Abstract
Exogenous polypeptides that self-assemble on biological membranes into pores are abundant and structurally diverse, functioning as transporters, toxins, ion channels, and antibiotics. A means for designing novel pore-forming sequences would unlock new opportunities for the development and engineering of protein function in membranes. Toward this goal, we designed a 9,604-member rational combinatorial peptide library based on the structural principles of known membrane-spanning β-sheets. When the library was screened under stringent conditions for sequences with pore-forming activity, a single active motif was found, which is characterized by aromatic residues at the lipid-exposed interfacial positions and basic residues in the pore-lining portion of the sequence. Peptides with this motif assembled on bilayer membranes into β-sheets and formed transient peptide/lipid pores of ≈1-nm diameter. The mechanism of action is very similar to that of natural, pore-forming peptides. These methods provide a powerful means for selecting and engineering novel pore-forming sequences and will open prospects for designing peptide antibiotics, biosensors, and new membrane protein structures.
Keywords: high throughput, antimicrobial, self-assembly
Pore-forming peptides and proteins are found in all kingdoms of life, where they are involved in pathogen virulence and host defense, for example, and also in the action of many toxins and venoms (1, 2). Methods for designing and engineering pore-forming peptides have potentially important biotechnology applications in the fields of antibiotics, biosensors, and drug delivery. However, our understanding of the fundamental principles of self-assembly, insertion, and folding of peptides in membranes is not advanced enough for rational design. In this work, the broadly defined structural principles of natural membrane β-sheets (3–5) served as a framework for the design of a rational combinatorial peptide library, which was screened to find a novel sequence motif that self-assembles into β-sheet pores in membranes.
Many pore-forming polypeptides, such as the vertebrate defensins and the anthrax toxin's PA subunit, use the β-sheet structural motif. Membrane-spanning β-sheets have an amphipathic dyad repeat (6) with hydrophobic residues at every second position. Constitutive β-barrel proteins are constructed of β-hairpins with adjacent 10-residue dyad repeats presenting a continuous 30-Å hydrophobic surface to the bilayer (Fig. 1a). In comparison, the natural pore-forming β-sheet peptides, although also amphipathic, have shorter dyad repeats that are often closely bracketed by basic residues (7) (Fig. 1 b and c). These peptides self-assemble on membranes into transient pores that depend on bilayer distortion and the formation of nonbilayer lipid–peptide complexes (8). We are interested in designing membrane β-sheets of both classes and delineating the structural determinants of each. A β-hairpin with an amphipathic dyad repeat sequence may be sufficient to drive membrane β-sheet formation (9), but these properties also promote nonspecific aggregation, a parallel and nonlinear self-assembly process. Here, we show that rational combinatorial chemistry is an especially powerful tool in this circumstance because libraries that conform to the composition and architecture of the target structure can be used to sample a large number of closely related sequences to find those that favor the target assembly process over alternatives.
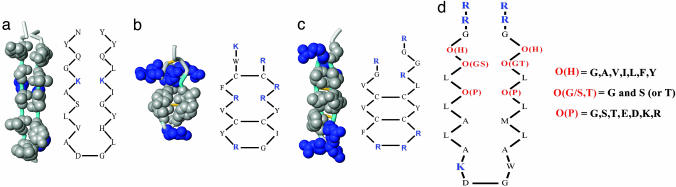
Fig. 1.
