Abstract
Glutamine synthetase catalyzes the ligation of glutamate and ammonia to form glutamine, with the resulting hydrolysis of ATP. The enzyme is a central component of bacterial nitrogen metabolism and is a potential drug target. Here, we report a high-yield recombinant expression system for glutamine synthetase of Mycobacterium tuberculosis together with a simple purification. The procedure allowed the structure of a complex with a phosphorylated form of the inhibitor methionine sulfoximine, magnesium, and ADP to be solved by molecular replacement and refined at 2.1-Å resolution. To our knowledge, this study provides the first reported structure for a taut form of the M. tuberculosis enzyme, similar to that observed for the Salmonella enzyme earlier. The phospho compound, generated in situ by an active enzyme, mimics the phosphorylated tetrahedral adduct at the transition state. Some differences in ligand interactions of the protein with both phosphorylated compound and nucleotide are observed compared with earlier structures; a third metal ion also is found. The importance of these differences in the catalytic mechanism is discussed; the results will help guide the search for specific inhibitors of potential therapeutic interest.
Keywords: enzyme catalysis, transition-state analogue, x-ray crystallography
The World Health Organization (www.who.org) estimates that 2 million people die each year of tuberculosis and that one-third of the world's population is infected by the causative pathogen, Mycobacterium tuberculosis. The current “short-term” treatment for tuberculosis lasts a minimum of 4 months. Given the expense and difficulties in successfully completing such cures and the increasing incidence of drug-resistant strains, additional targets for drug design are urgently needed, together with studies that bridge the enormous gaps in our understanding of the organism's biology.
Glutamine synthetase (GS) (EC 6.3.1.2, also known as γ-glutamyl:ammonia ligase) plays a central role in bacterial nitrogen metabolism by catalyzing the condensation of ammonia and glutamate to form glutamine, with the consequent hydrolysis of ATP. The extracellular localization of the GlnA1 enzyme from M. tuberculosis (MtGS) is associated with its involvement in the synthesis of a cell wall poly(l-glutamic acid/glutamine) component that is found only in pathogenic mycobacteria (1, 2). The enzyme is also thought to influence ammonia levels within infected host cells, thereby enabling the pathogen to inhibit phagosome–lysosome fusion and phagosome acidification (2, 3).
MtGS has been classified as essential for optimal growth based on Himar1-based transposon mutagenesis in strain H37Rv (4). Inhibition of the enzyme has a strong impact on growth of the bacterium in in vitro as well as in vivo systems, such as human macrophages and guinea pigs (5–8). The extracellular location means that drugs aimed at GS need not penetrate the formidable mycobacterial cell wall. The closest human analogue has very low similarity (≈23% amino acid sequence identity). The combined observations suggest that MtGS is an attractive subject for drug design. However, inhibitors would need to be tight-binding and selective to avoid unwanted interactions with human enzymes. Human GS is thus a possible source of complications, as are enzymes involved in glutathione synthesis, such as γ-glutamyl: cysteine synthetase (γ-GCS) (9). Structural studies are expected to play a vital role in finding solutions to such problems.
The pioneering work of Eisenberg and coworkers (10–13) has generated a number of GS structures from Salmonella typhimurium (StGS) in complex with relevant amino acids (substrates or inhibitors), metals (Mn2+ replacing the biologically relevant Mg2+ and thallium ions mimicking the ammonium substrate), and nucleotides (AMP or ADP), as well as one with the inhibitor phosphinothricin together with ADP and metals. These represent the “taut” (active) state that exists when multiple metal ions are bound. A more recent structure with bound AMP (14) represents a “relaxed” (inactive) form seen at lower concentrations of metal ions and provides the first GS structure from M. tuberculosis.
l-methionine-S-sulfoximine (MSO) can be phosphorylated by GS to form a transition-state analogue, l-methionine-S-sulfoximine phosphate (MSO-P) (15). Here, we report the structure of the “taut” form of MtGS in complex with MSO-P, ADP, and the physiologically relevant metal Mg2+ refined at a resolution of 2.1 Å. This complex was obtained by incubating the protein with MSO and ATP before crystallization and, as such, represents in situ production of the phosphorylated compound. The highest-resolution GS structure to date and the best-defined ligand complex, it offers several insights into the enzyme's specificity and catalytic activity and a more solid basis for drug design.
