Abstract
Because of the near geometric identity of Watson-Crick (W-C) G·C and A·T base pairs, a given DNA polymerase forms the four possible correct base pairs with nearly identical catalytic efficiencies. However, human DNA polymerase ι (Polι), a member of the Y family of DNA polymerases, exhibits a marked template specificity, being more efficient at incorporating the correct nucleotide opposite template purines than opposite pyrimidines. By using 7-deazaadenine and 7-deazaguanine as the templating residues, which disrupt Hoogsteen base pair formation, we show that, unlike the other DNA polymerases belonging to the A, B, or Y family, DNA synthesis by Polι is severely inhibited by these N7-modified bases. These observations provide biochemical evidence that, during normal DNA synthesis, template purines adopt a syn conformation in the Polι active site, enabling the formation of a Hoogsteen base pair with the incoming pyrimidine nucleotide. Additionally, mutational studies with Leu-62, which lies in close proximity to the templating residue in the Polι ternary complex, have indicated that both factors, steric constraints within the active site and the stability provided by the hydrogen bonds in the Hoogsteen base pair, contribute to the efficiency of correct nucleotide incorporation opposite template purines by Polι.
Keywords: translesion DNA synthesis, 7-deaza purines, syn and anti conformations
Virtually all DNA polymerases synthesize DNA with a strong preference for incorporating the nucleotide that forms the correct Watson-Crick (W-C) base pair with the template base, and moreover, a given polymerase incorporates the correct nucleotide opposite each of the four template bases with nearly equivalent efficiencies. Human DNA polymerase ι (Polι), a member of the Y family of DNA polymerases, is an exception to these rules. By contrast to other DNA polymerases, Polι incorporates nucleotides opposite the four template bases with very different efficiencies and fidelities, and it incorporates nucleotides opposite template purines with a much higher efficiency and fidelity than opposite template pyrimidines (1-5). Polι exhibits the highest efficiency and fidelity opposite template A, where it misincorporates nucleotides with frequencies of ≈10-4 to 10-5; opposite template G, Polι incorporates the correct nucleotide with an at least 10-fold reduced efficiency than opposite template A, and it misincorporates a T opposite template G with a frequency of ≈10-1. Polι is highly inefficient at incorporating the correct nucleotide opposite templates C and T, and, opposite template T, it misincorporates a G more often than an A (1-5).
Because the nearly equivalent efficiencies and fidelities of nucleotide incorporation opposite each of the four template bases by DNA polymerases result from the snug accommodation of the correct W-C base pair by geometric selection (6, 7), this fact raised the possibility that the unusual nucleotide incorporation specificity of Polι was not based upon W-C base pairing geometry. In keeping with this idea, the crystal structure of Polι bound to the template-primer junction of DNA with an A in the templating position and with an incoming dTTP has shown that Polι incorporates a T opposite template A by means of Hoogsteen base pairing (8). However, because of the fact that Hoogsteen base pairing has never before been observed to direct synthesis by DNA polymerases, we have carried out biochemical experiments to establish that DNA synthesis by Polι, in fact, requires Hoogsteen base pairing.
To provide evidence that the Hoogsteen base pairing seen in the crystal structure of Polι occurs in solution and thus is a valid reflection of its action mechanism during normal DNA synthesis, we determined whether DNA synthesis by Polι was inhibited by the 7-deazaadenine (7-deaza-A) or 7-deazaguanine (7-deaza-G) residues at the templating site in DNA. Importantly, we find that, unlike all of the other DNA polymerases we examined, DNA synthesis by Polι is severely inhibited by these N7-modified template bases. These observations provide strong evidence that nucleotide incorporation by Polι opposite both the template purine bases A and G involves Hoogsteen base pairing. Thus, in addition to confirming the structural observations of Hoogsteen base pairing for template A, the biochemical evidence presented here shows that Polι also utilizes Hoogsteen base pairing for incorporating nucleotides opposite template G.
To learn more about the mechanism of nucleotide incorporation by Polι opposite template purines, we have examined the effects of the Leu-62 → Ala mutational alteration on the efficiency and fidelity of nucleotide incorporation by Polι. In the ternary structure of Polι, Leu-62 is one of several residues in the finger domain that lie in close proximity to the templating base, and that could contribute to imposing a syn conformation upon the templating nucleotide. Interestingly, whereas the efficiencies of T incorporation opposite template A, and of C and T incorporation opposite template G, are lowered to varying degrees by the Ala-62 mutation, the misincorporation of purines opposite templates A and G is not as significantly affected. From these observations, we infer that the steric constraints imposed by the active site, as well as the stability provided by the hydrogen bonds involved in Hoogsteen base pair formation, contribute to the efficiency of T incorporation opposite template A, and of C and T incorporation opposite template G. Additionally, from our observations with the Ala-62 mutant protein, and also those with the 7-deaza-A and 7-deaza-G templates, we infer that, for purine misincorporation to occur opposite templates A and G, the templating residue adopts an “abasic-like” structure, opposite which purines are inserted preferentially over pyrimidines because of their better stacking ability.
