Abstract
Methylenetetrahydrofolate reductase (MTHFR) catalyzes the reduction of methylenetetrahydrofolate to methyltetrahydrofolate, the methyl donor for the conversion of homocysteine to methionine. Regulation of MTHFR activity is crucial for maintaining cellular concentrations of methionine and S-adenosylmethionine (AdoMet). Purified recombinant human MTHFR expressed in insect cells is multiply phosphorylated on an N-terminal extension of the protein that contains a highly conserved serine-rich region. Treatment by alkaline phosphatase removes seven phosphoryl groups from the enzyme. Thr-34 was identified as one of the seven phosphorylation sites by using a monoclonal antibody directed toward pThr-Pro. Mutation of Thr-34 to Ala completely blocks modification as judged by mass spectrometric analysis, suggesting that Thr-34 is the priming phosphorylation site. The Thr34Ala mutant was expressed in baculovirus-infected insect cells, and its enzymic properties were compared with wild-type enzyme. The mutant enzyme and alkaline phosphatase-treated wild-type enzyme are more active than untreated wild-type enzyme and less sensitive to inhibition by saturating AdoMet, indicating that phosphorylation at Thr-34 is critical for allosteric regulation of human MTHFR activity by AdoMet. The absence of methionine and the presence of adenosine in the cell culture medium, which lead to a low intracellular AdoMet/S-adenosylhomocysteine ratio, are associated with faster electrophoretic mobility of MTHFR, presumably because of less or no phosphorylation. Because the faster-mobility MTHFR is associated with the more active form of MTHFR, this response is likely to increase methionine production. Those observations suggest that AdoMet functions not only as an allosteric inhibitor but also to control phosphorylation of human MTHFR.
Keywords: homocysteine, posttranslational modification
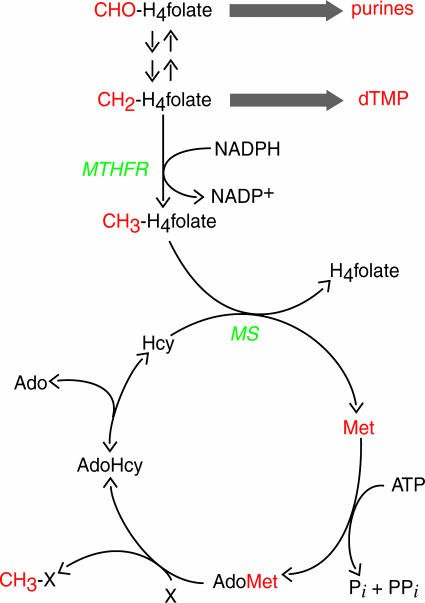
Methylenetetrahydrofolate reductase (MTHFR) is a flavoprotein that catalyzes reduction of methylenetetrahydrofolate to methyltetrahydrofolate by using NADPH as the source of reducing equivalents (1). As shown in Fig. 1, MTHFR controls the partitioning of tetrahydrofolate-bound one-carbon units between nucleotide biosynthesis and remethylation of homocysteine to form methionine. The product of the MTHFR reaction, methyltetrahydrofolate, can be used only by methionine synthase, which will remove the methyl group and transfer it to homocysteine to form tetrahydrofolate and methionine. Thus, the reaction of MTHFR commits tetrahydrofolate-bound one-carbon units to use in methionine regeneration. Regulation of MTHFR activity plays a critical role in controlling the level of homocysteine in plasma, and even mild MTHFR deficiencies are associated with elevated plasma homocysteine (2). Elevated plasma homocysteine in humans is associated with increased risk for cardiovascular disease (3) and, in women, with increased incidence of neural tube defects in their offspring (4, 5).
Fig. 1.
One-carbon metabolism in mammalian cells. One-carbon units and metabolites incorporating these one-carbon units are shown in red. CHO-H4folate, 10-formyltetrahydro-folate; dTMP, deoxythymidine monophosphate; Hcy, homocysteine; MS, methionine synthase, X, methyl acceptor.
Methionine is an essential amino acid in humans, serving both for protein synthesis and as the precursor to the active methyl donor S-adenosylmethionine (AdoMet). Although homocysteine can be converted to methionine by the action of methionine synthase, humans are unable to synthesize either compound de novo. AdoMet can donate its methyl group to many substrates and form S-adenosylhomocysteine (AdoHcy). AdoHcy is hydrolyzed by the function of adenosylhomocysteine hydrolase into adenosine and homocysteine. Because the hydrolysis of AdoHcy is reversible, elevated levels of homocysteine lead to reduced intracellular ratios of AdoMet/AdoHcy, inhibiting the activities of many AdoMet-dependent methyltransferases (6).
