Abstract
The phosphoprotein paramyosin is a major structural component of invertebrate muscle thick filaments. To investigate the importance of paramyosin phosphorylation, we produced transgenic Drosophila melanogaster in which one, three, or four phosphorylatable serine residues in the N-terminal nonhelical domain were replaced by alanines. Depending on the residues mutated, transgenic lines were either unaffected or severely flight impaired. Flight-impaired strains had decreases in the most acidic paramyosin isoforms, with a corresponding increase in more basic isoforms. Surprisingly, ultrastructure of indirect flight muscle myofibrils was normal, indicating N-terminal phosphorylation is not important for myofibril assembly. However, mechanical studies of active indirect flight muscle fibers revealed that phosphorylation site mutations reduced elastic and viscous moduli by 21–59% and maximum power output by up to 42%. Significant reductions also occurred under relaxed and rigor conditions, indicating that the phosphorylation-dependent changes are independent of strong crossbridge attachment and likely arise from alterations in thick filament backbone properties. Further, normal crossbridge kinetics were observed, demonstrating that myosin motor function is unaffected in the mutants. We conclude that N-terminal phosphorylation of Drosophila paramyosin is essential for optimal force and oscillatory power transduction within the muscle fiber and is key to the high passive stiffness of asynchronous insect flight muscles. Phosphorylation may reinforce interactions between myosin rod domains, enhance thick filament connections to the central M-line of the sarcomere and/or stabilize thick filament interactions with proteins that contribute to fiber stiffness.
Keywords: contraction, thick filament, sarcomere, mechanics, insect
Paramyosin is a major structural component of thick filaments in invertebrate muscle. The paramyosin to myosin ratio, which positively correlates with thick filament length and maximum active tension development (1), varies widely between different invertebrate phyla and even between different muscles within the same animal (2–5). For instance, this ratio is 1:34 in the fibrillar flight muscle of Drosophila melanogaster but is 1:6 in larval muscles (5). Sequence analysis of paramyosin isoforms isolated from different species reveal a rod-like molecule with a central α-helical region and two nonhelical terminal domains that can dimerize into a coiled-coil structure (6, 7). Like the myosin tail, amino acids of the paramyosin helical region have a periodic 28-residue positive and negative charge repeat distribution (6, 8). This charge arrangement on the surfaces of paramyosin and the myosin tail may regulate the packing of these two proteins in the thick filament (6, 9, 10).
Paramyosin is important for thick filament formation and for filament assembly into myofibrils (11, 12). A currently accepted model of invertebrate thick filament assembly is that paramyosin molecules, together with other thick filament structural proteins, form a thick filament core on which the motor protein myosin assembles (13). In Caenorhabditis elegans, the thick filament core is a tube-like structure composed of seven sleeves of paramyosin tetramers connected internally by filagenins spaced at regular distances (14, 15). Studies in various insect flight muscles also have indicated that paramyosin is in the core of the thick filaments. These results were derived by antibody labeling (16, 17) and by positively correlating paramyosin content to the number of thick filament core subfilaments (18). Examinations of specific regions in paramyosin have yielded insights into its function in assembly. A single charge change in the C. elegans paramyosin rod disrupts thick filament assembly (12). The assembly competence domain, a small, relatively neutral region in the rod domain near the C terminus found in both paramyosin and myosin (19), is believed to help initiate thick filament assembly (20). To our knowledge, previous to the current work, no investigations had been performed on the role of the nonhelical N-terminal domain of paramyosin in thick filament assembly.
Phosphorylation is an important means of regulating thick filament proteins such as vertebrate myosin binding protein-C (21), vertebrate and invertebrate myosin regulatory light chain (22–25), and Drosophila flightin (26, 27). Paramyosin is phosphorylated in vivo (5, 28) and can be phosphorylated in vitro (29–32), specifically in the nonhelical N-terminal domain (32). To investigate the function of the nonhelical N-terminal domain of paramyosin and the role of phosphorylation in muscle fiber assembly and performance, we took advantage of D. melanogaster genetics to generate phosphorylation site mutants. Transgenic flies were created in which one, three, or four putative phosphorylatable serine (Ser) residues in the N-terminal nonhelical domain of paramyosin were replaced with alanines (Alas). Some Ser mutations resulted in flies that exhibited flight impairment. Subsequent analysis of the mutant indirect flight muscle (IFM) structural and mechanical properties revealed that reduced elastic modulus, viscous modulus, and power output caused the flight impairment. Interestingly, no changes were observed in the mutants in thick filament assembly, myofibril organization, and fiber kinetics when compared with controls. These results indicate that phosphorylation of paramyosin is required for optimal force and power transduction, probably through a mechanism involving reinforcement of interactions within or with the thick filament.