Materials and Methods
Protein and DNA Substrates. Human DNA Polι was expressed and purified from yeast as described (1). Yeast DNA Polδ was a gift from Peter Burgers (Washington University, St. Louis). Yeast Polζ, human Polη, and human Polκ were purified as described (9-11). Escherichia coli DNA PolI and exonuclease-deficient Klenow fragment (KF exo-) were purchased from New England Biolabs. Oligonucleotides containing site-specific A, G, 7-deaza-A, or 7-deaza-G were obtained from Qiagen (Valencia, CA) and were PAGE purified before use. DNA primer:template substrates were composed of the oligodeoxynucleotide primer (32-mer), 5′-GTTTTCCCAG TCACGACGAT GCTCCGGTAC TC-3′, annealed to a 52-mer template 5′-TTCGTATAAT GCCTACACTX GAGTACCGGA GCATCGTCGT GACTGGGAAAAC-3′, where the X denotes either an A, a G, a 7-deaza-A, or a 7-deaza-G. DNA substrates consisted of the oligonucleotide primer, which was 5′-32P-end-labeled by using polynucleotide kinase (Roche Molecular Biochemicals) and [γ-32P]ATP (Amersham Pharmacia Biotech), annealed to the template by heating a mixture of primer:template at a 1:1.5 molar ratio to 95°C and allowing it to cool to room temperature over several hours.
Polι Leu-62 → Ala Mutation. The Leu-62 → Ala mutation was generated by using the quick change mutagenesis kit (Stratagene) and plasmid pBJ1142. Plasmid pBJ1142 harbors the Polι catalytic core and contains amino acids 1-420 of Polι. The N-terminal region of Polι from the ATG start codon to the BstZ17I site was sequenced to confirm the presence of the mutation, and the remaining region of Polι was replaced by the corresponding wild-type fragment from pBJ1142. Protein was expressed from plasmid pBJ1205 as a GST-fusion and purified as described (8).
DNA Polymerase Assays. The standard DNA polymerase reaction (5 μl) contained 25 mM Tris·HCl (pH 7.5), 5 mM MgCl2, 1 mM dithiolthreitol, 100 μg/ml BSA, 10% glycerol, 50 μM of each of the four deoxynucleotides (dGTP, dATP, dTTP, dCTP), and 10 nM DNA substrate. All reactions were carried out at 37°C, except for those containing yeast Polδ and Polζ, which were carried out at 30°C. For DNA synthesis assays in Fig. 2 A, reactions were carried out for 10 min and protein concentrations were as follows: Klenow fragment (KF exo-) [0.3 nM]; E. coli PolI [0.45 nM]; yeast Polδ [1 nM]; yeast Polζ [1 nM]; human Polη [1 nM]; human Polκ [2.5 nM]; and human Polι [2.5 nM]. For single nucleotide incorporation assays in Fig. 2B, only a single nucleotide (50 μM dGTP, dATP, dTTP, or dCTP) was included in the assay. Reactions were carried out for 5 min and the protein concentrations were as follows: KF exo- [0.3 nM], yeast Polζ [1.6 nM]; human Polη [1 nM]; human Polκ [1 nM]; and human Polι [1 nM]. Reactions were terminated by the addition of 5 vol of loading buffer (95% formamide/0.05% cyanol blue/0.05% bromophenol blue) before resolving products on 15% polyacrylamide gels containing 8 M urea. Gels were dried before autoradiography at -70°C. The primer was 5′-32P-end-labeled by using polynucleotide kinase (Roche Molecular Biochemicals) and [γ-32P]ATP (Amersham Pharmacia Biotech).
Fig. 2.
Replication through a 7-deaza-A (dzA) and 7-deaza-G (dzG) template by various DNA polymerases. (A) Inhibition of DNA synthesis by Polι on 7-deaza-A and 7-deaza-G templates. Each protein was incubated with DNA substrate (10 nM) and each of the 4 dNTPs (50 μM) for 10 min at 30°C or 37°C. The DNA polymerases examined were as follows: KF exo-, Klenow fragment lacking the 3′ → 5′ exonuclease activity; PolI, E. coli DNA polymerase I; and yeast Polδ, yeast Polζ, and human Polη, Polκ, and Polι. Note the poor synthesis by Polι opposite and beyond the template T. (B) Single nucleotide incorporation opposite 7-deaza-A and 7-deaza-G by various DNA polymerases. Reactions were carried out as in A but contained 50 μM a single dNTP (as indicated), except for KF exo-, where 2.5 μM dNTP was used. The sequence of the DNA substrate used is shown in A, where X denotes an A, a G, a 7-deaza-A, or a 7-deaza-G template base.
