Abstract
Previous studies of the pericentromeric region of the human X chromosome short arm (Xp) revealed an age gradient from ancient DNA that contains expressed genes to recent human-specific DNA at the functional centromere. We analyzed the finished sequence of this human genomic region to investigate its evolutionary history. Phylogenetic analysis of >1,500 alpha-satellite monomers from the region revealed the presence of five physical domains, each containing monomers from a distinct phylogenetic clade. The most distal domain contains long interspersed nucleotide element repeats that were active >35 million years ago, whereas the four proximal domains contain more recently active long interspersed nucleotide element repeats. An out-of-register, unequal recombination (i.e., crossover) detected at the edge of the X chromosome-specific alpha-satellite array (DXZ1) may reflect the most recent of a series of punctuating events during evolution that resulted in a proximal physical expansion of the X centromere. The first 18 kb of this array has 97–99% pairwise identity among all 2-kb repeat units. To perform more detailed evolutionary comparisons, we sequenced the junction between the ancient DNA of Xp and the primate-specific alpha satellite in chimpanzee, gorilla, orangutan, vervet, macaque, and baboon. The striking conservation found in all cases supports the ancestral nature of the alpha satellite at this location. These studies demonstrate that the primate X centromere appears to have evolved through repeated expansion events occurring within the central, active region of centromeric DNA, with the newly added sequences then conferring centromere function.
Keywords: evolution, alpha satellite, comparative genomics, genome sequencing
The centromeric regions of eukaryotic chromosomes represent an enigmatic example of conserved function in the face of rapidly evolving DNA sequences (1). The centromere is required for the proper segregation of chromosomes at mitosis and meiosis. This function is mediated by the kinetochore, which is a proteinaceous structure that assembles onto centromeric DNA (2, 3). Whereas the proteins of the kinetochore have largely been conserved throughout evolution (4), the DNA at the site of kinetochore formation (i.e., the centromere) has not (1, 5). Every species that has been studied appears to employ a different DNA sequence for attaining centromere function, with the precise role of that DNA being the subject of ongoing debate (6–9).
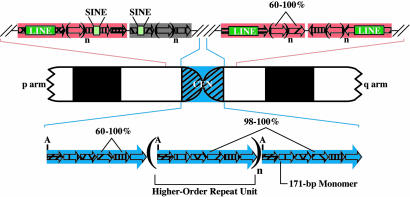
Based on a large body of evidence, a general model for the organization and content of human centromeric regions has emerged (Fig. 1). There is a large, chromosome-specific alpha-satellite array at the centromere of all human chromosomes (10, 11). Alpha-satellite DNA consists of 171-bp, AT-rich monomers arranged in a tandem, head-to-tail configuration (12, 13). A small group of these monomers comprises the higher-order repeat units that are then arranged in a tandem, head-to-tail configuration to form an array (10). Monomers within a higher-order repeat unit have average pairwise sequence identities of ≈72%; however, adjacent higher-order repeat units are typically 98–100% identical (14). Higher-order arrays can extend for 3–5 Mb (11, 15, 16) and are largely devoid of nonsatellite sequences (10, 17).
Fig. 1.
Structural organization of human centromeric and pericentromeric regions. A typical human chromosome is shown schematically, emphasizing the pericentromeric and centromeric (CEN; blue) regions above and below the ideogram, respectively. Each small arrow represents a single satellite monomer of n bp, with n being characteristic of a given satellite family (e.g., 171 for alpha satellite, such as that in the centromeric region; see Table 1, which is published as supporting information on the PNAS web site). In the pericentromeric regions, blocks of tandem satellite monomers from a single family (indicated by pink versus gray) occasionally contain embedded interspersed repetitive elements (e.g., LINEs and short interspersed nucleotide elements; not drawn to scale). Adjacent satellite blocks can exist in the same or opposite orientations. In the centromeric region, higher-order repeat units, which contain a characteristic number of monomers, are indicated with large blue arrows. The regular presence of a common restriction site (A) illustrates the periodic nature of centromeric DNA.
Global analyses of pericentromeric regions in the human genome sequence revealed a complex patchwork of intrachromosomal and interchromosomal duplication events (18). A major limitation of such analyses relates to the clone maps that were used for sequencing the human genome (19). Specifically, large-insert clones that join duplicon-rich pericentromeric regions with chromosome-specific alpha-satellite arrays are rare, and none provide continuity across a given array. As a result, the sequence assemblies of many human chromosome arms halt without ever reaching pericentromeric satellite DNA; only 11 of 43 such assemblies (excluding acrocentric short arms) contain higher-order alpha-satellite DNA (14), and no centromeric region has been sequenced in its entirety.
Regions flanking the chromosome-specific arrays contain a blend of different satellite families (both alpha satellite and other family types), often interrupted by transposable elements (20). The satellite DNA in these pericentromeric regions is in a monomeric form (21). Monomers within a given satellite family share average pairwise sequence identities of ≈72% and lie in tandem, head-to-tail configurations, but they generally lack higher-order structure (14). Among satellite families, alpha satellite is unique in several important ways. It has been detected in all primate species that have been studied, and it appears to be unique to the primate lineage (22). Also, it is the only satellite found at all human centromeres (10, 12), and it is the only one that is known to exist in two forms in the human genome (higher order at the centromere and monomeric in pericentromeric regions).
