Abstract
Systemic lupus erythematosus (SLE) is an autoimmune disease with a complex genetic basis that includes susceptibility gene(s) within the chromosome 1q41–1q42 region. Toll-like receptor 5 (TLR5), the innate immune receptor for bacterial flagellin, maps to chromosome 1q41 and contains a common stop codon polymorphism that abrogates signaling (allele C1174T) and is associated with an increased risk of infection. By using transmission disequilibrium testing in a cohort containing 199 affected patients and their 75 unaffected siblings and 326 parents, we found that allele 1174C, but not 1174T (with the stop codon), was preferentially transmitted to SLE-affected offspring (a 19:6 transmitted/not transmitted ratio, P = 0.009). In contrast, the alleles of the other three TLR5 SNPs did not exhibit preferential transmission. In addition, we found that allele 1174C was not preferentially transmitted to unaffected offspring (3:6 transmitted/not transmitted ratio, P value not significant). The allele frequency of 1174T in the probands was 3.2% compared with 5.8% in unaffected individuals, which was consistent with a protective association (odds ratio, 0.51; 95% confidence interval, 0.26–0.98; P = 0.041). Subjects with the TLR5 stop codon produced significantly lower levels of proinflammatory cytokines in comparison with individuals with the wild-type genotype. Together, these results indicate that the TLR5 stop codon polymorphism is associated with protection from the development of SLE. These data support a role for flagellated bacteria and the innate immune response in the development of SLE with implications for novel immunomodulatory treatment strategies.
Keywords: genetic markers, genetic predisposition to disease, immunity, inflammation
Systemic lupus erythematosus (SLE) is an autoimmune disease with a complex genetic basis that includes susceptibility gene(s) on multiple chromosomes (1–3). Linkage of chromosomal region 1q41–1q42 with susceptibility to SLE has been identified and confirmed in independent cohorts that included several different ethnic backgrounds (4–6). In addition, a syntenic murine region of chromosome 1, named Sle1, is also associated with lupus susceptibility (3). By using a multiallelic, transmission-disequilibrium test (TDT), we previously found preferential transmission in this region of a poly(ADP-ribose) polymerase allele to affected offspring and lack of transmission to unaffected offspring (7). However, a different study failed to confirm this association and raised the possibility that another locus in linkage disequilibrium with poly(ADP-ribose) polymerase was the causative gene (8). Subsequent genomic sequencing and mapping data revealed that Toll-like receptor (TLR)5, the innate immune receptor for bacterial flagellin, is located at 1q41 (9).
TLRs constitute a family of transmembrane proteins that differentially recognize pathogens and initiate inflammatory signaling pathways (10–13). TLRs are type I transmembrane proteins that contain an extracellular leucine-rich region involved in pathogen recognition and a conserved intracellular Toll/IL-1 receptor domain that activates a signaling pathway. Stimulation of the TLR pathway culminates in NF-κB activation and transcription of immune response genes, such as cytokines and chemokines. Because of their central role in the regulation of inflammation and the immune response to pathogens, TLRs are excellent candidate genes for genetic susceptibility studies for autoimmune diseases. In fact, a recent study indicated that TLR9 and MyD88 pathways, when stimulated by certain immune complexes, mediated production of autoantibodies (14). We also recently found that polymorphisms in TLR9 are associated with SLE and lupus nephritis.∥ We previously demonstrated that TLR5 recognizes bacterial flagellin, a potent inflammatory stimulus present in the flagellar structure of many bacteria (9). We also found that a common stop codon polymorphism in the ligand-binding domain of TLR5 (TLR5392STOP) is unable to mediate flagellin signaling, acts in a dominant fashion, and is associated with increased susceptibility to pneumonia caused by Legionella pneumophila (15).
Because TLR5 is a critical regulator of inflammatory pathways and maps to chromosome 1q41, we hypothesized that the stop codon variant is associated with susceptibility to SLE. To test this hypothesis, we used a TDT in a Caucasian SLE cohort and found that the TLR5 stop codon polymorphism, but not other TLR5 alleles, is associated with protection from developing SLE. We also found that this association was most pronounced in individuals who are seronegative for anti-dsDNA autoantibodies. These results suggest a role for the innate immune response in the development of SLE that involves flagellated bacterial infections.