Structure of transmembrane β-sheets. (a) Transmembrane β-hairpin from the outer membrane protein phospholipase OMPLA (24), a constitutive β-barrel protein in Escherichia coli. Hydrophobes, shown in gray, show the continuous 30-Å surface created by the dyad repeat pattern, which places alternate residues on the same side of the sheet. (b) The β-hairpin, pore-forming, antimicrobial peptide tachyplesin from the horseshoe crab, Tachypleus tridentatus. (c) The porcine β-hairpin, pore-forming, antimicrobial peptide protegrin. Hydrophobes, shown in gray, dominate the center of the structure, but the hydrophobic surface is smaller than for the hairpins of constitutive β-barrel membrane proteins and is bracketed by basic residues, shown in blue. (d) Sequence of the combinatorial library synthesized for this work. The 26-residue framework sequence has two dyad repeat sequences that are more hydrophobic near the turn sequence to facilitate binding and membrane crossing. The turn sequence, KDGW, is derived from a tight turn with the correct register between two TM β-strands in the β-barrel protein FepA (25). Six combinatorial sites were varied in the library, two hydrophobic sites, O(H), two polar sites, O(P), and two sites, O(G/S,T), that contained Gly and either Ser or Thr. The terminal Arg residues improve solubility and ease of purification, and the Trp residue provides a spectroscopic probe.
Materials and Methods
Peptide Synthesis. Peptides were synthesized with standard fluorenylmethoxycarbonyl chemistry (10) by using a combination of manual and automated synthesis with an Applied Biosystems Pioneer synthesizer. TentaGel NH2 macrobeads of 50–60 mesh pore size (≈0.3-mm diameter dry; 80,000 beads per g) were used for library synthesis. Active amino groups on the resin were first acylated with an acid-stable, photocleavable linker, RT1095 from Advanced ChemTech, followed by peptide synthesis. Combinatorial sites were varied by the split and pool method (11), giving a peptide library in which each bead contains ≈1.5 nmol of a single sequence from the library. Side-chain protecting groups were removed from the peptide with trifluoroacetic acid containing ethanedithiol, thioanisole, and anisole. Peptides synthesized in bulk (Table 1) were purified by reverse-phase HPLC. Microsequencing, HPLC, and mass spectrometry were routinely used to verify peptide sequences.
Table 1. Peptide sequences.
| Peptide | Pore-forming sequence* |
|---|---|
| FSSSTL | RRGFSLSLALAKDGWALMLSLTLGRR |
| FSGRGY† | RRGFSLGLALAKDGWALMLRLGYGRR |
| FSKGGY | RRGFSLKLALAKDGWALMLGLGYGRR |
| YGTKTF | RRGYGLTLALAKDGWALMLKLTFGRR |
| YGKRGY | RRGYGLKLALAKDGWALMLRLGYGRR |
| FSSRGY† | RRGFSLSLALAKDGWALMLRLGYGRR |
| LSRXXX | RRGLSLRLALAKDxxxxxxXxXXxxx |
| YSRRTF | RRGYSLRLALAKDGWALMLRLTFGRR |
| FSRKTY | RRGFSLRLALAKDGWALMLKLTYGRR |
| LSRRGF | RRGLSLRLALAKDGWALMLRLGFGRR |
| YGKRGF† | RRGYGLKLALAKDGWALMLRLGFGRR |
| FSKRGF | RRGFSLKLALAKDGWALMLRLGFGRR |
| FGRRTF | RRGFGLRLALAKDGWALMLRLTFGRR |
| Negative control sequences | |
| AGGKGF†‡ | RRGAGLGLALAKDGWALMLKLGFGRR |
| GGEDGA†§ | RRGGGLELALAKDGWALMLDLGAGRR |
| Composite positive sequences¶ | |
| FSKRGY† | RRGFSLKLALAKDGWALMLRLGYGRR |
| FGKRGY† | RRGFGLKLALAKDGWALMLRLGYGRR |
Sequences of the most potent pore-forming peptides selected form the combinatorial library. Residues in bold/underline are positions that were varied in the library (Fig. 1d). The C-terminal residues of FSRXXX are unknown due to sequencer malfunction
These sequences were synthesized and purified for detailed characterization of pore-forming activity
Negative control sequence from a well that had no visually detectible pore-forming activity at low stringency
Designed negative peptide that has none of the six features of the pore-forming motif. It was used in antimicrobial activity experiments
Composite positive sequences that conform to the pore-forming motif but were not actually observed in the library screen
High-Throughput Screening. For high-throughput screening, 100 mM lipid bilayer vesicles were prepared by extrusion (12) in the presence of 50 mM Tb3+/citrate. The external Tb3+/citrate was removed by gel filtration. Vesicles were then diluted to 200 μM, and 50 μM dipiccolinic acid (DPA) was added to the external medium to make the screening solution. Typical lipid composition was 90% palmitoyloleoylphostatidylcholine and 10% palmitoyloleoylphostatidylglycerol.