Functional studies of human chromosomes indicate that the kinetochore complex assembles within higher-order alpha-satellite arrays (23). Artificial chromosome assays have demonstrated de novo centromere formation with higher-order alpha satellite (24–27). Mapping of deletion chromosomes has delimited the functional centromere region of the human X chromosome to the higher-order alpha-satellite array, DXZ1 (26). Topoisomerase II cleavage activity is found at the active centromere but not the inactive centromere of dicentric chromosomes (28) and has been localized within higher-order arrays of the X (29) and Y (30) chromosome. Chromatin immunoprecipitation (31) and fiber-FISH (32) studies have found higher-order alpha-satellite sequences associated with essential kinetochore proteins.
These functional studies and previous genomic analyses of the pericentromeric region of the human X chromosome have yielded several important findings (26) that make this chromosome a model for centromere investigations. First, the presence of higher-order DXZ1 sequences, especially near the X chromosome short arm (Xp) (29), appeared to confer centromere function. Second, initial examination of embedded long interspersed nucleotide element (LINE) repeats provided preliminary insight about the recent evolution of the functional centromere sequences and the longer-term evolution of the monomeric alpha satellite immediately adjacent to the chromosome arm. Importantly, these data provided a valuable framework for comparative studies aiming to unravel the evolutionary history of centromere structure and function. Here, we present an analysis of the complete sequence of the pericentromeric transition from human Xp into the higher-order DXZ1 alpha-satellite array. Our results reveal important details about the history of the Xp centromere, and these findings are corroborated by comparative sequencing studies involving the Xp pericentromeric region of several primate species.
Materials and Methods
Sequence Analysis. We analyzed the most proximal 650 kb of the human Xp sequence (GenBank accession no. NT_011630), specifically, chrX:56,900,000–57,548,803 of NCBI Build 34 [July 2003; University of California, Santa Cruz (UCSC) Genome Browser, available at: http://genome.ucsc.edu] (Fig. 2). The content and organization of repetitive sequences were defined by using repeatmasker (59). Further analyses of repetitive sequences were performed by using perl scripts (developed by Jeff Bailey and John Dunn; available in Data Set 1, which is published as supporting information on the PNAS web site) that parsed alpha-satellite, HSAT4, and gamma-satellite monomers. Each script defines and extracts a monomer by searching for matches to beginning- and end-consensus sequences of each monomer family, requiring paired matches separated by the prototypic length of that monomer (see Table 1). The resulting list of monomers includes an indication of orientation, from which the overall directionality of each monomer block can be determined. A separate perl script (developed by John Dunn; available in Data Set 1) was used to search the alpha-satellite monomers for the presence of divergent centromere protein B (CENP-B) binding sites. The higher-order structure of the alpha satellite within the DXZ1 array was evaluated by using dotter (33).
Fig. 2.
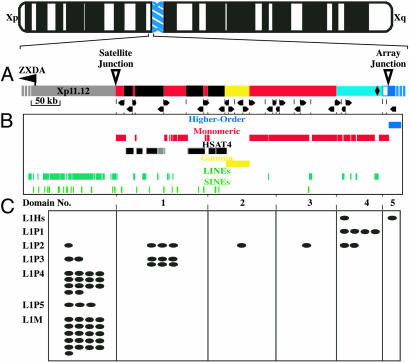
Sequence-based map of the pericentromeric region of human Xp. (A) The most proximal ≈650 kb of human Xp is depicted. The light gray bar at the left represents ≈150 kb of arm sequences, including the nearest expressed gene (ZXDA). The following blocks of satellite sequence are shown on the right: monomeric alpha satellite (red), HSAT4 (dark gray), and gamma satellite (yellow). The start of the X chromosome-specific higher-order alpha satellite (DXZ1) is shown in blue, with the light blue area reflecting monomers that lack higher-order structure but fall into the phylogenetic clade with DXZ1 monomers (see Fig. 3). The black diamond indicates the position of a single 880-bp block of nonsatellite sequence. Dotted lines indicate junctions between blocks of different satellite families (i.e., HSAT4 and alpha satellite) or junctions between blocks of the same family lying in opposite orientations. The arrowheads indicate the directionality of each satellite block. (B) Repetitive sequences are shown by using custom tracks from the UCSC Genome Browser (higher-order alpha satellite, blue; monomeric alpha satellite, red; HSAT4, dark gray; gamma satellite, yellow; LINEs, dark green; short interspersed nucleotide elements, light green). (C) The physical spans of the five satellite domains classified by phylogenetic analyses of alpha-satellite monomers (see Fig. 3) are indicated. Each L1 (LINE) repeat observed within proximal Xp and the alpha-satellite blocks is indicated as an oval, classified according to its subfamily [the currently active, human-specific elements (L1Hs) are shown at the top, with progressively older subfamilies listed below]. L1P subfamilies are primate-specific, whereas the L1M subfamily is quite large and composed of elements found throughout the mammalian radiation.
