Abstract
Protein conformational diseases, including Alzheimer's, Huntington's, and Parkinson's diseases, result from protein misfolding, giving a distinct fibrillar feature termed amyloid. Recent studies show that only the globular (not fibrillar) conformation of amyloid proteins is sufficient to induce cellular pathophysiology. However, the 3D structural conformations of these globular structures, a key missing link in designing effective prevention and treatment, remain undefined as of yet. By using atomic force microscopy, circular dichroism, gel electrophoresis, and electrophysiological recordings, we show here that an array of amyloid molecules, including amyloid-β(1–40), α-synuclein, ABri, ADan, serum amyloid A, and amylin undergo supramolecular conformational change. In reconstituted membranes, they form morphologically compatible ion-channel-like structures and elicit single ion-channel currents. These ion channels would destabilize cellular ionic homeostasis and hence induce cell pathophysiology and degeneration in amyloid diseases.
Keywords: atomic force microscopy, protein conformational disease, peptide ion channel, misfolding protein, 3D structure
Protein conformational diseases, including neurodegenerative (e.g., Alzheimer's, Huntington's, and Parkinson's diseases, prion encephalopathies, and familial British and Danish dementias), systemic (e.g., type II diabetes, light chain amyloidosis), and other (e.g., cystic fibrosis) diseases result from protein misfolding that alters their 3D conformations from native (often soluble) to nonnative (often insoluble) folded structures (1–4). Understanding such misfolding and the 3D conformations that induce pathophysiology and degeneration is one of the most important and yet challenging areas of research (1). One of the prevailing dogmas about these conformational diseases is that misfolded proteins assume a fibrillar feature termed amyloid that results into a gain-of-function and induce pathophysiological cellular response by altering cell-membrane composition and destabilizing cellular ionic homeostasis. Mechanisms underlying the formation of amyloid (amyloidosis) and its prevention have been studied extensively in the last few decades [for review, see Dobson (2)]. Recent studies, however, indicate that fibrillar aggregates could simply be a storage mechanism and/or even be protective and that only globular (not fibrillar) conformations of amyloid proteins are sufficient to induce cellular degeneration and pathophysiology (5–12).
Studies examining the mechanisms underlying globular peptide-induced cell dysfunction are available (1, 13–15). The deleterious effect of these globular proteins are proposed to be mediated either by means of their membrane poration as the key events followed by nonspecific membrane leakage (15, 16) or, most likely, by specific ionic transport through ion channels (refs. 14 and 17–21; for reviews, see refs 1, 13, and 22) that would destabilize ionic homeostasis. Indeed, amyloid peptides induce ionic conductances in both artificial membranes and in native cell plasma membrane (5, 12, 17–21). However, very little is known about the 3D structures of these globular peptides in the membrane. Lashuel et al. (23) have recently shown “pore-like” annular structure for amyloidogenic protofibrils. However, these protofibrils were never associated with membranes (i.e., neither isolated from membrane complexes nor reconstituted in membranes), and thus whether they form actual membrane pores is still a mystery. By using atomic force microscopy (AFM), circular dichroism (CD) spectrometry, gel electrophoresis, and electrophysiological recordings, we examined the 3D conformation and electrical activity of an array of amyloid molecules, including amyloid-β(1–40) [Aβ(1–40)], α-synuclein, ABri, ADan, serum amyloid A (SAA), and amylin in both native form as well as in reconstituted membranes.
Materials and Methods
1,2-Dioleoyl-sn-glycero-3-phosphocholine (DOPC) was purchased from Avanti Polar Lipids. Human α-synuclein recombinant protein (α-synuclein; molecular mass, 14.5 kDa) and human apo-serum amyloid A (SAA) were purchased from Alpha Diagnostics (San Antonio, TX) and PeproTech (Rocky Hill, NJ), respectively. Aβ(1–40), amylin, ADan, and ABri were synthesized in the W. M. Keck Facility (Yale University) by N-t-butyloxycarbonyl chemistry and purified by reverse-phase HPLC. Hepes was purchased from Sigma, and 16.5% Tris·N-tris(hydroxymethyl)methylglycine (Tricine)·SDS precast gel cassettes, SDS sample buffer, Tris·Tricine·SDS running buffer, and molecular mass standards were purchased from Bio-Rad. All solutions were prepared by using ultrapure water (resistivity > 18.2 MΩ·cm-1) from Milli-Q from Millipore purification system.
CD Spectrometry. Changes in the secondary structure were evaluated by monitoring the peptide species (typically 25–50 μg per 300 μl of 5 mM Tris, pH 7.4) spectrum in the far UV by using a J-720 spectropolarimeter (Jasco, Easton, MD) at 1-nm intervals over the wavelength range 190–260 nm at 24°C in a 0.1-cm path-length cell. Results are expressed in molar ellipticity (deg·cm2·mol-1).