In the high-throughput screens, library beads were separated into individual wells of a multiwell plate, and the peptides were cleaved into 10 μl of dry dimethyl sulfoxide for 5 h with UV light. Cleavage was carried out under a N2 atmosphere to prevent the hygroscopic DMSO from accumulating atmospheric water. This treatment releases ≈0.75 nmol (2.5 μg) of peptide from each bead, about half of the total peptide on the bead. To achieve different peptide-to-lipid ratios in the 96-well plate format, aliquots of 1–2.5 μl of the peptide/DMSO solution were added to 50–200 μl of Tb3+/DPA vesicles, and the solution was thoroughly mixed. After 30 min, the plates were photographed under short-wave UV light in a darkroom. Wells with the highest Tb3+/DPA luminescence were selected from the photographs, and the beads from those wells were sent to the Louisiana State University Protein Core Facility (New Orleans) for sequencing by Edman degradation.
Antimicrobial Assay. Staphylococcus aureus FDA 209 was grown to midlog phase and diluted to 103 colony-forming units per ml with liquid test medium (1% growth broth in PBS). In each row of a 96-well plate, 120 μl of cells were incubated with serial dilutions of selected peptides for 3 h at 37°C, followed by the addition of 125 μl of 2× concentrated growth broth. Cultures then were allowed to grow overnight, ≈18 h. Optical density at 600 nm was used to evaluate cell growth, and the lowest concentration of peptide that prevented cell growth was determined. A binary output was observed. For active peptides, wells with more than a minimum sterilizing concentration of peptide had no growth (A600 < 0.04). These wells were effectively sterilized because no bacterial colonies could be cultured from them on plates (<100 colony-forming units per ml). Wells with less than the minimum sterilizing concentration of peptide had stationary phase cultures, A600 > 0.5, and ≈107 colony-forming units per ml. Each test plate had control rows containing no peptide, no cells, or cells plus serial dilutions of ampicillin.
Permeability and Secondary Structure. Membrane permeability assays were performed with the 8-aminonaphthalene-1,3,6-trisulfonate/p-xylenebis(pyridinium)bromide f luorophore quencher pair and with self-quenching fluorescein-dextrans of 3 and 40 kDa molecular mass. In each case, vesicles were prepared as above in a solution of the probe molecule(s). External fluorophores were removed by gel filtration leaving a solution of vesicles with entrapped markers. Leakage of 8-aminonaphthalene-1,3,6-trisulfonate/p-xylenebis(pyridinium)bromide increases of 8-aminonaphthalene-1,3,6-trisulfonate fluorescence by the relief of p-xylenebis(pyridinium)bromide quenching. The fluorescence of the labeled dextrans increases upon leakage due to relief of self-quenching. In all experiments, the intensity for complete leakage was measured after the addition of 0.1% detergent Triton X-100 or by addition of 1 μM fungal pore-forming peptide alamethicin. Secondary structure was determined in a Jasco 810 circular dichroism (CD) spectrometer (Easton, MD) at room temperature.