Materials and Methods
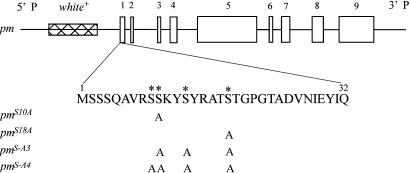
Construction of Transgenes. The normal paramyosin transgene (pm) was made from paramyosin genomic DNA, in which the miniparamyosin-specific exon was deleted (33). Four additional transgenes were created that mutated putative phosphorylation sites (Ser-9, -10, -13, and -18) in the N-terminal nonhelical domain of paramyosin. Either one (pmS10A and pmS18A), three (pmS-A3), or four (pmS-A4) Ser residues in the N-terminal region were mutated to Alas (Fig. 1). These sites were chosen because sequence analysis using the netphos program (www.cbs.dtu.dk/services/NetPhos) suggested that all four Ser residues have a >97% likelihood of being phosphorylated in vivo. To substitute the Sers with Alas, a 4.5-kb KpnI fragment from pm, containing a portion of exon 1 and most of the upstream promoter region, was subcloned into the pALTER-1 plasmid (Promega). Mutations in specific Ser codons were introduced into this plasmid by using in vitro mutagenesis. Single mutations at Ser-10 and -18 and a triple mutation at Ser-10, -13, and -18 were produced by using the Altered Sites II kit (Promega). Primers used to generate the mutations were: 5′-GTCCGCTCCGCGAAATACT-3′ (Ser-10), 5′-CGTGCCACGGCCACCGGAC-3′ (Ser-18), and 5′-TGTCCGCTCCGCGAAATACGCCTACCGTGCCACGGCCACCGGACC-3′ (Ser-10, -13, and -18). To mutate Ser-9, -10, -13, and -18, the plasmid containing a single mutation at Ser-18 served as a template for the QuikChange kit (Stratagene). The primer pair used was: 5′-GGCTGTCCGCGCCGCGAAATACGCCTACCG-3′ (forward) and 5′-CGGTAGGCGTATTTCGCGGCGCGGACAGCC-3′ (reverse). Each mutated KpnI fragment was subcloned back into pm. All constructs were sequenced for verification.
Fig. 1.
Structures of paramyosin transgenes used for germ-line transformation. Exons of the paramyosin gene are drawn as open boxes. The first 32 aa of the N-terminal nonhelical domain are emphasized, with four Ser residues that have high potential of phosphorylation marked by *. Locations of Ser residues replaced by Alas in mutant transgenes are indicated by “A.” All constructs are cloned in a CaSpeR vector, which has an eye color gene white+ used as a marker for selecting transgenic flies.
Generation of Drosophila Strains. Germ-line transformation of D. melanogaster embryos was performed as described in ref. 34. Mutated paramyosin transgenes were introduced into a paramyosin functional null mutant background, prm1, where endogenous paramyosin expression is <1% of wild-type levels (35).
Gel Electrophoresis and Western Blotting. 2D gel electrophoresis was performed by using the Immobiline DryStrip kit and the Multiphor II Electrophoresis system (Amersham Pharmacia Biotech). Briefly, proteins extracted from two upper thoraces of 2- to 3-day-old adult flies were loaded on Immobiline DryStrips (pH 4–7) and then electrically focused at 500 V for 5 h and then at 3,500 V for 13 h. After the first-dimensional separation, proteins on the strips were loaded onto a 12.5% ExcelGel (Amersham Pharmacia Biotech) for second-dimensional separation (300 V for 3 h). Proteins on the gel were then detected by silver staining (Amersham Pharmacia Biotech), and the relative amounts of paramyosin isoforms were determined by scanning densitometry of spots (Epson Expression 636 scanner, nih image 1.61, http://rsb.info.nih.gov/nih-image).
For Western blotting analysis, proteins separated on the ExcelGel were transferred to a nitrocellulose membrane and probed with a rabbit anti-Drosophila paramyosin antibody (36) and a secondary goat-anti-rabbit Ig conjugated with horseradish peroxidase (Bio-Rad). Chemical reaction and fluorescence signal detection were performed according to the SuperSignal system (Pierce).
Microscopy. Electron microscopy (EM) of adult IFMs was performed following procedures described in ref. 37. Briefly, dissected thoraces were fixed in glutaraldehyde/paraformaldehyde and postfixed in osmium tetraoxide. After dehydration with acetone, samples were infiltrated in Embed-812 resin (Electron Microscopy Sciences, Fort Washington, PA) under the following conditions: 50% resin/50% acetone (vol/vol) for 5 h, 75% resin/25% acetone overnight, and 100% resin overnight. Samples with fresh resin then were transferred into embedding molds, oriented within the molds, and polymerized at 60°C over 2 nights. After thick sectioning to expose the IFMs, superthin sections (70 nm) were collected on grids and stained with 1% uranyl acetate and Reynold's lead citrate. Images were collected with a Philips 410A transmission electron microscope. Sarcomere length, myofibril cross-sectional area, thick filaments per myofibril cross-sectional area, and myofibril area per fiber cross-sectional area were analyzed on scanned photos by using the imagej program (Version 1.26, National Institutes of Health). Light microscopy of devitellinized and fixed late embryos/young larvae was carried out on a Nikon Microphot microscope under polarized light. Samples were prepared by using the slow formaldehyde fix (substituting 4% paraformaldehyde) and the alternative rehydration procedures described by Rothwell and Sullivan (38).
Locomotory Performance. Flight testing was performed on 2- to 3-day-old female flies at room temperature (22°C) by releasing flies individually from the center of a Plexiglas flight chamber (39). Each fly was scored as flying up (U), horizontal (H), down (D), or not at all (N). The flight index for each line was determined by using the formula: 6 × U/T + 4 × H/T + 2 × D/T + 0 × N/T, where U, H, D, and N are the number of flies in each category of flight ability, and T is the total number of flies tested for that line (22). Crawling speed of third-instar larvae was assayed as the number of 0.5-cm grid boxes crossed in 5 min (40).