Polyacrylamide Gel Electrophoresis. Freshly dissolved ABri, ADan, SAA, and α-synuclein were electrophoresed on a 16.5% Tris·Tricine polyacrylamide gel under reducing conditions without cross-linking, whereas amylin and Aβ(1–40) were electrophoresed under the same conditions but after covalent cross-linking using glutaraldehyde as described below. Extraction of peptide oligomers reconstituted in DOPC liposomes was performed by freeze-thawing of the lipid–peptide mixture followed by pelleting through centrifugation. The pellet was washed by using 10 mM Hepes solution (pH 7.4) and subsequently resuspended in 10 mM Hepes. The procedure was repeated three times to ensure that no unincorporated peptides were left in the mixture. Afterward, liposomes were dissolved in SDS sample buffer (200 mM Tris·HCl/2% SDS/40% glycerol/0.04% Coomassie blue G-250, pH 6.8). SDS sample buffer was added to peptides freshly dissolved in water. The peptides were separated by electrophoresis on 16.5% Tris·Tricine·SDS polyacrylamide gels (SDS/PAGE). Molecular mass markers (from Bio-Rad) were run parallel to the samples. Peptides were fixed with 10% acetic acid and stained with Coomassie Blue G-250 (Invitrogen) or silver stain (Bio-Rad).
Cross-Linking of Aβ(1–40) and Amylin in DOPC Membrane and in Solution. Without cross-linking, the amount of multimers in the gels for Aβ(1–40) and amylin was very small, most likely because they fall apart when heated up to 90°C before running them through the gels. We cross-linked Aβ(1–40) and amylin oligomers reconstituted in DOPC membranes as described in ref. 5, by using 50 μl of glutaraldehyde, added to 400 μl of DOPC/Aβ(1–40) and DOPC/amylin mixtures, to a final concentration of 12 mM glutaraldehyde. The reaction was stopped after 10 min for amylin and 20 min for Aβ(1–40), respectively, with 100 μl of Tris solution (1 M). Six microliters of glutaraldehyde was added to 24 μl (1 mg/ml) of Aβ(1–40) or amylin solutions in ultrapure water to a final concentration of 12 mM glutaraldehyde, followed by the addition of 20 μl of Tris·SDS/PAGE sample buffer after 10 min for amylin and 20 min for Aβ(1–40), respectively. Cross-linked products were solubilized in 2% SDS solution and analyzed by SDS/PAGE. For comparison, we also cross-linked nonmembrane-associated peptides.
Ion-Channel Current Measurements. Planar phospholipid bilayer membranes were formed as described in ref. 24. A bubble of lipid dissolved in heptane was placed at the end of a small (100–300 μm) Teflon tube. Silver/silver chloride electrodes connected the aqueous components bounding the membrane to a voltage clamp. Ion-channel currents through the membrane were recorded by an Axopatch amplifier (Axon Instruments, Sunnyvale, CA). Data were filtered at 1 kHz and stored on VHS tape. Membrane capacitance and resistance were monitored continuously to ensure the formation and stability of reproducible membranes and the proper membrane thickness. Membranes that showed instability, abnormal capacitance, or abnormal resistance were not used. Control experiments with soluble proteins (e.g., BSA) showed that membranes did not interact with nonamyloid peptides. Peptide samples were introduced by perfusing the aqueous solution bounding one side of the membrane.
Sample Preparation for AFM Imaging. Planar lipid bilayers were prepared by means of liposome fusion followed by rupture on the mica surface by procedure modified from Lin et al. (5). Briefly, DOPC was dissolved in chloroform and dried under a flow of dry argon. DOPC pellet was vacuum-desiccated overnight and subsequently resuspended in 10 mM Hepes (pH 7.4) to a final concentration of 1 mg/ml. Lipids were hydrated for 1 h during which occasional vortexing was applied. Liposomes then were freeze-thawed and passed subsequently through a set of 400- and 200-nm pore size filters. Peptides were dissolved in ultrapure water and mixed with the DOPC liposomes at a 1:20 weight ratio. Lipid–protein mixture was bath-sonicated for 30 sec. Liposomes reconstituted with peptides then were deposited on freshly cleaved mica for 20 min and allowed to fuse and rupture upon contact with the mica surface forming planar lipid bilayers. The sample then was washed, and no additional amyloids were added so that no unincorporated amyloids were left before imaging.
AFM Imaging and Image Analysis. AFM images were acquired by using Nanoscope IIIa Multimode AFM with an Extender electronics module (Veeco, Santa Barbara, CA) as described in ref. 5. Oxide-sharpened silicon nitride cantilevers with a nominal spring constant of ≈0.06 N/m were used for most experiments. Imaging was carried out in both regular contact mode and in tapping mode (at oscillation frequencies between 9 and 15 kHz). Occasionally, higher-frequency resonance peaks (28–33 kHz) were used. The scan rates ranged between 1 and 12 Hz. All imaging was performed in 10 mM Hepes solution (pH 7.4) by using AFM liquid cell at room temperature. Through a continuous adjustment of the scanning parameters, it was ensured that imaging did not affect surface structure by routinely examining for damage by increasing the scan size at regular time intervals.
AFM images were processed and analyzed by using Veeco software. Some AFM images were low-pass filtered. Single ion channels images were passed through an additional low-pass Gaussian filter to reduce pixilation. Sizes of freshly dissolved peptide molecules as well as reconstituted channels in membrane were obtained by cross-sectional and bearing analyses software. The size of the structures observed in the cross-sections of height mode AFM images were measured at two-thirds of full height with respect to the substrate plane (mica surface for freshly dissolved nonmembrane-associated peptides; the bilayer membrane surface for amyloid channels) (5). Sizes and pore statistics for reconstituted channels were obtained from 50–200 channel-like features for each particle in amplitude-mode images. For the bilayers reconstituted with the peptide, often low gains in AFM imaging were required, rendering the amplitude image more reliable for analysis than the height images.