The activity of mammalian MTHFR is allosterically inhibited by AdoMet (7, 8). Mammalian MTHFR enzymes are homodimers in which each subunit contains an N-terminal catalytic domain of ≈40 kDa and a C-terminal regulatory domain of ≈37 kDa (9-11). The C-terminal domain has been shown to bind AdoMet (12). The two domains can be separated by partial digestion of the native enzyme with trypsin, and longer digestion results in further digestion of the smaller regulatory domain (11). After tryptic treatment, mammalian MTHFR is still active but has lost sensitivity to AdoMet. AdoHcy blocks the inhibitory effect of AdoMet on MTHFR activity (7) but does not itself alter enzyme activity (13). Thus, the activity of mammalian MTHFR is regulated by the AdoMet/AdoHcy ratio in the cell.
In this article, we describe the posttranslational modification of human MTHFR by phosphorylation and demonstrate that phosphorylation is regulated by the AdoMet/AdoHcy ratio in the cell. Phosphorylation leads to decreased activity and increased sensitivity to allosteric inhibition by AdoMet.
Methods
Reagents. Calf intestine alkaline phosphatase was purchased from Roche. Modified trypsin (sequencing grade) was obtained from Promega. Custom DMEM lacking one or more amino acids was prepared from cell culture-grade reagents purchased from Sigma. Lys-C was from Wako, and Glu-C was from Roche.
Expression Vectors and cDNA Constructs. pProExHT, pFastBacHT (Invitrogen), and pcDNA3HA (laboratory of K. L. Guan, University of Michigan) plasmids were used for expression of human MTHFR in Escherichia coli, insect, and human cells, respectively. The wild-type cDNA for human MTHFR was derived from pTrcAE (14). pProExHT(hMRwt) was constructed as follows. pProExHT and pTrcAE were digested by NcoI and HindIII, and the DNA fragments were separated by electrophoresis in 1% agarose gel. After the appropriate DNA fragments were purified, they were ligated by using T4 DNA ligase. The expression construct used with the baculovirus system was described in ref. 15. pcDNA3(hMRwt) was constructed as follows. pcDNA3HA was digested with BglII and HindIII or with BglII and NcoI (partial digestion). pTrcAE was digested with NcoI and HindIII. After purification by electrophoresis in an agarose gel, the appropriate DNA fragments were joined by using T4 DNA ligase. Because of this procedure, the expression vector lacks the hemagglutinin epitope present in the original vector and can be used to express untagged protein. Site-directed mutagenesis of human MTHFR cDNA was performed by using the QuikChange kit (Stratagene) according to the manufacturer's instructions. All constructs were confirmed by sequencing both strands of the MTHFR cDNA.
Protein Expression. Plasmid pProEXHT(hMRwt) was used to transform the E. coli K-12 strain XL1-Blue for expression of human MTHFR in E. coli. The strain was propagated in LB medium containing ampicillin but lacking isopropyl-β-d-thiogalactopyranoside. Cells were harvested and stored at -80°C.
Recombinant baculovirus containing the N-terminally histidine-tagged human MTHFR cDNA was generated by using the Bac-to-Bac expression system (Invitrogen) as described in ref. 15. Human MTHFR was expressed in Spodoptera frugiperda Sf9 cells that were maintained in the serum-free medium Sf900II SFM (Invitrogen) at 27°C. The histidine-tagged protein was purified by chromatography on a DE-52 column (Whatman) followed by a Hi-Trap Chelating HP column (Amersham Pharmacia) charged with NiSO4.
Expression of human MTHFR in human embryonic kidney (HEK) 293 cells was accomplished by transient transfection with pcDNA3(hMRwt) by using Lipofectamine (Invitrogen). The cells were maintained at 37°C under 5% CO2 in DMEM (Invitrogen) containing 10% FBS and a penicillin-streptomycin mixture (Invitrogen).
Western Blotting. Proteins were separated by electrophoresis under denaturing conditions (SDS/PAGE) (16) and transferred to a nitrocellulose membrane. Human MTHFR was visualized by using anti-human MTHFR rabbit antiserum, alkaline phosphatase-conjugated anti-rabbit IgG (Sigma), 5-bromo-4-chloro-3-indolyl phosphate, and nitroblue tetrazolium chloride (Invitrogen). The specific rabbit antiserum was generated by using histidine-tagged human MTHFR purified from baculovirus-infected insect cell cultures.
Enzyme Assay. MTHFR activity was determined by using NADPH-menadione oxidoreductase, NADPH-methylenetetrahydrofolate oxidoreductase, or methyltetrahydrofolate-menadione oxidoreductase assays as described in refs. 1 and 15.