Skinned Muscle Fiber Mechanics. Isolated single dorso-longitudinal muscle fibers were skinned (demembranated) in a relaxing solution plus a protease inhibitor mixture (Roche) with 0.5% Triton X-100 and 50% (wt/vol) glycerol for 1 h at 4°C. Relaxing solution [-log10 calcium concentration (pCa) 8.0] consisted of 20 mM N,N-bis[2-hydroxyethyl]-2-aminoethanesulfonic acid, 15 mM creatine phosphate, 240 units/ml creatine phosphokinase, 1 mM DTT, 5 mM EGTA, 1 mM free Mg2+, 5 mM MgATP, and 8 mM Pi (pH 7.0) at an ionic strength of 200 mEq adjusted with sodium methane sulfate. Fibers were split in half lengthwise (to ≈100 μm diameter) to reduce the cross-sectional area to facilitate quicker diffusion of solutions. Fibers were immediately used for mechanical experiments or were transferred to storage solution (relaxing solution plus protease inhibitor mixture and 50% glycerol) at -20°C and used within a few days of dissection. Skinned fibers, in relaxing solution, were attached to aluminum T-clips at both ends and mounted to a force transducer and a length driver (23, 41). The fiber was stretched until just taut (minimal stress), and the initial fiber length was measured. The fiber was subsequently stretched by 5% of the initial length in 1% increments. Fiber diameter was measured at the narrowest part to calculate cross-sectional area. Fibers were activated (pCa 5.0) by three exchanges of equal amounts of relaxing solution with activating solution (same as relaxing solution, except pCa 4.0) and then stretched until work production, evaluated by sinusoidal analysis, was maximized. Subsequently, fibers were returned to relaxing solution (pCa 8.0) and progressively Ca-activated (from pCa 8.0 to 4.5) with isometric tension measured and sinusoidal analysis performed at each Ca level. At the end of the experiment, measurements were performed in rigor solution (activation solution lacking MgATP, creatine phosphate, and creatine phosphokinase, with ionic strength maintained at 200 mEq).
To perform sinusoidal analysis, small-amplitude sinusoidal length changes (0.125% muscle length) were applied to the fiber at 47 frequencies (0.5–1,000 Hz) while measuring the force response (23). Length and force were normalized to determine strain (ΔL/L) and tension (F/CSA) by dividing the length change (ΔL) by total fiber length (L) and by dividing the force (F) by the fiber cross-sectional area (CSA). Elastic (Ee) and viscous (Ev) moduli were calculated from the tension transient by determining the magnitudes of the in-phase and out-of-phase components (0° and 90° with respect to strain, respectively) (42). Power (P in W·m-3) generated by the muscle fiber was calculated from P = πf(-Ev)(Lamp)2, where f is the frequency of the length perturbations (Hz), Ev is the viscous modulus (kN/m2), and the fractional change in length (Lamp) is 0.00125. Note that positive power output results from a negative viscous modulus. All measurements were conducted at 15°C.
Statistical Analysis. Statistical analyses were performed by using spss v.11.0 (SPSS, Chicago). Statistical tests were considered significant at the P < 0.05 level. For most of the data (flight index, EM, and most sinusoidal analysis data), one-way ANOVA was performed to determine the effects of the different strains. If differences were significant, the least significant difference post hoc test was used to determine which means differed. The only exceptions were variables, such as elastic or viscous moduli, that were examined across the different sinusoidal oscillation frequencies. In this case, repeated-measures ANOVA with frequency as the repeated measure was performed first to determine the effects of the different transgenic and control strains. If a significant frequency by interaction effect was found, then one-way ANOVAs were performed at each frequency to determine significant differences.
Results
Generation of Mutant Paramyosin Fly Lines. We used a reverse genetics approach to study phosphorylation of D. melanogaster paramyosin in vivo and to investigate the role of paramyosin phosphorylation in the structure and function of the IFM. To this end, we constructed paramyosin transgenes in which one, three, or four Ser codons were mutated to Ala. Mutations affected residues 9, 10, 13, and/or 18 in the N-terminal nonhelical domain of paramyosin (Fig. 1). We introduced these transgenes into a paramyosin functional null mutant background, prm1, to yield transgenic replacement of the endogenous paramyosin gene. All four mutant transgenes rescued the embryonic-lethal homozygous prm1 to adulthood.
Locomotory Performance. We observed no significant differences in flight ability in flies with the single Ser substitution at position 10 (pmS10A: flight index 3.8 ± 0.2, n = 157) compared with control (pm: 3.8 ± 0.2, n = 156). In contrast, the flight abilities of flies with Ser substitutions at position 18 (pmS18A: 0.7 ± 0.1, n = 242), positions 10, 13, and 18 (pmS-A3: 0.6 ± 0.1, n = 197), and positions 9, 10, 13, and 18 (pmS-A4: 0.6 ± 0.1, n = 259) were severely impaired. Two strains that exhibited severe defects in flight performance were chosen for further analysis (one with a single Ser substitution, pmS18A, and one with four Ser substitutions, pmS-A4). In contrast to the dramatic effects on flight, these mutations did not affect larval crawling speed (pm control, 13.2 ± 0.8; pmS18A, 13.0 ± 0.8; pmS-A4, 13.1 ± 0.7; n = 19 for each line).