Results and Discussion
Peptide Library Design. The library sequence used in this work (Fig. 1d) contained a 26-residue framework designed to promote β-sheet formation and disfavor α-helix formation in membranes. The framework sequence has two potential dyad repeat motifs separated by a four-residue turn sequence. There are six combinatorially varied sites (O) in the library sequence, three in each of the two dyad repeats (Fig. 1d). The six combinatorial sites were juxtaposed in the putative hairpin structure, allowing for the potential selection of specific cross-strand interactions. Because of the importance of interfacial interactions in peptide structure and folding in membranes (5), we selected the combinatorial sites to start at the ends of the two strands with a collection of hydrophobic residues. The hydrophobic sites are designated OH. To maintain a dyad repeat pattern, two additional sites, at one and three residues away, were selected to contain only polar or charged amino acids or Gly. The +1 polar sites contained Gly and either Ser or Thr (OG/S,T), the three most abundant amino acids on the inside of membrane-spanning β-barrels (6). These choices provide the potential for structural flexibility (Gly) and for polar interactions (Ser/Thr) in this interfacial polar site. The +3 polar sites are near the center of the putative β-strands and contain a full set of polar, acidic, and basic amino acids (Fig. 1d). These polar sites are designated OP. We assumed that these residues would be the key polar residues that would form the lining of the pore structure. By selectively varying only 6 of 26 residues within a framework designed to form β-sheets in membranes, we created a rational combinatorial library that could be used to explore a narrow region of sequence space for those peptides that favor self-assembly into pore-forming, transmembrane β-sheets over closely related sequences that favor nonspecific aggregation on the membrane or in the aqueous phase.
The 9,604-member peptide β-hairpin library was synthesized as a one-bead, one-peptide library by the “split and pool” method (11). Each 300-μm polystyrene microbead has ≈1.5 nmol, or 5 μg, of a single peptide sequence tethered to it with a photolabile linker. For screening, individual beads were sorted into multiwell plates, peptides were cleaved from the beads with UV light, and aliquots were assayed for pore-forming activity. A binary luminescence reporter system was adapted to create a high-throughput, visual screen for pore-formation in membranes (13). Interaction of the lanthanide metal terbium III (Tb3+) with the aromatic chelator DPA creates a complex that is visibly luminescent at low micromolar concentrations. By entrapping Tb3+ inside membrane vesicles and then adding DPA to the external medium, we created a visual reporter system for membrane pore formation that can detect as little as 5 pmol of pore-forming peptide (13).
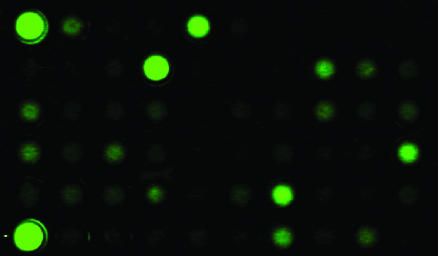
Under low stringency conditions of 1 peptide per 20 lipids (1:20 P:L), >25% of the library sequences caused detectable bilayer permeabilization (Fig. 2). Thus, the pore-forming potential of the framework sequence is substantial. In high-stringency screens, 1:500 P:L, <1% of all sequences produced a visible response. Peptides from positive and negative beads bound to membranes with similar affinity, and there was no difference in the amount of peptide released. Thus, our high-throughput screen is able to select from a library composed of membrane-binding peptides those sequences that readily assemble into membrane-spanning pores.
Fig. 2.
Example of high-throughput screen for pore-forming peptides. Beads with tethered peptides were sorted into a 96-well plate, and peptides were released by UV light as described in the text. Each well contains ≈0.75 nmol of a single peptide sequence along with 15 nmol of lipid in 200 μl of buffer. The lipid vesicles, prepared as described above, contain entrapped Tb3+ and external DPA, which combine to produce visible fluorescence only when the bilayers have been permeabilized. Under these low-stringency conditions (1:20 P:L), a significant proportion of the library sequences caused detectable leakage, indicated by green fluorescence. The high proportion of pore formers validates the library design. At high-stringency conditions (1:500 P:L), only 1% of sequences have detectable pore-forming activity (see text). Upper and lower left wells are positive controls for complete leakage induced by adding the detergent Triton X-100 (upper) or alamethicin, a natural pore-forming peptide (lower). The upper and lower right wells are negative controls containing only buffer or only DMSO with no peptide.