Results
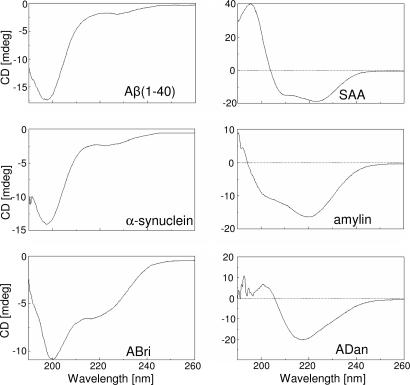
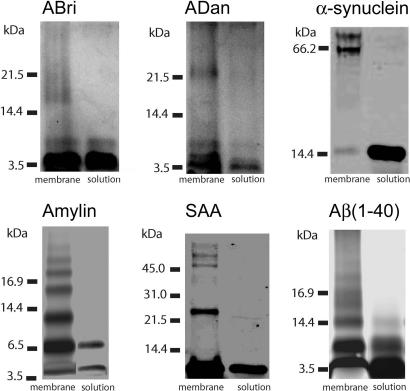
Secondary Structure and Membrane-Induced Oligomerization. The secondary structures of Aβ(1–40), α-synuclein, ABri, ADan, SAA, and amylin were evaluated by CD spectrometry. Various conformations were observed for the different amyloid peptides; Aβ(1–40) and α-synuclein showed predominantly unordered conformations, ADan and amylin were rich in β-structures, ABri was a mixture of β-sheet and random conformations, and SAA was basically α-helix (Fig. 1). The oligomeric nature of soluble globular amyloids before and after their reconstitution in bilayer membrane then was analyzed on SDS/PAGE. Freshly dissolved amyloid peptides appear predominantly monomeric with a strong band corresponding to their respective molecular masses (Fig. 2). Weaker bands corresponding to smaller amounts of dimers and higher-order oligomers are also present (Fig. 2). Conversely, amyloid peptides isolated after their reconstitution in liposomes appear as higher-order (trimers to octamers) oligomers at significantly higher concentration compared with their soluble counterparts (Fig. 1, left bands). The extent of membrane-induced oligomerization varied considerably among various peptides. Whereas amylin and Aβ(1–40) were predominantly trimeric to hexameric, α-synuclein and SAA were tetrameric to octameric, but ADan and ABri were only hexameric and tetrameric, respectively (Fig. 2).
Fig. 1.
CD spectra of mutant amyloid molecules in solution. CD spectrometry analysis of ABri, ADan, α-synuclein, amylin, SAA, and Aβ(1–40) in 5 mM Tris (pH 7.4) was carried out on a Jasco J-720 spectropolarimeter at 1-nm intervals over the wavelength range 190–260 nm at 24°C in a 0.1-cm path-length cell. Results are expressed in molar ellipticity (deg·cm2·mol-1).
Fig. 2.
Electrophoresis of ABri, ADan, α-synuclein, amylin, SAA, and Aβ(1–40) on 16.5% SDS/PAGE under reducing conditions. The right lane shows peptide in aqueous solution; the left lane shows peptide in DOPC membrane after solubilization in 2% SDS. Positions of molecular mass markers are indicated on the left. Amylin and Aβ(1–40) were cross-linked both in solution and in membrane. In solution, for all peptides the monomers are observed. Small amounts of dimers are observed in solution for Aβ(1–40), ABri, and amylin. In the membrane, besides monomers and dimers, trimers are observed for amylin and Aβ(1–40), and tetramers are observed for ABri, amylin, Aβ(1–40), and α-synuclein. Also observed are pentamers for amylin and Aβ(1–40); hexamers for α-synuclein, SAA, ADan, amylin, and Aβ(1–40); and heptamers and octamers for α-synuclein and SAA.
These results indicate that in lipid bilayers, a significantly higher percentage of these amyloids are oligomers (trimers and larger), while a small percentage of monomers and dimers are also present. On the contrary, soluble amyloid peptides are primarily monomers or dimers with a small percentage of higher-order oligomeric complexes. In the lipidic environment, thus, amyloid peptides undergo conformational changes favoring larger oligomeric complexes, although some large oligomeric complexes of soluble peptides can still retain their structure when inserted in a lipidic membrane (5, 23). A presence of large oligomeric complexes in membrane suggests that they could form supramolecular structures.