Phylogenetic Analysis. Alpha-satellite monomers were aligned by using clustalw (available at www.ebi.ac.uk/clustalw). An alpha-satellite monomer from African green monkey (34) was included as an outgroup. The resulting monomer alignment was then analyzed by the mega3 program (35). Given the large number (1,568 monomers) of small (171 bp), closely related taxa, the resulting unrooted neighbor-joining tree was evaluated with 500 replicates of both bootstrap and interior branch tests.
Comparative Sequencing. Bacterial artificial chromosome (BAC) libraries (available at: http://bacpac.chori.org) generated from chimpanzee (Pan troglodytes; CHORI-251), gorilla (Gorilla gorilla; CHORI-255), orangutan (Pongo pygmaeus; CHORI-253), vervet (Cercopithecus aethiops; CHORI-252), macaque (Macaca mulatto; CHORI-250), and baboon (Papio anubis; RPCI-41) were screened as described (36, 37). Probe sequences are available on request. After isolation and mapping (36, 37), a subset of BACs was analyzed by FISH (data not shown); only those clones found to derive from the pericentromeric region of the X chromosome of the originating species were sequenced. Comparative sequence analyses were performed by using vista (60).
Results
Analysis of Human Xp Sequence. Our established (26) BAC-based physical map of the pericentromeric region of human Xp was used as a framework for generating high-quality genomic sequence. Detailed analyses of the finished sequence for an ≈650-kb region, which links the most proximal portion of Xp with the higher-order satellite array (Fig. 2 A and B), provide a complete accounting of the sequence elements present in the region (26) and, importantly, the physical relationships of those elements. The sequence consists of data from up to four independent X chromosomes; restriction fragments that were expected based on the sequence were detected in each of three unrelated males (26). Consistent with the current model (Fig. 1), each satellite family is characterized by an underlying monomer length and sequence (see Table 1). Monomers are arranged in a tandem, head-to-tail configuration, providing directionality to each satellite block (Fig. 2A). The sequence of monomers within a given family varies by as much as 30% (14), resulting in considerable sequence variation; additional variation is provided by the presence of interspersed repetitive elements that have been associated with monomeric satellites (20) (Fig. 2B). In addition to these somewhat expected features, these analyses detail a high degree of sequence rearrangement. Sequence blocks composed of many monomers of a satellite family range in size from ≈3–60 kb (Fig. 2A). Careful scrutiny of the 25 junctions between these blocks reveals that changes in directionality occur both between blocks of the same satellite family and between blocks of different families (Fig. 2A).
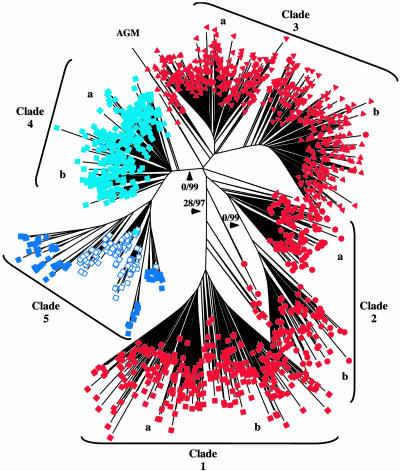
Analysis of Alpha-Satellite Sequences. Next, we analyzed the alpha-satellite monomers identified in the finished sequence. The monomers (1,568) were parsed from the finished sequence and subjected to neighbor-joining analysis by using the mega phylogenetic program (35); these were found to distribute across five major phylogenetic clades (Fig. 3). Importantly, monomers within each clade localize to a discrete physical domain in an exclusive fashion, such that clade 1 monomers reside only in domain 1, clade 2 monomers reside only in domain 2, and so forth (Fig. 2C). With the exception of clade 5, each clade is divided into at least two subclades (Fig. 3, a and b). Monomers from each pair of subclades are physically interspersed within the corresponding physical domain. The most proximal domain (domain 5, clade 5) contains 127 monomers (for a total length of ≈21 kb; Figs. 2C and 4) of the X chromosome higher-order repeat [type 1 and 2 DXZ1 (26); described below]. Monomers from a ≈13-kb region immediately distal to the array junction (Fig. 3, open squares) are also in clade 5; these monomers lack DXZ1 higher-order structure yet share sequence features with typical DXZ1 monomers and have been referred to as type 3 and 4 DXZ1 (26). Domain 4 extends distally for ≈74 kb (Fig. 2C) and is composed of monomers within clade 4 (type 4 DXZ1). Monomers within clades 3, 2, and 1 lie distally and sequentially in ≈107-, 40-, and 64-kb domains, respectively (Fig. 2C). These analyses provide the strongest evidence yet in support of the hypothesis that the alpha satellite in the region is physically separated into distinct homogenization zones (26). Indeed, five monomer clades corresponding to five physical domains are evident, whereas only two were distinguishable based on previous studies (26).
Fig. 3.