MS. Purified human MTHFR from baculovirus-infected insect cells was digested with trypsin, Lys-C, or Glu-C. The proteolytic fragments were desalted and partially fractionated by using C18-ZipTips (Millipore). Peptides were sequentially eluted with 20%, 40%, and 70% acetonitrile in aqueous 0.1% trifluoracetic acid (TFA). MALDI-MS and tandem MS (MS/MS) analyses were performed by using α-cyano 4-hydroxy cinnamic acid (2 mg/ml in 50% acetonitrile/50% aqueous 0.1% TFA) containing ammonium citrate (10 mM) as matrix. Spectra were acquired on a 4700 Proteomics Analyzer (Applied Biosystems). Peptide identifications were performed by using gps explorer (Version 2.0, Applied Biosystems), which acts as a front end for the mascot search engine (version 1.9, Matrix Science, London). Each MS/MS spectrum was searched against the human MTHFR sequence alone and against the NCBInr database (July 2004).
For determination of the molecular mass of intact human MTHFR before and after treatment with alkaline phosphatase, purified human MTHFR from baculovirus-infected insect cells was subjected to liquid chromatography-MS (LC-MS) as described for the analysis of cobalamin-independent methionine synthase (17). Mass spectral data were obtained in the positive mode by using a ThermoFinnigan LCQ mass spectrometer, and ESI spectra were deconvoluted by using biomass software provided by ThermoFinnigan.
Results
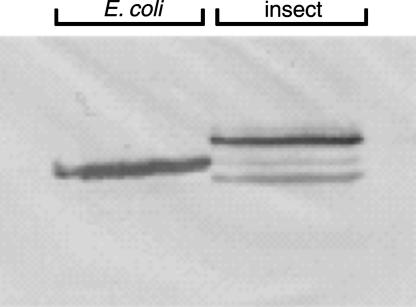
The Electrophoretic Mobility of Human MTHFR Depends on the Expression System. The apparent molecular weights of histidine-tagged human MTHFRs expressed in E. coli and insect cells were compared.§ Fig. 2 shows a Western blot using antiserum raised against human MTHFR. Because of the low expression level, the expressed human MTHFR in E. coli cells was partially purified by using Ni-affinity chromatography, and >100× diluted crude extract from Sf9 cells was used for this analysis. The enzyme expressed in E. coli shows clearly increased mobility, suggesting posttranslational modification of the human MTHFR expressed in insect cells. The minor band with greater mobility seen in the Sf9 cells may represent endogenous insect MTHFR, which would be expected to crossreact with antiserum against the human enzyme.
Fig. 2.
Expression of human MTHFR in baculovirus-infected insect cells results in a protein with decreased mobility as compared with expression of the same construct in E. coli. N-terminally histidine-tagged human MTHFR was expressed in the E. coli K-12 strain XL1-Blue (left) or in recombinant baculovirus-infected insect Sf9 cells (right). Polypeptides were detected by Western blotting with anti-human MTHFR rabbit antiserum.
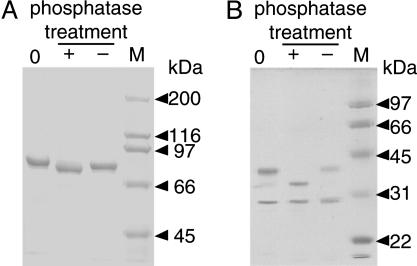
Alkaline Phosphatase Treatment Alters the Electrophoretic Mobility of Human MTHFR Expressed in Baculovirus Cultures. Factors that could change the apparent molecular weight upon electrophoresis under denaturing conditions were screened by using purified human MTHFR isolated from baculovirus-infected insect cells. Glycosylation of the purified human MTHFR was found to be unlikely by our preliminary analysis using periodic acid/Schiff stain (Sigma) and monoclonal antibody for O-linked N-acetylglucosamine (Covance). As shown in Fig. 3A, alkaline phosphatase treatment increased the mobility of the enzyme, indicating that the purified protein is phosphorylated.
Fig. 3.
Alkaline phosphatase treatment leads to increased mobility of human MTHFR. (A) SDS/PAGE analysis of purified full-length human MTHFR from Sf9 cells. (B) Analysis of human MTHFR after tryptic cleavage of the native enzyme (0), enzyme as isolated, or after incubation of enzyme with (+) or without (-) addition of alkaline phosphatase for 1 h at 37°C. Molecular mass markers are shown in the rightmost lane of each panel.