Steady-State Kinetic Analysis. Steady-state kinetic analyses for deoxynucleotide incorporation were done as described (12). The standard DNA polymerase assay was used except that only a single deoxynucleotide was included at various concentrations and reactions were carried out for 5-15 min. Protein concentrations used were as follows; KF exo- [0.07 nM]; yeast Polζ [1.6 nM]; human Polη [1 nM]; human Polκ [1 nM]; human Polι [1 nM or 5 nM]; human Polι (1-420) [0.5 nm or 5 nM]; and human Polι (1-420) L62A mutant protein [1 nM or 10 nM]. Gel band intensities of the substrate and products of the deoxynucleotide incorporation reactions were quantitated by using a Phosphor-Imager and imagequant software (Molecular Dynamics). The observed rate of deoxynucleotide incorporation, vobs, was determined by dividing the amount of product formed by the reaction time and protein concentration. We plotted the vobs as a function of the deoxynucleotide concentration and fit these data to the Michaelis-Menten equation describing a hyperbola: vobs = (kcat[E] × [dNTP])/(Km + [dNTP]), where [E] is enzyme concentration.
Results
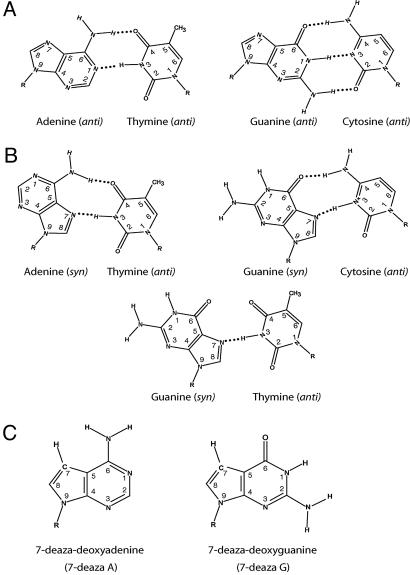
By contrast to a W-C base pair where the complementary purine and pyrimidine bases are both in the anti configuration (Fig. 1A), a Hoogsteen base pair forms when a purine base adopts the syn conformation in DNA and hydrogen bonds with the complementary pyrimidine nucleotide, which retains the anti configuration (Fig. 1B). In an A·T Hoogsteen base pair, two hydrogen bonds form between the “Hoogsteen edge” of adenine (N7 and N6) and the W-C edge of thymine (N3 and O4). The Hoogsteen edge of guanine (N7 and O6) and the W-C edge of cytosine (N3 and N4), which remains in the anti conformation, can also form two hydrogen bonds, one between O6 in G and N4 in C, and the other between N7 of G and N3 of C; the latter hydrogen bonding, however, requires the protonation of N3 in C (Fig. 1B). A geometrically similar Hoogsteen base pair can form between G and T by means of the single hydrogen bond between the N7 of G and N3 of T (Fig. 1B). The highest efficiency and fidelity of Polι at template A may then result from the efficient Hoogsteen base pairing of template A with a T; and the intermediate efficiency and fidelity opposite template G and the incorporation of both C and T opposite this template residue might then result from the relative efficiencies of Hoogsteen base pair formation of template G with a C vs. T (13). The highly inefficient incorporation of the correct nucleotide opposite templates T and C would then be due to the lack of hydrogen bonding possibilities when these template residues adopt a syn conformation.
Fig. 1.
Hydrogen bonding in W-C and Hoogsteen base pairs. (A) W-C A·T and G·C base pairs. (B) Hoogsteen base pair formation between adenine and thymine, guanine and cytosine, and guanine and thymine. (C) 7-Deaza purine analogs 7-deaza-A and 7-deaza-G. The R indicates the sugar moiety.
Inhibitory Effects of 7-Deaza-A and 7-Deaza-G on DNA Synthesis by Polι. To determine whether DNA synthesis by Polι depended upon Hoogsteen base pair formation, oligonucleotides containing a site-specific adenine (A), guanine (G), or their analogs 7-deaza-A or 7-deaza-G were used as a template in DNA synthesis assays (Fig. 1C). The 7-deaza purine nucleotides are modified at the N7 position, wherein the nitrogen has been replaced by carbon. This change would disrupt Hoogsteen base pairing because it removes the hydrogen bond acceptor nitrogen and, additionally, a hydrogen atom is added at this position (Fig. 1C). In fact, because W-C base pair formation is not affected by this modification (Fig. 1 A), 7-deaza-A and 7-deaza-G have been used to aid DNA sequencing, because these nucleotide triphosphates are readily incorporated into DNA, and they eliminate DNA secondary structure and polymerase arrest in GC-rich sequences, or in other sequences, where triplex DNA could form by means of Hoogsteen base pairing (14-16). The effects of 7-deaza-A and 7-deaza-G were tested on a variety of DNA polymerases in addition to Polι. Fig. 2A shows the DNA synthesis activities of several DNA polymerases in a standing start assay on templates containing an A or a G or their respective 7-deaza analogs. The A family DNA polymerases PolI and its 3′ → 5′ exonuclease deficient Klenow fragment derivative (KF exo-) were not adversely effected by the presence of 7-deaza-A or 7-deaza-G in the template. The yeast B family polymerases Polδ or Polζ (comprised of Rev3 and Rev7) were similarly unaffected by these analogs. The Y family of human DNA polymerases include Polη, Polκ, and Polι (17, 18). Although neither Polη or Polκ were affected by 7-deaza-A or 7-deaza-G, Polι displayed a severe defect in nucleotide incorporation opposite both these analogs (Fig. 2 A, lanes 26 and 28).