Materials and Methods
Materials. RPMI medium 1640, l-glutamine, and penicillin–streptomycin were from Life Technologies (Carlsbad, CA). Ultrapure lipopolysaccharide was from Salmonella minnesota R595 (List Biological Laboratories, Campbell, CA). Flagellin (FliC) was purified from Salmonella typhimurium strain TH4778, which is fljB-/fliC+, as described in refs. 9 and 16.
Human Subjects and Data Collection. Approval for human study protocols was obtained from the human subjects review boards at University of California, Los Angeles, the University of Washington, and the Western Institutional Review Board. All participants gave written informed consent. Genomic DNA was purified from peripheral blood leukocytes. The study design and enrolment criteria for this cohort have been described in ref. 7.
Genotyping. Genotyping was carried out with MassARRAY (Sequenom, San Diego), a chip-based MALDI-TOF mass spectrometer technique (17). Multiplex SNP assays were designed with spectrodesigner software (Sequenom); 384-well plates containing 5 ng of DNA in each well were amplified by PCR by following the manufacturer's (Sequenom) specifications. After PCR, arctic shrimp alkaline phosphatase (Sequenom) was added to samples to prevent future incorporation of unused dNTPs that could interfere with the primer extension assay. Allele discrimination reactions were conducted by adding the extension primer(s), DNA polymerase, and a mixture of dNTPs and dideoxynucleoside triphosphates to each well. MassEXTEND clean resin (Sequenom) was added to the mixture to remove extraneous salts that could interfere with MALDI-TOF analysis. Genotypes were determined by spotting 15 nl of each sample onto a 384 SpectroCHIP (Sequenom) which was subsequently read by the MALDI-TOF mass spectrometer.
Statistics. The TDT for individual SNPs and haplotypes was performed with genehunter and transmit. A contingency table Fisher exact two-tailed test was performed using prism 3.02 (GraphPad, San Diego). Linkage disequilibrium statistics were calculated with gold software.
Protein Analysis. Peripheral blood mononuclear cells (PBMCs) were isolated from 50 ml of blood from individuals with a Ficoll gradient separation as described in ref. 15. Cells were plated in RPMI medium 1640 supplemented with 10% FCS, penicillin, and streptomycin and stimulated for 18 h; supernatants were then harvested. Cytokine levels were determined with a sandwich ELISA technique (DuoSet, R & D Systems).
Results
To examine whether TLR5 polymorphisms are associated with susceptibility to SLE, we used the TDT in an extended Caucasian SLE cohort containing 199 affected patients and their 75 unaffected siblings and 326 parents (7). TLR5 is a type I transmembrane protein with a 642-aa, leucine-rich extracellular domain, an 18-aa transmembrane domain, and a 198-aa cytoplasmic Toll/IL-1 receptor homology signaling domain. The coding region of TLR5 has four common SNPs, including a cytosine-to-thymidine transition at base pair 1174 that changes an arginine at amino acid 392 to a stop codon and prematurely truncates TLR5 in the extracellular domain and causes the loss of the transmembrane domain and the entire signaling cytoplasmic tail (TLR5392STOP) (15). Two additional nonsynonymous SNPs [A1775G (amino acid N592S) and T1846C (F616L)] alter residues in the ectodomain, and a fourth synonymous SNP [A2523G (K841K)] is in the cytoplasmic tail. By using the TDT, we found that allele 1174C, but not 1174T (with the stop codon), was preferentially transmitted to SLE-affected offspring [a 19:6 transmitted/not transmitted (T/NT) ratio, P = 0.009), whereas the alleles of the other three TLR5 SNPs did not exhibit preferential transmission (Table 1). Thus, the stop codon allele (1174T) was significantly undertransmitted to SLE offspring. In addition, we found that allele 1174C was not preferentially transmitted to unaffected offspring (a 3:6 T/NT ratio; P value not significant). SNP 1174 exhibited linkage disequilibrium with SNPs 1775 and 1846 but not with 2523 (D′ = 1 for 1174–1775, D′ = 0.88 for 1174–1846, and D′ = 0.64 for 1174–2523; gold software). In light of this finding, we analyzed TLR5 haplotypes and their association with SLE (Table 1). Consistent with the TDT results, the 2-, 3- and 4-loci haplotypes containing SNP 1174 also showed association with SLE in affected siblings (global P = 0.0037, 0.0069, and 0.043, respectively; transmit 2.5.4 software) but not in unaffected siblings. The percent transmission was not significantly altered in haplotypes containing 1174C with other TLR5 SNPs when compared with analyzing allele 1174C alone. Together, these results suggest a strong association of the TLR5 1174 locus with susceptibility to lupus.