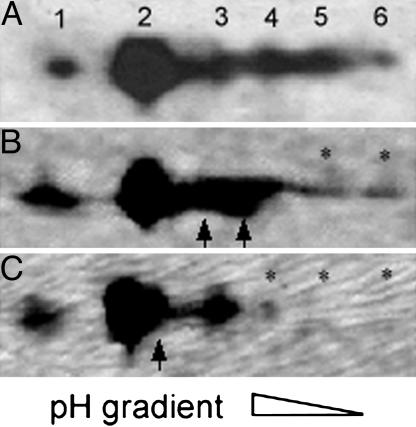
Paramyosin Phosphorylation. We characterized the paramyosin phosphovariant profile in the pmS18A and pmS-A4 lines. Paramyosin from upper thoraces of wild-type flies and prm1 rescued with the normal paramyosin transgene separated into six major spots on high-resolution 2D gels (Fig. 2A), compared with three spots observed by Vinos et al. (5). Substitution of Ser residues with Ala in the N-terminal nonhelical domain of paramyosin changed the phosphovariant profile in pmS18A and pmS-A4, as determined by measuring the proportion of paramyosin in each spot by densitometry. Substitution of Ser-18 decreased the amount of the two most acidic spots (spot 5 by 90%; spot 6 by 72%) and was accompanied by increases in spots 3 (to 128%) and 4 (to 170%) (Fig. 2B). Even more dramatic changes occurred in pmS-A4, where spots 5 and 6 disappeared and spot 4 decreased (by 95%), with a corresponding increase in spot 2 (to 131%) (Fig. 2C). These changes in spot distribution are consistent with the N terminus of paramyosin being phosphorylated, possibly at multiple sites.
Fig. 2.
Paramyosin isoform profiles of control and mutant flies on 2D gels. (A) On a Western blot using paramyosin antibody, six paramyosin isoforms are detected in homozygous prm1 rescued with the normal paramyosin transgene pm, as is the case for silver-stained gels and in yw control flies (data not shown). (B and C) In silver-stained gels, substitution of Ala for Ser-18 in pmS18A (B) or four Ser residues (Ser-9, -10, -13, and -18) in pmS-A4 (C) with Alas causes reduction in acidic isoforms and a corresponding increase in basic isoforms. * marks isoforms decreased; arrows mark isoforms increased.
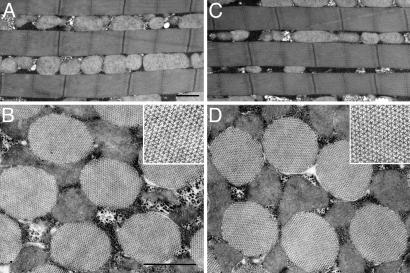
Muscle Ultrastructure. Because paramyosin is the major component of the core of invertebrate muscle thick filaments, we had speculated that subtle changes in this protein might affect thick filament assembly, resulting in structural abnormalities. To examine this possibility, we used transmission EM to study IFM myofibril ultrastructure in the pmS18A and pmS-A4 lines. Surprisingly, the mutant transgenes permitted normal assembly of IFM myofibrils. Even with the full complement of four Ser substitutions in pmS-A4, no defects in myofibril structure were observed (Fig. 3). As in wild-type IFM myofibrils, thick and thin filaments of mutant myofibrils interdigitated to form highly organized hexagonal arrays with a well-defined cylindrical shape. There were no significant differences between control and mutant flies in major structural parameters such as sarcomere length, thick filament number per myofibril cross-sectional area, and percentage of myofibril area per muscle cross-sectional area (Table 1). Mutant flies showed a 1.4–2.7% increase in myofibril cross-sectional area (Table 1). However, the calculated value of thick filaments per cross-sectional area (obtained by multiplying the thick filament number per myofibril area by the myofibril area per fiber area) was similar between control and mutant lines, indicating that the numbers of thick filaments per fiber were unchanged. Likewise, the structure of lateral body wall muscles of mutant and control first-instar larvae showed normal striation patterns under polarized light microscopy (data not shown).
Fig. 3.
EM of IFM. Substitution of four Ser residues in the N-terminal nonhelical domain of paramyosin does not affect the ultrastructure of IFMs. Compared with those of wild-type flies (A and B), IFMs of pmS-A4 flies (C and D) are normal. In longitudinal section (C), pmS-A4 myofibrils have sarcomeres of constant length and width. In cross section (D Inset), pmS-A4 thick and thin filaments interdigitate into regular hexagonal arrays, typical of IFMs (B Inset). (Bars: 1 μm.)
Table 1. IFM myofibril parameters of paramyosin transgenic flies.
| Strain | SL, μm | MF area, μm2 | TF per MF area, filaments·μm–2 | MF area per fiber area, % |
|---|---|---|---|---|
| pm | 3.06 ± 0.02 (25) | 0.964 ± 0.005 | 783 ± 7 | 55.4 ± 0.9 (5) |
| pmS18A | 3.06 ± 0.02 (21) | 0.978 ± 0.003* | 792 ± 5 | 55.5 ± 0.3 (5) |
| pmS-A4 | 3.06 ± 0.02 (25) | 0.990 ± 0.003* | 799 ± 4 | 55.2 ± 0.5 (5) |
SL, sarcomere length; MF, myofibril; TF, thick filament; area, cross-sectional area. Values are means ± SEM. Number of measurements (n-values) for pm, pmS18A, and pmS-A4 are 31, 33, and 31, respectively, except where noted in parentheses.
Indicates significant difference (P < 0.05) from pm
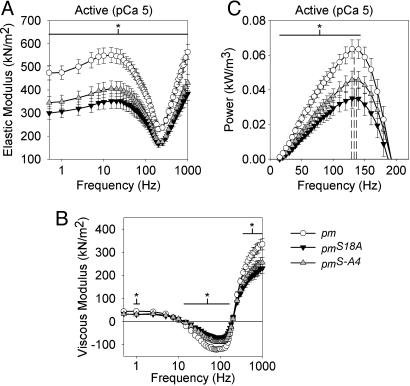
Skinned Muscle Fiber Mechanics. We investigated the effect of altering the paramyosin phosphorylation status on the mechanical properties of mutant IFM skinned fibers of the pmS18A and pmS-A4 lines. Passive, active, and crossbridge-dependent isometric force generation was significantly decreased by the phosphorylation site disruption (Table 2). However, the relationship between isometric tension and Ca concentration showed no significant differences in pCa50 values and Hill coefficients (Table 2). At peak Ca activation (pCa 5.0), over a wide range of oscillation frequencies, the magnitudes of the elastic modulus, viscous modulus, and power output for pmS18A and pmS-A4 fibers were dramatically reduced compared with those of the pm control (Fig. 4). The active elastic moduli of pmS18A and pmS-A4 fibers were significantly less (values ranged from 23% to 37% less at each frequency) than that of pm for all frequencies examined (Fig. 4A). The viscous moduli of active pmS18A and pmS-A4 fibers were significantly less for most frequency values of >20 Hz (values ranged from 21% to 59% less) (Fig. 4B). Consequently, pmS18A and pmS-A4 active fibers produced significantly less power compared with pm throughout most of the positive power-producing range (42% and 28% less at maximum power, respectively) (Fig. 4C).