After limit digestion of the native enzyme with trypsin, the electrophoretic mobility of the larger N-terminal catalytic domain was dramatically increased upon alkaline phosphatase treatment, whereas no effect on the mobility of the smaller regulatory domain was seen (Fig. 3B). N-terminal sequencing of a tryptic digest conducted under similar conditions established that the N-terminal fragment began at residue 7 whereas the C-terminal fragment began at residue 410. This experiment suggests that phosphorylation occurs on the N-terminal domain.
Attempts to Detect Phosphopeptides Using MALDI-MS. Intact and iodoacetamide-treated purified human MTHFR (wild type) from the insect cell expression system were analyzed. Protein samples were digested by trypsin, Lys-C, or Glu-C, and then the proteolytic fragments were analyzed by MALDI-MS. A summary of these analyses is shown in Fig. 4. These analyses did not reveal the phosphorylation sites, although 88% of the expected sequence was recovered. However, three large regions, indicated by yellow highlighting, were not detected.
Fig. 4.
MALDI-MS analysis of human MTHFR peptides limits possible phosphorylation sites. The amino acid sequence of histidine-tagged human MTHFR is shown. Digestions of human MTHFR were performed by using trypsin, Lys-C, or Glu-C. Regions of the protein that were not recovered are highlighted in yellow. Regions of the protein that were recovered constitute 88% of the sequence, and no modifications other than N-terminal acetylation were identified. Arrows indicate the start and end of the catalytic domain based on the crystal structure of E. coli MTHFR (24). The boxed sequence KRREED is thought to be the most sensitive position for initial tryptic cleavage of native enzyme, which separates the catalytic and regulatory domains. After extensive digestion of the native enzyme with trypsin, N-terminal amino acid analysis indicated sequences *GNSSL and *SKSPK, respectively.
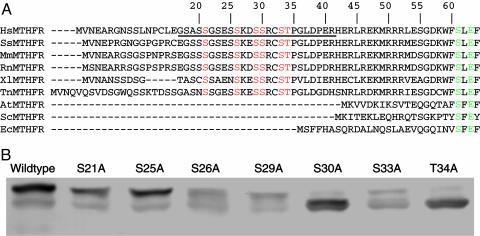
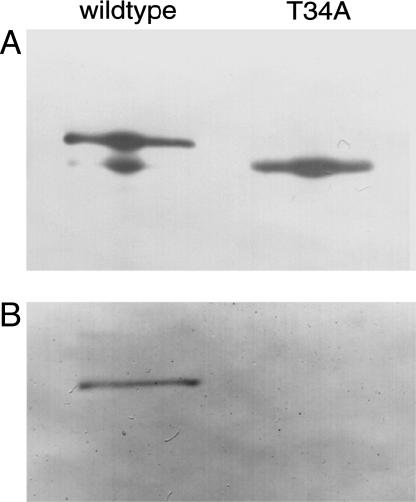
Mutation of Conserved Serines and Threonines in the N-Terminal Extension. Although the identification of phosphorylated peptides by using MALDI-MS failed, those analyses were still useful, because the phosphorylation sites presumably lie in regions undetected by MALDI-MS analysis. Because the results of the experiment shown in Fig. 3B indicated that phosphorylation was occurring in the N-terminal catalytic domain, we focused on the serine-rich region comprising amino acids 18-34 in the MTHFR sequence. This region is contained in an N-terminal extension found only in vertebrate MTHFRs. The sequence alignment of vertebrate sequences shown in Fig. 5A shows that there are six conserved serine and threonine residues in this region. Conserved serine and threonine residues in this region were mutated to Ala. The mutants were expressed in HEK293 cells and analyzed by Western blotting (Fig. 5B). A Thr34Ala mutation led to increased mobility of the protein as compared with the wild-type enzyme. Phosphorylation of Thr-34 (the only threonine residue in human MTHFR that is followed by a proline) was confirmed by using a commercially available monoclonal antibody against pThr-Pro (Cell Signaling Technology, Beverly, MA) as shown in Fig. 6.
Fig. 5.
Mutational analysis of conserved serine and threonine residues in the N-terminal extension of human MTHFR. (A) Alignment of vertebrate sequences of the N-terminal extension of MTHFR. Abbreviations are as follows, with GenBank accession nos. in parentheses: HsMTHFR, Homo sapiens MTHFR (U09806); SsMTHFR, Sus scrofa MTHFR (AW619446); MmMTHFR, Mus musculus MTHFR (BC051017); RnMTHFR, Rattus norvegicus MTHFR (U57049); XlMTHFR, Xenopus laevis MTHFR (BC046708); TnMTHFR, MTHFR from Tetraodon nigroviridis (CAF90576); AtMTHFR, Arabidopsis thaliana MTHFR (AF181966); ScMTHFR, Saccharomyces cerevisiae MTHFR (Z72647); EcMTHFR, E. coli MTHFR (AAN83327). Conserved serine and threonine residues in vertebrate MTHFRs are shown in red letters. Amino acids that are conserved in all MTHFR sequences are shown in green letters. The region of the human N-terminal extension that was not recovered in MALDI-MS analysis is underlined. (B) Mutant proteins were expressed in transiently transfected HEK293 cells cultured in DMEM, and the expressed human MTHFR proteins were detected by Western blotting after SDS/PAGE.