Table 1. Transmission disequilibrium test of TLR5 alleles and haplotypes in Caucasian SLE cohort.
| Affected offspring
|
Unaffected offspring
|
||||||
|---|---|---|---|---|---|---|---|
| TLR5 allele(s) | Allele(s) | T/NT | T, % | P value | T/NT | T, % | P value |
| Allele | |||||||
| C1174T | C | 19:6 | 76.0 | 0.009 | 3:6 | 33.3 | NS |
| A1775G | A | 37:26 | 58.7 | 0.166 | 11:11 | 50.0 | NS |
| T1846C | T | 61:57 | 51.7 | 0.713 | 20:22 | 47.6 | NS |
| A2523G | A | 34:26 | 56.7 | 0.302 | 7:9 | 43.8 | NS |
| Haplotype | |||||||
| 1-2 | CA | 45:22 | 67.2 | 0.005 | 11:14 | 44.0 | NS |
| TA | 4:17 | 19.0 | 0.005 | 5:3 | 62.5 | NS | |
| 1-2-3 | CAC | 48:29 | 62.3 | 0.030 | 15:14 | 51.7 | NS |
| TAC | 3:14 | 17.6 | 0.008 | 4:1 | 80.0 | NS | |
| 1-2-3-4 | CACA | 48:29 | 62.3 | 0.030 | 15:14 | 51.7 | NS |
| TACA | 3:14 | 17.6 | 0.008 | 4:1 | 80.0 | NS | |
Data presented are derived from TLR5 alleles transmitted and not transmitted from heterozygous parents to offspring in 199 Caucasian families. Data represent families that had complete genotyping data available for both parents and the affected or unaffected offspring. Data are shown as the percentage transmission (T) of each allele. The P value for each allele is the level of significance based on the deviation from the expected random (50%) allele transmission and was evaluated by using a χ2 statistic. NS, not significant.
We analyzed this association further by comparing allele frequencies of the four TLR5 SNPs in the affected probands with unaffected siblings and parents (Table 2). The allele frequency of 1174T in the probands was 3.0% compared with 5.8% in the total unaffected individuals (5.3% in the unaffected siblings and 5.9% in the unaffected parents) (Table 2). The odds ratio for this comparison was 0.51 (95% confidence interval, 0.26 to 0.98; P = 0.041; prism 3.02 software), consistent with a protective association. The allele frequency of the other three TLR5 SNPs did not show a significant difference between these two groups. Similar to the TDT analysis, these results suggested that the TLR5 stop codon allele (1174T) is associated with protection from SLE.
Table 2. TLR5 polymorphism genotype frequencies in cases and controls.
| No. of polymorphisms (frequency)
|
||||
|---|---|---|---|---|
| Base pair | Amino acid | Affected offspring | Unaffected siblings | Unaffected parents |
| 1174CC | 392RR | 187 (0.940) | 67 (0.893) | 257 (0.889) |
| 1174CT | 392R* | 12 (0.060) | 8 (0.107) | 30 (0.104) |
| 1174TT | 392** | 0 (0) | 0 (0) | 2 (0.007) |
| 1775AA | 592NN | 121 (0.742) | 51 (0.729) | 196 (0.737) |
| 1775AG | 592NS | 39 (0.239) | 18 (0.257) | 61 (0.229) |
| 1775GG | 592SS | 3 (0.018) | 1 (0.014) | 9 (0.034) |
| 1846TT | 616FF | 68 (0.407) | 29 (0.408) | 98 (0.364) |
| 1846TC | 616FL | 70 (0.419) | 33 (0.465) | 130 (0.483) |
| 1846CC | 616LL | 29 (0.174) | 9 (0.127) | 41 (0.152) |
| 2523AA | 841KK | 152 (0.813) | 64 (0.780) | 238 (0.799) |
| 2523AG | 841KK | 32 (0.171) | 18 (0.220) | 58 (0.196) |
| 2523GG | 841KK | 3 (0.016) | 0 (0) | 2 (0.007) |
TLR5 genotypes are presented for cases (SLE affected offspring) and controls (unaffected siblings and parents). Data are from families that had complete genotyping data available for both parents and the affected offspring.