Phylogenetic analysis of alpha-satellite monomers in the Xp pericentromeric region. A total of 1,568 alpha-satellite monomers (each represented by a colored shape) from the human Xp pericentromeric region and a single alpha-satellite monomer from the African green monkey (AGM) were subjected to neighbor-joining analysis with midpoint rooting, yielding the depicted phylogenetic tree. In separate analyses, several independent monomers of human, macaque, and African green monkey alpha satellite were used as the root without affecting clade topography (data not shown). Alpha-satellite monomers from each of the five physical domains within the Xp region are designated by a different shape or color: domain 1, filled red square; domain 2, filled red circle; domain 3, filled red triangle; domain 4, filled light-blue square; domain 5, dark-blue square. Numbers on the major branches of the tree are the results of 500 bootstrap replicates and 500 interior-branch-test replicates, respectively (i.e., 0/99 indicates 0% of 500 bootstrap replicates and 99% of 500 interior-branch-test replicates). Only major branches reaching a significant interior-branch-test value of >95% are labeled. The binary characteristic of domains 1–4 is indicated (a and b). Monomers of the higher-order alpha satellite DXZ1 (filled blue squares) constitute clade 5, which also contains monomers that are immediately adjacent to the higher-order array but lack higher-order structure (open blue squares).
Fig. 4.
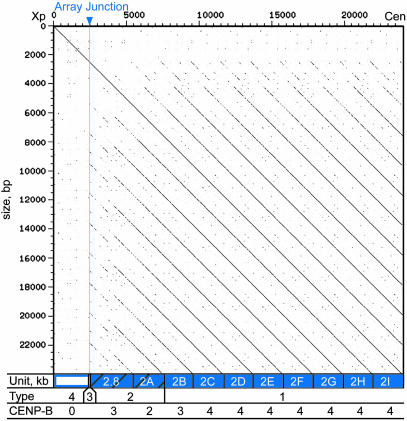
Characteristics of the edge of the DXZ1 array. A dotter plot displays a self–self alignment of the most proximal 24 kb of the generated Xp sequence (domain 5), ≈3 kb of pericentromeric alpha satellite (open rectangle), followed by ≈21 kb of centromeric higher-order repeats (blue rectangles) The coordinates of the analyzed region are depicted along the left and the top. The periodic nature of the higher-order repeats [solid blue rectangles along bottom, reflecting type 1 DXZ1 (26)] is demonstrated by the evenly spaced diagonal lines (14). Type 2 DXZ1 (hatched blue rectangles) contains one typical 2-kb repeat unit and an aberrant 2.8-kb higher-order repeat unit at the array junction. The size of each DXZ1 higher-order repeat unit is labeled (2.8 or 2 kb), and each 2-kb unit is given a letter to indicate its position relative to the others (2A and 2I being the most distal and proximal, respectively). The unusual nature of the aberrant repeat unit is revealed by reduced identity with the 2-kb units (see Table 2), distinct size, and the interrupted diagonal seen on the dotter plot. The region distal to the array junction (open rectangle) contains type 3 and 4 DXZ1 (open blue squares in Fig. 3). The number of CENP-B binding sites present in each higher-order repeat unit is indicated at the bottom.
The alpha-satellite monomers were also examined for the presence of a CENP-B box, a 17-bp recognition sequence found in a single copy in a subset of monomers within higher-order alpha satellite (38). Binding sites for CENP-B are present within human and great ape higher-order alpha satellite, but they are generally absent from monomeric alpha satellite (39, 40). Anti-CENP-B antibodies detect the protein at centromeres on most, but not all, primate chromosomes (41). Of the 1,568 Xp alpha-satellite monomers examined, only 36 contain a CENP-B box. Strikingly, all 36 of these monomers fall within the DXZ1 higher-order repeat units of domain 5, within the most proximal ≈21 kb (Figs. 4 and 6, which is published as supporting information on the PNAS web site).
Interspersed Repetitive Elements and LINE Dating. The presence of interspersed repetitive elements (specifically, LINE repeats) within the Xp pericentromeric satellite sequence can be used to gain insight about the evolution of the region. The interspersed elements interrupt monomers of a given satellite family, indicating the presence of that satellite at the time of active LINE transposition. Thus, the position of interspersed repeats can be used to infer the age of different satellite domains (42).
Based on analyses of the draft Xp sequence, we proposed (26) the presence of an age gradient that progresses from a very ancient distal region through the more recent proximal alpha-satellite sequences. Other data (13) suggest that alpha satellite near the junction between the chromosome arm and the distal edge of the satellite region is 35–40 million years old, whereas alpha satellite within the higher-order arrays is 6–8 million years old (43–45). Detailed analysis of the finished Xp sequence, including the identification of three additional LINEs, confirms the age-gradient hypothesis (Fig. 2C). Specifically, the distal domain (domain 1) contains LINEs (L1P3) that were active >35 million years ago (mya; 7% divergence), whereas the oldest LINEs (L1P2) present in the two central domains (domains 2 and 3) were active ≈25 mya (4% divergence). The oldest LINEs (L1P2) in domain 4 reside in the most distal block (13.9 kb) of monomeric alpha satellite present in this domain. Additional LINEs in domain 4 (all proximal to this 13.9-kb block) are L1P1 or L1Hs elements, which have both been active only since the emergence of the great ape lineage (3% divergence). The analyzed sequence contains ≈35 kb of alpha satellite that contains no interspersed LINE repeats (all within domain 5). However, previous attempts to identify LINEs within unanchored DXZ1 samples yielded a single L1Hs element (26). The complete inventory of L1 interspersed elements (Fig. 2C) demonstrates strict adherence to the proposed age gradient. Importantly, the absence of older elements (L1P3) in the proximal domains may indicate more recent addition of proximal monomeric alpha satellite to Xp, whereas the absence of newer LINEs (L1P1 and L1Hs) in the distal domains suggests a mechanism that prohibits L1 transposition into monomeric alpha satellite in this region.