Selections and Characterization of the Pore-Forming Motif. Approximately 10,000 peptides were screened at 1:500 P:L, a sequence-space coverage of 75%. Thirty peptides were found to have detectable pore-forming activity. The most potent of these were identified by direct Edman sequencing from the beads. A consistent motif was observed in the 13 highly active sequences, shown in Table 1. The hydrophobic combinatorial sites (OH) were occupied by aromatic residues in 88% of the sequences (22/25), a highly significant (P < 10-9) overabundance. In membrane proteins and membrane-spanning peptide pores, aromatic residues are abundant at the bilayer water interface and have important roles in folding and structure (3, 14). The abundance of aromatics at the putative interfacial, hydrophobic sites in these peptides supports the idea that they are assembling at the membrane interface into membrane-spanning structures. The polar sites (OP) in the pore-forming library members were found to have 20/25 (80%) basic residues (P < 10-6). No carboxyl-containing side chains were observed (P = 0.0003). The abundance of basic residues present in the transmembrane strands is reminiscent of the natural antibiotic pore-forming peptides (1, 15, 16) (Fig. 1 b and c). The selection of basic residues cannot be explained solely by the electrostatic interactions with the anionic bilayers used for selection because these peptides were also active in zwitterionic membranes. Instead, the ability of the basic residues to create a polar pore while also being able to interact with membranes through their long alkyl chains may drive the their selection. No significant preference for either available residue was seen in the OG/T and OG/S polar sites; however, there was a weak preference for having a single hydroxyl moiety, 19/25 (76%; P = 0.0075), in the adjacent hydrophobic, OH, and OG/S,T sites when taken together. In other words, YG, FS, and FT pairs were statistically overabundant at these potential interfacial sites relative to FG, YS, and YT. Within the framework sequence of this peptide library, the active pore-forming motif has an interfacial aromatic residue, a single interfacial hydroxyl group, and a basic residue on each of the two putative β-strands. Approximately 0.2% of the library sequences have all six of these features. With two exceptions, the pore-forming peptides selected contain either five or six of them.
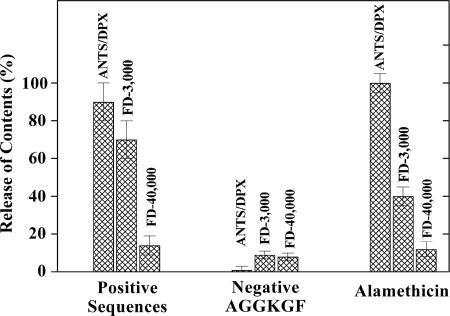
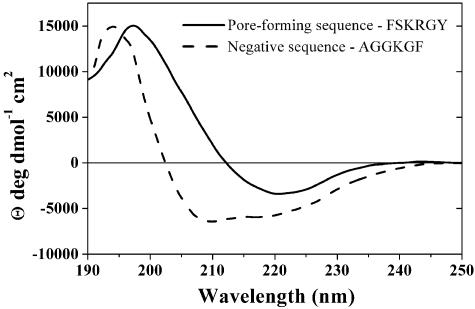
Mechanism of Pore Formation. To determine the mechanism by which the selected peptides permeabilize membranes, we synthesized and purified three peptides selected from the library (Table 1) and two other peptides conforming to the active motif but not actually observed in the screen. We also synthesized a negative control peptide with the residues AGGKGF in the combinatorial sites. AGGKGF was selected from the library for its lack of pore-forming activity under low stringency conditions. We also made a designed negative control peptide, GGEDGA, that contains none of the features of the pore-forming motif. In purified form, the positive sequences caused detectable leakage in Tb3+/DPA vesicles at stringent conditions of 1:500 P:L and also caused leakage when assayed with other fluorescence reporter systems (Fig. 3). The negative peptide AGGKGF did not cause substantial leakage in any system. The extent of membrane permeabilization was very similar for the five positive peptides. In real-time permeability assays, leakage began rapidly after peptide addition and then slowed during the first 10–15 min, ceasing completely before all vesicle contents were released. Additional peptide caused an additional burst of leakage. Requenching analysis (17) showed that partial leakage occurred by a graded mechanism in which all vesicles released some of their contents during the active period rather than an all-or-none process in which some of the vesicles release all of their contents. Taken together, these observations show that membrane assembly and pore formation by