Experimental Procedures
Cloning, Protein Expression, and Purification. The gene encoding MtGS (GlnA1; Rv2220) was amplified by PCR from M. tuberculosis DNA strain H37Rv (16) by using PfuTurbo DNA polymerase and the primer pair 5′-GTGACGGAAAAGACGCCCGAC-3′ (forward) and 5′-CCTAAACGTCGTAGTACAGCGCGAATTC-3′ (reverse). In a second amplification, the forward primer 5′-ATGGCTCATCATCATCATCATCATGGTACGGAAAAGACGCCCGAC-3′ and the above-mentioned reverse primer were used to add an N-terminal six-histidine tag to the protein sequence, using the product from the first PCR as a template. The PCR product was gel-purified, incubated with dNTPs and Taq polymerase to introduce 3′ A-overhangs, and cloned into the pCR T7/CT-TOPO vector by using the TOPO Cloning pCR kit (Invitrogen). Recombinant plasmids were isolated from Escherichia coli Top10F′ cells (Invitrogen). For protein production, E. coli BL21-AI strain was transformed with the pCRT7::Rv2220 plasmid. Cells were grown at 37°C in LB medium containing 100 μg/ml ampicillin to an A550 of 0.6, at which time expression was induced with 0.2% (wt/vol) l-arabinose. After 3 h, the cells were harvested by centrifugation, washed with lysis buffer (20 mM Tris·HCl, pH 8.0/500 mM NaCl/5 mM imidazole), and stored at -80°C.
For protein purification, thawed cells were resuspended in lysis buffer supplemented with 5 μg/ml DNase I and 10 μg/ml RNase A and then lysed by a passage through a constant cell disruptor (Constant Systems, Northants, U.K.). Although a large fraction of MtGS was produced as inclusion bodies, high soluble expression was also obtained. The soluble fraction was recovered after high-speed centrifugation and incubated with preequilibrated Ni-nitrilotriacetic acid agarose (Qiagen, Valencia, CA) for 1 h at 4°C; the resin was then washed, and MtGS was eluted with 20 mM Tris·HCl, pH 8.0/500 mM NaCl/250 mM imidazole. The eluted protein was applied to a size-exclusion chromatography column (HiLoad 16/60 Superdex 200, Amersham Pharmacia Biotech) using a buffer consisting of 20 mM Tris·HCl (pH 7.5), 150 mM NaCl, and 2 mM MgCl2. Fractions containing significant amounts of MtGS as judged by SDS/PAGE analysis were pooled and then concentrated to 30 mg/ml (A280, ε = 1.08 ml mg-1·cm-1) by using a Centriprep 30K concentrator (Millipore), flash-frozen in liquid nitrogen, and stored at -80°C.
Crystallization, Data Collection, and Processing. Crystals of MtGS in complex with magnesium, ADP, and MSO-P were obtained by vapor diffusion in hanging drops at room temperature. A mixture of 25 mg/ml enzyme, 10 mM NaATP, and 11 mM MSO (pure S enantiomer from Sigma-Aldrich) was incubated for 30 min at room temperature and then mixed with an equal volume of mother liquor consisting of 30% polyethylene glycol 400, 100 mM Mes (pH 6.8), and 200 mM MgCl2. Well shaped 3D crystals appeared within 48 h and were used for streak-seeding to further improve crystal quality under the same conditions. For x-ray data collection, crystals were directly flash-cooled in liquid nitrogen. Two data sets, one extending to 2.5-Å resolution and the other to 2.1-Å resolution, were collected at 100 K on an ADSC Q4 charge-coupled device detector at beamline ID14–2 at the European Synchrotron Radiation Facility in Grenoble, France. The data were integrated and scaled by using denzo and scalepack (17). Statistics for the higher-resolution data set are shown in Table 1.
Table 1. Statistics for data collection and refinement.
| Statistics | Value |
|---|---|
| Data collection* | |
| Cell axial lengths, Å | 133.3, 229.5, 203.2 |
| Space group | C2221 |
| Resolution range, Å | 20.00–2.10 (2.17–2.10) |
| No. of reflections measured | 568,452 |
| No. of unique reflections | 160,432 |
| Average multiplicity | 3.5 (2.1) |
| Completeness, % | 89.6 (69.1) |
| Rmerge | 0.059 (0.342) |
| I/σI | 13.2 (2.4) |
| Solvent content, % | 50.0 |
| Refinement | |
| Resolution range, Å | 20.00–2.10 |
| No. of reflections used in working set | 152,255 |
| No. of reflections for Rfree calculation | 7612 |
| R value, Rfree, % | 19.2, 23.0 |
| No. of nonhydrogen atoms | 23,469 |
| No. of solvent water molecules | 621 |
| Mean B factor, protein atoms, Å2† | 25.5 |
| Mean B factor, ligand atoms, Å2† | 21.5 |
| Mean B factor, magnesium ions, Å2† | 21.1 |
| Mean B factor, solvent atoms, Å2† | 21.4 |
| Ramachandran plot outliers, %‡ | 1.1 |
| rms deviation from ideal bond length, ŧ | 0.009 |
| rms deviation from ideal bond angle, °§ | 1.2 |
Structure Determination and Refinement. Structure determination and rebuilding were initially carried out using the 2.5-Å resolution data and later were extended to 2.1 Å. The structure was solved by molecular replacement in phaser (18) using the A protein chain of the MtGS/citrate/MnAMP structure [1HTO (14)] as a search model. Six solutions were clearly above background. After inspection in the graphics program o (19), crystallographic symmetry operators could be used to reconstruct one hexameric ring of the asymmetric unit. The model was refined by using rigid body refinement in refmac5 and ncsref (20) as implemented in the ccp4i suite (21, 22), which reduced the R factor to 29%. A total of 100 cycles of automated model building in arp/warp (23) resulted in improved electron density maps and a partially traced model; the model was used to aid rebuilding of one of the subunits in o. Copies of that subunit were subsequently placed at the positions of the other molecules in the hexamer by using the noncrystallographic symmetry (NCS) operators. Initially, the structure was refined with tight NCS restraints, which were later loosened to account for the differences between the subunits. Ligands and water were added only after protein structure was accounted for. Statistics for the final refined model are given in Table 1.