, stop codon
We next considered whether the TLR5 stop codon showed preferential association with particular clinical manifestations of SLE. Congenic analyses in mice have shown that Sle1 is composed of several loci that are associated with a loss of tolerance to chromatin and the production of anti-chromatin antibodies (Sle1a, Sle1b, or Sle1c) (3). These three loci require an additional locus, Sle1d, to increase susceptibility to nephritis. Mapping studies indicate that TLR5 is in the Sle1d region. These findings in the murine system suggest that different SLE clinical phenotypes may be preferentially associated with alleles in the 1q41 region. To explore this possibility, we used a stratified analytic approach with TDT to determine whether transmission of allele 1174 was associated with different SLE phenotypes. Although our sample size was limited for this analysis, the results were intriguing. For most of the phenotypes, there was a similar transmission frequency of allele 1174C for those with the phenotype in comparison with those without it (Table 3). However, for anti-dsDNA antibody, 100% of the seronegative individuals transmitted allele 1174C compared with only 60% of seropositive individuals (an 8:0 seronegative T/NT ratio, P = 0.00087; a 9:6 seropositive T/NT ratio, P = 0.44). In addition, the stop codon heterozygous genotype frequency (1174CT) was lower in the seronegative subjects (3.2%) in comparison with the seropositive subjects (8.5%) (Table 3). Although this latter comparison suggested a trend toward a difference, it was not statistically significant because of the small sample size (P = 0.21). Together, these results suggest that transmission of TLR5 1174C is more strongly associated with SLE in anti-dsDNA seronegative as opposed to seropositive individuals. Titers of autoantibodies to dsDNA are found in many patients with SLE and often correlate with the level of disease activity. The selective association of TLR5392STOP with anti-dsDNA seronegative individuals suggests that the clinical heterogeneity of SLE may be caused by distinct molecular mechanisms.
Table 3. Transmission disequilibrium test and genotype frequency of TLR5 stop codon in SLE clinical subgroups.
| SLE characteristic | T/NT for 1174C | T, % | P value | 1174CC, n | 1174CT; n (%) | P value | |
|---|---|---|---|---|---|---|---|
| Renal | + | 9:2 | 81.8 | 0.035 | 72 | 4 (5.3) | 0.76 |
| – | 8:4 | 66.7 | 0.25 | 96 | 7 (6.8) | ||
| Arthritis | + | 13:5 | 72.2 | 0.059 | 147 | 11 (7.0) | 0.61 |
| – | 4:1 | 80.0 | 0.17 | 19 | 0 (0) | ||
| Hematologic | + | 10:5 | 66.7 | 0.20 | 83 | 8 (8.8) | 0.22 |
| – | 6:1 | 85.7 | 0.047 | 77 | 3 (3.8) | ||
| α-CL Ab | + | 8:2 | 80.0 | 0.058 | 54 | 4 (6.9) | 1.00 |
| – | 6:3 | 66.7 | 0.31 | 66 | 6 (8.3) | ||
| α-dsDNA Ab | + | 9:6 | 60.0 | 0.44 | 97 | 9 (8.5) | 0.21 |
| – | 8:0 | 100.0 | 0.00087 | 61 | 2 (3.2) | ||
| α-Sm Ab | + | 2:2 | 50.0 | 1 | 19 | 2 (9.5) | 0.67 |
| – | 14:4 | 77.8 | 0.019 | 110 | 9 (7.6) |
Transmission (T) rates and genotype frequency of TLR5 SNP C1174T were evaluated in 199 SLE patients with different available clinical features. Rates indicate transmission of 1174C (wild-type allele). For the genotype frequency analysis, only SLE patients who had the indicated clinical data available were included. α-CL, anticardiolipin; Sm, Smith antigen.