Table 2. Summary of isolated IFM isometric data.
| Tension, kN/m2
|
Hill fit parameters
|
||||||
|---|---|---|---|---|---|---|---|
| Strain | Active (pCa = 5.0) | Passive (pCa = 8.0) | Crossbridge dep. (active–passive) | n | pCa50 | Hill coefficient | n |
| pm | 2.8 ± 0.3 | 1.2 ± 0.2 | 1.6 ± 0.2 | 10 | 5.9 ± 0.1 | 2.8 ± 0.4 | 6 |
| pmS18A | 1.9 ± 0.1* | 0.8 ± 0.1* | 1.0 ± 0.1* | 12 | 5.8 ± 0.1 | 2.8 ± 0.5 | 5 |
| pmS-A4 | 2.2 ± 0.2* | 0.9 ± 0.1† | 1.3 ± 0.1‡ | 12 | 6.1 ± 0.1 | 1.8 ± 0.3 | 9 |
Isometric tension and Hill fit data from isolated IFM. Values are means ± SEM. Crossbridge dep., crossbridge dependent. Temperature = 15 °C. †, P = 0.054; ‡, P = 0.061.
Indicates significant difference (P < 0.05) from pm
Fig. 4.
Elastic modulus (A), viscous modulus (B), and power production (C) values for active IFM across muscle oscillation frequencies for paramyosin mutants (pmS18A and pmS-A4) and pm control. Values are means ± SEM. Dashed lines in C represent fPmax (frequency of maximum power) from Table 3. * indicates a span of frequencies over which there is a significant difference (P < 0.05) between pm and the pmS18A and pmS-A4 lines. Temperature = 15°C. Note that the elastic and viscous moduli frequency values were plotted on a log scale over the entire sinusoidal analysis frequency range, whereas the power data were plotted on a linear scale over the power-producing range (where the viscous modulus is negative).
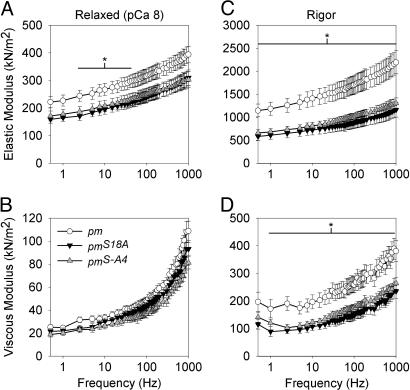
To determine whether the decreased elastic and viscous moduli of active mutant fibers depends on the attachment of myosin crossbridges, we examined IFM fibers while relaxed (pCa 8, where no myosin crossbridges are strongly attached to actin) or in rigor (no ATP, where myosin crossbridges are strongly attached). In the relaxed condition, pmS18A and pmS-A4 fibers showed a significant reduction in magnitude of elastic modulus between 2.5 and 40 Hz (ranging from 25% to 37% less) compared with pm control fibers (Fig. 5A). In rigor conditions, pmS18A and pmS-A4 fibers showed dramatic reductions in elastic modulus (ranging from 40 to 49% less) over the entire frequency range and in viscous modulus (ranging from 28 to 51% less) at all frequencies except at 0.5 and 1,000 Hz (Fig. 5 C and D). These results indicate that mutation of paramyosin phosphorylation sites in the N-terminal nonhelical domain significantly reduces the moduli of myofibril components, regardless of whether crossbridges are strongly attached (active and rigor conditions) or not (passive conditions).
Fig. 5.
Elastic and viscous modulus values for relaxed (A and B) and rigor (C and D) IFM across muscle oscillation frequencies for paramyosin mutants (pmS18A and pmS-A4) and pm control. Values are means ± SEM. * indicates a span of frequencies over which there is a significant difference (P < 0.05) between pm and the pmS18A and pmS-A4 lines. Temperature = 15°C.
Although active stiffness and power output showed dramatic differences between the pm control and mutants, sinusoidal analysis revealed no significant differences in active (pCa 5.0) muscle fiber kinetics. The oscillatory frequency at which maximum power occurred was similar between the strains tested (Table 3; Fig. 4C, dashed lines). In addition, the oscillatory frequency at the minimum value of complex modulus amplitude, a conventional index for kinetic changes, showed no significant differences (208 ± 7 Hz for pm, 197 ± 8 Hz for pmS18A, and 207 ± 6 Hz for pmS-A4). These results strongly suggest that myosin crossbridge kinetics are not affected by the changes in paramyosin phosphorylation.