Next, we identified the nucleotides that are incorporated by the various DNA polymerases opposite 7-deaza-A and 7-deaza-G. The nucleotide incorporation profiles for KF exo-, Polζ, Polη, and Polκ opposite A and 7-deaza-A, and opposite G and 7-deaza-G, were nearly identical, indicating that these polymerases are relatively insensitive to the modification of the N7 of template purines (Fig. 2B). Polι, however, shows little propensity to incorporate a nucleotide opposite 7-deaza-A or 7-deaza-G (Fig. 2B).
Polι Incorporates the Correct Nucleotide Opposite 7-Deaza-A and 7-Deaza-G with a Much Reduced Catalytic Efficiency. To quantitatively assess the effects of 7-deaza purine templates on DNA synthesis by Polι and other DNA polymerases, we determined the efficiency (kcat/Km) of nucleotide incorporation opposite templates A, G, 7-deaza-A, and 7-deaza-G under steady-state conditions (Table 1). Whereas 7-deaza-A and 7-deaza-G imparted only a little or no reduction in the efficiency of nucleotide incorporation by KF exo-, or Pols ζ, η, and κ, the incorporation of T opposite 7-deaza-A by Polι was reduced >200-fold relative to incorporation opposite A, and the efficiency of C incorporation opposite 7 deaza-G by Polι was reduced by ≈30-fold compared with incorporation opposite G. However, because Polι is ≈10 fold less efficient for the incorporation of a C opposite template G than for the incorporation of T opposite template A to begin with, the actual efficiency of C incorporation opposite 7-deaza-G and of T incorporation opposite 7-deaza-A is nearly the same (3.3 × 10-4 vs. 4.3 × 10-4, respectively).
Table 1. Steady-state kinetic parameters for correct nucleotide incorporation opposite 7-deaza purines by Polι and other DNA polymerases.
| Enzyme | Template base | Incoming nucleotide | kcat, min–1 | Km, μM | kcat/Km | Relative efficiency | Fold reduction in efficiency |
|---|---|---|---|---|---|---|---|
| KF exo– | A | dTTP | 0.7 ± 0.05 | 0.005 ± 0.001 | 140 | 1 | – |
| 7-deaza A | dTTP | 1.6 ± 0.09 | 0.005 ± 0.001 | 320 | 2.3 | – | |
| G | dCTP | 1.4 ± 0.1 | 0.016 ± 0.005 | 88 | 1 | – | |
| 7-deaza G | dCTP | 1.9 ± 0.1 | 0.004 ± 0.001 | 475 | 5.4 | – | |
| Polζ | A | dTTP | 0.11 ± 0.007 | 0.20 ± 0.08 | 0.55 | 1 | – |
| 7-deaza A | dTTP | 0.08 ± 0.003 | 0.53 ± 0.08 | 0.15 | 0.27 | 3.7 | |
| G | dCTP | 0.17 ± 0.009 | 0.14 ± 0.05 | 1.2 | 1 | – | |
| 7-deaza G | dCTP | 0.21 ± 0.014 | 0.47 ± 0.13 | 0.45 | 0.38 | 2.6 | |
| Polη | A | dTTP | 0.78 ± 0.05 | 2.7 ± 0.7 | 0.29 | 1 | – |
| 7-deaza A | dTTP | 0.55 ± 0.01 | 3.2 ± 0.2 | 0.17 | 0.59 | 1.7 | |
| G | dCTP | 0.91 ± 0.05 | 2.1 ± 0.5 | 0.43 | 1 | – | |
| 7-deaza G | dCTP | 0.78 ± 0.07 | 3.0 ± 1.0 | 0.26 | 0.6 | 1.7 | |
| Polκ | A | dTTP | 1.5 ± 0.05 | 0.37 ± 0.06 | 4.1 | 1 | – |
| 7-deaza A | dTTP | 1.1 ± 0.03 | 0.99 ± 0.1 | 1.1 | 0.27 | 3.7 | |
| G | dCTP | 1.6 ± 0.2 | 0.72 ± 0.3 | 2.2 | 1 | – | |
| 7-deaza G | dCTP | 1.2 ± 0.04 | 2.3 ± 0.3 | 0.5 | 0.23 | 4.3 | |
| Polι | A | dTTP | 1.0 ± 0.05 | 11.5 ± 1.2 | 0.09 | 1 | – |
| 7-deaza A | dTTP | 0.08 ± 0.002 | 185 ± 16 | 4.3 × 10–4 | 4.8 × 10–3 | 208 | |
| G | dCTP | 0.18 ± 0.009 | 18.8 ± 4.1 | 9.6 × 10–3 | 1 | – | |
| 7-deaza G | dCTP | 0.08 ± 0.004 | 241 ± 45 | 3.3 × 10–4 | 3.4 × 10–2 | 29 |
Efficiency of Incorrect Nucleotide Incorporation Opposite 7-Deaza-A and 7-Deaza-G by Polι. Next, we determined the efficiency of incorrect nucleotide incorporation by Polι opposite templates 7-deaza-A and 7-deaza-G in comparison with normal templates A and G under steady-state conditions. Table 2 gives the kinetic parameters for Polι for the incorporation of dGTP, dATP, and dCTP opposite A and 7-deaza-A, and for the incorporation of dGTP, dATP, and dTTP opposite G and 7-deaza-G. Interestingly, whereas the efficiency of dCTP incorporation opposite 7 deaza-A was reduced by 20-fold compared with that opposite A, the incorporation of dATP opposite 7 deaza-A was only ≈3-fold lower, and of dGTP was nearly the same as that opposite A. A similar pattern was seen for the incorporation of nucleotides opposite 7-deaza-G, where the incorporation of dTTP opposite 7-deaza-G was 25-fold lower than opposite normal G, and the incorporation of purine nucleotides was either unchanged, as for dATP, or actually much higher, ≈33-fold, for dGTP. Thus, whereas the incorporation of pyrimidines, whether correct or incorrect, is adversely affected by the modification of N7 of adenine or guanine, the misincorporation of purines remains unaffected, or is even enhanced.