Fig. 6.
Confirmation of phosphorylation at Thr-34. Wild-type and Thr34Ala MTHFR were expressed in HEK293 cells and purified by immunoprecipitation using antiserum against human MTHFR and protein G-Sepharose. The expressed human MTHFR proteins were detected by Western blotting after SDS/PAGE. Five times as much sample was loaded in B as in A to ensure sufficient sensitivity. Antiserum against human MTHFR and anti-pThr-Pro antibody were used for A and B, respectively.
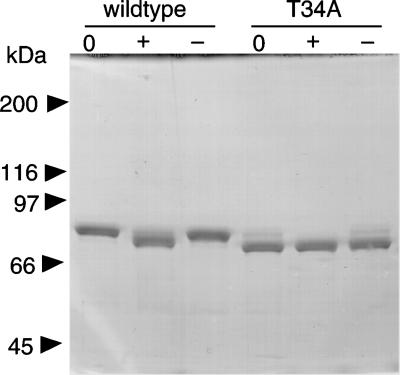
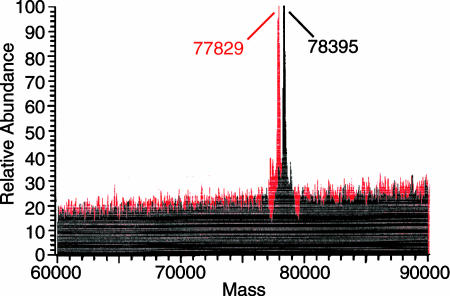
Comparison of the Molecular Weights of Purified Wild-Type and Thr34Ala Mutant Enzymes. Histidine-tagged Thr34Ala human MTHFR was expressed in baculovirus-infected insect cells. A comparison of the effect of alkaline phosphatase treatment on electrophoretic mobility of the mutant and wild-type proteins is shown in Fig. 7. We determined the molecular masses of the proteins by LC-MS: the molecular masses of the purified and phosphatase-treated wild-type enzyme were 78,395 and 77,829 Da, respectively (Fig. 8), and the calculated mass for the unmodified protein was 77,602 Da. Because the mass change due to removal of a phosphoryl group is 80 Da, we calculated that seven phosphate groups were hydrolyzed by alkaline phosphatase from the purified wild-type enzyme. The molecular mass of the alkaline phosphatase-treated wild-type enzyme was still larger than expected, and the mass difference between wild-type MTHFR treated with alkaline phosphatase and the predicted value remains to be explained. The mass of the untreated wild-type enzyme was determined four times, on two different LC-MS instruments, and from three different preparations; the masses were 78,372 ± 21, indicating that the accuracy was ≈0.03%.
Fig. 7.
Effect of phosphatase treatment on the mobility of the Thr34Ala mutant of human MTHFR. Purified human MTHFR was incubated at 37°C with or without alkaline phosphatase and then subjected to SDS/PAGE, as described in the legend to Fig. 3.
Fig. 8.
Alkaline phosphatase treatment leads to removal of seven phosphoryl groups from human MTHFR as analyzed by LC-MS. Peaks obtained by deconvolution of the electrospray spectra of purified human MTHFR (wild-type) with (red) or without (black) alkaline phosphatase treatment are shown.
The mobility of the Thr34Ala mutant was not changed by phosphatase treatment, and the measured molecular mass was 77,552 Da, within 0.03% of the predicted mass of 77,572 Da (data not shown). Thus, we believe that Thr-34 may be the first residue in MTHFR to be phosphorylated, and, once phosphorylated, it may serve as a priming site for recognition by subsequent kinases.