Table 3. Summary of isolated IFM sinusoidal analysis data at the frequency of maximum power output (pCa = 5.0).
| Strain | Pmax, W·m–3 | fPmax, Hz | Ee, kN·m–2 | Ev, kN·m–2 | Complex modulus, kN·m–2 | n |
|---|---|---|---|---|---|---|
| pm | 65 ± 5 | 133 ± 4 | 323 ± 17 | –114 ± 8 | 343 ± 18 | 10 |
| pmS18A | 38 ± 5* | 129 ± 8 | 226 ± 20* | –71 ± 8* | 237 ± 21* | 12 |
| pmS-A4 | 47 ± 5* | 137 ± 3 | 242 ± 19* | –82 ± 9* | 256 ± 21* | 12 |
Averaged sinusoidal analysis data from isolated IFM at the frequency of maximum power. Values are means ± SEM. Maximum power (Pmax) occurs at specific oscillatory frequencies (fPmax). Ee refers to the elastic (in-phase) modulus. Ev refers to the viscous (out-of-phase) modulus. Complex modulus is the vector sum of Ee and Ev. Temperature = 15 °C.
Indicates significant difference (P < 0.05) from pm
Discussion
By using a reverse genetics approach, we investigated the role of Drosophila paramyosin phosphorylation in regulating muscle structure and function. We showed that Drosophila paramyosin can be phosphorylated in its N-terminal nonhelical domain and that this posttranslational modification is not critical for normal IFM assembly but is important for flight muscle power output and normal flight ability. In comparison with the phosphorylation mutants, phosphorylated paramyosin yields enhanced elastic modulus, viscous modulus, and power output in IFM fibers. As discussed below, this result may arise from paramyosin strengthening the sarcomere by linking myosin α-helical rod domains, by connecting the thick filaments to the M-line, and/or by attaching the thick filament to other proteins important for generating passive stiffness. Importantly, our studies show that paramyosin N-terminal phosphorylation plays a key role in producing the high passive stiffness observed in asynchronous IFMs (41, 43).
Paramyosin Phosphorylation Isoforms. Even though paramyosin is encoded by a single gene in D. melanogaster (7, 36), the protein migrates as several isoelectrically discrete isoforms in 2D gels. Phosphorylation of Ser residues in the N-terminal domain is responsible for much of the isoform variability, as the number and amount of acidic isoforms decrease in the Ser → Ala mutant flies. Ala substitutions at all four phosphorylatable Ser residues in the N-terminal domain yield more than a single paramyosin spot on 2D gels, indicating that paramyosin is likely to be posttranslationally modified at other residues in addition to the mutated sites.
Muscle Ultrastructure. Paramyosin is important for formation of thick filaments and for their assembly into myofibrils (11, 12). The zones of positive and negative charge on the α-helical region of paramyosin and their interaction with the myosin tail appear to play a key role in these processes (6, 9, 10, 12).
This work is an investigation of the previously undescribed role of the nonhelical N-terminal domain of paramyosin in thick filament assembly. We initially speculated that phosphorylation regulates paramyosin packing during thick filament assembly by modifying the charge distribution at the ends of paramyosin dimers. Unexpectedly, our ultrastructural examination of mutant fly IFMs showed that thick filament assembly and myofibril organization are not affected by mutating the N-terminal phosphorylation sites. Thus, the mutated residues (Ser-9, -10, -13, and -18) do not serve to direct the assembly of the α-helical regions of paramyosin and myosin into thick filaments.
Locomotory Performance. The changes in flight index provide insight into the role of paramyosin N-terminal phosphorylation sites in muscle function. The Ser-18 phosphorylation site in particular appears to be important for proper IFM function because its removal, individually (pmS18A) or jointly (pmS-A3 and pmS-A4), causes a significant decrease in flight index. In contrast, removing the Ser-10 phosphorylation site (pmS10A) has no effect on flight index when compared with the pm control, suggesting a less significant role for Ser-10 in muscle function. Establishing the relative importance of Ser-9 or -13 to flight performance is difficult because the pmS18A (Ser-18 switched to Ala), pmS-A3 (Ser-10, -13, and -18 switched) and pmS-A4 (Ser-9, -10, -13, and -18 switched) have equivalent flight indices and all contain the Ser-18 → Ala switch, which may drive the flight performance decrease.
Although our work indicates that specific Ser residues in paramyosin are critical for IFM function, we detected no effects of the phosphorylation site mutations on larval locomotion or myofibrillar structure. Thus, either phosphorylation serves a specific regulatory function in the IFMs, or the less-ordered architecture of larval body wall muscle myofibrils precludes detection of phenotypic defects. In contrast, C. elegans unc-82 mutants have reduced paramyosin phosphorylation and exhibit defects in body wall myofibril assembly and function (32, 44). However, unc-82 mutations may affect additional paramyosin phosphorylation sites and/or phosphorylation of other muscle proteins.
Skinned Muscle Fiber Mechanics. Surprisingly, the reduced phosphorylations in pmS18A (a single site mutation) and pmS-A4 (four-site mutation) have similar magnitude effects on all measured skinned fiber mechanics. Reductions in isometric force as well as elastic (in-phase) and/or viscous (out-of-phase) modulus values occur regardless of whether crossbridges are strongly attached (active and rigor conditions) or not strongly attached (passive conditions). Because the reduction in modulus values does not depend on crossbridge attachment, phosphorylation-dependent changes in moduli are most likely due to alterations in the thick filament structural properties rather than alterations within the crossbridge itself. The stiff connections bridging Z-lines and thick filaments in the IFM (45) permit the changes in thick filament properties to be observed in the relaxed fiber. The lack of measurable changes in the oscillatory frequency at which maximum power occurred, oscillatory frequency at which the minimum value of complex modulus amplitude occurred, pCa50, and Hill coefficient further validate the idea that the myosin motor domain is unchanged. These results indicate that the decreased power-generating capability of active fibers with disrupted phosphorylation sites is due to a reduction in the ability of the thick filament to transmit the force and oscillatory power produced by the myosin heads. As with the flight performance, the observed changes in muscle mechanics suggest that the Ser-18 phosphorylation site is very important for normal muscle performance because this site is mutated in both lines examined, and both are similar in all mechanical measurements.