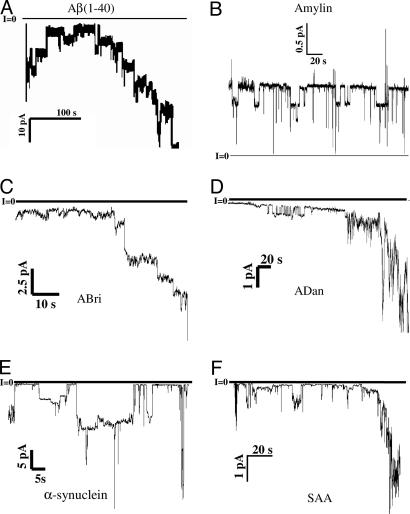
Amyloid Peptides Induce Single Ion-Channel Currents When Reconstituted in Lipid Membrane. We examined the activity of these oligomeric complexes in reconstituted bilayers by using a single-channel electrophysiological recording technique. All six amyloid peptides induced single-channel ion conductances when reconstituted in appropriate lipid bilayers. Fig. 3 shows an example of single-channel currents as a function of time across planar lipid bilayer membranes for each of these six peptides. With the exception of amylin (islet amyloid polypeptide, or IAPP), all amyloids formed channels with heterogeneous single-channel conductances, suggesting that several distinct oligomeric species formed channel structures (5). Single-channel recording could often be seen to merge into macroscopic conductances, implying that the former are responsible for the latter. One-sided addition of amyloid peptide to the solution bounding a bilayer membrane was sufficient to induce channel activity. Multiple sizes of single channels usually could be observed that depended on the peptide aggregation state. Channels were never observed in the absence of added peptide. Channel-forming activity could sometimes vary depending on the aggregation state of the peptide; e.g., disaggregation with DMSO followed by brief reaggregation in water often could enhance channel-forming ability.
Fig. 3.
Single-channel records of amyloid channels. Current traces as a function of time under voltage-clamp conditions are shown. Current jumps corresponding to the opening or closing of individual ion channels can be observed for all six amyloid peptides. Solutions contained 100 mM KCL (except B, which contained 10 mM KCl, and F, which contained 1 M KCl), buffered to pH 7.4. Peptide concentrations were as follows. (A) Aβ(1–40): 21 μg/ml, V = -30mV. (B) Amylin: 3 μg/ml, V =+50 mV. (C) ABri: 50 μg/ml, V =-50 mV. (D) ADan: 100 μg/ml, V =-50 mV. (E) NAC: 15 nM, V =-50mV. (F) SAA: 1 μg/ml, V = -60 mV.
A complete electrical characterization of amyloid channels was not the main focus of the work; rather, the goal was a confirmatory element to support the results that the 3D membrane structures of various amyloids that we report in this work indeed elicit ion-channel conductance and currents. Previous electrophysiological studies of amyloid peptides [Aβ(1–40) (20), amylin (25), SAA (21), and NAC (α-synuclein 60–95)**] have characterized electrophysiological properties in details that include multiple conductances, ion selectivity, and roles of specific agonists, antagonists, and antibodies. In general, results obtained in the present work are consistent with earlier studies of these peptides. Previously unidentified ABri and ADan channel conductances are reported here: they both exhibit heterogeneous single-channel conductances and macroscopic conductance increases strikingly similar to those of other amyloid peptide channels (27).
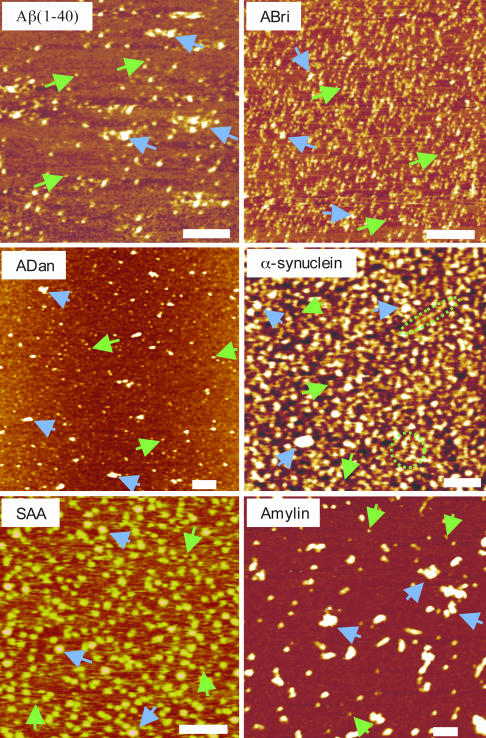
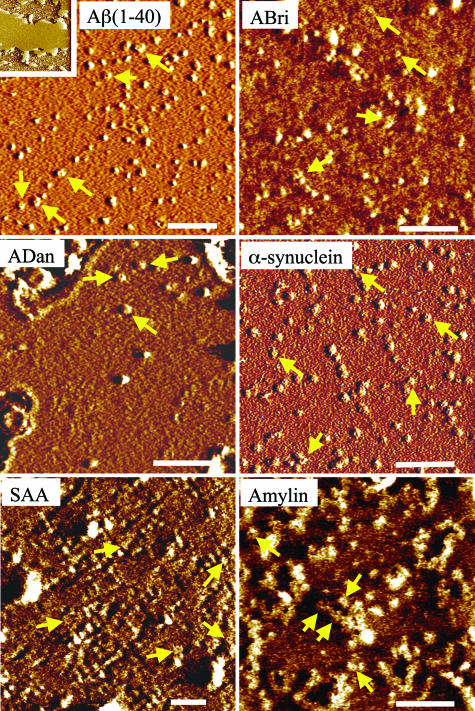
Amyloid Peptides Reconstituted in Bilayer Membrane Form Channel-Like Structures. To understand the structural features of membrane-induced conformational changes, we used AFM to image 3D structures of these amyloids present in both native (soluble, nonmembranous) form and when reconstituted in a lipid bilayer. AFM images of freshly dissolved peptides show globular features with average diameters of 1–10 nm (Fig. 4). Based on their size distributions in the AFM images and their comparison with images of other similar peptides (5–7, 17, 18, 23, 28–31), these globular structures appear to be mostly monomers and dimers, although higher-order oligomeric complexes cannot be ruled out. Significantly, unlike earlier reports indicating amyloids' tendency to form large fibrillar aggregates in solution, by using real-time AFM imaging we confirmed that these peptides retained their globular structure over a period of several (>4) hours with no significant change in the size distributions and without significant aggregation, even at physiologically high concentrations (>1 mg/ml) (data not shown). The complexity of their surface physiochemical properties was reflected in their adsorption to the mica substrate. It was difficult to adsorb soluble ADan and amylin on mica surface, which reflects the presence of very few monomeric or oligomeric complexes and mostly large aggregates in AFM images (Fig. 4). Conversely, ABri, α-synuclein, and SAA were highly attractive to the mica surface, which reflects the presence of both monomers as well as their high-density clusters.
