Those of you, like this author, who have managed to stay alive for close to eight decades or more, will have had the experience of observing increasing degrees of phenotypic discordances among our identical twin friends as we age together. They may succumb to the same disease, but often the age of onset is years or even decades apart. An interesting example is a report of twins, both of whom developed histologically confirmed dementia of the Alzheimer's type; one had the diagnosis in her late 60s but the other was not diagnosed until age 83 (1). The conventional wisdom is that the bulk of these differences can be attributable to good or bad luck with one's environmental exposures, to the quality of medical and spousal care (or abuse), or to learned behavioral differences in how we exercise (perhaps mentally and physically) and what we eat, drink, or smoke, among myriad other possibilities. Many have assumed that, apart from some rare point mutations or aberrant chromosomal segregations and their impacts upon the emergence of neoplastic diseases, the differences we see are not due to heritable changes in cells. There is growing interest, however, in that class of potentially heritable change in DNA that is not associated with alterations in the primary nucleotide sequence or copy number but rather is modulated by what Joshua Lederberg (2) long ago referred to as “epinucleic.” Modern molecular genetics now points to two major classes of such alterations: methylations of cytosine at cytosine-guanine dinucleotides and covalent modifications of DNA-bound histones, notably acetylations and methylations. Cytosine methylations at regions of gene promoters rich in CpG islands are generally associated with the silencing of genes, whereas histone acetylations are generally associated with the activation of genes.
Gene Expression Studies Reflect Increase in Variation Between Twin Pairs as They Age
In this issue of PNAS, a group of Spanish, Swedish, Danish, English, and American investigators report their findings of both global and locus-specific differences in DNA methylation and histone acetylation in identical twins of various ages (3). Their general conclusion is that whereas young identical twin pairs are essentially indistinguishable in their epigenetic markings, older identical twin pairs show substantial variations. Moreover, and of considerable importance, were their studies of gene expression in these aging twin pairs. Differences in gene expression among older twin pairs were some four times greater than those observed in young twin pairs.
There is widespread “epigenetic drift” associated with aging.
The studies reported by Fraga et al. (3) in this issue of PNAS mostly used peripheral blood lymphocytes, but a smaller number of samples of buccal mucosal epithelial cells, skeletal muscle biopsies, and aspirations of s.c. fat provided results consistent with the conclusions from the research with lymphocytes. The authors therefore concluded that there is indeed widespread “epigenetic drift” associated with aging. Their article cites other lines of evidence consistent with an important role of epigenetic alterations in aging mammalian tissues. A particularly nice example is one reported from the David Burke laboratory at the University of Michigan. Their studies of age-associated activation of epigenetically repressed genes in aging mouse tissues indicate that they are perhaps two orders of magnitude greater than somatic mutations (4). The present study is by far the most comprehensive and detailed study of age-associated epigenetic changes so far reported, certainly for the case of human subjects. It is a technical tour de force with the utilization of a battery of powerful molecular genetic methodologies coupled with competitive chromosomal hybridizations.
The reported epigenetic shifts in these aging identical twins could have arisen through endogenous, stochastic mechanisms, independent of environmental perturbations, or could have resulted from such environmental perturbations. The fact that there was an association between the extent of environmental differences between twins (lifestyles, time spent together, etc.) and the degree of epigenetic shifts cannot definitively answer that question. Moreover, one cannot know from this data the extent to which specific gene alterations were adaptive or nonadaptive. Although the classic evolutionary biological theory of why aging exists argues that senescent phenotypes are nonadaptive (5), compensatory changes in gene expression can continue for some decades after the peak of reproductive activity; I have referred to such compensatory changes in gene expression as “sageing” (6). One can imagine various degrees of the efficiency of such compensations, thus leading to widening discordances among aging identical twins.
Definitive Analyses Require Special Methodologies to Account for Shifts in Cellular Population Heterogeneity
I have one technical concern about Fraga et al.'s (3) interpretations of their results, however. Shifts in cellular population heterogeneity within mammalian tissues during aging are exceedingly well established and have, in fact, been one of the major stumbling blocks of biochemical investigations of aging for many decades. For the case of lymphocytes, the major cell types used for this study, it is clear that, as people age, samples of peripheral blood have proportionately more memory T cells and fewer naïve T cells (7). There are also nonmalignant clonal expansions of classes of memory T cells in aged individuals (8). Each type of cell can be expected to have a characteristic profile of gene expression. Thus, when admixtures change during aging, the results of studies that use DNA and RNA from the mass population may not reflect a change in expression for a given specific cell type but only a shift in the proportions of different cell types in the sample. Similar arguments can be made for the case of the other tissues examined. Older individuals often have periodontal disease. Scrapings of their buccal mucosa may therefore include variable amounts of inflammatory cells and, perhaps, vascular cells, as well as buccal mucosal epithelial cells; the proportion of basal stem cells and stages of differentiated cells within the epithelium may be altered in aged, inflamed mucosa (9). As a pathologist, I have observed variable admixtures of fibrous connective tissue and adipocytes in the skeletal muscles of aging human subjects. s.c. fat tissue is also heterogeneous, with variable admixtures of histiocytes, fibroblasts, and vascular tissues; there are also distinct classes of preadipocytes (10). More definitive analyses of epigenetic shifts in aging twins would therefore have to use such techniques as flow cytometric separations of specific cell types; these methods are particularly well developed for the case of human peripheral blood lymphocytes (11).