Transverse EM sections of Drosophila IFM labeled with an anti-paramyosin antibody indicate that paramyosin is uniformly distributed along the length of the thick filaments (16, 17). Cross-sectional EM sections of various insect flight muscles show a direct correlation between paramyosin content and the number of thick filament core subfilaments, with low paramyosin to myosin ratios (Phormia terrae-novae or fleshfly) resulting in hollow filaments and higher ratios (Apis mellifica or honey bee) in solid filaments (18). Drosophila IFM has a low paramyosin to myosin ratio of 1:34 (3, 5) and a hollow core, which fits the former category.
Drosophila paramyosin most likely interacts with the rod portion of the myosin filaments at sites along the length of the thick filament, as in C. elegans (10). Thus, we postulate that paramyosin lines the inner wall of the thick filament and anchors portions of the myosin rod domains in place through interactions with its phosphorylated N-terminal. Altering the phosphorylation sites of paramyosin would weaken the connections between paramyosin and the myosin rod domains, causing a decrease in the ability of the thick filament to transmit the force and oscillatory power produced by the myosin heads.
Paramyosin also may reinforce sarcomere stiffness by stabilizing the connection between thick filaments and the M-line. This model is bolstered by the observation that a potential interacting partner, miniparamyosin, is located in the M-line (16). Miniparamyosin contains the same C-terminal region as paramyosin (7), permitting the coiled-coil formation of heterodimers. Thus, it is possible that the phosphorylated N termini of paramyosin initiate or stabilize an interaction between paramyosin and miniparamyosin. Because paramyosin is differentially phosphorylated (16), phosphovariants may be differentially localized along the length of the thick filament (5). Phosphovariant-specific localization directed at the M-line may be one mechanism by which an interaction of paramyosin with miniparamyosin is enhanced. In addition, paramyosin may stabilize thick filament interactions with proteins such as projectin (46) or kettin (47), which contribute to fiber properties, especially passive stiffness.
Acknowledgments
We thank Dr. Steve Barlow for advice regarding microscopy. This work was supported by National Institutes of Health Grants R01 HL68034 (to D.W.M.) and R01 AR43396 (to S.I.B.) and a predoctoral fellowship from the Western States Affiliate of the American Heart Association (to H.L.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: IFM, indirect flight muscle; pCa, -log10 calcium concentration; pm, paramyosin transgene; pmS10A, pm with Ser-10 replaced by Ala; pmS18A, pm with Ser-18 replaced by Ala; pmS-A3, pm with Ser-10, -13, and -18 replaced by Ala; pmS-A4, pm with Ser-9, -10, -13, and -18 replaced by Ala; prm1, paramyosin functional null mutant.
References
- 1.Levine, R. J., Elfvin, M., Dewey, M. M. & Walcott, B. (1976) J. Cell Biol. 71, 273-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Winkelman, L. (1976) Comp. Biochem. Physiol. B 55, 391-397. [DOI] [PubMed] [Google Scholar]
- 3.Beinbrech, G., Meller, U. & Wolfgang, S. (1985) Cell Tissue Res. 241, 607-614. [Google Scholar]
- 4.Bullard, B., Luke, B. & Winkelman, L. (1973) J. Mol. Biol. 75, 359-367. [DOI] [PubMed] [Google Scholar]
- 5.Vinos, J., Domingo, A., Marco, R. & Cervera, M. (1991) J. Mol. Biol. 220, 687-700. [DOI] [PubMed] [Google Scholar]
- 6.Kagawa, H., Gengyo, K., McLachlan, A. D., Brenner, S. & Karn, J. (1989) J. Mol. Biol. 207, 311-333. [DOI] [PubMed] [Google Scholar]
- 7.Becker, K. D., O'Donnell, P. T., Heitz, J. M., Vito, M. & Bernstein, S. I. (1992) J. Cell Biol. 116, 669-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McLachlan, A. D. & Karn, J. (1982) Nature 299, 226-231. [DOI] [PubMed] [Google Scholar]
- 9.Hoppe, P. E. & Waterston, R. H. (1996) J. Cell Biol. 135, 371-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoppe, P. E. & Waterston, R. H. (2000) Genetics 156, 631-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waterston, R. H., Fishpool, R. M. & Brenner, S. (1977) J. Mol. Biol. 117, 679-697. [DOI] [PubMed] [Google Scholar]
- 12.Gengyo-Ando, K. & Kagawa, H. (1991) J. Mol. Biol. 219, 429-441. [DOI] [PubMed] [Google Scholar]
- 13.Nonomura, Y. (1974) J. Mol. Biol. 88, 445-455. [DOI] [PubMed] [Google Scholar]