these peptides is a transient, burst phenomenon, a mode of action that is very similar to the natural pore-forming antimicrobial peptides (2, 8). The most widely accepted model for transient pore formation by antimicrobial peptides is the “carpet model” (2, 18) in which amphipathic, pore-forming peptides bind to the outer surface of a membrane and self-assemble on that surface to create an unfavorable mass, charge, or surface tension asymmetry across the bilayer. The transbilayer asymmetry is relieved by the formation of membrane-spanning, peptide/lipid pore complexes and subsequent movement of peptides across the membrane. The pores formed by the peptides in Table 1 released molecules as large as 3 kDa but did not substantially release a 40-kDa dextran (Fig. 3) even at 1:50 P:L, indicating a pore size of ≈1-nm diameter. CD spectroscopy was used to show that all of the pore-forming sequences have a variable amount of β-sheet content in solution but assembled into structures with substantially more β-sheet content in the presence of lipid bilayers (Fig. 4). The mean residue ellipticity values for the pore-forming peptides in bilayers are smaller than expected for fully ordered β-sheets, suggesting that the peptides contain a mixture of β-sheet and random coil structure. Interestingly, the negative peptides AGGKGF and GGEDGA contained partial α-helical and random coil structures, respectively, in bilayers (Fig. 4). We speculate that library members that are partially helical do not assemble into pores because they can stably interact with membrane interfaces while exposing their polar residues to the aqueous phase. Thus, the rational design of the combinatorial library leads to preferential selection of β-sheet pore formers.
Fig. 3.
Characterization of peptide pores in large unilamellar vesicles. Vesicles containing entrapped solutes of various molecular masses were made from palmitoyloleoylphostatidylcholine with 10% palmitoyloleoylphostatidylglycerol. Release of 8-aminonaphthalene-1,3,6-trisulfonate/p-xylenebis-(pyridinium)bromide (425 D), and fluorescein-labeled dextrans (3 and 40 kDa) were determined fluorimetrically at 1:50 P:L. Complete (100%) leakage was determined by solubilizing the vesicles with the detergent Triton X-100. The five positive peptides tested (Table 1) caused very similar levels of leakage. Values shown are mean ± SE of all experiments taken together (n = 10–15). The negative peptide, AGGKGF (Table 1), binds well to membranes but causes negligible leakage. The values shown are mean ± SE (n = 3). Alamethicin, a helical pore-forming toxin from Trichoderma viride, is a positive control that forms small discrete pores in membranes.
Fig. 4.
Examples of secondary structure of positive and negative peptides in lipid bilayers. CD spectra of pore-forming peptide FSGRGY (solid line) and negative peptide AGGKGF (dashed line) in the presence of 90% palmitoyloleoylphostatidylcholine/10% palmitoyloleoylphostatidylglycerol lipid bilayer vesicles. Before CD measurements, peptides were dissolved in buffer, then 1 mM lipid vesicles was added, and the sample was allowed to equilibrate. The secondary structure of the pore-forming FSGRGY peptide in the membrane contains a significant amount of β-sheet, consistent with the design principles of the library framework. The non-pore-forming AGGKGF contains some α-helical secondary structure, indicative of partially ordered structure.
Membrane-Spanning Structures. The hypothesis that a hydrophobic dyad repeat motif is sufficient to drive β-hairpin peptides to self-assemble into membrane-spanning pores is supported by the promiscuity of pore formation by library members under low-stringency conditions of 1:20 P:L (Fig. 2). At high-stringency conditions, we selected for those sequence motifs that strongly favor membrane pore formation over other possible structural pathways. The sequence motif selected under these conditions (Table 1) is active in the same concentration range as the natural pore-forming peptides and has a similar mechanism of action in synthetic lipid vesicles.