Table 2. Efficiency of incorrect nucleotide incorporation by Polι opposite purines versus 7-deaza purines.
| Template base | Incoming nucleotide | kcat, min–1 | Km, μM | kcat/Km | Relative efficiency |
|---|---|---|---|---|---|
| A | dGTP | 0.029 ± 0.001 | 1140 ± 160 | 2.5 × 10–5 | – |
| 7-deaza-A | dGTP | 0.007 ± 0.0003 | 203 ± 49 | 3.4 × 10–5 | 1.4 |
| A | dATP | 0.030 ± 0.003 | 1,420 ± 310 | 2.1 × 10–5 | – |
| 7-deaza-A | dATP | 0.008 ± 0.0005 | 1,390 ± 220 | 5.8 × 10–6 | 0.28 |
| A | dCTP | 0.03 ± 0.002 | 2,210 ± 380 | 1.4 × 10–5 | – |
| 7-deaza-A | dCTP | ND* | ≥2500 | 6.8 × 10–7† | 0.05 |
| G | dGTP | 0.02 ± 0.002 | 3,430 ± 730 | 5.8 × 10–6 | – |
| 7-deaza-G | dGTP | 0.02 ± 0.001 | 104 ± 23 | 1.9 × 10–4 | 33 |
| G | dATP | 0.01 ± 0.001 | 905 ± 95 | 1.1 × 10–5 | – |
| 7-deaza-G | dATP | 0.01 ± 0.001 | 626 ± 130 | 1.6 × 10–5 | 1.5 |
| G | dTTP | 0.07 ± 0.002 | 63 ± 10 | 1.1 × 10–3 | – |
| 7-deaza-G | dTTP | 0.04 ± 0.003 | 860 ± 197 | 4.7 × 10–5 | 0.04 |
ND, Not determined
Because nucleotide incorporation remained linear throughout the dNTP concentration, the efficiency was determined from the slope of the line
Effects of Leu-62 → Ala Mutation on the Efficiency and Fidelity of Nucleotide Incorporation by Polι. In the ternary structure of Polι with a templating A and an incoming dTTP, a number of amino acid residues are apparently involved in conferring a syn conformation upon the templating nucleotide. Three amino acids, Leu-62, Val-64, and Gln-59 in the fingers domain of Polι bear down upon the templating A (Fig. 3), tilting and rotating it to the syn configuration with a significant restructuring of the sugar-phosphate backbone. The syn conformation is also stabilized by the sugar of templating A fitting into a small hydrophobic cavity formed by Gln-59 and Lys-60 (Fig. 3).
Fig. 3.
Surface representation of the Polι active site with the nascent base pair. In the A·T base pair, the templating A adopts a syn conformation and forms a Hoogsteen base pair with the incoming T, which retains the anti conformation. The amino acid residues that impact the template base and the incoming nucleotide are shown.
To begin to delineate the contributions of different residues in the Polι active site to conferring a syn conformation upon the template purines, we have begun a mutational analysis of residues that could effect this ability of Polι. Here, we report our results with mutational alteration of Leu-62 to Ala. We made this change because Leu-62 in Polι corresponds to Ala-42 in Dpo4, and because, in other Y family polymerases also, the structural equivalent of Leu-62 is a smaller residue, as for example, a Ser in yeast and human Polη.
We examined the efficiency and fidelity of nucleotide incorporation by the Polι Ala-62 mutant protein opposite template A and opposite template G by steady-state kinetic analyses. As shown in Table 3, compared with wild-type Polι, the catalytic efficiency of T incorporation opposite template A by the mutant protein is reduced by ≈7-fold, and the efficiency of C and T incorporation opposite template G by the mutant protein is reduced by over 6- and 30-fold, respectively. The misincorporation of purine nucleotides opposite templates A and G, however, is not as significantly affected by the Ala-62 mutation: the mutant protein incorporates a G opposite templates A and G, and also incorporates an A opposite template G, with almost the same efficiency as the wild-type protein, whereas A is incorporated opposite template A ≈4-fold less well. These observations with the Ala-62 mutation have allowed us to draw some inferences regarding the contributions of steric constraints imposed by Polι active site upon the templating base, and of hydrogen bonding involved in Hoogsteen base pair formation, to the incorporation of correct nucleotide opposite template purines. Additionally, studies with the mutant protein and those with the 7-deaza-A and 7-deaza-G templates have suggested that purine misincorporation opposite template purines occurs opposite an “abasic-like” template intermediate. We elaborate upon these points in Discussion.