Properties of the Array Junction. The most proximal 24 kb of the human Xp pericentromeric sequence (chrX: 57,524,803–57,548,803) includes a transition from the satellite region, characterized by monomeric satellites and interspersed repeats, to the higher-order alpha-satellite array (Fig. 4). A typical DXZ1 higher-order repeat unit consists of twelve 171-bp monomers that together span ≈2 kb (46). An aberrant 2.8-kb higher-order repeat unit that signifies the beginning of the DXZ1 array resides at the array junction (Figs. 2 A and 4). The percent identity between this 2.8-kb unit and a typical 2-kb unit is reduced (see Table 2, which is published as supporting information on the PNAS web site), and the structure is discontinuous (Fig. 4). Analysis of the monomers that comprise the aberrant 2.8-kb unit reveals evidence for an out-of-register exchange between two misaligned 2-kb units (see Fig. 6A). Also, a 15-bp insertion (46) observed in all other DXZ1 higher-order repeats is absent from this 2.8-kb unit. Interestingly, this 15-bp segment is also absent in the available sequence of chimpanzee and gorilla X chromosome higher-order repeats (see Fig. 6B), suggesting that the repeat unit without the 15-bp insertion is the ancestral form.
Proximal to this 2.8-kb unit are nine 2-kb units, all in the tandem, head-to-tail arrangement that creates the hallmark periodicity of alpha-satellite arrays (Figs. 1 and 4). The 2-kb unit immediately proximal to the 2.8-kb aberrant unit (rectangle 2A in Fig. 4) is only 96% identical to other DXZ1 units (Table 2); these two units have been termed type 2 DXZ1, because they share higher-order structure with typical DXZ1 but exhibit reduced identity (26), indicative of a transition zone. The remaining eight 2-kb units (rectangles 2B–2I in Fig. 4) have the typical 98–100% pairwise identity seen with higher-order alpha-satellite arrays, and they are considered type 1 DXZ1 (Table 2). Interestingly, the number of CENP-B boxes found in each 2-kb higher-order repeat unit increases from two in the most distal 2-kb unit to three in the next proximal unit, followed by four in each of the seven remaining 2-kb units (Fig. 4). The 2.8-kb aberrant unit at the array junction (Fig. 4; also see Fig. 6A) contains three CENP-B boxes within an apparent combination of two different higher-order repeats: one unit containing a CENP-B box in monomer 10 and one unit without a CENP-B box in monomer 10 (see Fig. 6A). Distal to the array junction reside three monomers that share sequence features with specific DXZ1 monomers (26); these monomers share reduced identity with type 1 DXZ1 but fall into the DXZ1 phylogenetic clade, thus defining type 3 (Fig. 3, clade 5).
Conservation of the Satellite Junction Among Multiple Primates. An essential component of the age-gradient hypothesis is that the satellite region immediately proximal to the euchromatic chromosome arm is a remnant of the ancestral primate X centromere (26). To search for conservation of this junction, as predicted by the model, we sequenced the orthologous region in six other primate genomes. vista analysis (Fig. 5) reveals sequence-identity levels throughout this region consistent with the estimated divergence times between human and chimpanzee, gorilla, orangutan, baboon, macaque, and vervet, respectively (47). Moreover, within the alpha-satellite region, an inserted L1P3 element (specifically, a L1PA7 element) is present at the orthologous position in all six primates (Fig. 5; also see Fig. 7, which is published as supporting information on the PNAS web site). Members of the L1P3 subfamily of LINEs are estimated to have been active in the primate genome over 35 mya (42), indicating that this segment of alpha satellite was present in an early primate ancestor. Within the same stretch of alpha satellite, an LTR5 (ERVK) element is present only in the great apes, revealing the continuing evolution of the region. The absence of this element in all three old world monkeys suggests its insertion occurred between 12 and 25 mya, after the origin of the old world monkey lineage (≈25 mya) but before the split creating the great ape lineage (≈12 mya).
Fig. 5.
Comparative sequence analysis of the Xp satellite junction. An adapted vista plot depicts alignments of the 10 kb of sequence encompassing the Xp satellite junction from each of six primate species with the human reference sequence (in each case, the overall percent identity shared with the human sequence is shown in parentheses). The repeat content of the human sequence is shown along the top (see key at bottom; αsat, alpha satellite). For each species, a percent identity plot is displayed ranging from 50% to 100% sequence identity, as indicated. Each plot was generated by searching for 100-bp windows of ≥90% identity. An arrowhead and dotted line depict the position of the satellite junction. L1P3 and LTR5 elements embedded within alpha satellite are highlighted.
Discussion
Our understanding of centromere dynamics (specifically, the role of DNA sequence in centromere function, the evolution of centromere sequences, and the impact of sequence changes on speciation) suffers from inadequate molecular details (14). The human X chromosome has emerged as a model for the study of centromere structure and function, in part because it was the first chromosome for which a sequence-ready BAC contig was constructed that spanned the transition between the chromosomal arm and the higher-order satellite array (26). Here, we report key details about the X-chromosome centromere based on analyses of the complete sequence of the Xp pericentromeric transition.