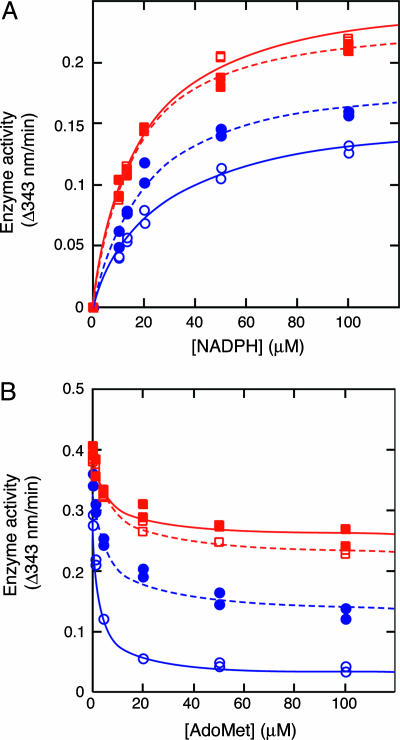
Effects of Phosphorylation on Human MTHFR Activity. Basic kinetic parameters were measured by using the NADPH-menadione oxidoreductase assay (Fig. 9). Alkaline phosphatase-treated wild-type enzyme was 1.2 times more active at saturating NADPH than untreated enzyme (Fig. 9A). The Thr34Ala mutant was more active at saturating NADPH than either treated or untreated wild-type enzyme. Alkaline phosphatase treatment and the mutation did not affect the apparent affinity for NADPH (Fig. 9A) or the apparent Ki for AdoMet (Fig. 9B). However, the degree of inhibition at saturating AdoMet was dramatically reduced. The purified wild-type enzyme was ≈80% inhibited by AdoMet. The phosphatase-treated wild-type enzyme was only ≈60% inhibited by AdoMet, and the Thr34Ala mutant was only ≈40% inhibited.
Fig. 9.
Effects of alkaline phosphatase treatment or mutation on human MTHFR activity. The NADPH-menadione oxidoreductase assay was used to measure activity after preincubation of the enzyme with 100 μM NADPH and AdoMet at the indicated concentration. (A) Effect of alkaline phosphatase treatment on the activity of purified wild-type and Thr34Ala human MTHFR. (B) Effect of alkaline phosphatase treatment and/or mutation of Thr34Ala on inhibition of NADPH-menadione oxidoreductase activity by AdoMet. Shown are wild-type MTHFR with (•) or without (○) alkaline phosphatase treatment and Thr34Ala mutant MTHFR with (▪) or without (□) alkaline phosphatase treatment.
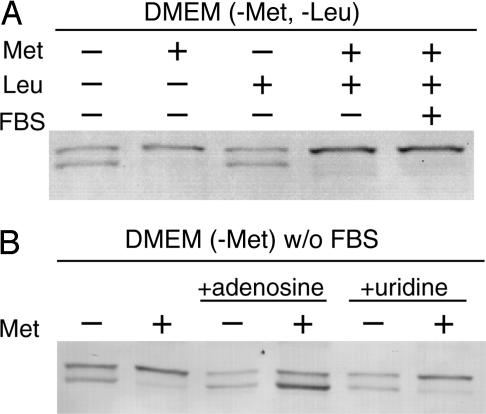
Alteration of Modification Status by Methionine Restriction and the Addition of Adenosine to the Medium. We have screened extracellular factors that might alter the modification status of human MTHFR using HEK293 cells. Modification status was judged by SDS/PAGE and Western blotting as shown in Fig. 10. The modification status was not altered by serum depletion or by removal of leucine from the medium (Fig. 10A). However, lack of methionine in the medium led to decreased phosphorylation as judged by increased mobility of human MTHFR. We hypothesized that this was a response to a low intracellular AdoMet/AdoHcy ratio. To test our hypothesis, adenosine was added to the medium. Addition of adenosine decreases the intracellular AdoMet/AdoHcy ratio (18). MTHFR exhibited a mobility characteristic of unmodified enzyme when adenosine was added to the medium, even when methionine was present (Fig. 10B). However, there was no additional effect of adenosine in the absence of methionine.
Fig. 10.
Effects of methionine and adenosine on the mobility of human MTHFR in transfected HEK293 cells. (A) Wild-type human MTHFR was expressed in HEK293 cells cultured in modified DMEM containing or lacking methionine or leucine and containing or lacking FBS. Cells were harvested after 12 h of nutrient limitation. (B) Wild-type human MTHFR was expressed in HEK293 cells cultured in modified DMEM containing or lacking methionine, and the effect of additions of adenosine (1 mM) or uridine (1 mM) was examined. Cells were harvested 12 h after the initiation of methionine limitation and/or addition of adenosine or uridine.
Discussion
In this article, we provide evidence for dual regulation of human MTHFR activity by allosteric inhibition and by phosphorylation. Earlier studies of the allosteric inhibition of porcine MTHFR suggested a two-state model in which AdoMet led to alterations in the fractions of active (R) and inactive (T) states by preferentially binding to enzyme in the T state (19). Our present data are consistent with the notion that the primary effect of phosphorylation is alteration of the intrinsic equilibrium between the R and T states in the absence of ligands, so as to favor the R state. The differential free energy of binding of AdoMet is now insufficient to drive the equilibrium fully over to the T state, so that, even at saturating levels of AdoMet, the enzyme is only partially inhibited.