Table 3. Efficiency of nucleotide incorporation by Polι Ala62 mutant.
| Template base | Incoming nucleotide | Enzyme | kcat, min–1 | Km, μM | kcat/Km | Relative efficiency |
|---|---|---|---|---|---|---|
| G | dGTP | WT | 0.037 ± 0.005 | 264 ± 87 | 1.4 × 10–4 | – |
| Ala62 | 0.018 ± 0.002 | 224 ± 66 | 8.5 × 10–5 | 0.6 | ||
| dATP | WT | 0.081 ± 0.004 | 705 ± 82 | 1.1 × 10–4 | – | |
| Ala62 | 0.038 ± 0.003 | 632 ± 106 | 6.0 × 10–5 | 0.5 | ||
| dTTP | WT | 0.15 ± 0.005 | 19.2 ± 3.8 | 7.8 × 10–3 | – | |
| Ala62 | 0.06 ± 0.005 | 273 ± 63 | 2.2 × 10–4 | 0.03 | ||
| dCTP | WT | 0.74 ± 0.032 | 14.5 ± 2.8 | 5.1 × 10–2 | – | |
| Ala62 | 0.3 ± 0.02 | 36 ± 5.8 | 8.3 × 10–3 | 0.16 | ||
| A | dGTP | WT | 0.069 ± 0.004 | 322 ± 54 | 2.1 × 10–4 | – |
| Ala62 | 0.05 ± 0.003 | 228 ± 30 | 2.2 × 10–4 | 1 | ||
| dATP | WT | 0.14 ± 0.005 | 175 ± 21 | 8.0 × 10–4 | – | |
| Ala62 | 0.06 ± 0.003 | 287 ± 43 | 2.1 × 10–4 | 0.26 | ||
| dTTP | WT | 3.1 ± 0.06 | 2.7 ± 0.2 | 1.1 | – | |
| Ala62 | 0.85 ± 0.04 | 5.5 ± 0.9 | 0.15 | 0.14 | ||
| dCTP | WT | 0.08 ± 0.006 | 888 ± 173 | 9.0 × 10–5 | – | |
| Ala62 | ND* | > 1,500† | 8.4 × 10–6 | 0.09 |
ND, Not determined
Because nucleotide incorporation remained linear throughout the dNTP concentration, the efficiency was determined from the slope of the line
Discussion
The inhibitory effects of 7-deaza-A and 7-deaza-G templates on DNA synthesis by Polι, as well as the large reduction in the efficiency of correct nucleotide incorporation opposite both these modified templates, provide strong support to the proposal that DNA synthesis by Polι requires Hoogsteen base pairing rather than normal W-C base pairing. The much higher efficiency and fidelity of nucleotide incorporation opposite template purines than opposite pyrimidines could then be ascribed to the templates A and G adopting a syn conformation in the polymerase active site and forming two hydrogen bonds by means of their Hoogsteen edge with the respective correct incoming pyrimidine, which retains the anti conformation. By contrast, the lack of a Hoogsteen edge on template pyrimidines would preclude hydrogen bond formation with the incoming nucleotide, accounting thereby for the highly inefficient incorporation of the correct nucleotide opposite templates T and C.
These studies provide biochemical evidence in support of the recently determined crystal structure of Polι bound to a template-primer and an incoming nucleotide (8). In this structure, the templating A adopts a syn conformation and forms two hydrogen bonds by means of its Hoogsteen edge (N7 and N6) with the W-C edge of the incoming dTTP (N3 and O4), which remains in the anti conformation. Our finding that the efficiency of dTTP incorporation opposite template 7-deaza-A is reduced by >200-fold than opposite template A provides strong biochemical evidence that the Hoogsteen base-pairing geometry seen in the structure occurs in solution, and thus is a valid reflection of its action mechanism. Additionally, these studies provide strong evidence that Polι utilizes Hoogsteen base pairing for replicating through a template G also. Based upon these observations, we conclude that Polι replicates through template purines by means of Hoogsteen base pairing, wherein the templating A or G residue adopts a syn conformation and the incoming pyrimidine nucleotide remains in the anti conformation.