Compared with the strict periodicity found in the centromeric alpha-satellite arrays, the long-range structure of pericentromeric regions is disorganized (14). Despite its highly repetitive nature, the sequence of such regions is actually stratified (analogous to the layers of earth laid down over time). Our previous analysis of a smaller sample of alpha-satellite monomers (<500) (26) indicated the presence of two major types of alpha satellite; notably, this sample did not include monomers from the ≈200-kb region between the gamma satellite and the DXZ1 array. Examination of the finished sequence shows the presence of both monomeric and higher-order alpha satellite and, in addition, confirms their spatial separation. Moreover, phylogenetic analysis of the complete set of alpha-satellite monomers demonstrates five physical domains, including four domains that are populated with monomeric alpha satellite and one domain with a higher-order structure.
Previous dating of L1 elements (42) in the Xp pericentromic region provided important evolutionary insights (42). Our identification of LINE repeats within the alpha satellite between gamma satellite and DXZ1 clearly defines the age gradient that reflects the evolutionary history of the region. The strict correlation between LINE ages and the phylogenetically defined physical domains revealed by the complete sequence indicates a progressive proximal expansion of centromere content. Specifically, the most distal alpha-satellite domain is the oldest, with an age gradient progressing proximally through the satellite region by an apparent series of events culminating in the addition of the higher-order alpha-satellite array. Our analyses indicate that a similar age gradient exists within the Xq pericentromeric region (data not shown). Together, these findings suggest that the primate X centromere evolved through repeated expansion events involving the central, functional alpha-satellite domain, such that ancestral centromeric sequences were split and displaced distally onto each arm.
Whereas absence of older LINEs within the proximal domains indicates the successive additions of alpha satellite to the X centromere, the absence of newer LINEs from distal domains is not accounted for by this hypothesis. This observed bias toward the insertion of active LINEs into a discrete region of alpha satellite, which occurs at the exclusion of adjacent (and very similar) sequences, suggests that changes in chromatin composition that accompany centromere activation/inactivation (48) may influence transposition events. The ongoing homogenization of centromeric satellite sequences by unequal crossover (49–52) and clustering of crossover breakpoints at or near CENP-B binding sites (53) may be mechanistically related to this phenomenon. The CENP-B protein is closely related to the pogo superfamily of transposases and may retain single-strand nicking capability (54). CENP-B binds a subset of alpha-satellite DNA (55) and has been shown to accumulate below the kinetochore (56), within the region of the active centromere. In addition to CENP-B, topoisomerase II enzymatic activity has been localized within the active centromere site (29, 30). The unique chromatin environment created by the centromere-specific histone, CENP-A, combined with the activity of CENP-B and topoisomerase II may contribute to the apparent promiscuity of centromeric regions by confining the proposed expansion events and insertion of transposable elements to functionally active centromeric DNA.
Possible negative consequences of the dynamic nature of the region may be offset by the advantages of having a fluid centromere environment. The sites of human kinetochore assembly (29, 30) are largely devoid of interspersed repeats (10, 17), span large intervals (15, 57), and have an underlying sequence periodicity, all of which facilitate the formation of higher-order structures that might be necessary for kinetochore interactions (4). Insertion of active LINEs would disrupt that periodicity, potentially compromising centromere activity and requiring a compensatory expansion of centromeric DNA (58). Consistent with this model is the notion that the higher-order alpha satellite at the centromere evolved as a replacement for the monomeric alpha satellite (13), providing a more efficient mechanism for homogenization. The increased rate of chromosome-specific homogenization of higher-order alpha satellite in higher primates (51) may represent an important step in evolutionary efforts to adapt centromeric DNA to the needs of its protein partners.
Last, the generated comparative sequence data allowed refinement of our model of Xp centromere structure and evolution. Examination of multiple primate sequences confirms that Xp orthology spans from the chromosome arm, across the satellite junction, and deep into the satellite region. Our model predicts that the most distal alpha satellite on human Xp (domain 1) is likely to be present in very early primates, with the remaining domains appearing sequentially later in the primate lineage. Meanwhile, one would expect that concerted evolution of centromeric sequences would lead to the accumulation and fixation of species-specific changes in each genome. The ongoing sequencing of primate genomes should provide valuable insights about the timing of these events and their influence on centromere function and speciation.
Supplementary Material
Acknowledgments
We thank all members of the NISC Comparative Sequencing Program for generating the comparative sequence data. M.R. and L.V. were supported by Centro di Eccellenza Geni in campo Biosanitario e Agroalimentare, Ministero Italiano della Universitá e della Ricerca (Cluster C03, Program L.488/92), and European Commission INPRI-MAT Grant QLRI-CT-2002-01325.
Abbreviations: LINE, long interspersed nucleotide element; mya, million years ago; BAC, bacterial artificial chromosome; Xp, X chromosome short arm; CENP-B, centromere protein B.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AC140661 (chimpanzee), AC147693 (gorilla), AC147722 (orangutan), AC147591 (baboon), AC134314 (macaque), and AC147690 (vervet)].