Structural Comparisons and Structure Analysis. Structures were compared by using the programs o (19) and lsqman (24). Figures were prepared with o, molray (25), chemdraw (CambridgeSoft, Cambridge, MA), and canvas (ACD Systems, Saanichton, British Columbia, Canada).
Results and Discussion
Expression. In the first reported study of MtGS expression, the enzyme was purified directly from the highly virulent Erdman strain (2), an extremely unsuitable procedure for most laboratories. Previously reported work with recombinant expression in E. coli required either large culture volumes (14) or refolding from inclusion bodies (26). The approach reported here is much more effective, providing soluble material in good yield (10 mg of highly pure MtGS per liter culture) and a simple purification protocol. It is anticipated that this approach will provide a boost for GS work in many laboratories.
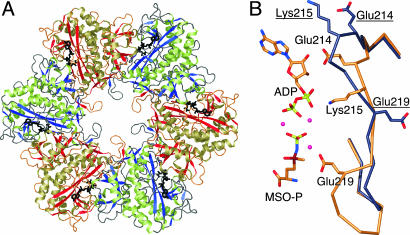
Overall Structure. The structure of MtGS in complex with MSO-P and MgADP was solved by molecular replacement using a subunit from the MtGS/citrate/MnAMP model reported earlier (14) as a probe; it was ultimately refined to 2.1-Å resolution. The final model consists of six subunits that constitute one hexameric ring (Fig. 1A). The biologically relevant dodecamer is formed as the result of a crystallographic twofold axis that places the second hexamer ring behind the first. In the course of model building, the MtGS/MSO-P/MgADP structure was renumbered according to the actual MtGS sequence, instead of by reference to the Salmonella structure as used earlier. In the final refined model, all residues of each subunit are supported by good electron density, beginning with Lys-4 and ending with Val-478 of the native sequence. Pairwise comparisons of the various subunits show them to be very similar, with rms distances in the range of 0.1–0.3 Å.
Fig. 1.
Overall MtGS structure. (A) The subunits of the MtGS hexamer are colored either gold/red or green/blue. Ball-and-stick representations of MSO-P and ADP (black) show the location of the active sites at subunit–subunit interfaces. (B) The β-strand register shift that distinguishes the current taut structure of MtGS (gold carbons) from the previous relaxed one (dark gray carbons) is shown. Residues of the relaxed structure are underlined.
In vivo, GS activity is controlled in a number of ways, including reversible covalent modification. Adenylylation catalyzed by one domain of GlnE reduces GS activity, whereas deadenylylation performed by the other domain of GlnE activates the enzyme (27). The electron density of the relevant side chain in the present MtGS structure (Tyr-406) clearly shows that it is unmodified, allowing it to occupy a position that is buried within the protein, inaccessible to GlnE. Thus, the structure represents an active enzyme. Interestingly, the only significant conformational differences between the six subunits in the asymmetric unit are rearrangements in the adjacent 407–415 loop. We conclude that this portion of the structure has some inherent flexibility, as would be needed if the tyrosine were to be available to modifying enzymes with the required frequency.