Dual regulation is surprisingly common for mammalian proteins, and the first mammalian enzyme shown to be regulated by phosphorylation (20), glycogen phosphorylase, exhibits exactly such dual regulation. Studies from the laboratory of C. F. Cori and G. T. Cori (Washington University School of Medicine, St. Louis) first showed that glycogen phosphorylase existed in two forms, phosphorylase a and b, which differed primarily in their sensitivity to allosteric effectors (21). Phosphorylase b requires AMP for activation, whereas phosphorylase a is active in the absence of AMP. Phosphorylation of glycogen phosphorylase occurs on Ser-14, forming phosphorylase a, and leads to a shift in the allosteric equilibrium between active and inactive forms, so as to favor the active form in the absence of the allosteric activator AMP. The structural basis for the activating effect of phosphorylation was revealed by a comparison of x-ray structures of the inactive forms of phosphorylase b and a with their active forms (reviewed in ref. 22). AMP activates phosphorylase a by binding at the interface between subunits of the homodimer, leading to a conformational change that exposes the active site. The phosphoryl group introduced into phosphorylase a has a similar effect, interacting with arginine residues on the opposite subunit so as to initiate the conformational change leading to exposure of the active site. The conversion of inactive to active forms of phosphorylase leads to a dramatic rearrangement of the N terminus of phosphorylase, which interacts with residues of its own subunit in the inactive state, and with residues of the opposite subunit in the active state. The well studied dual regulation of glycogen phosphorylase can serve as a model to understand the dual regulation of human MTHFR, where only limited structural information is available, with the important caveat that phosphorylation of MTHFR favors the inactive rather than the active state.
A pertinent question is what physiological advantage is conferred by dual regulation of human MTHFR. Generally, three advantages have been proposed to account for modulation of allosteric enzymes by reversible covalent modification: response to a greater number of allosteric stimuli, greater flexibility in control patterns, and amplification in responses to variations in effector concentrations (23). The amplification arises if the activities of the pertinent kinase and/or phosphatase are also regulated by covalent modification, allowing an amplification cascade. In the case of human MTHFR, our investigations indicate that the allosteric stimuli resulting in covalent modification are the same as those required for allosteric modulation, although once the pertinent kinases and phosphatases are identified other factors may be identified that modulate the level of phosphorylation of the enzyme. Clearly, a full understanding of the significance of human MTHFR phosphorylation will require that we identify the kinase(s) and phosphatase(s) responsible for reversible modification.
Taken together, our results suggest that, when the cellular AdoMet/AdoHcy ratio is low, MTHFR modification is decreased, leading to a more active enzyme that is less susceptible to inhibition by AdoMet. Increasing the AdoMet/AdoHcy ratio results in a somewhat less active enzyme that is much more completely inhibited by AdoMet. The mechanism by which the AdoMet/AdoHcy ratio effects altered modification of human MTHFR is of interest, but one possibility is that it does so by altering the exposure of the N-terminal extension of MTHFR where phosphorylation occurs.
Acknowledgments
We thank Kun Liang Guan and Ken Inoki (University of Michigan) for the gift of plasmid pcDNA3HA and for helpful advice throughout the course of this work, Elise Hondorp for assistance with the LC-MS analysis of intact MTHFR, and Robert Pejchal for preparing the samples for N-terminal amino acid analysis after limited digestion of human MTHFR with trypsin. This work was supported in part by National Institutes of Health Grant GM24908 from the National Institute of General Medical Sciences (to K.Y. and R.G.M.) and by National Resource for Proteomics and Pathways National Center for Research Resources Grant P41-18627 (to J.R.S. and P.C.A.).
Abbreviations: AdoHcy, S-adenosylhomocysteine; AdoMet, S-adenosylmethionine; HEK, human embryonic kidney; LC-MS, liquid chromatography-MS; MTHFR, methylenetetrahydrofolate reductase.
Footnotes
The expression vectors pFASTBacHT (baculovirus) and pProExHT (E. coli) specify the same sequence for the histidine tag that is added to the N terminus of the protein.