Fig. 4.
AFM images of freshly dissolved peptide molecules. Green arrows indicate surface-adsorbed peptides with a width of 8–12 nm and a height of 1–2 nm (consistent with the size of monomers). Blue arrows indicate higher-order oligomers and clusters. For ABri, most observed features are monomers and dimers; for other peptides, larger multimers and aggregates are observed. For ADan and amylin, the amount of monomers adsorbed is small, and mostly multimers and clusters are observed. In one experiment for α-synuclein, fiber-like structures were observed, indicated by green dotted lines. Peptides were adsorbed on mica for 20–40 min and then washed to remove unadsorbed peptides and imaged in buffer (see Materials and Methods). (Scale bars: 500 nm for ADan and 100 nm for all other peptides.)
We then investigated the possibility that the observed ionic currents (Fig. 3) are indeed due to the pores formed by the globular oligomeric amyloids when they are reconstituted in bilayer membranes. We reasoned that freshly dissolved peptide molecules would adsorb on the liposome surface and, after undergoing partial folding, oligomerize in the lipid-bilayer membrane (5). Alternately, preformed oligomeric complexes with defined pores (23, 28, 29) could insert directly in the membrane. Bilayers obtained from fusion of large liposomes were resolved in ≈75% of all reconstitution experiments. An example of a planar lipid-bilayer membrane reconstituted with Aβ(1–40) and immobilized on the mica surface is shown in Fig. 5 Inset. Consistent with earlier studies (5), the lipid bilayer shows flat patches, irregular in shape in the range of a few square micrometers in size and ≈5- to 5.5-nm thick [the thickness of a single bilayer membrane (5)]. When examined at higher resolution using AFM, the surface of the planar lipid-bilayer patch formed by only lipid vesicles without any amyloid peptides showed no distinguished features (data not shown). The average roughness of these surfaces varied by <0.1 nm.
Fig. 5.
AFM images of amyloid peptides reconstituted in membrane bilayers. Inset shows lipid bilayer with thickness of ≈5 nm. For Aβ(1–40), ADan, and α-synuclein, channel-like structures with a central pore can be easily resolved. For ABri, SAA, and amylin, the central pore is only resolved on some multimer structures. Arrows indicate locations where annular structures can be observed clearly. For those indicated channels, channel sizes (outer diameter) are 16 nm for Aβ(1–40), 14 nm for ABri, 14 nm for ADan, 16 nm for α-synuclein, 12 nm for SAA, and 15 nm for amylin. (Scale bars: 100 nm.)
Surfaces of lipid bilayers show that, once reconstituted in the lipid membranes, predominantly monomeric and dimeric globular peptides appear to coexist with stable higher-order multimers. At medium-resolution imaging (scan size 500–1,000 nm, 512 × 512 pixels), multimeric peptide complexes have disk-like shapes with an outer diameter of 8–12 nm and often contain a central pore-like concavity with a diameter of 1–2 nm (Fig. 5). These structures protrude ≈1 nm above the surrounding flat lipid-bilayer membrane. The presence of channel-like features varied among various amyloids, perhaps reflecting diversity in their interactions with lipids and their eventual stable insertion in the bilayer. On average, 66–75% of all reconstituted bilayers show globular multimeric complexes with the diameter of 10–12 nm, suggesting that they form supramolecular structures. In ≈20% of these structures, a central pore-like feature could be resolved, indicating the formation of channel-like structures. The presence of distinct central pore-like features could be an underestimation due to the AFM tip morphology (blunt tip) or local movement of subunits (due to the imaging in hydrated condition), or it could reflect the low rate of channel formation in reconstituted membrane. In most bilayer samples, larger structures also could be observed on or in the membrane but had no central pore-like feature.