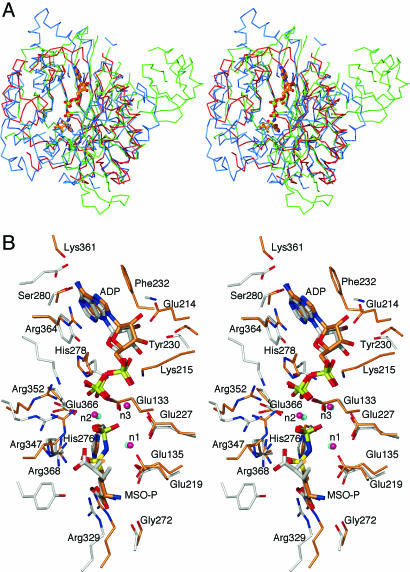
Comparison with Other GS Structures. Several GS structures are compared in Table 2. The A subunit of the MtGS/MSO-P/MgADP structure can be superimposed on that of the MtGS/citrate/MnAMP complex (14) with an rms distance of 1.0 Å. Ninety-five percent of the Cα atoms match within a 3.5-Å cutoff, with only residues 56–58, 65–66, 180–185, 219–223, 294, 335, 359, 406–407, and 460 falling outside. The most striking difference between the two structures is a three-residue register shift of the β-strand starting at Glu-214; thus, the preceding loop is smaller and the following loop is larger than in MtGS/citrate/MnAMP (Fig. 1B). As a result, the observed sequence identity for the matched regions in the structural comparison is <100% (Table 2). Similar results are obtained in other pairwise comparisons of subunits. Because the experimental data for the earlier MtGS structure were deposited in the Protein Data Bank, the relevant map could be obtained from the Uppsala Electron Density server (http://eds.bmc.uu.se/eds) (28). Density for the loops preceding and following the shifted strand is very clear in this map and unambiguously supports the previous researchers' conclusions. However, residues 54–69, 98–99, 332–335, and 406–415 appear to be disordered (using the current numbering scheme). One of these segments (residues 54–69) includes important components of the active site. Another includes tyrosine (Tyr-406), which is not ordered and buried as in the MtGS/MSO-P/MgADP structure; greater exposure of the tyrosine side chain is expected to make it more accessible to GlnE. Alignments of the hexameric units show 96% of all Cα atoms matching with a 1.2-Å rms distance, indicating that there are no major changes in the relative positions of the various subunits in the two structures.
Table 2. Structural comparisons.
| Model | MtGS/MSO-P/MgADP | MtGS/citrate/MnAMP | StGS/phosphinothricin/MnADP | YbdK | γ-GCS/TS analogue/MgADP |
|---|---|---|---|---|---|
| MtGS/MSO-P/MgADP | 475 | 1.0 | 1.1 | 2.0 | 1.9 |
| MtGS/citrate/MnAMP | 452 (99) | 474 | 1.1 | 1.9 | 1.9 |
| StGS/phosphinothricin/MnADP | 462 (50) | 442 (50) | 468 | 2.1 | 1.9 |
| YbdK | 139 (17) | 141 (16) | 140 (14) | 352 | 2.0 |
| γ-GCS/TS analogue/MgADP | 114 (18) | 121 (16) | 114 (19) | 176 (20) | 507 |
Numbers at top right are rms distances obtained by using a cutoff of 3.5 Å in the superposition in lsqman (fast-force algorithm, default settings). At bottom left are the numbers of Cα atoms matching (with percent amino acid sequence identity in parentheses), and those on the diagonal (in boldface) are the number of residues in the chains being compared. The structures compared represent PDB entries 1HTO, 1FPY, 1R8G, and IVA6, respectively; A molecules are used in each case. TS, transition state.
In contrast, comparisons of MtGS/MSO-P/MgADP with the StGS/phosphinothricin/MnADP structure (13) reveal that ≈97% of the residues match with a similar rms distance; in this case, there is no difference in the register of the relevant β-strand, an observation that could be confirmed by inspection of the relevant electron density. The rms distance is in the range expected for the core regions of two proteins related by 50% amino acid sequence identity (29). Thus, our structure represents a taut (active) GS conformation as seen for StGS, rather than the relaxed (inactive) form observed in the MtGS/citrate/MnAMP complex, confirming that the two forms reflect a true structural transition rather than species differences.
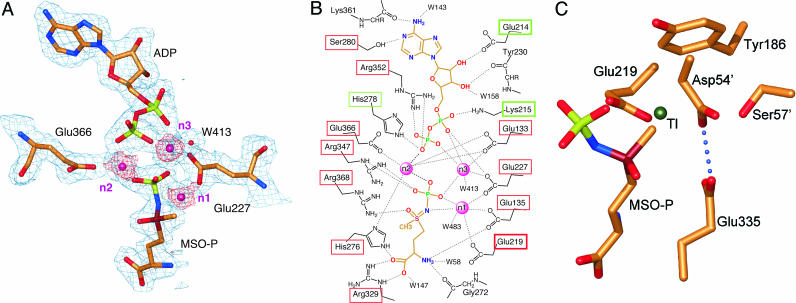
Active Site. The MtGS/MSO-P/MgADP complex has the highest resolution for any GS structure reported to date, and each of the bound ligands is well defined in the electron density maps (Fig. 2A). Fig. 2B outlines the observed polar protein–ligand interactions.