References
- 1.Matthews, R. G. (1986) Methods Enzymol. 122, 372-381. [DOI] [PubMed] [Google Scholar]
- 2.Jacques, P. F., Bostom, A. G., Williams, R. R., Ellison, R. C., Eckfeldt, J. H., Rosenberg, I. H., Selhub, J. & Rozen, R. (1996) Circulation 93, 7-9. [DOI] [PubMed] [Google Scholar]
- 3.Graham, I. M., Daly, L. E., Refsum, H. M., Robinson, K., Brattstrom, L. E., Ueland, P. M., Palma-Reis, J., Boers, G. H. J., Sheahan, R. G., Israelsson, B., et al. (1997) J. Am. Med. Assoc. 277, 1775-1781. [DOI] [PubMed] [Google Scholar]
- 4.Steegers-Theunissen, R. P. M., Boers, G. H. J., Trijbels, F. J. M. & Eskes, T. K. A. B. (1991) New. Engl. J. Med. 324, 199-200. [DOI] [PubMed] [Google Scholar]
- 5.Mills, J. L., McPartlin, J. M., Kirke, P. N., Lee, Y. J., Conley, M. R., Weir, D. G. & Scott, J. M. (1995) Lancet 345, 149-151. [DOI] [PubMed] [Google Scholar]
- 6.Hoffman, D. R., Comatzer, W. E. & Duerre, J. A. (1979) Can. J. Biochem. 57, 56-65. [DOI] [PubMed] [Google Scholar]
- 7.Kutzbach, C. & Stokstad, E. L. R. (1971) Biochim. Biophys. Acta 250, 459-477. [DOI] [PubMed] [Google Scholar]
- 8.Daubner, S. C. & Matthews, R. G. (1982) in Flavins and Flavoproteins, eds. Massey, V. & Williams, C. H. (Elsevier, New York), pp. 165-172.
- 9.Frosst, P., Blom, H. J., Milos, R., Goyette, P., Sheppard, C. A., Matthews, R. G., Boers, G. J., den Heijer, M., Kluijtmans, L. A., van den Heuvel, L. P. & Rozen, R. (1995) Nat. Genet. 10, 111-113. [DOI] [PubMed] [Google Scholar]
- 10.Goyette, P., Sumner, J., Milos, R., Duncan, A. M. V., Rosenblatt, D. S., Matthews, R. G. & Rozen, R. (1994) Nat. Genet. 7, 195-200. [DOI] [PubMed] [Google Scholar]
- 11.Matthews, R. G., Vanoni, M. A., Hainfeld, J. F. & Wall, J. (1984) J. Biol. Chem. 259, 11647-11650. [PubMed] [Google Scholar]
- 12.Sumner, J., Jencks, D. A., Khani, S. & Matthews, R. G. (1986) J. Biol. Chem. 261, 7697-7700. [PubMed] [Google Scholar]
- 13.Matthews, R. G. (1990) in Chemistry and Biochemistry of Flavoenzymes, ed. Muller, F. (CRC, Boca Raton, FL), Vol. I, pp. 371-387. [Google Scholar]
- 14.Weisberg, I. S., Jacques, P. F., Selhub, J., Bostom, A. G., Chen, Z., Curtis Ellison, R., Eckfeldt, J. H. & Rozen, R. (2001) Atherosclerosis 156, 14853-14858. [DOI] [PubMed] [Google Scholar]
- 15.Yamada, K., Chen, Z., Rozen, R. & Matthews, R. G. (2001) Proc. Natl. Acad. Sci. USA 98, 14853-14858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laemmlie, U. K. (1970) Nature 227, 680-685. [DOI] [PubMed] [Google Scholar]
- 17.Hondorp, E. R. & Matthews, R. G. (2004) PLoS Biol. 2, 1738-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Møller, M. T. N., Samari, H. R., Fengsrud, M., Strømhaug, P. E., Østvold, A. C. & Seglen, P. O. (2003) Biochem. J. 373, 505-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jencks, D. A. & Matthews, R. G. (1987) J. Biol. Chem. 262, 2485-2493. [PubMed] [Google Scholar]
- 20.Fischer, E. H., Graves, D. J., Snyder Crittenden, E. R. & Krebs, E. G. (1959) J. Biol. Chem. 234, 1698-1704. [PubMed] [Google Scholar]
- 21.Cori, C. F., Cori, G. T. & Green, A. A. (1943) J. Biol. Chem. 151, 39-55. [Google Scholar]
- 22.Johnson, L. N. & Barford, D. (1993) Annu. Rev. Biophys. Biomol. Struct. 22, 199-232. [DOI] [PubMed] [Google Scholar]
- 23.Voet, D. & Voet, J. G. (2004) Biochemistry (Wiley, New York), 3rd Ed.
- 24.Guenther, B. D., Sheppard, C. A., Tran, P., Rozen, R., Matthews, R. G. & Ludwig, M. L. (1999) Nat. Struct. Biol. 6, 359-365. [DOI] [PubMed] [Google Scholar]