Williams. 2001. Ability of laboratories to detect emerging antimicrobial resistance: proficiency testing and quality control results from the World Health Organization's external quality assurance system for antimicrobial susceptibility testing. J. Clin. Microbiol. 39:241-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tripodi, M. F., V. Attanasio, L. E. Adinolfi, A. Florio, P. Cione, S. Cuccurullo, R. Utili, and G. Ruggiero. 1994. Prevalence of antibiotic resistance among clinical isolates of methicillin-resistant staphylococci. Eur. J. Clin. Microbiol. Infect. Dis. 13:148-152. [DOI] [PubMed] [Google Scholar]
- 73.Tsiodras, S., H. S. Gold, G. Sakoulas, G. M. Eliopoulos, C. Wennersten, L. Venkataraman, R. C. Moellering, and M. J. Ferraro. 2001. Linezolid resistance in a clinical isolate of Staphylococcus aureus. Lancet 358:207-208. [DOI] [PubMed] [Google Scholar]
- 74.Tuazon, C. U., and H. Miller. 1983. Clinical and microbiologic aspects of serious infections caused by Staphylococcus epidermidis. Scand. J. Infect. Dis. 15:347-360. [DOI] [PubMed] [Google Scholar]
- 75.Turano, A., G. Ravizzola, L. Peroni, T. Ceruti, L. M. Greco, E. Pitzus, G. Santini, S. Cresti, and G. Satta. 1994. A multicentre study: Staphylococcus and Enterococcus susceptibility to antibiotics. Eur. J. Epidemiol. 10:567-572. [DOI] [PubMed] [Google Scholar]
- 76.Turco, T. F., G. P. Melko, and J. R. Williams. 1998. Vancomycin intermediate-resistant Staphylococcus aureus. Ann. Pharmacother. 32:758-760. [DOI] [PubMed] [Google Scholar]
- 77.Udo, E. E., L. E. Jacob, and T. D. Chugh. 1995. Antimicrobial resistance of coagulase-negative staphylococci from a Kuwait hospital. Microb. Drug Resist. 1:315-320. [DOI] [PubMed] [Google Scholar]
- 78.Veach, L. A., M. A. Pfaller, M. Barrett, F. P. Koontz, and R. P. Wenzel. 1990. Vancomycin resistance in Staphylococcus haemolyticus causing colonization and bloodstream infection. J. Clin. Microbiol. 28:2064-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.von Eiff, C., R. R. Reinert, M. Kresken, J. Brauers, D. Hafner, and G. Peters. 2000. Nationwide German multicenter study on prevalence of antibiotic resistance in staphylococcal bloodstream isolates and comparative in vitro activities of quinupristin-dalfopristin. J. Clin. Microbiol. 38:2819-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Watanakunakorn, C. 1984. Mode of action and in-vitro activity of vancomycin. J. Antimicrob. Chemother. 14(Suppl. D):7-18. [DOI] [PubMed] [Google Scholar]
- 81.Wenzel, R. P., and M. B. Edmond. 1998. Vancomycin-resistant Staphylococcus aureus: infection control considerations. Clin. Infect. Dis. 27:245-251. [DOI] [PubMed] [Google Scholar]
- 82.Wong, S. S., P. L. Ho, P. C. Woo, and K. Y. Yuen. 1999. Bacteremia caused by staphylococci with inducible vancomycin heteroresistance. Clin. Infect. Dis. 24:760-767. [DOI] [PubMed] [Google Scholar]
- 83.Ziegler, D. W. 1956. Vancomycin, a new antibiotic II. In vitro antibacterial studies, p. 612-618. In Antibiotic annual 1955-1956. Medical Encyclopedia, New York, N.Y. [PubMed]