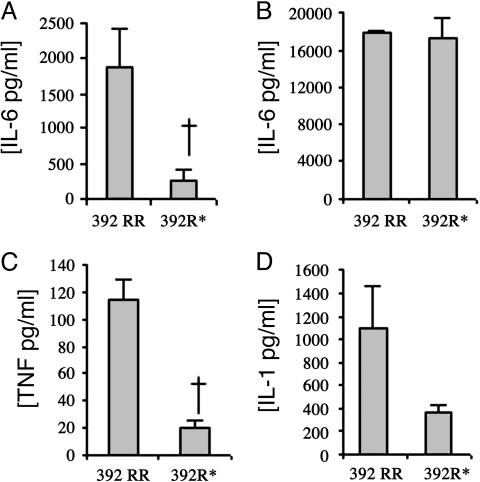
Previous investigators have found dysregulated levels of proinflammatory cytokines in SLE individuals, including IL-6, TNF-α, and IL-1 (18). To better understand the cellular function of the TLR5 stop codon and its ability to mediate inflammation during SLE pathogenesis, we examined whether primary cells from individuals with the stop codon produced similar levels of these proinflammatory cytokines in comparison with subjects with the wild-type TLR5 gene. We have previously shown that IL-6 production is impaired in flagellin-stimulated PBMCs from individuals who are heterozygous for the stop codon (15). To further understand the ability of the TLR5 stop codon to mediate inflammation, we examined additional cytokines. We isolated PBMCs from individuals who were TLR5 wild type homozygotes (base pair 1174CC, amino acids 392RR) or TLR5 stop codon heterozygotes (base pair 1174CT, AA392R*). We stimulated the PBMCs with purified flagellin (125 ng/ml) or lipopolysaccharide (10 ng/ml) for 18 h, collected culture supernatants, and determined IL-6, TNF-α, and IL-1β levels by ELISA. Flagellin-stimulated PBMCs from wild-type individuals (392RR) secreted IL-6, TNF-α, and IL-1β (Fig. 1 A, C, and D). In contrast, PBMCs from individuals with the TLR5 stop codon (392R*) produced less of each of these cytokines in comparison with wild type [392RR vs. 392R* average ± SEM: IL-6, 1,893 ± 528 pg/ml vs. 249 ± 164 pg/ml (P < 0.05); TNF-α, 114 ± 15 pg/ml vs. 20 ± 6 pg/ml (P < 0.05); IL-1β, 1,098 ± 364 pg/ml vs. 358 ± 70 pg/ml (P = 0.08)]. As a control, the PBMCs were stimulated with lipopolysaccharide, and no significant differences were detected between TLR5 wild-type and TLR5 stop codon individuals for each of the cytokines (Fig. 1B and data not shown). Together, these results suggest that individuals who are heterozygous for the TLR5 stop codon have a significantly impaired ability to produce several cytokines in response to flagellin stimulation. This blunted inflammatory response may protect these individuals from developing SLE.
Fig. 1.
Flagellin stimulation of cytokine production in PBMCs. PBMCs were harvested from individuals who were wild type TLR5 homozygotes (392RR, n = 7–8) or stop codon TLR5 heterozygotes (392R*, n = 3–4). Cells were stimulated with flagellin at 125 ng·ml-1 (A, C, and D) or lipopolysaccharide at 10 ng·ml-1 (B) for 18 h, and supernatants were assayed by ELISA for IL-6 (A and B), TNF-α (C), or IL-1β (D). The mean level and SEM of each cytokine were derived by averaging the responses of different individuals' cells stimulated in triplicate. †, P < 0.05 by Student's t test.
Discussion
Linkage of chromosome region 1q41–1q42 region with SLE susceptibility has been found in multiple independent cohorts with different ethnic backgrounds, including European-Americans, African-Americans, and Asians (4–6). In addition, linkage of region 1q44 with SLE has been found in Mexican-American families (19). The causative gene(s) and polymorphism(s) in this region remain(s) unknown. We have found a strong genetic association of a stop codon polymorphism in TLR5 with protection from SLE. Although these results suggest an association of TLR5 with lupus, we cannot exclude the possibility that these SNPs are in linkage disequilibrium with a nearby causative gene. However, the lack of transmission of allele 1174C to unaffected siblings and the lack of an association of the three other TLR5 SNPs with SLE argue against this possibility. Furthermore, because of its central role in regulating inflammatory pathways, the biologic plausibility of TLR5's association with SLE is compelling.