A similarity of action in vitro between the pore-forming peptides and known antimicrobial peptides (2) prompted us to conduct a test of antimicrobial activity. Pore-forming peptides were assessed for bactericidal activity based on the ability of the peptide to sterilize a solution of 103 cells per ml of S. aureus. In these assays, we used a negative control peptide with the residues GGEDGA in the combinatorial sites (Table 1). This negative peptide is 77% identical (20/26 residues) to the pore-forming sequences because it has the framework sequence but contains none of the six recognizable features of the pore-forming motif, as discussed above. In serial dilution experiments, the minimum sterilizing concentration for the pore-forming peptides was consistently in the low micromolar range (Table 2), very similar to the activity of naturally occurring antimicrobial peptides (Fig. 1) (2, 19). The negative peptide GGEDGA was unable to kill Staphylococcus at concentrations up to 15 μM.
Table 2. Antimicrobial activity.
Peptides that were examined for antimicrobial activity against Staphylococcus aureus. The six residues shown are the amino acids in the combinatorially varied sites within the 26-residue framework sequence shown in Table 1
Minimum sterilizing concentration is the average concentration in a serial dilution experiment at which the peptides sterilized a culture containing 103 S. aureus cells per ml. The numbers listed are the mean minimum sterilizing concentration with SE calculated from three to six separate experiments
A pore-forming peptide that was found during the in vitro high-throughput screen
A “composite positive” peptide (Table 1) designed to contain the six features of the pore-forming motif but not actually observed in the screen
A designed negative peptide that contains none of the six features of the pore-forming motif
An interesting structure–function question that arises from this work is whether the mechanism by which peptides assemble into carpet model pores in membranes is on a pathway toward structured, protein-like β-barrel pores, another highly desirable protein-engineering goal (20). Although both classes share interfacial aromatic residues and the amphipathic dyad repeat motif, peptide–lipid pores probably do not require a specific, hydrogen-bonded structure but require a balance of physicochemical interactions with each other and the membrane lipids (7). This idea is supported by our observation that all peptides tested that conform to the active motif have comparable pore-forming activity. Similarly, other authors have shown that antimicrobial activity of pore-forming peptides can be surprisingly insensitive to alterations in sequence, length, or even secondary structure (21, 22). New libraries need to be screened with framework sequences based on the pore-forming motif identified in this work. Carpet model pores, which require a minimum mass of peptide on the bilayer surface, have not been observed at P:L < 1:500. In contrast, protein-like pores can cause vesicle permeabilization with as few as 8–10 peptides, or one pore complex, per vesicle (23) (1:10,000 P:L). If the peptide–lipid pores achieved in this first library iteration are on a mechanistic path toward a stable, transmembrane β-barrel, then additional iterations of library design and selection at increasing stringencies may yield peptides that assemble into stable transmembrane pores.
Rational combinatorial chemistry and high-throughput screening have been used to design β-sheet peptides that self-assemble into membrane-spanning pores. The potent sequence motif selected from a 9,604-member library assembles on bilayer surfaces at low peptide concentration to create β-sheet pores, which are similar to those formed by naturally occurring antibiotic peptides, and they have similar antimicrobial activity. The pore-forming motif described here will form the basis for the discovery of more potent sequences selected from additional iterations of library design and screening. These methods provide a powerful means for selecting and engineering novel pore-forming sequences and will open new prospects for designing peptide antibiotics and biosensors and engineering membrane protein structures.
Acknowledgments
We thank the New Orleans Protein Folding Intergroup for many invaluable discussions and Samuel H. Gellman, Arthur E. Johnson, and Samuel J. Landry for carefully critiquing this manuscript. This work was supported by National Institutes of Health Grant GM60000 and by the Louisiana Board of Regents Support Fund.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: DPA, dipiccolinic acid; n:m P:L, n peptide per m lipids.