- 14.Epstein, H. F., Lu, G. Y., Deitiker, P. R., Oritz, I. & Schmid, M. F. (1995) J. Struct. Biol. 115, 163-174. [DOI] [PubMed] [Google Scholar]
- 15.Muller, S. A., Haner, M., Ortiz, I., Aebi, U. & Epstein, H. F. (2001) J. Mol. Biol. 305, 1035-1044. [DOI] [PubMed] [Google Scholar]
- 16.Maroto, M., Arredondo, J., Goulding, D., Marco, R., Bullard, B. & Cervera, M. (1996) J. Cell Biol. 134, 81-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Royuela, M., Garcia-Anchuelo, R., Arenas, M. I., Cervera, M., Fraile, B. & Paniagua, R. (1996) Histochem. J. 28, 247-255. [DOI] [PubMed] [Google Scholar]
- 18.Beinbrech, G., Ashton, F. T. & Pepe, F. A. (1992) Biophys. J. 61, 1495-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen, C. & Parry, D. A. (1998) J. Struct. Biol. 122, 180-187. [DOI] [PubMed] [Google Scholar]
- 20.Sohn, R. L., Vikstrom, K. L., Strauss, M., Cohen, C., Szent-Gyorgyi, A. G. & Leinwand, L. A. (1997) J. Mol. Biol. 266, 317-330. [DOI] [PubMed] [Google Scholar]
- 21.Winegrad, S. (2003) Adv. Exp. Med. Biol. 538, 31-40. [DOI] [PubMed] [Google Scholar]
- 22.Tohtong, R., Yamashita, H., Graham, M., Haeberle, J., Simcox, A. & Maughan, D. (1995) Nature 374, 650-653. [DOI] [PubMed] [Google Scholar]
- 23.Dickinson, M. H., Hyatt, C. J., Lehmann, F. O., Moore, J. R., Reedy, M. C., Simcox, A., Tohtong, R., Vigoreaux, J. O., Yamashita, H. & Maughan, D. W. (1997) Biophys. J. 73, 3122-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levine, R. J., Yang, Z., Epstein, N. D., Fananapazir, L., Stull, J. T. & Sweeney, H. L. (1998) J. Struct. Biol. 122, 149-161. [DOI] [PubMed] [Google Scholar]
- 25.Adhikari, B. B., Somerset, J., Stull, J. T. & Fajer, P. G. (1999) Biochemistry 38, 3127-3132. [DOI] [PubMed] [Google Scholar]
- 26.Vigoreaux, J. O. (1994) Biochem. Genet. 32, 301-314. [DOI] [PubMed] [Google Scholar]
- 27.Vigoreaux, J. O. & Perry, L. M. (1994) J. Muscle Res. Cell Motil. 15, 607-616. [DOI] [PubMed] [Google Scholar]
- 28.Cooley, L. B., Johnson, W. H. & Krause, S. (1979) J. Biol. Chem. 254, 2195-2198. [PubMed] [Google Scholar]
- 29.Watabe, S., Tsuchiya, T. & Hartshorne, D. J. (1989) Comp. Biochem. Physiol. B 94, 813-821. [DOI] [PubMed] [Google Scholar]
- 30.Achazi, R. K. (1979) Pflügers Arch. 379, 197-201. [DOI] [PubMed] [Google Scholar]
- 31.Chen, G. X., Tan, R. Y., Gong, Z. X., Huang, Y. P., Wang, S. Z. & Cao, T. G. (1988) Biophys. Chem. 29, 147-153. [DOI] [PubMed] [Google Scholar]
- 32.Schriefer, L. A. & Waterson, R. H. (1989) J. Mol. Biol. 207, 451-454. [DOI] [PubMed] [Google Scholar]
- 33.Mardahl-Dumesnil, M. (1998) Ph.D. dissertation (San Diego State Univ./Univ. of California, San Diego).
- 34.Rubin, G. M. & Spradling, A. C. (1982) Science 218, 348-353. [DOI] [PubMed] [Google Scholar]
- 35.Liu, H. (2003) Ph.D. dissertation (San Diego State Univ./Univ. of California, San Diego).
- 36.Maroto, M., Arredondo, J. J., San Roman, M., Marco, R. & Cervera, M. (1995) J. Biol. Chem. 270, 4375-4382. [DOI] [PubMed] [Google Scholar]
- 37.O'Donnell, P. T. & Bernstein, S. I. (1988) J. Cell Biol. 107, 2601-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rothwell, W. F. & Sullivan, W. (2000) in Drosophila Protocols, eds. Sullivan, W., Ashburner, M. & Hawley, R. S. (Cold Spring Harbor Lab. Press, Woodbury, NY), pp. 141-157.
- 39.Drummond, D. R., Hennessey, E. S. & Sparrow, J. C. (1991) Mol. Gen. Genet. 226, 70-80. [DOI] [PubMed] [Google Scholar]
- 40.Naimi, B., Harrison, A., Cummins, M., Nongthomba, U., Clark, S., Canal, I., Ferrus, A. & Sparrow, J. C. (2001) Mol. Biol. Cell 12, 1529-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peckham, M., Molloy, J. E., Sparrow, J. C. & White, D. C. (1990) J. Muscle Res. Cell Motil. 11, 203-215. [DOI] [PubMed] [Google Scholar]
- 42.Kawai, M. & Brandt, P. W. (1980) J. Muscle Res. Cell Motil. 1, 279-303. [DOI] [PubMed] [Google Scholar]
- 43.Josephson, R. K., Malamud, J. G. & Stokes, D. R. (2000) J. Exp. Biol. 203, 2713-2722. [DOI] [PubMed] [Google Scholar]
- 44.Waterston, R. H., Thomson, J. N. & Brenner, S. (1980) Dev. Biol. 77, 271-302. [DOI] [PubMed] [Google Scholar]
- 45.White, D. C. (1983) J. Physiol. 343, 31-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moore, J. R., Vigoreaux, J. O. & Maughan, D. W. (1999) J. Muscle Res. Cell Motil. 20, 797-806. [DOI] [PubMed] [Google Scholar]
- 47.Kulke, M., Neagoe, C., Kolmerer, B., Minajeva, A., Hinssen, H., Bullard, B. & Linke, W. A. (2001) J. Cell Biol. 154, 1045-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]