References
- 1.Henikoff, S., Ahmad, K. & Malik, H. S. (2001) Science 293, 1098-1102. [DOI] [PubMed] [Google Scholar]
- 2.Cleveland, D. W., Mao, Y. & Sullivan, K. F. (2003) Cell 112, 407-421. [DOI] [PubMed] [Google Scholar]
- 3.Amor, D. J., Kalitsis, P., Sumer, H. & Choo, K. H. (2004) Trends Cell. Biol. 14, 359-368. [DOI] [PubMed] [Google Scholar]
- 4.Sullivan, B. A., Blower, M. D. & Karpen, G. H. (2001) Nat. Rev. Genet. 2, 584-596. [DOI] [PubMed] [Google Scholar]
- 5.Malik, H. S. & Henikoff, S. (2002) Curr. Opin. Genet. Dev. 12, 711-718. [DOI] [PubMed] [Google Scholar]
- 6.Karpen, G. H. & Allshire, R. C. (1997) Trends Genet. 13, 489-496. [DOI] [PubMed] [Google Scholar]
- 7.Csink, A. K. & Henikoff, S. (1998) Trends Genet. 14, 200-204. [DOI] [PubMed] [Google Scholar]
- 8.Willard, H. F. (1998) Curr. Opin. Genet. Dev. 8, 219-225. [DOI] [PubMed] [Google Scholar]
- 9.Choo, K. H. (2000) Trends Cell Biol. 10, 182-188. [DOI] [PubMed] [Google Scholar]
- 10.Willard, H. F. & Waye, J. S. (1987) Trends Genet. 3, 192-198. [Google Scholar]
- 11.Lee, C., Wevrick, R., Fisher, R. B., Ferguson-Smith, M. A. & Lin, C. C. (1997) Hum. Genet. 100, 291-304. [DOI] [PubMed] [Google Scholar]
- 12.Manuelidis, L. (1978) Chromosoma 66, 23-32. [DOI] [PubMed] [Google Scholar]
- 13.Alexandrov, I., Kazakov, A., Tumeneva, I., Shepelev, V. & Yurov, Y. (2001) Chromosoma 110, 253-266. [DOI] [PubMed] [Google Scholar]
- 14.Rudd, M. K. & Willard, H. F. (2004) Trends Genet. 20, 529-533. [DOI] [PubMed] [Google Scholar]
- 15.Wevrick, R. & Willard, H. F. (1989) Proc. Natl. Acad. Sci. USA 86, 9394-9398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahtani, M. M. & Willard, H. F. (1998) Genome Res. 8, 100-110. [DOI] [PubMed] [Google Scholar]
- 17.Schindelhauer, D. & Schwarz, T. (2002) Genome Res. 12, 1815-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.She, X., Horvath, J. E., Jiang, Z., Liu, G., Furey, T. S., Christ, L., Clark, R., Graves, T., Gulden, C. L., Alkan, C., et al. (2004) Nature 430, 857-864. [DOI] [PubMed] [Google Scholar]
- 19.International Human Genome Sequencing Consortium (2001) Nature 409, 860-921. [DOI] [PubMed] [Google Scholar]
- 20.Wong, L. H. & Choo, K. H. (2004) Trends Genet. 20, 611-616. [DOI] [PubMed] [Google Scholar]
- 21.Alexandrov, I. A., Medvedev, L. I., Mashkova, T. D., Kisselev, L. L., Romanova, L. Y. & Yurov, Y. B. (1993) Nucleic Acids Res. 21, 2209-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willard, H. F. (1990) Trends Genet. 6, 410-416. [DOI] [PubMed] [Google Scholar]
- 23.Porter, A. C. & Farr, C. J. (2004) Chromosome Res. 12, 569-583. [DOI] [PubMed] [Google Scholar]
- 24.Harrington, J. J., Van Bokkelen, G., Mays, R. W., Gustashaw, K. & Willard, H. F. (1997) Nat. Genet. 15, 345-355. [DOI] [PubMed] [Google Scholar]
- 25.Ikeno, M., Grimes, B., Okazaki, T., Nakano, M., Saitoh, K., Hoshino, H., McGill, N. I., Cooke, H. & Masumoto, H. (1998) Nat. Biotechnol. 16, 431-439. [DOI] [PubMed] [Google Scholar]
- 26.Schueler, M. G., Higgins, A. W., Rudd, M. K., Gustashaw, K. & Willard, H. F. (2001) Science 294, 109-115. [DOI] [PubMed] [Google Scholar]
- 27.Grimes, B. R., Rhoades, A. A. & Willard, H. F. (2002) Mol. Ther. 5, 798-805. [DOI] [PubMed] [Google Scholar]
- 28.Andersen, C. L., Wandall, A., Kjeldsen, E., Mielke, C. & Koch, J. (2002) Chromosome Res. 10, 305-312. [DOI] [PubMed] [Google Scholar]
- 29.Spence, J. M., Critcher, R., Ebersole, T. A., Valdivia, M. M., Earnshaw, W. C., Fukagawa, T. & Farr, C. J. (2002) EMBO J. 21, 5269-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Floridia, G., Gimelli, G., Zuffardi, O., Earnshaw, W. C., Warburton, P. E. & Tyler-Smith, C. (2000) Chromosoma 109, 318-327. [DOI] [PubMed] [Google Scholar]
- 31.Vafa, O. & Sullivan, K. F. (1997) Curr. Biol. 7, 897-900. [DOI] [PubMed] [Google Scholar]