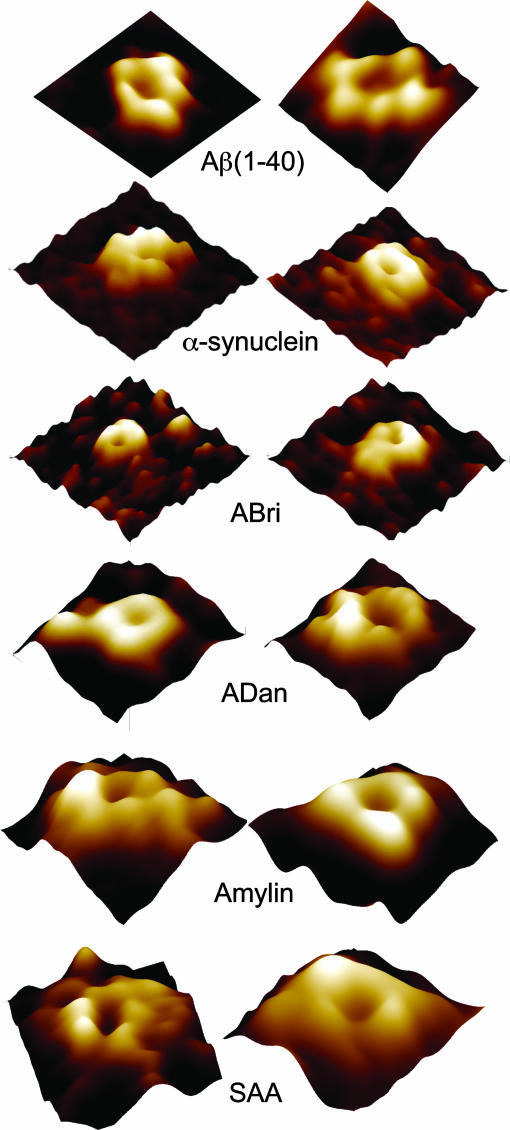
Upon closer examination of individual channel-like structures at higher resolution, several possible subunit arrangements were revealed: rectangular with four subunits, pentagonal with five subunits, hexagonal with six subunits, and octahedral with eight subunits (Fig. 6). Individual subunits' extramembranous protrusion varied by 0.2–0.5 nm. Aβ(1–40) and ADan were mainly four- and six-subunit structures. Up to eight subunits were observed only for SAA and α-synuclein. Pentamers were mainly observed for amylin. For ABri, resolution was only good enough to resolve substructures on few channels. The four-subunit channels, and in some cases the six-subunit channels, show an overall twofold rotational symmetry. It is possible that lower-order oligomers (e.g., dimers, trimers) could form higher-order complexes (tetramers, hexamers, etc.), although we did not ascertain their presence. Occasionally, subunits seem dislocated, breaking the symmetry of arrangement of the subunits. Pore sizes were smallest (≈1 nm) for α-synuclein and ABri and larger (≈2 nm) for Aβ(1–40), ADan, amylin and SAA. The differing multimeric structures and substructures of various amyloid peptides are consistent with data obtained from SDS/PAGE (Fig. 2) and size-exclusion chromatography (data not shown) and are consistent with the model of amyloid–membrane interactions (1, 32, 33), the 3D structure proposed by Durell et al. (34). The subunit variation is also consistent with the multiple electrical conductances observed in this work (Fig. 3) as well as with those described in refs. 5 and 17–21.
Fig. 6.
Individual channel-like structures at high resolution. Two examples are shown for each molecule, in which a central pore can be observed. The number of subunits observed protruding from the surface varies from four to eight subunits. Resolution of AFM images is not enough to resolve individual subunit structures. [Image sizes are 25 nm for Aβ(1–40), 25 nm for α-synuclein, 35 nm for ABri, 20 nm for ADan, 25 nm for amylin, and 20 nm for SAA.]
Discussion
Despite the substantial progress made in understanding the mechanisms underlying the formation of amyloid (amyloidosis) and its prevention, very little can be attributed to amyloidosis as the prime initiator of protein conformational diseases. Our present results show that soluble amyloid subunits, regardless of their initial secondary structure (Fig. 1), assume a supramolecular 3D structure when reconstituted in membrane bilayer (Figs. 4, 5, 6). Conformations of soluble amyloids depend on several factors, including solvents, pH, and metals (Zn, Cu) (32). In our study, we avoided any alcohol derivatives, samples were prevented from going through multiple phase transitions, and AFM imaging was performed in appropriate physiological conditions. Thus, the structures imaged in our study truly represent the supramolecular 3D features of globular amyloidogenic peptides. Globular amyloidogenic peptides have been reported to partition membrane in two phases (16), and large globular complexes, usually termed as ADDLs, have induced nonspecific membrane disruption and leakiness (15). In our earlier studies on calcium uptake in liposomes reconstituted with amyloids, we never observed any nonspecific calcium leakage (17, 18). Membrane-partition studies (16) had used preformed membranes adsorbed to a substrate before addition of peptides. Such supported bilayers have limited ability for refolding/restructuring when incubated with amyloids and are inappropriate for ion channel reconstitutions; to our knowledge, in all published studies of ion-channel reconstitutions, either in artificial membranes (as in the present work) or in vivo in cell plasma membranes, bilayer membranes were accessible to peptides from the both sides.