Fig. 2.
Active site. (A) Two of the glutamates of the MtGS active site that interact directly with metal ions (magenta spheres) are shown. The water molecule (W413) interacting directly with n3 is shown as a red sphere. Electron density of ligands, ions, and water is illustrated by using the final sigmaa-weighted [2mFo-dFc (36)] map contoured at 1σ (blue; σ = 0.29 electrons Å-3) and 3σ (magenta). (B) Polar interactions between ligands and protein are indicated by dotted lines. Metals are shown as magenta spheres. Atoms of ADP and MSO-P are colored as in A. Green boxes highlight side-chain interactions of residues not conserved in eukaryotes; red boxes indicate that the residues are conserved in GSs from all organisms; residues of the shifted β-strand are indicated by boxes with thicker lines. (C) The position occupied by thallium in an earlier structure of StGS (30), proposed to represent the site that would normally bind ammonium during the reaction, is shown together with the inhibitor and surrounding residues in the MtGS/MSO-P/MgADP structure. The methyl group of the inhibitor and the thallium are only ≈1 Å apart after superposition as described in Table 2.
In each amino acid-binding site is a molecule of MSO-P, an activated version of the prodrug MSO; the preferred S enantiomer is clearly supported by electron density. As MSO was added together with magnesium and ATP before crystallization, phosphorylation at the sulfoximido nitrogen was carried out in situ by an active enzyme. In the StGS/phosphinothricin/MnADP complex (13), the phospho compound was placed in a very similar way in the active site. However, the methyl group of phosphinothricin was rotated ≈180° away from the orientation we observe for the equivalent atoms of MSO-P. The clear density for the phosphate group indicates that the mode we suggest for MSO-P must be correct; at 2.9-Å resolution, such details of phosphinothricin docking cannot really be established. Furthermore, the polar and nonpolar groups of the inhibitor are now placed in more favorable local environments.
Two manganese ions (n1 and n2) have been reported per subunit in the taut StGS structures. The β-strand shift that distinguishes taut and relaxed forms has been attributed to a difference in occupancy of one of the metal ion sites (14). In the relaxed MtGS/citrate/MnAMP structure, the site assumed by Mg502 here (n1) is empty; the other metal site (Mg503, n2) is half-occupied. The lower metal occupancy in the relaxed MtGS structure was ascribed to the chelating properties of the citrate in the crystallization solutions. In MtGS/MSO-P/MgADP, we see a third ion as well (Mg504, n3). All three of the metals have almost perfect octahedral coordination geometry, with metal–ligand distances appropriate to magnesium (≈2.1 Å). Inspection of the surroundings strongly suggests that this third metal will help counter the high local negative charge in both protein and ligands/substrates. Such charge balancing would be expected to assist in binding of substrates as well as in stabilizing the transition state. Concentrations of metal in previous crystallization experiments (2–3 mM), although physiologically reasonable, may have been too low to allow saturation of all three sites in the crystal (subunit concentration = 1–2 mM). It is not clear whether any differences in the metal sites might be due to the substitution of manganese for the biological magnesium in previous work.
Binding of the nucleotide is different from that reported earlier for both MtGS and StGS with regard to the orientation of the nucleotide base. The plane of the adenosyl ring is similar, although, in the present structure, this group is rotated by 180° around the glycosidic bond. As a result, three previously unobserved hydrogen bonds are seen (Fig. 2B). N6 interacts with 361-O and an active-site water. The third previously unobserved hydrogen bond links N1 and Ser-280-OG, a completely conserved residue for which no function has previously been suggested. These interactions shed light on data concerning GS's nucleotide specificity (2). ATP is preferred, although GTP can also serve as a phosphate donor with somewhat lower specific activity. Both nucleotides would be expected to make a hydrogen bond with Ser-280, although this residue will accept a hydrogen bond from, rather than donate it to, GTP. The lower activity with GTP can be explained by the loss of the interaction with 361-O. A direct interaction of the α-phosphate group of the nucleotide with Lys-215 was also not reported previously. The equivalently placed histidine (because of the β-strand rearrangement) does not interact in MtGS/citrate/MnAMP; this hydrogen bond is also lacking in the Salmonella structures because of an alanine replacement in that sequence. As with the extra metal ion, the positively charged amino group would help compensate for the large local negative charge and so would be expected to assist in ligand binding. The inherent flexibility of this side chain could also allow changes in its conformation as needed during phosphoryl transfer. The ring pucker and interactions made by the ribosyl unit in the MtGS/MSO-P/MgADP complex are essentially identical to those reported earlier.