A three-step hypothetical model has been previously proposed to illustrate the role of different genes in SLE pathogenesis (3). Step 1 includes genes that trigger the loss of tolerance to nuclear autoantigens. In step 2, disruption of the immune system occurs, leading to immune dysregulation. Finally, step 3 includes genes that mediate autoimmune destruction of specific organs. Previous studies in congenic mice suggest that the Sle1 region includes at least four different loci and that three of them (Sle1a, Sle1b, and Sle1c) mediate loss of tolerance to nuclear antigens (step 1) (20, 21). In contrast, Sle1d, the fourth locus, which is syntenic with human chromosome 1q41–1q42, is involved in the development of nephritis (step 3). It is hypothetically plausible that TLR5 and flagellin could affect any of the three steps in the SLE pathogenesis model, including step 3, for which murine genetic studies suggest its strongest effect. In addition to stimulating cytokine production and innate effector mechanisms, flagellin promotes maturation of dendritic cells and influences formation of the adaptive immune response (22). In fact, flagellin is a powerful adjuvant that promotes a T helper-2-type T cell response that stimulates antibody production (23, 24). Activation of TLR5 triggers production of proinflammatory cytokines, such as IL-6, which, in turn, can stimulate B cells to proliferate, differentiate, and secrete antibodies. Dysregulation of this process may lead to excessive production of cytokines as well as autoantibodies (18). We previously demonstrated that TLR5392STOP is nonfunctional in reconstitution assays and associated with markedly decreased IL-6 production in response to flagellin stimulation of primary cells from heterozygous individuals (15). In the current study, we have extended these studies and found that production of additional proinflammatory cytokines (TNF-α and IL-1β) is also impaired. TLR5392STOP may provide protection from SLE by decreasing production of proinflammatory cytokines during infection, which may influence formation of the adaptive immune response and production of autoantibodies.
These findings suggest a provocative hypothesis that flagellated bacteria trigger the development of SLE. Flagellated bacteria cause a number of medically important infections from pathogens, such as Escherichia coli, Pseudomonas aeruginosa, Salmonella species, Listeria monocytogenes, Treponema pallidum, and Borrelia burgdorferi. A recent study found that flagellins are immunodominant antigens that may trigger autoimmune intestinal pathology in patients with Crohn's disease (25, 26). We hypothesize that flagellin could similarly stimulate systemic pathology in SLE. Overall, these genetic findings suggest a role for TLR5 in the pathogenesis of SLE and a potential treatment strategy directed at control of flagellated bacterial infections. The high population frequency of TLR5392STOP raises the question of whether there is an evolutionary advantage to having this genetic variant. Our data suggests that TLR5392STOP provides protection against autoimmune disease as one of those evolutionary pressures.
Acknowledgments
We thank all participating patients and their family members and many physicians for referring patients and verifying their diagnoses. We also thank Marta Janer and Sarah Li for genotyping work. This work was supported by grants from the National Institutes of Health, the Southern California Chapter of the Arthritis National Research Foundation, and the Paxson Family Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: SLE, systemic lupus erythematosus; TDT, transmission-disequilibrium test; TLR, Toll-like receptor; T/NT, transmitted/not transmitted; PBMC, peripheral blood mononuclear cell.
Footnotes
Wu, H., Cantor, R. M., Park, E., Rumbin, A. A., Wallace, D. J., Hahn, B. H. & Tsao, B. P. (2004) Arthritis Rheum. 50, S459–S460 (abstr.).
References
- 1.Nath, S. K., Kilpatrick, J. & Harley, J. B. (2004) Curr. Opin. Immunol. 16, 794-800. [DOI] [PubMed] [Google Scholar]
- 2.Tsao, B. P., Cantor, R. M., Kalunian, K. C., Wallace, D. J., Hahn, B. H. & Rotter, J. I. (1998) Proc. Assoc. Am. Physicians 110, 113-117. [PubMed] [Google Scholar]