In the Polι ternary structure, a number of amino acid residues (Leu-62, Val-64, Gln-59, and Lys-60) in the fingers domain that lie in close proximity to the templating base could have a role in imposing the syn conformation upon the templating nucleotide. We have examined the effects of mutational alteration of Leu-62 to Ala upon the efficiency and fidelity of nucleotide incorporation opposite the templates A and G. Interestingly, whereas the Ala-62 mutation lowers the efficiency of T incorporation opposite template A ≈7-fold, and of C and T incorporation opposite template G by over 6- and 30-fold, respectively, the efficiency of purine misincorporation opposite these templates is not as significantly affected. These observations taken together with our studies with the 7-deaza-A and 7-deaza-G templates have allowed us to draw some inferences about the mechanism of nucleotide incorporation by Polι opposite template purines, and they are discussed below.
Because the Ala-62 mutation has a much less debilitating effect on the ability of Polι to incorporate a correct pyrimidine nucleotide opposite the templating purine base than do the N7-modified purine bases, amino acid residues other than Leu-62 must additionally contribute to the adoption of a syn conformation by the templating base. Although a number of amino acid residues are apparently involved in conferring a syn conformation upon the templating nucleotide, our observations with the Ala-62 mutation suggest that the somewhat less constrained active site afforded by this mutational change reduces the proficiency of Polι for accommodating the template purines in a syn conformation, lessening thereby their ability for Hoogsteen base pair formation with the incoming pyrimidine nucleotide. However, the degree of adverse effect of the mutation upon pyrimidine nucleotide incorporation seems to be dependent upon the number of hydrogen bonds that could form within a Hoogsteen base pair. Thus, whereas the A·T Hoogsteen base pair has two hydrogen bonds between the N7 and N6 of template adenine and N3 and O4 of thymine, respectively, the G·C Hoogsteen base pair has two hydrogen bonds between the N7 and O6 of template guanine and N3 and N4 of cytosine, respectively, and the G·T Hoogsteen base pair has only one hydrogen bond that forms between the N7 of G and N3 of T. Because, in the absence of steric constraints that would be imposed by Leu-62 upon the templating base, the ability of Polι to incorporate a T opposite template A and to incorporate a C opposite template G is reduced only by 6- to 7-fold, we presume that the stability provided by the two hydrogen bonds in these base pairs compensates for the loss of steric constraints in the Ala-62 mutant protein. The >30-fold reduction in the efficiency of T incorporation opposite template G in the Ala-62 mutant protein could then be ascribed to the increased dependence upon the steric constraints of the active site, because, for this Hoogsteen base pair, hydrogen bonding will not provide the same measure of stability as for the A·T or G·C Hoogsteen base pair. From these observations with the Ala-62 mutant protein, we infer that both factors, the stability provided by the hydrogen bonding and the steric constraints of the active site, contribute to the efficiency of correct nucleotide incorporation opposite template purines by Polι.
Interestingly, because the misincorporation of an A oraGbyPolι is not reduced in the presence of 7-deaza-A or 7-deaza-G templates, this observation suggests that the inability of these template bases to engage in Hoogsteen base pairing has no adverse effect upon purine misincorporation. Further, because the Ala-62 mutation procures no significant effect upon purine misincorporation opposite template purines, this result suggests that the geometric constraints introduced by Leu-62 on the templating base are also of little consequence for this misincorporation. From these observations, we infer that, for purine misincorporation to occur opposite template purines, the templating nucleotide does not adopt a syn conformation in the Polι active site, but, instead, it assumes an extrahelical abasic-like structure, opposite which the polymerase then inserts an A or a G, because of their much better stacking ability than those of the two pyrimidines (19-21).
How could Hoogsteen base pairing contribute to lesion bypass ability of Polι? DNA polymerases form the four possible correct base pairs with nearly equal efficiencies because of the optimal geometrical alignment of the W-C A·T and G·C base pairs in their active sites. In addition to this geometric selection, DNA polymerases sense the correct W-C base pair by means of hydrogen bonding between the polymerase and the N3 (purine) and O2 (pyrimidine) atoms in the minor groove region of the duplex DNA (22-24), which occupy the same position for all four correct W-C base pairs but are in different geometric positions for mismatched base pairs. Hence, distortions of the DNA minor groove are inhibitory to synthesis by DNA polymerases. Hoogsteen base pairing by Polι provides for a mechanism to displace minor groove adducts of purines, such as those covalently attached to the N3 of AortheN2 group of G, into the major groove, where there is much less contact of the polymerase with the DNA. In support of this idea, we have recently shown that Polι is highly adept at incorporating a C opposite the γ-HOPdG adduct (25), which results from the reaction of acrolein, an α,β-unsaturated aldehyde, with the N2 of G in DNA followed by ring closure at N1. As a templating residue, the ring closed form of γ-HOPdG would be in the syn conformation, forming a Hoogsteen base pair with the C (26-28). The proficient ability of Polι to incorporate the correct nucleotide opposite such a lesion further suggests that Hoogsteen base pairing can additionally serve to promote replication through purine adducts that impinge upon the W-C base pairing geometry, but which do not affect the Hoogsteen base pairing geometry.
In conclusion, in its dependence upon Hoogsteen base pairing for DNA synthesis, Polι differs from all other known DNA polymerases. We suggest that this unusual replication mode endows Polι with the ability to incorporate nucleotides opposite highly distorting DNA lesions that impinge upon the DNA minor groove or that disturb the W-C edge of the templating purine.