The ammonium site identified by binding thallium in the Salmonella enzyme (13, 30) is blocked by the methyl group of the inhibitor in the present structure (Fig. 2C). This site lies in close proximity to Glu-335 as well as Asp-54 from the neighboring subunit of GS; we note that these residues share a proton, forming a previously unreported short, tight (2.6-Å) hydrogen bond.
Transitions between the taut and relaxed forms of MtGS have a number of direct and indirect effects on the active site. For example, the ammonium site includes Glu-335 as well as residues 54 and 57 of the adjacent subunit (Fig. 2C), all of which are disordered in the relaxed structure. Tyr-406, which stabilizes the Glu-335 loop in the taut structure, also becomes disordered. The strand shift itself creates a very different environment in the nucleotide, ammonium, and metal-binding sites, both directly (Figs. 1B and 2B) and by means of effects on nearby Tyr-186 (Fig. 2C). Sequence conservation in this strand is good in comparisons of prokaryotic GSs, and so such changes are expected to be relevant for these enzymes; however, the sequence is not well conserved in eukaryotic GSs, which may reflect fundamental differences in both active sites and regulatory properties.
Comparison with Other Proteins. Searches using a subunit of MtGS as the probe in dali (31) indicated that the most similar structures were YbdK [Z score of 12.4 to PDB entry 1R8G (32)] and γ-GCS [Z score of 11.3 to 1V4G (33)], both from E. coli, supporting earlier suggestions of distant relationships among the three proteins (32, 34). In each case, the structure-based sequence identity was very low (11–12%) and the rms distances very high (3.6–3.7 Å) in the equivalent regions (≈240 residues, 50–70% of the structures). More detailed comparisons are summarized in Table 2. The results indicate that YbdK and γ-GCS are more similar to each other than to any of the GS structures; although the rms distances are similar, more residues are matched in the comparison of the YbdK and γ-GCS pair, with a higher sequence identity. Superposition (Fig. 3A) shows that the conserved elements include the highly curved β-sheet that wraps around the active site, together with several long helices that cradle the sheet. One wall of the GS active site is supplied by a neighboring subunit (Fig. 1 A). In the γ-GCS structure, the same function is performed by three noncontiguous segments of the same subunit. Although YbdK is a dimer, the second subunit is placed elsewhere, and nearby loops that could form a lid are disordered, giving this apo enzyme a more open active site than either of the other two enzymes.
Fig. 3.
Comparisons of MtGS with YbdK and γ-GCS. (A) The Cα backbones of the complex form of γ-GCS (blue) and the apo form of the YbdK protein (red) were superimposed on that of the A molecule of MtGS/MSO-P/MgADP (green) by using the superposition described in Table 2. For γ-GCS and MtGS, ADP and phosphorylated inhibitor are shown. (B) The active sites of MtGS (gold carbons) and γ-GCS (steel-gray carbons) are compared. Metal ions of the two structures are shown in magenta and blue, respectively. Interacting residues of MtGS are labeled.
The physiological substrate of YbdK is not yet known, although it has been shown to have low γ-glutamyl:cysteine ligase activity. The sites for nucleotide and amino acid are similar, but changes near the site that binds ammonia in GS (13) suggest that YbdK will not be a γ-glutamyl:ammonia ligase.
γ-GCS is known to be the rate-limiting enzyme in glutathione biosynthesis and important in glutathione homeostasis. This enzyme has been considered as a target for therapeutic agents in the treatment of parasite infections and cancer. The structure of γ-GCS in complex with a phosphorylated transition-state analogue ((2S)-2-amino-4-[(2S)-2-carboxybutyl-(R)-sulfonimidoyl]butanoic acid) was solved at 2.1-Å resolution (33). The striking similarity of the γ-GCS active site to that of MtGS/MSO-P/MgADP is shown in Fig. 3B. The nucleotide base is oriented in the same way as described here for MtGS/MSO-P/MgADP. The interaction of N6 with a main-chain carbonyl oxygen is, however, replaced by one with the side chain of Glu-325, and Ser-154 takes on the role of Ser-280 in its interaction with N1 of ATP. The glutamic acid-binding site is also highly similar. Most interestingly, the arrangement near the β-phosphate group of ADP and the phosphate of the transition-state analogue is essentially identical, including the presence of all three magnesium ions. The placement of the sulfoximido nitrogen is exactly as described above for the MtGS/MSO-P/MgADP structure, suggesting that this orientation is indeed the correct one for the group in this type of enzyme. Most of the residues in the area are conserved, although the roles of Arg-347 and Arg-352 in MtGS will be fulfilled by Arg-304 and Lys-306 of γ-GCS. We conclude that the reaction mechanisms of the two enzymes will be extremely similar, although the ammonia site in GS has been redesigned to accommodate cysteine in γ-GCS.