- 3.Wakeland, E. K., Liu, K., Graham, R. R. & Behrens, T. W. (2001) Immunity 15, 397-408. [DOI] [PubMed] [Google Scholar]
- 4.Graham, R. R., Langefeld, C. D., Gaffney, P. M., Ortmann, W. A., Selby, S. A., Baechler, E. C., Shark, K. B., Ockenden, T. C., Rohlf, K. E. & Moser, K. L. (2001) Arthritis Res. 3, 299-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsao, B. P., Cantor, R. M., Kalunian, K. C., Chen, C. J., Badsha, H., Singh, R., Wallace, D. J., Kitridou, R. C., Chen, S. L. & Shen, N. (1997) J. Clin. Invest. 99, 725-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moser, K. L., Neas, B. R., Salmon, J. E., Yu, H., Gray-McGuire, C., Asundi, N., Bruner, G. R., Fox, J., Kelly, J., Henshall, S., et al. (1998) Proc. Natl. Acad. Sci. USA 95, 14869-14874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsao, B. P., Cantor, R. M., Grossman, J. M., Shen, N., Teophilov, N. T., Wallace, D. J., Arnett, F. C., Hartung, K., Goldstein, R., Kalunian, K. C., et al. (1999) J. Clin. Invest. 103, 1135-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Criswell, L. A., Moser, K. L., Gaffney, P. M., Inda, S., Ortmann, W. A., Lin, D., Chen, J. J., Li, H., Gray-McGuire, C., Neas, B. R., et al. (2000) J. Clin. Invest. 105, 1501-1502. [PubMed] [Google Scholar]
- 9.Hayashi, F., Smith, K. D., Ozinsky, A., Hawn, T. R., Yi, E. C., Goodlett, D. R., Eng, J. K., Akira, S., Underhill, D. M. & Aderem, A. (2001) Nature 410, 1099-1103. [DOI] [PubMed] [Google Scholar]
- 10.Aderem, A. & Ulevitch, R. J. (2000) Nature 406, 782-787. [DOI] [PubMed] [Google Scholar]
- 11.Beutler, B., Hoebe, K., Georgel, P., Tabeta, K. & Du, X. (2004) Microbes Infect. 6, 1374-1381. [DOI] [PubMed] [Google Scholar]
- 12.Iwasaki, A. & Medzhitov, R. (2004) Nat. Immunol. 5, 987-995. [DOI] [PubMed] [Google Scholar]
- 13.Takeda, K., Kaisho, T. & Akira, S. (2003) Annu. Rev. Immunol. 21, 335-376. [DOI] [PubMed] [Google Scholar]
- 14.Leadbetter, E. A., Rifkin, I. R., Hohlbaum, A. M., Beaudette, B. C., Shlomchik, M. J. & Marshak-Rothstein, A. (2002) Nature 416, 603-607. [DOI] [PubMed] [Google Scholar]
- 15.Hawn, T. R., Verbon, A., Lettinga, K. D., Zhao, L. P., Li, S. S., Laws, R. J., Skerrett, S. J., Beutler, B., Schroeder, L., Nachman, A., et al. (2003) J. Exp. Med. 198, 1563-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ibrahim, G. F., Fleet, G. H., Lyons, M. J. & Walker, R. A. (1985) J. Clin. Microbiol. 22, 1040-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Storm, N., Darnhofer-Patel, B., van den Boom, D. & Rodi, C. P. (2003) Methods Mol. Biol. 212, 241-262. [DOI] [PubMed] [Google Scholar]
- 18.Dean, G. S., Tyrrell-Price, J., Crawley, E. & Isenberg, D. A. (2000) Ann. Rheum. Dis. 59, 243-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shai, R., Quismorio, F. P., Jr., Li, L., Kwon, O. J., Morrison, J., Wallace, D. J., Neuwelt, C. M., Brautbar, C., Gauderman, W. J. & Jacob, C. O. (1999) Hum. Mol. Genet. 8, 639-644. [DOI] [PubMed] [Google Scholar]
- 20.Morel, L., Blenman, K. R., Croker, B. P. & Wakeland, E. K. (2001) Proc. Natl. Acad. Sci. USA 98, 1787-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morel, L., Croker, B. P., Blenman, K. R., Mohan, C., Huang, G., Gilkeson, G. & Wakeland, E. K. (2000) Proc. Natl. Acad. Sci. USA 97, 6670-6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Means, T. K., Hayashi, F., Smith, K. D., Aderem, A. & Luster, A. D. (2003) J. Immunol. 170, 5165-5175. [DOI] [PubMed] [Google Scholar]
- 23.Didierlaurent, A., Ferrero, I., Otten, L. A., Dubois, B., Reinhardt, M., Carlsen, H., Blomhoff, R., Akira, S., Kraehenbuhl, J. P. & Sirard, J. C. (2004) J. Immunol. 172, 6922-6930. [DOI] [PubMed] [Google Scholar]
- 24.McSorley, S. J., Ehst, B. D., Yu, Y. & Gewirtz, A. T. (2002) J. Immunol. 169, 3914-3919. [DOI] [PubMed] [Google Scholar]
- 25.Lodes, M. J., Cong, Y., Elson, C. O., Mohamath, R., Landers, C. J., Targan, S. R., Fort, M. & Hershberg, R. M. (2004) J. Clin. Invest. 113, 1296-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sitaraman, S. V., Klapproth, J. M., Moore, I. D., Landers, C., Targan, S., Williams, I. R. & Gewirtz, A. T. (2005) Am. J. Physiol. 288, G403-G406. [DOI] [PubMed] [Google Scholar]