The supramolecular 3D structure of reconstituted amyloid peptides in our work is similar to an ion channel (5, 23, 28, 29). We see a heterogeneous population of multimeric channels that vary for different amyloid peptides. Structural heterogeneity of amyloid channels [tetrameric to hexameric and higher-order structures (Fig. 6)] is consistent with the higher-order oligomeric transformation of monomeric and dimeric soluble peptides after their membrane insertion and correlates with the nature of the peptides (Fig. 2). This result is further supported by size-exclusion chromatography and spectral analysis of peptides isolated after insertion in the reconstituted membrane (data not shown). Moreover, such heterogeneity conforms to the original varying secondary structure, charge distribution, and tissue and disease specificity of the amyloid peptides that we have examined in this work. Structural and biochemical findings are supported by electrophysiological data that show heterogeneous single-channel conductances for these amyloids and are also consistent with previous studies that ion channels formed by various amyloid lengths exhibit multilevel channel conductances. These multilevel conductances could be due to the multiple conformational changes in the amyloid channel structure or could simply reflect the difference in the number of subunits that form a single channel (19, 20, 34, 35). Channel-forming activity also could vary with the nature of lipid and lipid mixtures (36, 37). A detailed study of such complexity is beyond the scope of this work. Nevertheless, our data show strongly that all these peptides induce ion-channel activity when reconstituted in bilayer membranes.
Amyloid ion-channels would provide the most direct pathway for inducing pathophysiological and degenerative effects when cells encounter amyloidogenic peptides; these channels would mediate specific ion transport (5, 17–21) and thus destabilize the cell ionic homeostasis. A loss of ionic homeostasis would increase the cell calcium to toxic levels, which is the common denominator for the early cellular event leading to pathophysiology and degeneration (5–7, 19, 26). In vivo and in vitro studies have shown that amyloid molecules can form stable small oligomers at physiological concentrations (low nanomolar) as well as up to micromolar levels. The production, oligomerization, and degradation of these amyloids is a dynamic process. Under normal conditions, soluble amyloids are bound to various amyloid-binding proteins and are usually cleared from cerebrospinal fluid into the bloodstream, most likely via receptor transport mechanisms across the blood–brain barrier. In the diseased brain, the level of soluble amyloids is significantly elevated. This elevation could result in an excessive accumulation of amyloid in the cerebrospinal fluid and the formation of calcium-permeable amyloid channels in the cell plasma membrane. Continued accumulation of amyloid channels over an extended time period would eventually increase the disruptive level of cellular free calcium in a dose-dependent manner. With other cellular weaknesses as yet unidentified, toxic calcium level would lead to cellular dysfunction and degeneration. The cellular toxicity data from several recent studies support such a scenario.
In summary, our data provide clear evidence that various amyloid molecules indeed form pore-like structures and elicit channel activity in membrane. Our results provide the structural identity of globular amyloid complexes that would induce pathophysiological cellular activity and degeneration resulting from protein misfolding; amyloid ion channels would allow ionic exchange across the plasma membrane and thus disrupt the cellular ionic homeostasis. Overwhelming electrophysiological evidence suggests that such ionic exchange ultimately leads to cellular calcium loading, the common denominator of the amyloidogenic cellular pathophysiology and degeneration.
Acknowledgments
This work was supported by National Institutes of Health Grants AG08721, AG05891, and GM056290, the Alzheimer's Association of America, and Alzheimer's Disease Program of the Department of Health (California) Grant 04-35524.
This work was presented in part at the 48th Annual Meeting of the Biophysical Society, Baltimore, February 14–18, 2004.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Aβ(1–40), amyloid-β(1–40); AFM, atomic force microscopy; DOPC, 1,2-dioleoyl-sn-glycero-3-phosphocholine; SAA, serum amyloid A; Tricine, N-tris(hydroxymethyl)methylglycine.
Footnotes
Kagan, B. L. & Azimova, R. K. (2003) Biophys. J. 84, 53a (abstr.).
References
- 1.Temussi, P. A., Masino, L. & Pastore, A. (2003) EMBO J. 22, 355-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dobson, C. M. (2003) Nature 426, 884-890. [DOI] [PubMed] [Google Scholar]