Implications for GS Mechanism. The first step in GS catalysis is the ordered binding of ATP and glutamate (35). Our results suggest some differences in the binding of the nucleotide base compared with previous reports and give a better understanding of the observed nucleotide specificity. However, these differences are not expected to have direct consequences for the reaction mechanism.
The next step is the formation of the activated intermediate γ-glutamyl phosphate by means of the transfer of the terminal phosphate of ATP to glutamate. Two major observations are in contrast to previous suggestions for GS but in agreement with structural studies of γ-GCS (33). First, the arrangement of the phosphorylated compound shows a different placement of the terminal phosphate group than was proposed earlier (13). Second, three magnesium ions are intimately involved in balancing the overwhelming negative charge of both substrates and neighboring protein. The residues coordinating these ions are completely conserved throughout the known GS sequences (Fig. 2B). It has been proposed that magnesium ion coordination is a common catalytic motif among ATP-dependent ligases, increasing the reactivity of the γ-phosphate group of ATP and stabilizing the γ-glutamyl intermediate (33).
Subsequent binding of ammonium is thought to occur near the side chain of Glu-219 (14); the position is occupied by the methyl group of MSO-P in our present structure (Fig. 2C) and by the cysteine mimic in the γ-GCS complex. Thus, the inhibitors act by blocking the ammonium- and cysteine-binding sites, respectively, which prevents further reaction. Multiple interactions with active-site metals, as well as protein, ensure that ADP and phosphorylated inhibitor remain tightly bound.
The tight hydrogen bond between Glu-335 and Asp-54′ would stabilize subunit–subunit interactions and shield MSO-P from hydrolysis by solvent. Both of these residues are completely conserved and are believed to be involved in the next steps of the reaction, which include deprotonation of ammonium to form ammonia (33, 34). Their interaction is likely to affect their catalytic properties by perturbing the relevant pKas. Ammonia is then in an ideal position for attack on the γ-glutamyl phosphate intermediate; the MtGS/MSO-P/MgADP complex most closely mimics the tetrahedral adduct before the generation of glutamine and inorganic phosphate products. Local conformation changes are believed to be crucial at various stages of the reaction.
The present structure thus sheds light on both substrate binding and catalysis, and the complex with a tight-binding transition-state mimic provides a good starting point for future drug design. Because the nucleotide-binding site is rather different in prokaryotic and eukaryotic GSs (Fig. 2B), an improved understanding of its properties may be particularly useful in the search for inhibitors that bind specifically to the bacterial enzymes.
Acknowledgments
We thank Evalena Andersson and Nina Bäckbro for data collection, Martin Högbom for help with data processing, and Erling Wikman for patiently recompiling ever-larger versions of the programs needed. This work was supported by the Swedish Foundation for Strategic Research and the Swedish Research Council.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: GS, glutamine synthetase; MtGS, GlnA1 enzyme from M. tuberculosis; StGS, GS from S. typhimurium; MSO, l-methionine-S-sulfoximine; MSO-P, l-methionine-S-sulfoximine phosphate; γ-GCS, γ-glutamyl:cysteine synthetase.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2BVC).
References
- 1.Hirschfield, G. R., McNeil, M. & Brennan, P. J. (1990) J. Bacteriol. 172, 1005-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harth, G., Clemens, D. L. & Horwitz, M. A. (1994) Proc. Natl. Acad. Sci. USA 91, 9342-9346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gordon, A. H., Hart, P. D. & Young, M. R. (1980) Nature 286, 79-80. [DOI] [PubMed] [Google Scholar]
- 4.Sassetti, C. M., Boyd, D. H. & Rubin, E. J. (2003) Mol. Microbiol. 48, 77-84. [DOI] [PubMed] [Google Scholar]
- 5.Tullius, M. V., Harth, G. & Horwitz, M. A. (2003) Infect. Immun. 71, 3927-3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harth, G. & Horwitz, M. A. (1999) J. Exp. Med. 189, 1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harth, G., Zamecnik, P. C., Tang, J. Y., Tabatadze, D. & Horwitz, M. A. (2000) Proc. Natl. Acad. Sci. USA 97, 418-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harth, G. & Horwitz, M. A. (2003) Infect. Immun. 71, 456-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richman, P. G., Orlowski, M. & Meister, A. (1973) J. Biol. Chem. 248, 6684-6690. [PubMed] [Google Scholar]
- 10.Yamashita, M. M., Almassy, R. J., Janson, C. A., Cascio, D. & Eisenberg, D. (1989) J. Biol. Chem. 264, 17681-17690. [DOI] [PubMed] [Google Scholar]
- 11.Liaw, S. H., Pan, C. & Eisenberg, D. (1993) Proc. Natl. Acad. Sci. USA 90, 4996-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liaw, S. H., Jun, G. & Eisenberg, D. (1994) Biochemistry 33, 11184-11188. [DOI] [PubMed] [Google Scholar]
- 13.Gill, H. S. & Eisenberg, D. (2001) Biochemistry 40, 1903-1912. [DOI] [PubMed] [Google Scholar]
- 14.Gill, H. S., Pfluegl, G. M. & Eisenberg, D. (2002) Biochemistry 41, 9863-9872. [DOI] [PubMed] [Google Scholar]
- 15.Ronzio, R. A. & Meister, A. (1968) Proc. Natl. Acad. Sci. USA 59, 164-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cole, S. T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., Gordon, S. V., Eiglmeier, K., Gas, S., Barry, C. E., III, et al. (1998) Nature 393, 537-544. [DOI] [PubMed] [Google Scholar]
- 17.Otwinowski, Z. (1993) in Data Collection and Processing, eds. Sawyer, L., Issacs, N. & Bailey, S. (Science and Engineering Research Council Daresbury Laboratory, Daresbury, U.K.), pp. 56-62.