References
- 1.Zasloff, M. (2002) Nature 415, 389-395. [DOI] [PubMed] [Google Scholar]
- 2.Yeaman, M. R. & Yount, N. Y. (2003) Pharmacol. Rev. 55, 27-55. [DOI] [PubMed] [Google Scholar]
- 3.Wimley, W. C. (2003) Curr. Opin. Struct. Biol. 13, 404-411. [DOI] [PubMed] [Google Scholar]
- 4.Wimley, W. C. & White, S. H. (1996) Nat. Struct. Biol. 3, 842-848. [DOI] [PubMed] [Google Scholar]
- 5.White, S. H. & Wimley, W. C. (1999) Annu. Rev. Biophys. Biomol. Struct. 28, 319-365. [DOI] [PubMed] [Google Scholar]
- 6.Wimley, W. C. (2002) Protein Sci. 11, 301-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yount, N. Y. & Yeaman, M. R. (2004) Proc. Natl. Acad. Sci. USA 101, 7363-7368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang, H. W., Chen, F. Y. & Lee, M. T. (2004) Phys. Rev. Lett. 92, 198304. [DOI] [PubMed] [Google Scholar]
- 9.Bishop, C. M., Walkenhorst, W. F. & Wimley, W. C. (2001) J. Mol. Biol. 309, 975-988. [DOI] [PubMed] [Google Scholar]
- 10.Grant, G. A. (1992) Synthetic Peptides: A User's Guide (Freeman, New York).
- 11.Lam, K. S., Lehman, A. L., Song, A., Doan, N., Enstrom, A. M., Maxwell, J. & Liu, R. (2003) Methods Enzymol. 369, 298-322. [DOI] [PubMed] [Google Scholar]
- 12.Mayer, L. D., Hope, M. J. & Cullis, P. R. (1986) Biochim. Biophys. Acta 858, 161-168. [DOI] [PubMed] [Google Scholar]
- 13.Rausch, J. M. & Wimley, W. C. (2001) Anal. Biochem. 293, 258-263. [DOI] [PubMed] [Google Scholar]
- 14.Schiffer, M., Chang, C. H. & Stevens, F. J. (1992) Protein Eng. 5, 213-214. [DOI] [PubMed] [Google Scholar]
- 15.Wang, Z. & Wang, G. (2004) Nucleic Acids Res. 32, D590-D592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brahmachary, M., Krishnan, S. P., Koh, J. L., Khan, A. M., Seah, S. H., Tan, T. W., Brusic, V. & Bajic, V. B. (2004) Nucleic Acids Res. 32, D586-D589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ladokhin, A. S., Wimley, W. C., Hristova, K. & White, S. H. (1997) Methods Enzymol. 278, 474-486. [DOI] [PubMed] [Google Scholar]
- 18.Shai, Y. & Oren, Z. (2001) Peptides 22, 1629-1641. [DOI] [PubMed] [Google Scholar]
- 19.White, S. H., Wimley, W. C. & Selsted, M. E. (1995) Curr. Opin. Struct. Biol. 5, 521-527. [DOI] [PubMed] [Google Scholar]
- 20.Bayley, H. & Jayasinghe, L. (2004) Mol. Membr. Biol. 21, 209-220. [DOI] [PubMed] [Google Scholar]
- 21.Papo, N. & Shai, Y. (2004) Biochemistry 43, 6393-6403. [DOI] [PubMed] [Google Scholar]
- 22.Blazyk, J., Wiegand, R., Klein, J., Hammer, J., Epand, R. M., Epand, R. F., Maloy, W. L. & Kari, U. P. (2001) J. Biol. Chem. 276, 27899-27906. [DOI] [PubMed] [Google Scholar]
- 23.Parente, R. A., Nir, S. & Szoka, F. (1990) Biochemistry 29, 8720-8728. [DOI] [PubMed] [Google Scholar]
- 24.Snijder, H. J., Ubarretxena-Belandia, I., Blaauw, M., Kalk, K. H., Verheij, H. M., Egmond, M. R., Dekker, N. & Dijkstra, B. W. (1999) Nature 401, 717-721. [DOI] [PubMed] [Google Scholar]
- 25.Buchanan, S. K., Smith, B. S., Venkatramani, L., Xia, D., Esser, L., Palnitkar, M., Chakraborty, R., van der Helm, D. & Deisenhofer, J. (1999) Nat. Struct. Biol. 6, 56-63. [DOI] [PubMed] [Google Scholar]