- 3.Selkoe, D. J. (2003) Nature 426, 900-904. [DOI] [PubMed] [Google Scholar]
- 4.Revesz, T., Ghiso, J., Lashley, T., Plant, G., Rostagno, A., Frangione, B. & Holton, J. L. (2003) J. Neuropathol. Exp. Neurol. 62, 885-898. [DOI] [PubMed] [Google Scholar]
- 5.Lin, H., Bhatia, R. & Lal, R. (2001) FASEB J. 15, 2433-2444. [DOI] [PubMed] [Google Scholar]
- 6.Zhu, Y. J., Lin, H. & Lal, R. (2000) FASEB J. 14, 1244-1254. [DOI] [PubMed] [Google Scholar]
- 7.Bhatia, R., Lin, H. & Lal, R. (2000) FASEB J. 14, 1233-1243. [DOI] [PubMed] [Google Scholar]
- 8.Walsh, D. M., Klyubin, I., Fadeeva, J. V., Cullen, W. K., Anwyl, R., Wolfe, M. S., Rowan, M. J. & Selkoe, D. J. (2002) Nature 416, 535-539. [DOI] [PubMed] [Google Scholar]
- 9.Gibson, G., Gunasekera, N., Lee, M., Lelyveld, V., El-Agnaf, O. M. A., Wright, A. & Austen, B. (2004) J. Neurochem. 88, 281-290. [DOI] [PubMed] [Google Scholar]
- 10.Bucciantini, M., Giannoni, E., Chiti, F., Baroni, F., Formigli, L., Zurdo, J., Taddei, N., Ramponi, G., Dobson, C. & Stefani, M. (2002) Nature 416, 507-511. [DOI] [PubMed] [Google Scholar]
- 11.Koistinaho, M., Ort, M., Cimadevilla, J. M., Vondrous, R., Cordell, B., Koistinaho, J., Bures, J. & Higgins, L. S. (2001) Proc. Natl. Acad. Sci. USA 98, 14675-14680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Etcheberrigaray, R., Ito, E., Kim, C. S. & Alkon, D. L. (1994) Science 264, 276-279. [DOI] [PubMed] [Google Scholar]
- 13.Pollard, H. B., Arispe, N. & Rojas, E. (1995) Cell. Mol. Neurobiol. 15, 513-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kourie, J. I. & Henry, C. L. (2002) Clin. Exp. Pharmacol. Physiol. 29, 741-753. [DOI] [PubMed] [Google Scholar]
- 15.Kayed, R., Sokolov, Y., Edmonds, B., MacIntire, T. M., Milton, S. C., Hall, J. E. & Glabe, C. G. (2004) J. Biol. Chem. 279, 46363-46366. [DOI] [PubMed] [Google Scholar]
- 16.Green, J. D., Kreplak, L., Goldsbury, C., Blatter, X. L., Stolz, M., Cooper, G. S., Seelig, A., Kist-Ler, J. & Aebi, U. (2004) J. Mol. Biol. 342, 877-887. [DOI] [PubMed] [Google Scholar]
- 17.Lin, H., Zhu, Y. W. J. & Lal, R. (1999) Biochemistry 38, 11189-11196. [DOI] [PubMed] [Google Scholar]
- 18.Rhee, S. K., Quist, A. P. & Lal, R. (1998) J. Biol. Chem. 273, 13379-13382. [DOI] [PubMed] [Google Scholar]
- 19.Kawahara, M., Kuroda, Y., Arispe, N. & Rojas, E. (2000) J. Biol. Chem. 275, 14077-14083. [DOI] [PubMed] [Google Scholar]
- 20.Arispe, N., Pollard, H. B. & Rojas, E. (1993) Proc. Natl. Acad. Sci. USA 90, 10573-10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirakura, Y., Carreras, I., Sipe, J. D. & Kagan, B. L. (2002) Amyloid 9, 13-23. [DOI] [PubMed] [Google Scholar]
- 22.Kourie, J. I. & Shorthouse, A. A. (2000) Am. J. Physiol. 278, C1063-C1087. [DOI] [PubMed] [Google Scholar]
- 23.Lashuel, H. A., Hartley, D., Petre, B. M., Walz, T. & Lansbury, P. T. (2002) Nature 418, 291. [DOI] [PubMed] [Google Scholar]
- 24.Mirzabekov, T., Silberstein, A. & Kagan, B. L. (1999) Methods Enzymol. 294, 61-74. [DOI] [PubMed] [Google Scholar]
- 25.Mirzabekov, T. A., Lin, M. C. & Kagan, B. L. (1996) J. Biol. Chem. 271, 1988-1992. [DOI] [PubMed] [Google Scholar]
- 26.Palop, J. J., Jones, B., Kekonius, L., Chin, J., Yu, G. Q., Raber, J., Masliah, E. & Mucke, L. (2003) Proc. Natl. Acad. Sci. USA 100, 9572-9577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kagan, B. L., Azimov, R. & Azimova, R. (2004) J. Membr. Biol. 202, 1-10. [DOI] [PubMed] [Google Scholar]
- 28.Lashuel, H. A., Hartley, D. M., Petre, B. M., Wall, J. S., Simon, M. N., Walz, T. & Lansbury, P. T. (2003) J. Mol. Biol. 332, 795-808. [DOI] [PubMed] [Google Scholar]
- 29.Lashuel, H. A., Petre, B. M., Wall, J., Simon, M., Nowak, R. J., Walz, T. & Lansbury, P. T. (2002) J. Mol. Biol. 322, 1089-1102. [DOI] [PubMed] [Google Scholar]
- 30.Srinivasan, R., Jones, E. M., Liu, K., Ghiso, J., Marchant, R. E. & Zagorski, M. G. (2003) J. Mol. Biol. 333, 1003-1023. [DOI] [PubMed] [Google Scholar]
- 31.Ding, T. T., Lee, S. J., Rochet, J. C. & Lansbury, P. T. (2002) Biochemistry 41, 10209-10217. [DOI] [PubMed] [Google Scholar]
- 32.Curtain, C. C., Ali, F. E., Smith, D. G., Bush, A. I., Masters, C. L. & Barnham, K. J. (2003) J. Biol. Chem. 278, 2977-2982. [DOI] [PubMed] [Google Scholar]
- 33.Mobley, D. L., Cox, D. L., Singh, R. R. P., Maddox, M. W. & Longo, M. L. (2004) Biophys. J. 86, 3585-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Durell, S. R., Guy, H. R., Arispe, N., Rojas, E. & Pollard, H. B. (1994) Biophys. J. 67, 2137-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirakura, Y., Lin, M-C., Kagan, B.L. (1999) J. Neurosci. Res. 57, 458-466.10440895 [Google Scholar]
- 36.Arispe, N. & Doh, M. (2002) FASEB J. 16, 1526-1536. [DOI] [PubMed] [Google Scholar]
- 37.Lin, M. & Kagan, B. L. (2002) Peptides 23, 1215-1228. [DOI] [PubMed] [Google Scholar]