Acknowledgments
We thank Aneel Aggarwal for discussions. This work was supported by U.S. National Institute of Environmental Health Sciences Grant ES012411.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: W-C, Watson-Crick; Polι, polymerase ι; 7-deaza-A, 7-deazaadenine; 7-deaza-G, 7-deazaguanine; KF exo-, exonuclease-deficient Klenow fragment.
References
- 1.Johnson, R. E., Washington, M. T., Haracska, L., Prakash, S. & Prakash, L. (2000) Nature 406, 1015-1019. [DOI] [PubMed] [Google Scholar]
- 2.Tissier, A., McDonald, J. P., Frank, E. G. & Woodgate, R. (2000) Genes Dev. 14, 1642-1650. [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang, Y., Yuan, F., Wu, X. & Wang, Z. (2000) Mol. Cell. Biol. 20, 7009-7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haracska, L., Johnson, R. E., Unk, I., Phillips, B. B., Hurwitz, J., Prakash, L. & Prakash, S. (2001) Proc. Natl. Acad. Sci. USA 98, 14256-14261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Washington, M. T., Johnson, R. E., Prakash, L. & Prakash, S. (2004) Mol. Cell. Biol. 24, 936-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodman, M. F. (1997) Proc. Natl. Acad. Sci. USA 94, 10493-10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kool, E. T. (2002) Annu. Rev. Biochem. 71, 191-219. [DOI] [PubMed] [Google Scholar]
- 8.Nair, D. T., Johnson, R. E., Prakash, S., Prakash, L. & Aggarwal, A. K. (2004) Nature 430, 377-380. [DOI] [PubMed] [Google Scholar]
- 9.Johnson, R. E., Yu, S.-L., Prakash, S. & Prakash, L. (2003) Genes Dev. 17, 77-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haracska, L., Johnson, R. E., Unk, I., Phillips, B., Hurwitz, J., Prakash, L. & Prakash, S. (2001) Mol. Cell. Biol. 21, 7199-7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haracska, L., Unk, I., Johnson, R. E., Phillips, B. B., Hurwitz, J., Prakash, L. & Prakash, S. (2002) Mol. Cell. Biol. 22, 784-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Creighton, S., Bloom, L. B. & Goodman, M. F. (1995) Methods Enzymol. 262, 232-256. [DOI] [PubMed] [Google Scholar]
- 13.Leontis, N. B., Stombaugh, J. & Westhof, E. (2002) Nucleic Acids Res. 30, 3497-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Innis, M. A., Myambo, K. B., Gelfand, D. H. & Brow, M. A. D. (1988) Proc. Natl. Acad. Sci. USA 85, 9436-9440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baran, N., Lapidot, A. & Manor, H. (1991) Proc. Natl. Acad. Sci. USA 88, 507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jung, A., Ruckert, S., Frank, P., Brabletz, T. & Kirchner, T. (2002) J. Clin. Pathol. Mol. Pathol. 55, 55-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prakash, S. & Prakash, L. (2002) Genes Dev. 16, 1872-1883. [DOI] [PubMed] [Google Scholar]
- 18.Prakash, S., Johnson, R. E. & Prakash, L. (2005) Annu. Rev. Biochem. 74, 317-353. [DOI] [PubMed] [Google Scholar]
- 19.Bommarito, S., Peyret, N. & Santa Lucia, J. (2000) Nucleic Acids Res. 28, 1929-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guckian, K. M., Ren, R. X. F., Chaudhuri, M. C., Tahmassebi, D. C. & Kool, E. T. (2000) J. Am. Chem. Soc. 122, 2213-2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kool, E. T. (2001) Annu. Rev. Biophys. Biomol. Struct. 30, 1-22. [DOI] [PubMed] [Google Scholar]
- 22.Doublie, S., Tabor, S., Long, A. M., Richardson, C. C. & Ellenberger, T. (1998) Nature 391, 251-258. [DOI] [PubMed] [Google Scholar]
- 23.Li, Y., Korolev, S. & Waksman, G. (1998) EMBO J. 17, 7514-7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, Y. & Waksman, G. (2001) Protein Sci. 10, 1225-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Washington, M. T., Minko, I. G., Johnson, R. E., Wolfle, W. T., Harris, T. M., Lloyd, R. S., Prakash, S. & Prakash, L. (2004) Mol. Cell. Biol. 24, 5687-5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de los Santos, C., Zaliznyak, T. & Johnson, F. (2001) J. Biol. Chem. 276, 9077-9082. [DOI] [PubMed] [Google Scholar]
- 27.Singh, U. S., Moe, J. G., Reddy, G. R., Weisenseel, J. P., Marnett, L. J. & Stone, M. P. (1993) Chem. Res. Toxicol. 6, 825-836. [DOI] [PubMed] [Google Scholar]
- 28.Weisenseel, J. P., Reddy, G. R., Marnett, L. J. & Stone, M. P. (2002) Chem. Res. Toxicol. 15, 127-139. [DOI] [PubMed] [Google Scholar]