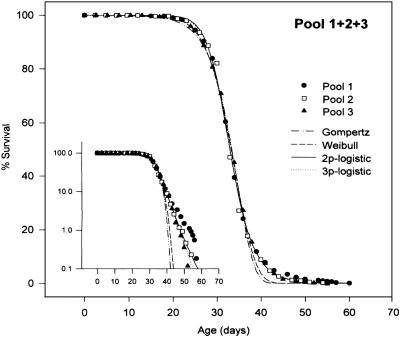
The last word has yet to be written on the range of somatic mutational events of relevance to the pathobiology of aging. Stochastic events such as retrotranspositions by long interspersed nuclear element-1 (12), point mutations of microRNAs (13), and segmental duplications and haploinsufficiencies (14, 15) could also contribute to our observations that some identical twins seem to age more rapidly than their siblings. My guess, however, is that epigenetic shifts will prove to be the most significant class of altered genetic expression, despite the caveats expressed in the preceding paragraph. We should also keep in mind that biogerontologists work with hundreds of genetically identical individuals in their experiments. Moreover, they do a very good job at controlling environmental variations. Perhaps the most striking example is the aging of Caenorhabditis elegans grown in axenic media in suspension cultures (16) (Fig. 1). In all these experiments, we see survival curves that reflect marked variations in ages of death. Are stochastic epigenetic shifts responsible for the generation of worms, fruit flies, mice, and humans destined to have comparatively short or long lives? Could these shifts be taking place during early development, thus setting the stage for differential phenotypic expressions in late life?
Fig. 1.
Genetically identical animals aged under rigorously controlled environments (in this case, C. elegans worms in spinner cultures) (16) exhibit wide ranges of lifespans. Such survival curves have been observed for inbred and outbred strains of fruit flies, mice, rats, and hamsters, among many other species whose lifespans have been observed under controlled experimental conditions. Such curves are also typical of outbred humans. The data in this figure (and in many other recent studies employing very large numbers of animals) (17) do not fit the classic Gompertz model of a continuous exponential increase in mortality. [Reproduced with permission from ref. 16 (Copyright 1998, The Gerentological Society of America).]
See companion article on page 10604.
References
- 1.Cook, R. H., Schneck, S. A. & Clark, D. B. (1981) Arch. Neurol. 38, 300-301. [DOI] [PubMed] [Google Scholar]
- 2.Lederberg, J. (1958) J. Cell. Comp. Physiol. 52, 383-401. [Google Scholar]
- 3.Fraga, M. F., Ballestar, E., Paz, M. F., Ropero, S., Setien, F., Ballestar, M. L., Heine-Suñer, D., Cigudosa, J. C., Urioste, M., Benitez, J., et al. (2005) Proc. Natl. Acad. Sci. USA 102, 10604-10609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett-Baker, P. E., Wilkowski, J. & Burke, D. T. (2003) Genetics 165, 2055-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirkwood, T. B. (2005) Cell 120, 437-447. [DOI] [PubMed] [Google Scholar]
- 6.Martin, G. M. (1997) Philos. Trans. R. Soc. London B 352, 1773-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linton, P. J., Li, S. P., Zhang, Y., Bautista, B., Huynh, Q. & Trinh, T. (2005) Immunol. Rev. 205, 207-219. [DOI] [PubMed] [Google Scholar]
- 8.Clambey, E. T., van Dyk, L. F., Kappler, J. W. & Marrack, P. (2005) Immunol. Rev. 205, 170-189. [DOI] [PubMed] [Google Scholar]
- 9.Celenligil-Nazliel, H., Ayhan, A., Uzun, H. & Ruacan, S. (2000) J. Periodontol. 71, 1567-1574. [DOI] [PubMed] [Google Scholar]
- 10.Tchkonia, T., Tchoukalova, Y. D., Giorgadze, N., Pirtskhalava, T., Karagiannides, I., Forse, R. A., Koo, A., Stevenson, M., Chinnappan, D., Cartwright, A., et al. (2005) Am. J. Physiol. 288, E267-E277. [DOI] [PubMed] [Google Scholar]
- 11.O'Gorman, M. R. & Nicholson, J. K. (2000) Clin. Diagn. Lab. Immunol. 7, 333-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muotri, A. R., Chu, V. T., Marchetto, M. C., Deng, W., Moran, J. V. & Gage, F. H. (2005) Nature 435, 903-910. [DOI] [PubMed] [Google Scholar]
- 13.Bentwich, I., Avniel, A., Karov, Y., Aharonov, R., Gilad, S., Barad, O., Barzilai, A., Einat, P., Einav, U., Meiri, E., et al. (2005) Nat. Genet. 37, 776-770. [DOI] [PubMed] [Google Scholar]
- 14.Sebat, J., Lakshmi, B., Troge, J., Alexander, J., Young, J., Lundin, P., Maner, S., Massa, H., Walker, M., Chi, M., et al. (2004) Science 305, 525-528. [DOI] [PubMed] [Google Scholar]
- 15.Sharp, A. J., Locke, D. P., McGrath, S. D., Cheng, Z., Bailey, J. A., Vallente, R. U., Pertz, L. M., Clark, R. A., Schwartz, S., Segraves, R., et al. (2005) Am. J. Hum. Genet. 77, 78-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vanfleteren, J. R., De, V. A. & Braeckman, B. P. (1998) J. Gerontol. A Biol. Sci. Med. Sci. 53, B393-B403. [DOI] [PubMed] [Google Scholar]
- 17.Vaupel, J. W., Carey, J. R., Christensen, K., Johnson, T. E., Yashin, A. I., Holm, N. V., Iachine, I. A., Kannisto, V., Khazaeli, A. A., Liedo, P., et al. (1998) Science 280, 855-860. [DOI] [PubMed] [Google Scholar]