- 18.Storoni, L. C., McCoy, A. J. & Read, R. J. (2004) Acta Crystallogr. D 60, 432-438. [DOI] [PubMed] [Google Scholar]
- 19.Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 20.Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240-255. [DOI] [PubMed] [Google Scholar]
- 21.Collaborative Computing Project No. 4 (1994) Acta Crystallogr. D 50, 760-763.15299374 [Google Scholar]
- 22.Potterton, E., Briggs, P., Turkenburg, M. & Dodson, E. (2003) Acta Crystallogr. D 59, 1131-1137. [DOI] [PubMed] [Google Scholar]
- 23.Perrakis, A., Sixma, T. K., Wilson, K. S. & Lamzin, V. S. (1997) Acta Crystallogr. D 53, 448-455. [DOI] [PubMed] [Google Scholar]
- 24.Kleywegt, G. J. & Jones, T. A. (1997) Methods Enzymol. 277, 525-545. [DOI] [PubMed] [Google Scholar]
- 25.Harris, M. & Jones, T. A. (2001) Acta Crystallogr. D 57, 1201-1203. [DOI] [PubMed] [Google Scholar]
- 26.Singh, J., Joshi, M. C. & Bhatnagar, R. (2004) Biochem. Biophys. Res. Commun. 317, 634-638. [DOI] [PubMed] [Google Scholar]
- 27.Jaggi, R., van Heeswijk, W. C., Westerhoff, H. V., Ollis, D. L. & Vasudevan, S. G. (1997) EMBO J. 16, 5562-5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kleywegt, G. J., Harris, M. R., Zou, J. Y., Taylor, T. C., Wahlby, A. & Jones, T. A. (2004) Acta Crystallogr. D 60, 2240-2249. [DOI] [PubMed] [Google Scholar]
- 29.Gan, H. H., Tropsha, A. & Schlick, T. (2001) Proteins 43, 161-174. [DOI] [PubMed] [Google Scholar]
- 30.Liaw, S. H., Kuo, I. & Eisenberg, D. (1995) Protein Sci. 4, 2358-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holm, L. & Sander, C. (1993) J. Mol. Biol. 233, 123-138. [DOI] [PubMed] [Google Scholar]
- 32.Lehmann, C., Doseeva, V., Pullalarevu, S., Krajewski, W., Howard, A. & Herzberg, O. (2004) Proteins 56, 376-383. [DOI] [PubMed] [Google Scholar]
- 33.Hibi, T., Nii, H., Nakatsu, T., Kimura, A., Kato, H., Hiratake, J. & Oda, J. (2004) Proc. Natl. Acad. Sci. USA 101, 15052-15057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abbott, J. J., Pei, J., Ford, J. L., Qi, Y., Grishin, V. N., Pitcher, L. A., Phillips, M. A. & Grishin, N. V. (2001) J. Biol. Chem. 276, 42099-42107. [DOI] [PubMed] [Google Scholar]
- 35.Eisenberg, D., Gill, H. S., Pfluegl, G. M. & Rotstein, S. H. (2000) Biochim. Biophys. Acta 1477, 122-145. [DOI] [PubMed] [Google Scholar]
- 36.Read, R. J. (1986) Acta Crystallogr. A 42, 140-149. [Google Scholar]
- 37.Kleywegt, G. J. (1997) J. Mol. Biol. 273, 371-376. [DOI] [PubMed] [Google Scholar]
- 38.Kleywegt, G. J. & Jones, T. A. (1996) Structure 4, 1395-1400. [DOI] [PubMed] [Google Scholar]
- 39.Engh, R. A. & Huber, R. (1991) Acta Crystallogr. A 47, 392-400. [Google Scholar]