- 32.Haaf, T. & Ward, D. C. (1994) Hum. Mol. Genet. 3, 697-709. [DOI] [PubMed] [Google Scholar]
- 33.Sonnhammer, E. L. & Durbin, R. (1995) Gene 167, GC1-GC10. [DOI] [PubMed] [Google Scholar]
- 34.Rosenberg, H., Singer, M. & Rosenberg, M. (1978) Science 200, 394-402. [DOI] [PubMed] [Google Scholar]
- 35.Kumar, S., Tamura, K. & Nei, M. (2004) Brief. Bioinform. 5, 150-163. [DOI] [PubMed] [Google Scholar]
- 36.Thomas, J. W., Prasad, A. B., Summers, T. J., Lee-Lin, S. Q., Maduro, V. V., Idol, J. R., Ryan, J. F., Thomas, P. J., McDowell, J. C. & Green, E. D. (2002) Genome Res. 12, 1277-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas, J. W., Touchman, J. W., Blakesley, R. W., Bouffard, G. G., Beckstrom-Sternberg, S. M., Margulies, E. H., Blanchette, M., Siepel, A. C., Thomas, P. J., McDowell, J. C., et al. (2003) Nature 424, 788-793. [DOI] [PubMed] [Google Scholar]
- 38.Muro, Y., Masumoto, H., Yoda, K., Nozaki, N., Ohashi, M. & Okazaki, T. (1992) J. Cell Biol. 116, 585-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ikeno, M., Masumoto, H. & Okazaki, T. (1994) Hum. Mol. Genet. 3, 1245-1257. [DOI] [PubMed] [Google Scholar]
- 40.Haaf, T., Mater, A. G., Wienberg, J. & Ward, D. C. (1995) J. Mol. Evol. 41, 487-491. [DOI] [PubMed] [Google Scholar]
- 41.Earnshaw, W. C., Sullivan, K. F., Machlin, P. S., Cooke, C. A., Kaiser, D. A., Pollard, T. D., Rothfield, N. F. & Cleveland, D. W. (1987) J. Cell Biol. 104, 817-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smit, A. F., Toth, G., Riggs, A. D. & Jurka, J. (1995) J. Mol. Biol. 246, 401-417. [DOI] [PubMed] [Google Scholar]
- 43.Durfy, S. J. & Willard, H. F. (1990) J. Mol. Biol. 216, 555-566. [DOI] [PubMed] [Google Scholar]
- 44.Haaf, T. & Willard, H. F. (1997) Chromosoma 106, 226-232. [DOI] [PubMed] [Google Scholar]
- 45.Haaf, T. & Willard, H. F. (1998) Mamm. Genome 9, 440-447. [DOI] [PubMed] [Google Scholar]
- 46.Waye, J. S. & Willard, H. F. (1985) Nucleic Acids Res. 13, 2731-2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goodman, M. (1999) Am. J. Hum. Genet. 64, 31-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sullivan, B. A. & Karpen, G. H. (2004) Nat. Struct. Mol. Biol. 11, 1076-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith, G. P. (1976) Science 191, 528-535. [DOI] [PubMed] [Google Scholar]
- 50.Durfy, S. J. & Willard, H. F. (1989) Genomics 5, 810-821. [DOI] [PubMed] [Google Scholar]
- 51.Warburton, P. & Willard, H. (1996) in Human Genome Evolution, eds. Jackson, M., Strachan, T. & Dover, G. (BIOS Scientific, Oxford), pp. 121-145.
- 52.Mashkova, T., Oparina, N., Alexandrov, I., Zinovieva, O., Marusina, A., Yurov, Y., Lacroix, M. H. & Kisselev, L. (1998) FEBS Lett. 441, 451-457. [DOI] [PubMed] [Google Scholar]
- 53.Warburton, P. E., Waye, J. S. & Willard, H. F. (1993) Mol. Cell. Biol. 13, 6520-6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kipling, D. & Warburton, P. E. (1997) Trends Genet. 13, 141-145. [DOI] [PubMed] [Google Scholar]
- 55.Masumoto, H., Masukata, H., Muro, Y., Nozaki, N. & Okazaki, T. (1989) J. Cell Biol. 109, 1963-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cooke, C. A., Bernat, R. L. & Earnshaw, W. C. (1990) J. Cell Biol. 110, 1475-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mahtani, M. M. & Willard, H. F. (1990) Genomics 7, 607-613. [DOI] [PubMed] [Google Scholar]
- 58.Laurent, A. M., Puechberty, J. & Roizes, G. (1999) Chromosome Res. 7, 305-317. [DOI] [PubMed] [Google Scholar]
- 59.Smit, A. F. A., Hubley, R. & Green, P. (1996) repeatmaster open (Inst. Syst. Biol., Seattle), Version 3.0.
- 60.Frazer, K. A., Pachter, A., Poliakou, A., Rubin, E. M., Duback, I. (2004) Nucleic Acids Res. 32, W273-W279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.